Abstract
Most currently available glycan structure databases use their own proprietary structure representation schema and contain numerous annotation errors. These cause problems when glycan databases are used for the annotation or mining of data generated in the laboratory. Due to the complexity of glycan structures, curating these databases is often a tedious and labor-intensive process. However, rigorously validating glycan structures can be made easier with a curation workflow that incorporates a structure-matching algorithm that compares candidate glycans to a canonical tree that embodies structural features consistent with established mechanisms for the biosynthesis of a particular class of glycans. To this end, we have implemented Qrator, a web-based application that uses a combination of external literature and database references, user annotations and canonical trees to assist and guide researchers in making informed decisions while curating glycans. Using this application, we have started the curation of large numbers of N-glycans, O-glycans and glycosphingolipids. Our curation workflow allows creating and extending canonical trees for these classes of glycans, which have subsequently been used to improve the curation workflow.
Keywords: curation, glycan, ontology, Qrator, structure matching
Introduction
Glycobiology is an emerging discipline aimed at understanding the diverse biological functions of complex glycans and the relationships between glycan structure, abundance, biosynthesis and function. Glycans participate in a broad range of cellular processes including cell–cell recognition and maintenance of cellular integrity (Packer et al. 2008). As a sub-discipline of glycobiology, glycomics seeks to identify glycan structures and determine how their abundance changes in various tissues, cells and organelles or as a function of cell development or pathology. The availability of robust and accurate collections of glycan structural data is a key element required for the success of this emerging field. Glycans are composed of monosaccharide residues that can be linked together in several different ways, often resulting in branched structures. This topological complexity distinguishes them from proteins and polynucleic acids (DNA/RNA), which are basically linear structures (Herget et al. 2009, 2008; Laine 1994; Werz et al. 2007). Unlike proteins and polynucleic acids, glycans are not synthesized using a template-based mechanism, but are generated by glycosyltransferases and glycosyl hydrolases that modify glycan structure by catalyzing the addition or removal of specific monosaccharide residues (Varki et al. 2009). This structural and biosynthetic complexity makes the determination and accurate representation of glycan structures a challenging endeavor.
Collecting and storing data are an essential part of every field of scientific research, with databases and ontologies, among other methods, being used to capture scientific data. Ontologies are formal, shared vocabularies of concepts and relations that represent knowledge within a domain, and are increasingly utilized for capturing scientific knowledge, as evidenced by the popularity of sites like OBO Foundry (http://www.obofoundry.org). Yet, the amount of information that has been recorded in databases and ontologies has not kept pace with the recent surge of data acquisition in biological and biomedical research. Furthermore, the quality of archived data is often compromised as a result of errors in data exchange, translation and annotation. When glycan structures are recorded in databases, they are translated into database specific, non-standard formats (Ranzinger and York 2012), often leading to inaccuracies ranging from simple typographical errors to fundamental inconsistencies in the structural representation. Thus, standardization and curation of glycan structural representations are important issues that must be addressed for the effective interpretation and utilization of laboratory data and associated metadata, particularly when populating databases or ontologies.
The Complex Carbohydrate Structural Database (CCSD), also called CarbBank (Doubet et al. 1989), established at the Complex Carbohydrate Research Center (CCRC), was the first major international effort to systematically archive structural and meta information of complex glycans. After the discontinuation of funding for CarbBank, other glycan structural databases, including GLYCOSCIENCES.de (Lütteke et al. 2006), the Consortium for Functional Glycomics Glycan Structure Database (Raman et al. 2006), KEGG Glycan (Hashimoto et al., 2006), the Bacterial Carbohydrate Structure Database (BCSDB) (Toukach 2011) and GlycoSuiteDB (Cooper et al. 2001) were established partially using the CCSD as a source of core data (Packer et al. 2008). Unfortunately, CCSD also contained its share of errors, which then propagated to the databases that make use of its data (Egorova and Toukach 2012). More recent bioinformatics efforts at the CCRC have emphasized the establishment and population of ontologies, such as GlycO (Thomas et al. 2006), to represent knowledge pertaining to glycan structures. However, the set of glycans to be represented is potentially very large, encompassing many complex, branched structures that are composed of many residues linked together in distinct ways. Therefore, manual data entry is prone to the introduction of errors, which can be mitigated by the development and implementation of effective curatorial tools.
An effective curation tool, whether used for populating ontologies or more traditional databases with glycan structures, requires a highly intuitive interface for reviewing glycan structures. Such an application should assist scientists in identifying and eliminating errors, and also provide provenance information when possible. Subtle changes in the linkage between two monosaccharide residues can completely change the physical and biological properties of the glycan. As the human eye can easily miss subtle errors of this nature, computational methods are extremely helpful in highlighting potential errors in the representation of the glycan.
