Abstract
The DNA damage response triggers cell-cycle checkpoints, DNA repair and apoptosis using multiple post-translational modifications as molecular switches. However, how ubiquitination regulates ATR signaling in response to replication stress and single-strand break is still unclear. Here, we identified the deubiquitination enzyme (DUB) USP20 as a pivotal regulator of ATR-related DDR pathway. Through screening a panel of DUBs, we identified USP20 as critical for replication stress response. USP20 is phosphorylated by ATR, resulting in disassociation of the E3 ubiquitin ligase HERC2 from USP20 and USP20 stabilization. USP20 in turn deubiquitinates and stabilizes Claspin and enhances the activation of ATR-Chk1 signaling. These findings reveal USP20 to be a novel regulator of ATR-dependent DNA damage signaling.
INTRODUCTION
Maintenance of genomic stability is critical for the well-being of organisms. To maintain genomic stability, cells have developed a network of signaling pathways, collectively known as the DNA damage response (DDR) pathway, to sense and repair DNA damage (1–4). The DDR pathway elicits various responses, including cell-cycle checkpoint activation, DNA repair, aging and apoptosis (5,6). Dysfunction in the DDR pathway results in genomic instability, which is one of the driving forces of tumorigenesis (2,7). The major regulators of the DDR are the phosphoinositide 3-kinase-related protein kinases (PIKKs), including ataxia-telangiectasia mutated (ATM) and ATM and Rad3 related (ATR). Following different type of DNA damage, these two kinases phosphorylate and activate downstream signaling networks (8,9). ATM is mainly activated by DNA double-stranded breaks (10), while ATR is activated in response to a broad variety of DNA damage, such as single-stranded breaks and replication stress (11,12). Studies in yeast and mammals suggest that ATR activation involves multiple steps. ATR and its partner ATR-interacting protein are recruited to DNA damage sites and stalled replication forks by RPA-coated ssDNA following DNA damage or replication stress (13–16). The Rad17-RFC complex recognizes the junctions between ssDNA and double-stranded DNA and loads the 9-1-1 complex (Rad9, Hus1 and Rad1) to the junctions (17–19). The 9-1-1 complex in turn recruits a crucial ATR activator TopBP1 to DNA damage sites through the interaction between C-terminal tail of Rad9 and N-terminal tandem BRCT domains in TopBP1, leading to ATR activation and the phosphorylation of downstream kinase Chk1 (20–26). In addition, a mediator protein named Claspin is important for Chk1 activation (27). Claspin is phosphorylated by ATR and directly binds to Chk1, which is important for Chk1 activation (28,29). On the other hand, activated Chk1 can also stabilize Claspin, suggesting a positive feedback loop for checkpoint activation (30).
Ubiquitination has proven to be an important regulatory mechanism of the DDR, especially in response to interstrand crosslinks and double strand breaks (4,31–34). However, how ubiquitination regulates ATR signaling in response to replication stress and single-strand breaks is largely unknown. In this study, we identified USP20 as a critical regulator of the ATR signaling pathway. USP20 deubiquitinates and stabilizes Claspin, which in turn facilitate the activation of cell-cycle checkpoint following DNA damage. USP20 itself is phosphorylated by ATR, resulting in its stabilization and further activating ATR-Chk1 signaling following replication stress.
MATERIALS AND METHODS
Cell culture, plasmids and antibodies
A549 and HEK293 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) supplemented with 10% fetal calf serum (FBS). USP20+/+ and USP20−/− mouse embryonic fibroblasts (MEFs) were culture in Dulbecco's modified Eagle's medium supplemented with 15% FBS. HA-USP20 was purchased from Addgene (Plasmid #22573, provided by Dr. Wade Harper) and subcloned into PGEX-4T-2 vector (Clontech). pIRES-SFB-Claspin were kindly provided by Larry Karnitz (Mayo Clinic). Deletion mutants were generated by site-directed mutagenesis (Stratagene).
Rabbit anti-USP20 antibodies were raised by immunizing rabbits with GST-USP20 (amino acids 1-200). The antisera were affinity-purified with AminoLink Plus immobilization and purification kit (Pierce). Anti-USP20 antibodies were also purchased from Abcam and Bethyl laboratories. Anti-HERC2 antibody was purchased from BD Biosciences. Anti-Claspin was purchased from Bethyl laboratories. Anti-FLAG (m2) and anti-HA antibodies were purchased from Sigma.
RNA interference
USP20 shRNAs were purchased from Sigma (NM_006676.2-2549s1c1 and NM_006676.2-4079s1c1). Lentiviruses for USP20 shRNAs were made according to the standard protocol.
Tandem affinity purification
Cells stably expressing FLAG-tagged USP20 were lysed with high salt NETN buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF and 1 μg/ml each of pepstatin A and aprotinin on ice for 25 min. Cell lysates were 1:1 diluted with NET buffer (NETN buffer without NaCl) and incubated with anti-HA beads (Sigma) overnight at 4°C. After washing with NETN buffer for three times, the bound proteins were eluted with HA peptide for 1 h at 4°C. Protein samples were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie blue. The eluted proteins were detected by mass spectrometry performed by the Taplin Biological Mass Spectrometry Facility at Harvard.