The curatorial framework described in this paper takes advantage of canonical trees, each of which represents the emergent structural features of a set of related glycan structures. This concept was first implemented as a “GlycoTree” used to predict the retention times of glycan structures (Takahashi and Kato 2003). Such trees are also formally implemented within the schema of the GlycO ontology and as “composite structure maps” in the KEGG database. These are powerful canonical representations assembled by overlaying many glycan structures of a particular class (e.g., N-glycans) to generate a superstructure containing all of the different residues and residue-to-residue linkages included in the glycan structures that were used to generate the tree. Canonical trees are generated using a set of naturally occurring, biosynthetically related glycans, rather than chemically synthesized glycans. Each glycan corresponds to a subset or subtree of the canonical tree. Moreover, the structure of the GlycO ontology allows us to place canonical trees within the context of other information that we use to support the curation process. This information consists of all the individual structures that make up the tree, as mentioned previously, as well as any references or other meta-information they contain. As curation progresses the amount of information increases, which in turn supports further curation. To date, the application of canonical trees for the curation of glycan structures has not been extensively explored, and software applications that use such trees to aid in the curation process are not currently available.
Most of the curation efforts published in the literature (not limited to glycan structure curation) rely on completely manual curation. Curation in GlycoSuiteDB or BCSDB is performed manually by trained glycobiologists, and data are based on scientific literature. The developers of these databases assert that disallowing direct data entry by researchers ensures consistency and integrity in the data, but Qrator offers users the option to upload their own structures for curation. Moreover, their meaning for curation seems somewhat different from ours since they curate structures by extracting them from literature results, while Qrator allows a scientist to submit a structure directly, verify its structural correctness against a canonical tree, and then present the structure to experts for their approval or rejection. GlycoBank (Berry 2004) is a system that has been used for the curation of glycosaminoglycans (GAGs). The system contains a repository of GAGs, their references, and classification material. Proposed entries or modifications are reviewed and approved manually by GlycoBank appointed curators with no apparent computer assistance, except for the actual display of pending additions. WikiPathways (Pico et al. 2008) offers wiki-based pathway curation by a community of scientists, allowing domain experts from all over the world to directly collaborate on improving pathway diagrams. However, this system is geared towards signaling pathways and pathways leading to cellular metabolites rather than glycan structures and does not feature any algorithms to aid scientists in curating glycan-specific pathways.
Henceforth, we describe a novel approach for curating glycan structures with the help of a web-based application, Qrator, which assists researchers in the curation of new structures by using existing knowledge of previously curated glycans. After passing through a two-stage human curation process, approved structures are available for download, and stored in the GlycO ontology.
Materials and methods
Knowledge representation
Glycan structures approved at the end of the curation workflow may be deposited into the GlycO ontology, which contains knowledge about many types of biomolecules including glycans. A key feature of GlycO is the representation of complex glycans as collections of canonical residues that are defined in terms of their local structures and context within a canonical tree. Within GlycO, each canonical tree corresponds to a particular class of glycans (e.g., N- or O-glycan), such that all known structures of that particular class are represented as subtrees of the tree. These trees are constructed by identifying the union of residues and links contained in a validated collection of structures of a particular class. When the tree is generated using a set of biosynthetically related glycans, it not only provides a convenient method to represent glycan structures but also constitutes a concise basis for inferring a subset of the rules for glycan biosynthesis. That is, adjacent structures in the overall biosynthetic pathway often correspond to valid subtrees that differ by the presence of a single residue; structures that cannot be mapped to a specific subtree cannot be generated by the biosynthetic mechanisms that give rise to the canonical tree. However, canonical trees are not static constructions. Although they grow over time, canonical trees tend to become more stable as structures are added to GlycO, as subsequent inclusion of new structures is less likely to require extension of the tree by the addition of new residues.
To supplement the structural knowledge contained in GlycO, we have created another ontology named ReferO that contains meta-information for each glycan structure in GlycO. This includes references to other databases that contain the same glycan structure, publications that describe or cite the structure, biological source information and provenance information that is collected at each stage of the curation workflow.
Structure matching algorithm
After a glycan is submitted to Qrator, one of the early steps in the curation process involves matching the glycan against a suitable canonical tree to establish its conformance to existing structural knowledge. To establish how well a candidate glycan matches within a canonical tree, we check if all of its paths are included within the canonical tree. These paths are enumerated by starting at leaf residues (residues that have no successor residues) and following the linkages to their predecessors, all the way back to the root residue of the glycan. At present, Qrator does not match glycan fragments.
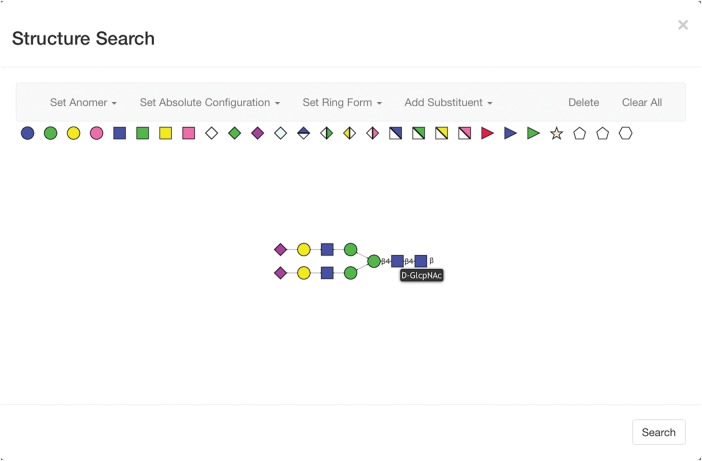
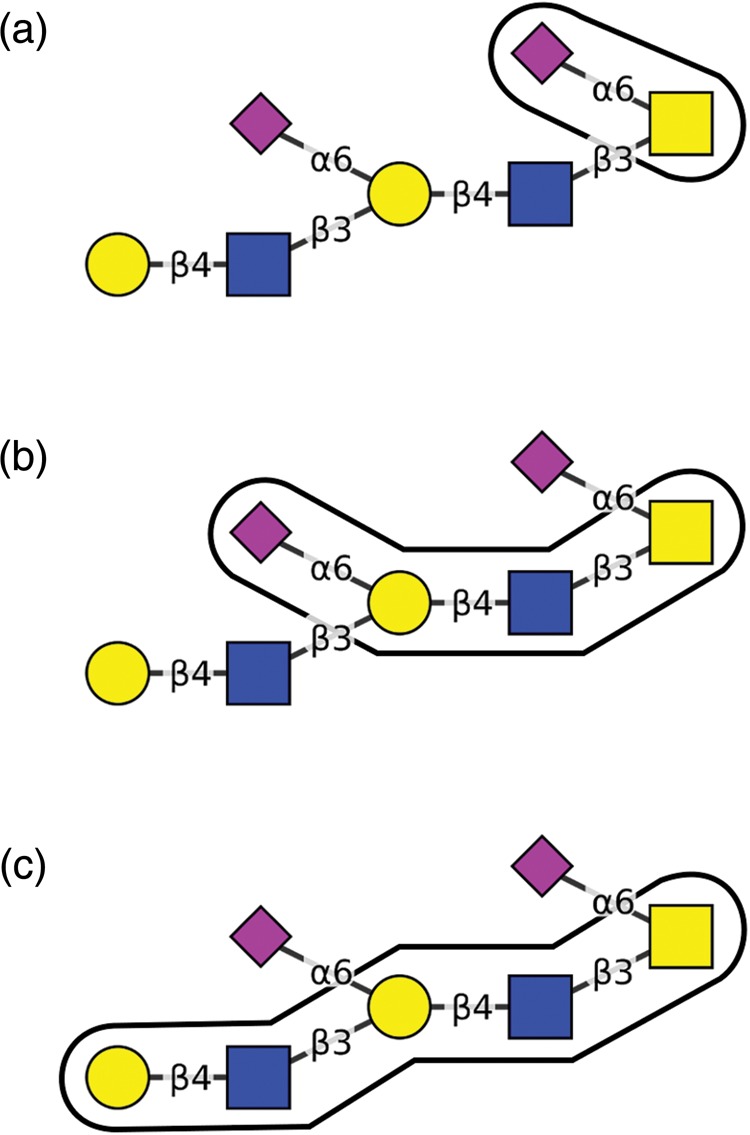
A path in a candidate structure is fully included in a canonical tree if all of its residues and linkages have the same corresponding residues and linkages in the canonical tree. For example, consider the O-glycan paths shown in Figure 1. This glycan has three paths, each starting with a leaf of the structure tree on the left and leading back to its root on the right. Two of the paths (Figure 1A and C) are fully included in the canonical tree shown in Figure 2. The third one (Figure 1B) is included only partially, since the α-linked sialic acid residue connected by a 1–6 linkage (the leaf residue) is not present in the canonical tree.
Fig. 1.
An example of possible paths within a single O-glycan structure.
Fig. 2.
The current canonical tree for GalNAc-initiated O-glycans.
If any path in a candidate structure is not fully included in the canonical tree, incorporation of the candidate structure will extend the canonical tree, providing new information about residues and linkages that have not been previously reviewed. The structure depicted in Figure 1 is one such example, and will extend the canonical tree if approved.
On the other hand, it is possible that a candidate structure contains errors. Their existence also results in partially included paths. One possibility is to treat non-included residues as correct and assume that they extend the canonical tree. However, our matching algorithm attempts to generate additional alignments in which the partially included paths correspond to potential errors in the representation of the candidate structure, and are returned to curators for consideration.
To measure the quality of a glycan's match within a canonical tree, we compute the match score between the glycan and its inclusion in the canonical tree. Pairs of corresponding residues in the candidate glycan and the canonical tree are compared with respect to their (1) residue types, e.g., mannose, glucose, galactose, etc.; (2) absolute configurations, i.e., d or l; (3) anomeric configurations, i.e., α or β; (4) ring forms, i.e., furanose and pyranose and (5) parent attachment site. A perfect residue match has a score of 5, which means the residue agrees with the canonical tree on all considered factors. Thus, the match score of an entire glycan is the total score of all residue assignments of all residues in the candidate structure. A perfect candidate glycan match consists entirely of residues perfectly matched to the canonical tree.
Our matching algorithm enumerates the list of all possible matches of the candidate glycan sorted by score in decreasing order and shows the 10 best matches to the user. The user has the option to request more matches as needed, though in practice this feature is rarely used. The list may contain perfect matches, matches that contain residues that disagree with the canonical tree, or matches that contain residues not currently found in the canonical tree to which it was compared. The matching algorithm, and consequently the user interface, gives indication of where and how each of these matches differ with respect to a canonical tree to assist a human curator in evaluating the structure.