Co-immunoprecipitation (Co-IP) assay
Cells were lysed with NETN buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) containing 50 mM β-glycerophosphate, 10 mM NaF and 1 mg/ml each of pepstatin A and aprotinin. Whole cell lysates obtained by centrifugation were incubated with 2 μg of antibody and protein A or protein G Sepharose beads (Amersham Biosciences) for 4 h at 4°C. The immunocomplexes were then washed with NETN buffer three times and separated by SDS-PAGE. Immunoblotting was performed following standard procedures.
Protein stability assay
Cycloheximide (CHX) was purchased from Sigma. For protein turnover analysis, CHX was added to cell culture medium at the final concentration of 0.1 mg/ml and cells were harvested at the indicated time points. Cells were then lysed and cell lysates were resolved by SDS-PAGE and analyzed by western blot.
Deubiquitination assay in vivo and in vitro
For the in vivo deubiquitination assay, cells stably expressing Ctrl or USP20 shRNAs were transfected with HA-USP20 wild-type (WT) or Mutant Cys 154 to Ala (CA mutant). Cells were then treated for 4 h with a proteasome inhibitor, MG132 (50 μM), before being harvested. The cells were lysed in 120 μl 62.5 mM Tris-HCl (PH 6.8), 2% SDS, 10% glycerol, 20 mM NEM and 1 mM iodoacetamide, boiled for 15 min, diluted 10 times with NETN buffer containing protease inhibitors, 20 mM NEM and 1 mM iodoacetamide and centrifuged to remove cell debris. The cell extracts were subjected to immunoprecipitation with the indicated antibodies, and blotted with anti-Ub antibody.
For the preparation of a large amount of ubiquitinated proteins as the substrate for the deubiquitination assay in vitro, HEK293 cells were transfected together with the FLAG-Claspin and HA-UB expression vectors. After treatment with MG132, ubiquitinated proteins were purified from the cell extracts with anti-FLAG-affinity column in FLAG-lysis buffer (50 mM Tris-HCl pH 7.8, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 1 mM DTT, 10% glycerol and fresh proteinase inhibitors). After extensive washing with the FLAG-lysis buffer, the proteins were eluted with FLAG-peptides (Sigma). The recombinant GST-USP20 and USP20CA were expressed in BL21 cells and purified following the standard protocol. For the deubiquitination assay in vitro, ubiquitinated proteins were incubated with recombinant USP20 in a deubiquitination buffer (50 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol) for 4 h at 37°C.
Colony formation assay
Cells were exposed to Hydroxyurea (HU) or ultraviolet (UV) at the indicated dosage and left for 10–14 days at 37°C to allow colony formation. Colonies were stained with 5% GIEMSA and counted. Results were normalized to plating efficiencies.
Soft agar colony-formation assays
The soft agar colony-formation assay was performed as described (35). Briefly, primary MEF cells were infected with retrovirus encoding c-Myc together with lentivirus encoding control, USP20shRNA, or USP20shRNA together with FLAG-tagged Claspin. Cells were then plated in 0.3% top agarose in 35 mm dishes and cultured for 2 weeks. Colonies were counted at room temperature under a light microscope (ECLIPSE 80i; Nikon) using a 4× NA 0.10 objective lens (Nikon). Images were captured with a camera (SPOT 2 Megasample; Diagnostic Instruments) and processed using SPOT 4.6 software (Diagnostic Instruments). Adobe Photoshop and Illustrator were used to generate figures.
Athymic nude mice tumor formation assay
Primary MEF cells infected with retrovirus encoding c-Myc together with lentivirus encoding control, USP20 shRNA or USP20 shRNA plus FLAG-tagged Claspin were injected subcutaneously and bilaterally into the dorsal left and right scapular areas of 5-week-old male athymic nude mice NCr nu/nu (NCI/NIH) using 19-gauge needles. Each mouse received two injections of a 200 μl mixture of 2 × 106 cells in 100 μl of 1 × PBS and 100 μl of growth factor reduced MATRIGEL (BD Biosciences). Tumor growth was monitored for 4 weeks and tumor volume was calculated as 0.5 × L × H × W. The tumors were surgically removed, weighed and processed.
Statistics
Values are presented as mean ± SEM of three experiments, unless otherwise indicated. The significance of differences between means was assessed by two-tailed Student's t-test, as indicated. A P-value less than 0.01 was considered significant.