Curation workflow
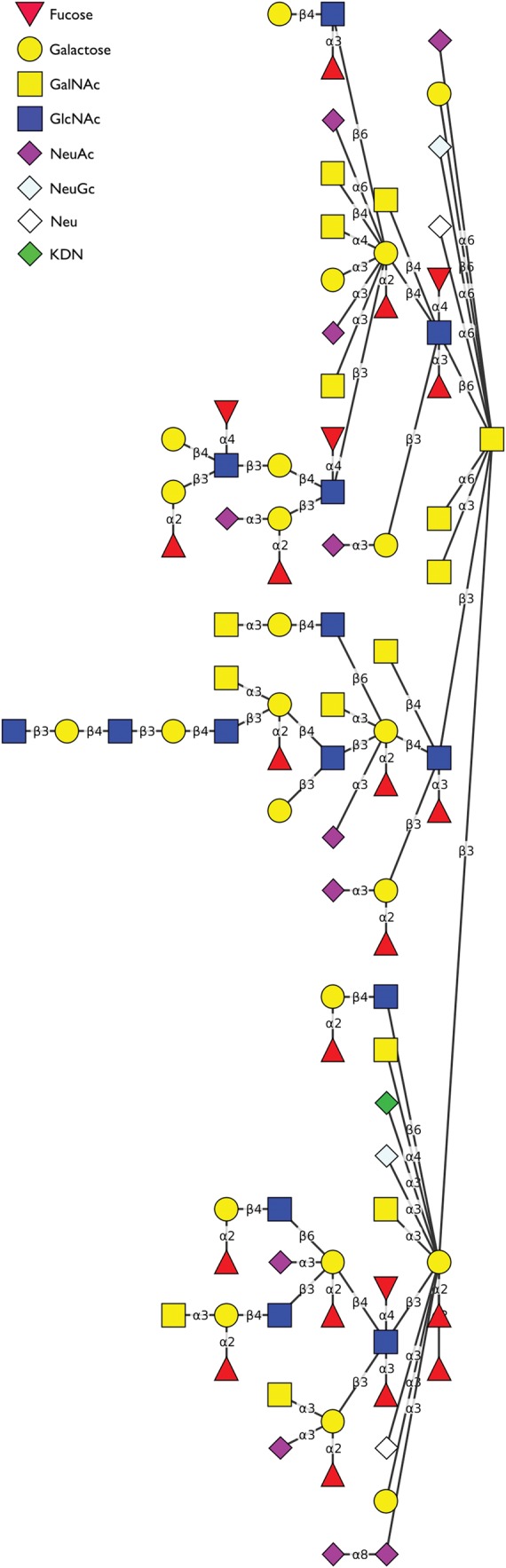
The curation workflow (Figure 3) is a multistep process that requires approval from curators at key points to minimize the possibility of incorrect structures making their way into the GlycO ontology. Combined with computer-aided structure validation, this approach provides an effective means to reduce both the amount of manual labor involved, and the amount of human error. Note that “incorrect” structures, in this context, mean structures that are not consistent with curators’ knowledge of biosynthesis.
Fig. 3.
The curation workflow. Solid lines represent the primary path that a structure takes to be included in GlycO. Dashed lines indicate secondary paths that a structure could take, such as being deferred or rejected. Once a structure is submitted to Qrator, it goes through separate review and curation before being added to the GlycO ontology. User roles for different workflow tasks are also shown.
The process for structure approval is purposely multi-stage to give ample opportunities for reviewers and curators to reject incorrect structures. In Qrator, reviewers are identified as users who initially examine a pending structure and match it against a canonical tree, as opposed to curators who have the power to approve or reject reviewed structures. In a typical usage scenario, anyone may register to use the Qrator for structure submission and review, but only known and trusted experts are allowed to make final approvals or rejections of structures. Interested labs may download and install their own Qrator software using source code hosted on Google Code (https://code.google.com/p/qrator/). In such cases, the decision of who should be allowed to submit, review, and curate structures, or whether the application should be restricted at all, is up to the lab director. The downloadable version of Qrator will facilitate the implementation of autonomous databases or ontologies that focus on glycan structures that are relevant in a specific context, such as those produced by a particular organism or those related to a particular disease.
During structure submission, Qrator can process a single file, or an archive containing several files. File formats accepted by Qrator are discussed in the section entitled “Submitting structures”. The structures parsed from these files are automatically classified using a set of core motifs to identify the canonical tree that most closely fits each one (e.g., the N-glycan tree), and its subtype (e.g., complex N-glycans), if applicable. In general, motifs are substructures that are shared among all glycans of a particular class, but only motifs that contain the root residue of the glycan are used for this classification. After classification, a reviewer determines whether a structure is consistent with his or her knowledge of biosynthesis. Structures that are consistent are computationally compared with the canonical tree that matches their classification. Then, depending on the judgment of the reviewer, one of the matches generated by Qrator may be chosen for a second review stage. However, if a structure is determined to be incorrect (e.g., it contains a structural feature that is biologically improbable), it is not compared with a canonical tree and is moved to the rejected status and kept for future reference. This means that the structure does not make it to the second stage of curation. Retaining rejected structures prevents identical structures from being uploaded in the future, as well as allowing previously rejected structures, along with comments and references, to still be viewed. In certain cases, the decision to reject a structure may be reversed, and structures can be brought out of the rejected state and reviewed again.