RESULTS
USP20 is involved in replication stress and DDR
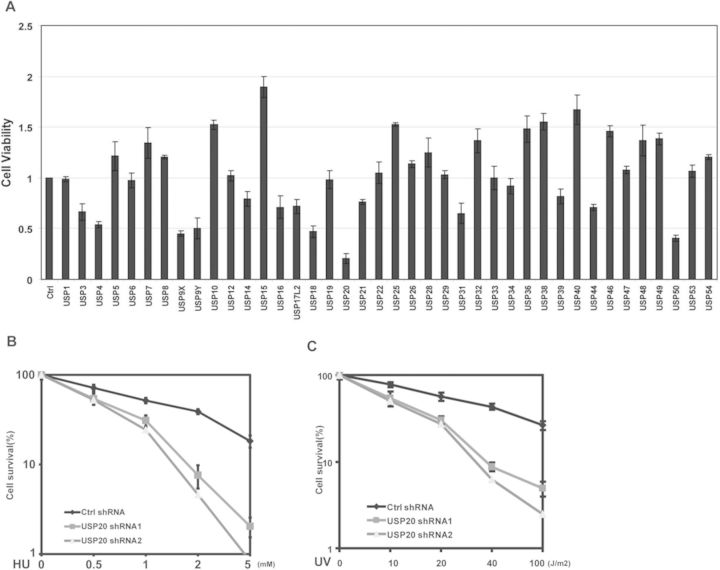
Deubiquitinating enzymes (DUBs) are a group of proteases that regulate ubiquitin-dependent pathways by cleaving ubiquitin-protein bonds (36). To identify DUBs that are important for replication stress response, we knocked-down DUBs individually in cells and checked cell viability following HU treatment. Depletion of several DUBs caused either resistance or hypersensitivity to HU treatment (Figure 1A); among them, USP20 knockdown showed most significant hypersensitivity to HU treatment, while knockdown of USP20 slightly decreased cell proliferation in untreated cells (Supplementary Figure S1A). We decided to further study how USP20 regulates replication stress response. First, to confirm the role of USP20 in replication stress response, we used two different shRNAs targeting USP20 and examined cell sensitivity to HU or UV treatment. Consistent with our initial screen, downregulation of USP20 resulted in hyper-sensitivity to HU (Figure 1B) or UV (Figure 1C). Interestingly, knockdown of USP20 also cause hypersensitivity to infrared (Supplementary Figure S1B). These results suggest that the USP20 is involved in replication stress and DDR.
Figure 1.
USP20 is involved in replication stress and DDR. (A) A549 cells were infected with lentivirus encoding Ctrl shRNA or 40 DUB shRNAs individually. Cells were left untreated or treated with 2mM HU. After 2 weeks, cell viability was determined by colony-formation assay. Error bars represent the SEM of three independent experiments. (B and C) A549 cells stably infected with the indicated shRNA were treated with different dosages of HU (B) or UV (C). After 2 weeks, cell viability was determined by colony-formation assay. Error bars represents the SEM of three independent experiments.
USP20 deubiquitinates and stabilizes Claspin
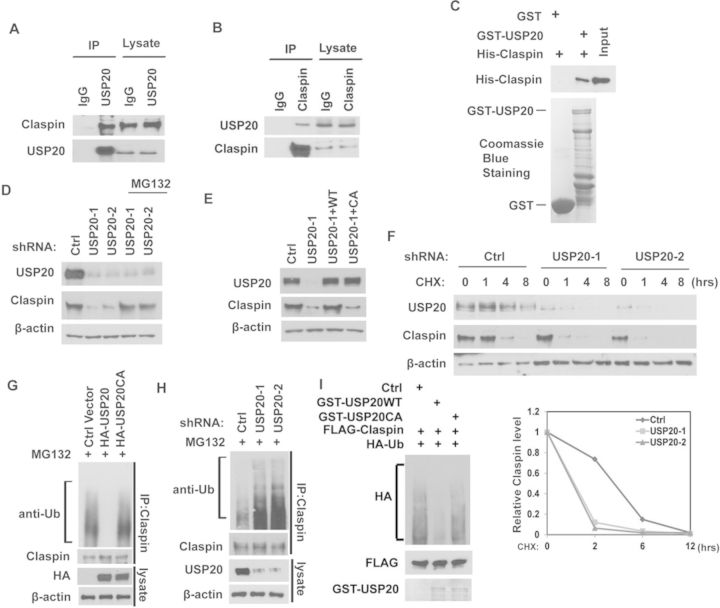
To elucidate the potential molecular mechanism that mediates the effect of USP20 effect on replication stress, we used cells stably expressing FLAG-USP20 to perform tandem affinity purification and mass spectrometry analysis. We identified Claspin as a major USP20-associated protein (Supplementary Figure S1C). We confirmed the USP20-Claspin interaction by Co-IP. As shown in Figure 2A, USP20 co-immunoprecipitated with Claspin in cells. Reciprocal Co-IP with anti-Claspin also pulled down USP20 (Figure 2B). To determine whether the interaction between USP20 and Claspin is direct, we generated and purified recombinant USP20 and Claspin. As shown in Figure 2C, GST-USP20 was able to interact with His-Claspin under cell-free conditions, suggesting a direct interaction between USP20 and Claspin.
Figure 2.
USP20 deubiquitinates and stabilizes Claspin. (A and B) HEK293T cell lysates were subjected to immunoprecipitation with control IgG or anti-USP20 (A) or anti-Claspin (B) antibodies. The immunoprecipitates were then blotted with the indicated antibodies. (C) Purified His-Claspin was incubated with GST or GST-USP20 coupled to GSH-Sepharose. Proteins retained on Sepharose were then blotted with the indicated antibodies. (D) A549 cells infected with lentivirus encoding the indicated shRNAs were left untreated or treated with MG132 for 4 h. Cells were lysed and the cell lysates were then blotted with the indicated antibodies. (E) A549 cells stably expressing control or USP20 shRNA were transfected with empty vector, shRNA resistant HA-USP20WT or its CA mutant. After 48 h, cells were lysed and cell lysates were then blotted with the indicated antibodies. (F) A549 cells stably expressing control shRNA or USP20 shRNAs were treated with CHX (0.1 mg/ml), and harvested at the indicated times. Cells were lysed and cell lysates were then blotted with the indicated antibodies. Lower panel: quantification of the Claspin protein levels relative to β-actin. (G) Cells transfected with the indicated constructs were treated with MG132 for 4 h before harvest. Claspin was immunoprecipitated and immunoblotted with the indicated antibodies. (H) Cells stably expressing Ctrl or USP20 shRNAs were treated with MG132 for 4 h before harvest. Claspin was immunoprecipitated and immunoblotted with the indicated antibodies. (I) Deubiquitination of Claspin in vitro by USP20. Ubiquitinated Claspin was incubated with purified USP20 or USP20CA in vitro, and then blotted with the indicated antibodies.
Since USP20 functions as an ubiquitin-specific protease, we next tested whether USP20 regulates Claspin in a proteasome-dependent manner. As shown in Figure 2D, the decrease in Claspin levels after depletion of USP20 was reversed by the addition of the proteasome inhibitor MG132, while the transcription level of Claspin did not change after depletion of USP20 (Supplementary Figure S1D), suggesting that USP20 regulates Claspin in a proteasome-dependent manner. Furthermore, reconstitution of WT USP20 but not catalytically inactive mutant of USP20 (USP20CA) in USP20-depleted cells restored Claspin protein levels (Figure 2E). These results confirmed the specificity of our USP20 shRNA and suggested USP20 deubiquitination enzyme activity is essential for regulation of Claspin. To further establish that USP20 affects Claspin stability, we treated cell with CHX and determined the half-life of Claspin. We found that Claspin stability was significantly decreased in cells stably expressing USP20 shRNAs (Figure 2F). These results demonstrate that USP20 stabilizes Claspin in cells. Next, we tested whether USP20 deubiquitinates Claspin. As shown in Figure 2G, overexpressing USP20 resulted in a significant decrease in polyubiquitination of Claspin, while overexpressing USP20CA mutant had no such effect. Conversely, knocking-down USP20 increased polyubiquitination of Claspin (Figure 2H). These results suggest that USP20 deubiquitinates Claspin in cells. To determine whether USP20 directly deubiquitinates Claspin, we performed an in vitro deubiquitination assay. As shown in Figure 2I, WT USP20 but not the USP20CA mutant dramatically deubiquitinated Claspin in vitro. Taken together, these results suggest that USP20 deubiquitinates Claspin both in vitro and in vivo.
USP20 is upregulated following DNA damage or replication stress and regulates cell-cycle checkpoint
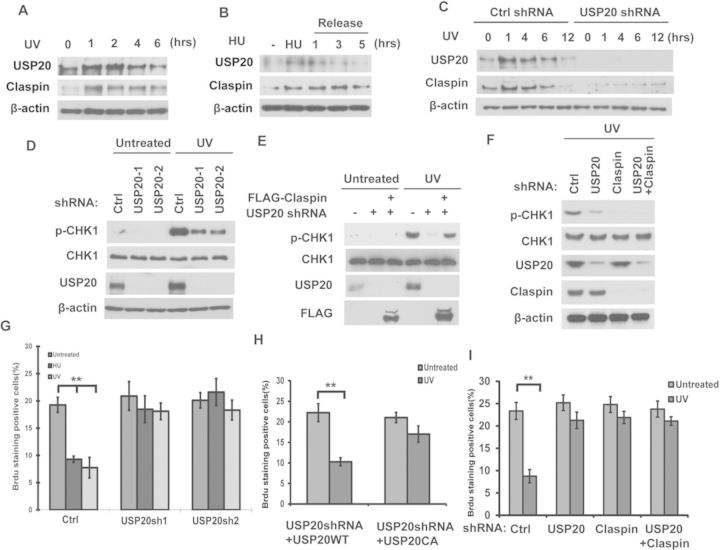
We have shown that USP20 regulates Claspin stability in unstressed cells. Claspin function as a key mediator in ATR-dependent DNA damage signaling and becomes upregulated following DNA damage (37–39). We further examined whether USP20 is important for Claspin upregulation following HU or UV treatment. As shown in Figure 3A and B, Claspin protein levels increased at early time points following UV or HU treatment and decreased at later time points. Interestingly, the protein levels of USP20 followed the same trend. There was no change in Claspin and USP20 mRNA levels (Supplementary Figure S2A and B). Furthermore, downregulation of USP20 blocked UV-induced Claspin upregulation (Figure 3C). These results suggest that USP20 is important for Claspin upregulation following genotoxic stress.
Figure 3.