In the second stage of curation, a curator examines structures in the reviewed state to make sure no errors are introduced to the ontology. In the curation stage, structures may be sent to approved status or to rejected status. There is no distinction in status between structures that were rejected during curation and those rejected during review. However, the provenance for such a structure will show that it passed the review stage, but was rejected during curation. Approved structures are eventually moved to committed status by an administrator, and added to the GlycO ontology. Reviewers and curators are able to make comments on structures (e.g., correctness, other concerns) at any stage of the curation process, even after a structure has achieved committed status.
Submitting structures
Glycan structures can be uploaded to Qrator using GLYDE-II, an XML-based glycan structure format (http://glycomics.ccrc.uga.edu/core4/informatics-glyde-ii.html) that has been accepted as a standard data exchange format (Packer et al. 2008). Glycans may also be uploaded as a GlycoWorkbench Structure file (GWS files) (Damerell et al. 2012), or zipped archives of many GLYDE-II or GWS files. Once logged in, a scientist may upload a structure or review structures already imported from a database. After a structure is imported or uploaded, it is parsed and converted into a Glycan Object Model (GOM) object, and then converted into a simplified structure representation in JavaScript Object Notation (JSON) (http://www.json.org), which is then stored in the database. GOM is an application programming interface (API) used for parsing GLYDE-II XML files for use by Java applications, while JSON is an information exchange format that is often used as an alternative to XML. Afterwards, both SHA-1 and MD5 hashes of the structure representation are computed to ensure uniqueness among structures. SHA-1 and MD5 are cryptographic hash functions that take potentially large strings of data as input, such as the aforementioned glycan representations, and generate fixed-length (shorter) hashed strings as output. These are useful as unique structure keys in the Qrator's database. After being imported, all new structures are given pending status (Figure 3).
Additionally, scientists may use the included structure builder interface to construct glycans for review. The interface allows users to rapidly construct glycans by clicking residues in a graphical menu to chain residues together. Default values for ring form and absolute configuration are provided for each residue type, but users may modify them. Users may also select a linkage position for each residue by clicking the link between two residues and selecting the desired position from the provided menu. Parent residues are checked to make sure that they do not have multiple child residues connecting to the same linkage position. Additionally, substituents are created in much the same way as creating a child residue, though users pick substituent types and set their positions from a drop-down menu. Moreover, the structure builder doubles as a substructure search interface. It does not utilize the canonical tree to guide construction, however, as users may want to build structures that are not yet subsumed by a canonical tree.
Structures in Qrator are rendered in CFG Nomenclature. When a structure contains residues with no CFG equivalent, they are rendered as dark circles. However, the user may mouse over a residue at any point to view the textual description of the residue. Other notations for rendering glycans may be considered in future versions.
Reviewing and matching structures
During the structure review process, a reviewer decides whether a structure is consistent with well-established biosynthetic pathways according to the reviewer's knowledge. Many biosynthetic pathways are summarized and accessible as public resources (e.g., http://www.ccrc.uga.edu/~moremen/glycomics/) but expert knowledge is still required, especially when considering isomeric complexities that may be known to interfere with specific elongation reactions (Varki et al. 2009). Also, no ambiguities are allowed within a structure's definition, since curation is meant to produce a collection of specific, completely defined structures. If, in the judgment of the curator, each linkage and residue is not supported by known biosynthetic capabilities or clear analytic data, the structure may be rejected (see Curation workflow).
After initial review, the structure may then be compared against one of the canonical trees present in GlycO using the previously discussed matching algorithm. A list of possible matching structural configurations (alignments) is then presented to the reviewer, with differences between the alignment and the submitted structure highlighted as colored circles. For example, the candidate structure shown in Figure 4A has eight residues, meaning the maximum score can be 40 (five possible points per residue and its linkage). The alignment shown in Figure 4B has a score of 36/40 because the linkage position, absolute and anomeric configuration and residue type for the β-galactose residue in the target structure do not match that of the canonical tree. The only matching criterion between the β-galactose and the α-fucose is the ring form (pyranose). In cases where two structures have the same score, they are ranked equivalently and it is left to the reviewer to determine which is correct. However, in practice the number of structures with identical scores is small and does not impair the curation process. A perfect match (an alignment with no differences) appears exactly as the submitted structure does, with no highlights around residues. It should be noted that a perfect match merely means that a structure is completely subsumed by the canonical tree to which it was compared, and that attaining perfect matches is not the ultimate goal of curation. However, perfect matches do indicate a higher probability that a structure is correct, since the curation process has previously validated other structures containing the same residues and linkages. However, in an imperfect alignment, candidate structure residues may exist that do not completely match a canonical tree residue, or simply are not present in the canonical tree to which the candidate structure was mapped. As shown in Figure 4B, a single residue in the candidate structure has been mapped to a topologically equivalent, yet structurally different, residue in the canonical tree and is highlighted with a red circle. Selection of a match highlighted thus indicates the candidate structure should be edited to correct a mistake in the indicated node to match the corresponding node in the canonical tree. Reviewers examining structures manually, or using software that does not attempt to detect errors may overlook such mistakes, since they are not pointed out automatically and are difficult to detect. Residues in the candidate that are not matched topologically or structurally to any residues in the canonical tree are highlighted with blue circles, as shown in Figure 4C. Selection of a match highlighted in this way should be carefully considered, as each highlighted residue requires the addition of a new residue to the canonical tree, fundamentally changing the tree's information content. Alignments such as these are most frequently selected when a canonical tree is initially being built. As a canonical tree becomes larger and more robust, addition of new residues is less often necessary, as the mature canonical tree is more likely to contain all of the residues in each new candidate structure. Structures that have been successfully matched by the reviewer against a canonical tree are assigned reviewed status.