USP20 is upregulated following DNA damage or replication stress and regulates cell-cycle checkpoint. (A and B) Cells treated as indicated were lysed and cell lysates were then blotted with the indicated antibodies. (C) Cells stably expressing control shRNA or USP20 shRNAs were left untreated or treated with UV (30 J/m2) and harvested at the indicated times. Cells were lysed and cell lysates were then blotted with the indicated antibodies. (D) A549 cells stably expressing control shRNA or USP20 shRNAs were left untreated or treated with UV (30 J/m2). One hour later, cells were lysed and cell lysates were then blotted with the indicated antibodies. (E) A549 cells stably expressing control shRNA or USP20 shRNAs were transfected with the indicated constructs. After 48 h, cells were left untreated or treated with UV (30 J/m2). One hour later, cells were lysed and cell lysates were then blotted with the indicated antibodies. (F) Cells stably expressing Ctrl, USP20shRNA, Claspin shRNA or USP20shRNA together with Claspin shRNA were treated with UV (30 J/m2). One hour later, cells were lysed and cell lysates were then blotted with the indicated antibodies. (G) Cells from (D) were left untreated or treated with HU (10 mM) or UV (30 J/m2). After 2 h, cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. (H) Cells stably expressing USP20 shRNA were transfected with shRNA resistant HA-USP20WT or its CA mutant. Cells were left untreated or treated with UV (30 J/m2). After 2 h, cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. (I) Cells stably expressing Ctrl, USP20shRNA, Claspin shRNA or USP20shRNA together with Claspin shRNA were left untreated or treated with UV (30 J/m2). After 2 h, cells were incubated with 20 μM BrdU; 30 min later, cells were fixed and stained with anti-BrdU antibody. (G–I) Error bars represent the SEM of three independent experiments. **P < 0.01.
Because Claspin is a key regulator of the ATR pathway, we next studied the biological significance of USP20 in this pathway. As shown in Figure 3D, depletion of USP20 in cells dramatically reduced Chk1 phosphorylation following DNA damage. Interestingly, ectopically expression of Claspin in USP20-depleted cells restored Chk1 phosphorylation (Figure 3E). Furthermore, depletion of USP20 cannot further reduce Chk1 phosphorylation in Claspin knocking-down cells (Figure 3F). These results suggested that USP20 regulates ATR signaling mainly through Claspin. Next, we examined whether USP20 regulates cell-cycle checkpoints in response to replication stress. Claspin is important for the intra-S-phase cell-cycle checkpoint (38,39). When the intra-S-phase checkpoint is activated, DNA synthesis will be suppressed. This could be revealed by a decrease in BrdU incorporation, which acts as a marker for DNA synthesis. As shown in Figure 3G, UV or HU treatment resulted in a decrease in BrdU-positive cells in control cells. Downregulation of USP20 largely abolished this decrease, suggesting a defect in the intra-S-phase checkpoint. Reconstitution of USP20-depleted cells with WT USP20, but not USP20 CA, rescued the intra-S-phase checkpoint activation (Figure 3H). Furthermore, depletion of USP20 in cells cannot cause further defect in the intra-S-phase checkpoint in Claspin knocking-down cells (Figure 3I). These results demonstrate that USP20 plays an important role in ATR-dependent signaling and cell-cycle checkpoint activation through targeting Claspin.
USP20 is regulated by HERC2
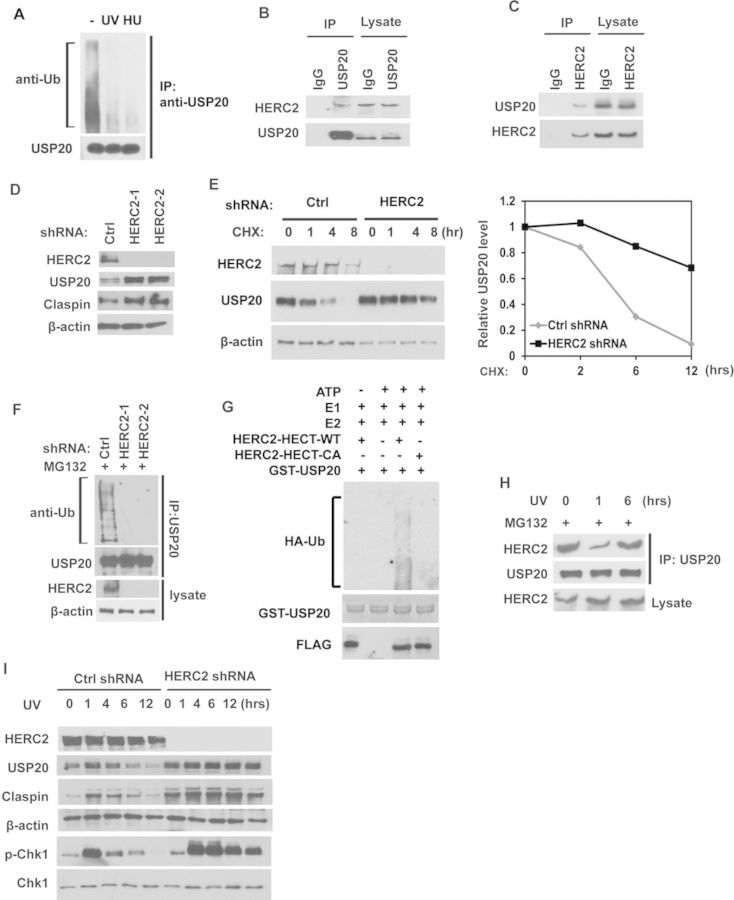