Fig. 4.
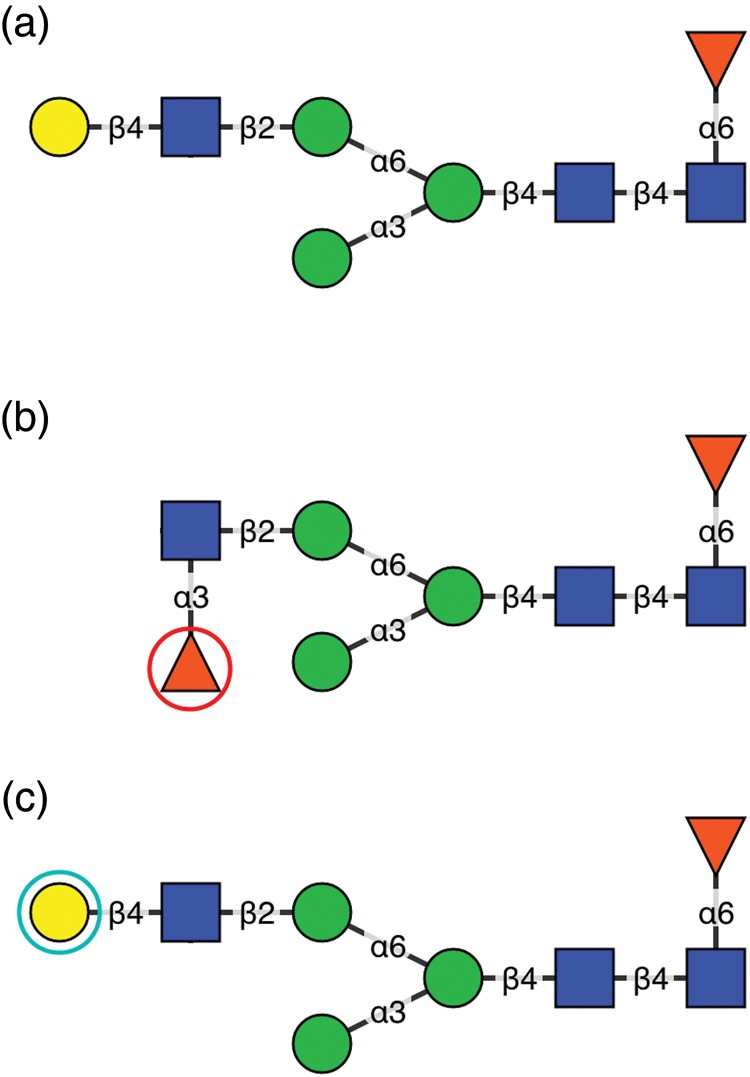
An example of an N-glycan structure that has been matched against a canonical tree. The originally reviewed structure is depicted in (A). An example of an alignment that differs from the original is shown in (B). In this case, Qrator suggests changing the β-galactose to an α-fucose, and the linkage from 1–4 to 1–3. An example of an alignment that will cause a new residue (in this case, β-galactose) to be added to a canonical tree is shown in (C).
The computer-assisted approach for structure review presented here reduces the incidence of human error and the amount of manual labor required in the overall curation process.
Curating structures
After a structure has been reviewed and matched, a second human curator further assesses the matched structure to determine whether it should be included in GlycO. When viewing a matched structure, all of the references and provenance data associated with it are immediately available to the curator, along with the aforementioned visual cues highlighting possible discrepancies between the structure and the canonical tree it was matched against. With this information readily accessible, the curator can make a well-informed decision as to whether the structure should be approved or rejected.
Another important feature of Qrator is its capability to construct canonical trees for new classes of glycans, provided that the root residue is present and an appropriate set of representative glycan structures is available for each class. In this context, we have built upon early work (Takahashi and Kato 2003) by expanding the initial N-glycan canonical tree that we had manually imported into the GlycO ontology. We subsequently regenerated and extended this tree, and generated several O-glycan and glycosphingolipid trees from scratch by defining appropriate root residues (i.e., reducing end residues) and adding new residues using the Qrator application.
Committing structures
After a number of structures have been subjected to the curation workflow and have been approved, they are given committed status. If Qrator is not in standalone mode (i.e., it is not configured to add structures to GlycO), they are written as GLYDE-II XML files and sent to a separate web application that manages the GlycO and ReferO ontologies. This application parses the XML and makes the necessary modifications to GlycO. All metadata about the structures are sent in much the same manner and added to ReferO. The local copies of the canonical trees are then updated for future structure matching.