Our results in Figure 3 suggest that USP20 itself is upregulated post-transcriptionally following replication stress. We next examined whether USP20 was regulated by post-translational modifications following HU or UV treatment. We found that USP20 was ubiquitinated in unstressed cells and USP20 ubiquitination was dramatically decreased after UV or HU treatment (Figure 4A), suggesting that USP20 is regulated by the ubiquitin-proteasome pathway. Our IP-MS data showed an E3 ubiquitin ligase HERC2 (HECT and RLD domain containing E3 ubiquitin protein ligase 2), might interact with USP20 (Supplementary Figure S1C). HERC2 has previously been implicated in regulating XPA, BRCA1 and RNF168 levels (40–42). We confirmed the interaction between USP20 and HERC2 by endogenous Co-IP (Figure 4B and C). Further mapping of the USP20-HERC2 interaction showed that the enzymatic domain of USP20, is required for the interaction between USP20 and HERC2 (Supplementary Figure S3A). We next investigated whether HERC2 is a regulator of USP20. As shown in Figure 4D and E, downregulation of HERC2 resulted in upregulation of USP20 levels and protein stability. These results suggest that HERC2 might be an E3 ubiquitin ligase of USP20. To confirm this, we examined the ubiquitination of USP20 in cells depleted of HERC2. Downregulation of HERC2 resulted in significant decrease in USP20 ubiquitination in vivo (Figure 4F). To determine whether HERC2 directly ubiquitinates USP20, we performed an in vitro ubiquitination assay. The HECT domain of WT HERC but not the catalytic inactive mutant (CA) ubiquitinated USP20 in vitro (Figure 4G). Taken together, these results suggested that HERC2 functions as an E3 ligase of USP20 and negatively regulates USP20 in unstressed cells. As USP20 ubiquitination decreases following replication stress, we next studied whether the USP20-HERC2 interaction is subjected to regulation by the DDR pathway. As shown in Figure 4H, the interaction between HERC2 and USP20 decreased at early time point following DNA damage and then increased at later time point. This is consistent with the level change of USP20 following DNA damage (Figure 3A–C). In HERC2-depleted cells, USP20 and its substrates Claspin were significantly upregulated and the phosphorylation of CHK1 is more sustained, even at later time points (Figure 4I). However, depletion of HERC2 did not affect the binding between USP20 and Claspin (Supplementary Figure S3B). These results suggest that HERC2 disassociates from USP20 following genotoxic stress, resulting in the upregulation of USP20 and its targets Claspin. On the other hand, when cells recover from DNA damage, HERC2 mediates USP20 ubiquitination, turning off the DDR signaling. Therefore, timely regulation of USP20 would ensure proper cellular response and recovery to genotoxic stress.
Figure 4.
USP20 is regulated by HERC2 following DNA damage or replication stress. (A) Cells treated with MG132. After 1 h, cells were left untreated or treated with HU (10 mM) or UV (30 J/m2). After 2 h, USP20 was immunoprecipitated and immunoblotted with the indicated antibodies. (B and C) HEK293T cell lysates were subjected to immunoprecipitation with control IgG, anti-USP20 (B) or anti-HERC2 (C) antibodies. The immunoprecipitates were then blotted with the indicated antibodies. (D) Cells stably expressing control shRNA or HERC2 shRNAs were lysed and cell lysates were then blotted with the indicated antibodies. (E) Cells stably expressing control shRNA or HERC2 shRNA were treated with CHX (0.1 mg/ml), and harvested at the indicated times. Cells were lysed and cell lysates were then blotted with the indicated antibodies. Right panel: quantification of the USP20 protein levels relative to β-actin. (F) Cells stably expressing Ctrl or HERC2 shRNAs were treated with MG132 for 4 h before harvest. USP20 was immunoprecipitated and immunoblotted with the indicated antibodies. (G) FLAG-HERC2-HECT-WT and CA mutant were purified from HEK-293T cells and incubated with E1, E2, ubiquitin (HA-Ub), ATP and GST-UPS20 or in the absence of the indicated reagents. Ubiquitinated products were detected by immunoblot with anti-HA antibody. (H) Cells treated with MG132 for 1 h were left untreated or treated with UV and cells were harvested at the indicated times. Cell lysates were subjected to immunoprecipitation with anti-USP20 antibody. The immunoprecipitates were then blotted with the indicated antibodies. (I) Cells stably expressing control shRNA or HERC2 shRNA were left untreated or treated with UV. Cells were lysed and cell lysates were then blotted with the indicated antibodies.