If Qrator is configured for standalone operation, committing structures to GlycO is not enabled. However, approved structures (or structures in other stages of the curation workflow) can always be downloaded as a zipped archive of GLYDE-II files from the Status panel. This allows scientists to utilize Qrator to curate structures for use in a specific biological context, or for situations where utilizing the semantic capabilities of the GlycO ontology is not needed.
Results
The Qrator web application has been thoroughly tested by scientists at the CCRC, and the application has evolved significantly based on the feedback we were given. Workflow changes were implemented, making certain stages of the workflow appear under-populated, as in the case of deferred structures. However, this disparity is expected to decrease over time as more curation is done. In all, over 2500 glycans from various classes including N-glycans, O-glycans, and glycosphingolipids have been reviewed thus far, and are in various stages of the curation workflow. These structures were all imported from the GlycomeDB meta-database, which provides access to structural information from several different databases (Ranzinger et al. 2008, 2009), including CCSD, BCSDB and the CFG database. Acquiring structures from GlycomeDB has distinct advantages, including the availability of numerous external references to the other databases that have been integrated within GlycomeDB. In our past curation efforts, the focus was on the curation of mammalian structures, which led to a temporary deferment/rejection of valid glycan structures that are not present in Mammalia. Such structures will be reviewed again at a later date. Moreover, Qrator has been designed for curating glycan structures, not glycoconjugates or aglycons, and thus we only import glycans for review.
It is important to note that not all classes of glycans are amenable to curation by Qrator. Each canonical tree used must consist of structurally related glycans (such as N-glycans) that are produced by variations of the same biosynthetic pathway. Thus, curation of N-glycans using Qrator was undertaken before other classes of glycans. This was due in part to the abundance of available structures to curate, as well as the availability of an existing canonical tree (GlycoTree) manually generated by Takahashi and Kato (2003). This tree, containing 74 residues, was initially utilized for curation efforts. After several hundred structures were curated, the N-glycan canonical tree was completely rebuilt using only the curated structures. This newly rebuilt tree has been further extended by continuous structure curation to 144 residues, as of May 2014. Of the 1911, N-glycan structures submitted for curation, experts have approved 879 for inclusion in GlycO (Table I). All numbers are current as of August 2014.
Table I.
Current curation status for different classes of glycans
| Canonical tree | Imported | Pending | Reviewed | Deferred/rejected | Approved/committed |
|---|---|---|---|---|---|
| N-Glycan lipid-linked precursor | 14 | 0 | 0 | 4 | 10 |
| N-Glycan | 1911 | 4 | 297 | 731 | 879 |
| O-GalNAc | 383 | 36 | 0 | 195 | 152 |
| O-Mannose | 466 | 456 | 0 | 1 | 9 |
| O-Fucose | 113 | 113 | 0 | 0 | 0 |
| Gal-initiated glycosphingolipid | 12 | 2 | 0 | 8 | 2 |
| Glc-initiated glycosphingolipid | 428 | 110 | 2 | 36 | 280 |
Currently, curation of O-glycans has been primarily focused on GalNAc-initiated O-structures, with limited curation on mannose-initiated structures and plans to start curation of fucose-initiated structures in the near future. Of the 383 GalNAc-initiated structures input for review, 347 have been curated thus far, with 152 structures approved for inclusion in GlycO. Currently, 9 mannose-initiated structures have been curated and approved, of 466 candidate structures with a mannose residue at the reducing end.
We have also added 440 glycosphingolipid structures including both glucose-initiated and galactose-initiated varieties. In all, 330 glycosphingolipid structures have been curated thus far, with 282 structures approved for addition to GlycO.
All structures are available for download at any stage of curation, either in batches from the status page or individually. Also, the latest version of GlycO is freely available from the CCRC's Ontology Web API website (http://glycomics.ccrc.uga.edu/ontologywebapi/showOntology.action).
Conclusion
Qrator takes a unique approach for glycan curation that leverages information embedded in canonical tree representations of glycan structures, matching new structures against these trees to facilitate the identification of possible mistakes and to assist reviewers in making more informed curation decisions. Algorithmic structure matching reduces the burden placed on reviewers to meticulously examine every structural detail, thereby decreasing the possibility of introducing errors due to human oversight. We have so far committed 1323 approved glycan structures to the GlycO ontology, including N-glycans, O-glycans and glycolipids. These structures can now be used for the interpretation of experimental data with trustworthy glycan structures, and the curation of related biological information, such as glycosylation reactions (Figure 5).
Fig. 5.
A screenshot of the Structure Builder interface. A sialylated bi-antennary N-glycan has been constructed, but not fully specified (i.e., all linkages, anomers, etc., have not been filled in). This interface can be used for both constructing new glycans and searching over existing glycans. Fully specifying a glycan is not necessary for search, but is necessary for adding new structures.
In upcoming versions, we plan to attach species information to each glycan, which will provide essential information that allows glycobiologists to more effectively interpret and mine data generated in the laboratory. For example, this will facilitate selection of glycan structures present in the taxonomic species under study. Furthermore, the structure-building interface will also likely be modified so that scientists could use an existing canonical tree to guide construction of new structures, or build structures based on existing template forms. The first approach has been described previously in our work on GlycoBrowser (Eavenson et al. 2008).