USP20 phosphorylation by ATR is important for its stabilization and checkpoint activation
We next studied how HERC2-USP20 interaction is regulated by the DDR pathway. We found that treating cells with the pan PIKK inhibitor caffeine blocked the disassociation of USP20 and HERC2 after UV treatment (Figure 5A), suggesting that PIKKs might be involved in the regulation of USP20-HERC2 interaction. When we performed a GST pull-down assay using GST-USP20, we found GST-USP20 brought down equal amounts of HERC2 from cells with or without UV treatment (Supplementary Figure S4A). This result led us to hypothesize that USP20 is subjected to regulation by the DDR pathway that results in HERC2 disassociation.
Figure 5.
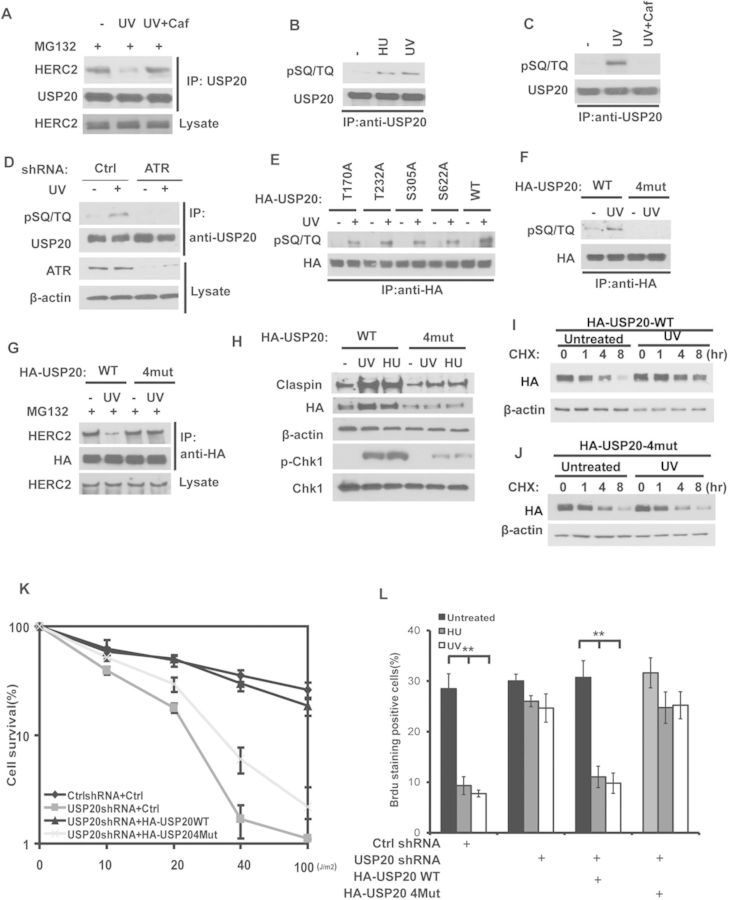
USP20 phosphorylation by ATR is important for its stabilization and checkpoint activation. (A) Cells treated with MG132 or caffeine as indicated for 1 h were left untreated or treated with UV. Cell lysates were subjected to immunoprecipitation with anti-USP20 antibody. The immunoprecipitates were then blotted with the indicated antibodies. (B) Cells were left untreated or treated with UV or HU. After 1 h, USP20 was immunoprecipitated with anti-USP20 antibody and immunoblotted with phospho-SQ/TQ antibody. (C) Cells were pretreated with Dimethyl sulfoxide (DMSO) or 3 mM caffeine. After 2-h incubation, cells were left untreated or treated with UV (30 J/m2). After an additional 1 h, USP20 was immunoprecipitated with anti-USP20 antibody and immunoblotted with phospho-SQ/TQ antibody. (D) Cells stably expressing control shRNA or ATR shRNA were left untreated or treated with UV. After 1 h, USP20 was immunoprecipitated with anti-USP20 antibody and immunoblotted with phospho-SQ/TQ antibody. (E and F) Cells transfected with indicated constructs were left untreated or treated with UV (30 J/m2). After 1 h, HA-USP20 was immunoprecipitated with anti-HA antibody and immunoblotted with phospho-SQ/TQ antibody. (B–F) The immunoprecipitated USP20 loading levels were equalized. (G) Cells stably expressing USP20 shRNA were transfected with shRNA resistant HA-USP20 WT or USP20-4mut mutant. After 48 h, cells were treated with MG132. 2 h later, cells were left untreated or treated with UV (30 J/m2) and after an additional 1 h, cell lysates were subjected to immunoprecipitation with anti-HA antibody. The immunoprecipitates were then blotted with the indicated antibodies. (H) Cells from (G) were left untreated or treated with UV (30 J/m2) or HU (10 mM). Cells were lysed and cell lysates were then blotted with the indicated antibodies. (I and J) Cells expressing the indicated constructs were pretreated for 15 min with CHX (0.1 mg/ml) followed by UV (50 J/m2) treatment. Cells harvested at the indicated times were lysed and cell lysates were then blotted with the indicated antibodies. (K) Cells stably expressing control or USP20 shRNA were stably transfected with empty vector, shRNA-resistant HA-USP20WT or HA-USP20-4mut mutant. Cells were treated as indicated. After 2 weeks, cell viability was determined by colony-formation assay. Error bars represents the SEM of three independent experiments. (L) Cells the same as (K) were left untreated or treated with HU (10 mM) or UV (30 J/m2). After 2 h, cells were incubated with 20 μM BrdU, 30 min later, cells were fixed and stained with anti-BrdU antibody. Error bar represents the SEM of three independent experiments. **P < 0.01.