Conflict of interest statement
None declared.
Funding
This work was supported by the National Institute of General Medical Sciences, a part of the National Institutes of Health, funding the National Center for Biomedical Glycomics (8P41GM103490).
Abbreviations
API, application programming interface; BCSDB, Bacterial Carbohydrate Structure Database; CCRC, Complex Carbohydrate Research Center; CCSD, Complex Carbohydrate Structural Database; GAGs, glycosaminoglycans; GOM, Glycan Object Model; JSON, JavaScript Object Notation
Supplementary Material
Acknowledgements
The authors thank the numerous glycan structure database providers that allow free access to their structural information and have been included in GlycomeDB. We also thank our colleagues at the Complex Carbohydrate Research Center who assisted with the design of Qrator, and continue to use this software to curate structures for inclusion in GlycO.
References
- Berry EZ. Bioinformatics and Database Tools for Glycans. Massachusetts Institute of Technology; 2004. [Google Scholar]
- Cooper CA, Harrison MJ, Wilkins MR, Packer NH. GlycoSuiteDB: A new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. 2001;29:332–335. doi: 10.1093/nar/29.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: Updates and new developments. Biol Chem. 2012;393:1357–1362. doi: 10.1515/hsz-2012-0135. [DOI] [PubMed] [Google Scholar]
- Doubet S, Bock K, Smith D, Darvill A, Albersheim P. The complex carbohydrate structure database. Trends Biochem Sci. 1989;14:475–477. doi: 10.1016/0968-0004(89)90175-8. [DOI] [PubMed] [Google Scholar]
- Eavenson M, Janik M, Nimmagadda S, Miller JA, Kochut KJ, York WS. GlycoBrowser – A tool for contextual visualization of biological data and pathways using ontologies. 4th International Symposium on Bioinformatics Research and Applications (ISBRA 2008); Atlanta, Georgia. 2008. [Google Scholar]
- Egorova KS, Toukach PV. Critical analysis of CCSD data quality. J Chem Inf Model. 2012;52:2812–2814. doi: 10.1021/ci3002815. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- Herget S, Ranzinger R, Thomson R, Frank M, von der Lieth CW. Introduction to Carbohydrate Structure and Diversity. Bioinformatics for Glycobiology and Glycomics: An Introduction. Chichester, UK: John Wiley & Sons, Ltd; 2009. pp. 21–47. [Google Scholar]
- Herget S, Toukach PV, Ranzinger R, Hull WE, Knirel YA, von der Lieth CW. Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): Characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct Biol. 2008;8:35. doi: 10.1186/1472-6807-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine RA. A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05×10(12) structures for a reducing hexasaccharide: The Isomer Barrier to development of single-method saccharide sequencing or synthesis systems. Glycobiology. 1994;4:759–767. doi: 10.1093/glycob/4.6.759. [DOI] [PubMed] [Google Scholar]
- Lütteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, Lieth C-Wvd. GLYCOSCIENCES.de: An Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- Packer NH, Lieth C-Wvd, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: Bioinformatics and biomarkers in disease an NIH White Paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11–13, 2006) Proteomics. 2008;8:8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: Pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. Advancing glycomics: Implementation strategies at the Consortium for Functional Glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Frank M, Lieth C-Wvd, Herget S. Glycome-DB.org: A portal for querying across the digital world of carbohydrate sequences. Glycobiology. 2009;19:1563–1567. doi: 10.1093/glycob/cwp137. [DOI] [PubMed] [Google Scholar]
- Ranzinger R, Herget S, Wetter T, Lieth C-Wvd. GlycomeDB – Integration of open-access carbohydrate structure databases. BMC Bioinformatics. 2008;9:384. doi: 10.1186/1471-2105-9-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranzinger R, York WS. Glyco-Bioinformatics Today (August 2011) – Solutions and Problems; Potsdam, Germany: 2012. 2nd Beilstein Symposium on Glyco-Bioinformatics, Cracking the Sugar Code by Navigating the Glycospace. [Google Scholar]
- Takahashi N, Kato K. GALAXY (glycoanalysis by the three axes of MS and chromatography): A web application that assists structural analyses of N-glycans. Trends Glycosci Glycotechnol. 2003;15:235–251. [Google Scholar]
- Thomas CJ, Sheth AP, York WS. Modular Ontology Design Using Canonical Building Blocks in the Biochemistry Domain. International Conference on Formal Ontology in Information Systems (FOIS).2006. [Google Scholar]
- Toukach PV. Bacterial carbohydrate structure database 3: Principles and realization. J Chem Inf Model. 2011;51:159–170. doi: 10.1021/ci100150d. [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. NY: Cold Spring Harbor; 2009. [PubMed] [Google Scholar]
- Werz DB, Ranzinger R, Herget S, Adibekian A, von der Lieth CW, Seeberger PH. Exploring the structural diversity of mammalian carbohydrates (“glycospace”) by statistical databank analysis. ACS Chem Biol. 2007;2:685–691. doi: 10.1021/cb700178s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.