Using an antibody against consensus ATM/ATR phosphorylation sites (anti-phospho-SQ/TQ), we found that phosphorylation of USP20 at SQ/TQ motifs following HU or UV treatment and USP20 phosphorylation was blocked by caffeine (Figure 5B and C). Furthermore, USP20 phosphorylation increased at the early time point in response to UV treatment and decreased when the cells were recovered from DNA damage (Supplementary Figure S4B). Since ATR is a major kinase activated by replication stress, we further confirmed the role of ATR in USP20 phosphorylation using cells depleted of ATR by shRNA. As shown in Figure 5D and Supplementary Figure S4C, ATR depletion compromised USP20 phosphorylation at the SQ/TQ motifs and resulted in USP20 downregulation, suggesting that ATR is the major kinase responsible for USP20 phosphorylation and upregulation following replication stress. We next set out to determine the ATR phosphorylation sites of USP20. There are four potential SQ/TQ motifs in USP20: T170Q, T232Q, S305Q and S662Q. We found that individual mutation at these sites partially affected USP20 phosphorylation, while mutating all four sites (4 mut) abolished USP20 phosphorylation (Figure 5E and F). Furthermore, USP20-4 mut failed to disassociate from HERC2 following UV treatment and failed to stabilize following replication stress or UV treatment (Figure 5G–J and Supplementary Figure S4D). These results suggest that ATR-mediated phosphorylation of USP20 is required for USP20 stabilization through inducing disassociation of HERC2 and USP20. In cells depleted of endogenous and reconstituted with USP20-4 mut, Claspin upregulation and Chk1 phosphorylation following UV and HU treatment were compromised (Figure 5H and Supplementary Figure S4E). When we overexpressed WT USP20 or USP20-4 mut, we found that Claspin ubiquitination was equally decreased (Supplementary Figure S4F), suggesting that mutation at ATR phosphorylation sites does not affect USP20 enzymatic activity. Furthermore, reconstitution of WT USP20 but not USP20-4 mut rescued the cell viability after UV treatment and restored UV or HU induced intra-S-phase checkpoint (Figure 5K and L). These results establish the important role of USP20 phosphorylation in ATR-dependent DDR.
USP20 functions as a tumor suppressor
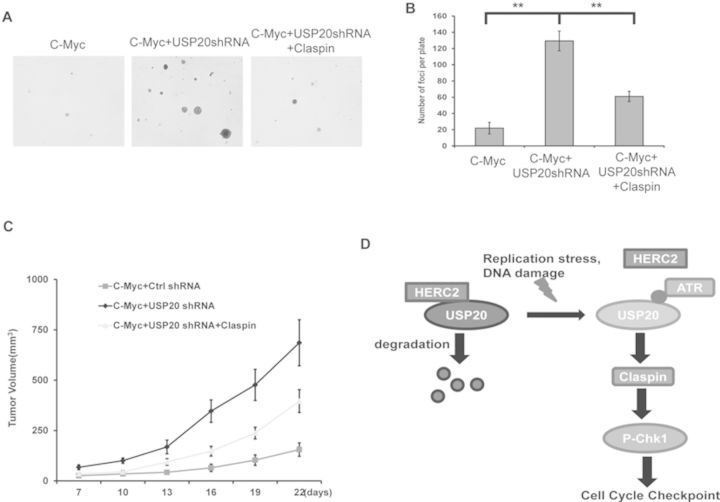
The DDR pathway is critical for maintaining genomic stability. Dysfunction of this pathway results in genomic instability and cancer predisposition (2,7). Therefore, many components of the DDR pathway are identified as tumor suppressors. We next explored a potential role of USP20 in tumorigenesis. We infected the primary MEFs cells with lentiviruses encoding c-Myc and USP20 shRNA and reconstituted the cells with Claspin (Supplementary Figure S5A). We found that in the c-Myc overexpression background, knocking-down USP20 significantly induced cell transformation in soft agar (Figure 6A and B). Ectopic expression of Claspin dramatically inhibited cell transformation induced by USP20 depletion (Figure 6A and B). Furthermore, knocking-down USP20 significantly induced tumorigenesis in xenograft mice models in the c-Myc overexpression background and ectopic expression of Claspin significantly inhibited tumorigenesis induced by USP20 depletion (Figure 6C). These results indicated that USP20 functions as a tumor suppressor.
Figure 6.
USP20 functions as a tumor suppressor. (A and B) Soft agar colony-formation assay was performed using primary MEF cells stably expressing the indicated constructs. Quantification of colonies formed in soft agar. Error bars represent SEM of three independent experiments. **P < 0.01. (C) Primary MEF cells stably expressing the indicated constructs were implanted into nude mice, and tumor formation was monitored. (n = 5; mean tumor volume ± SD.). (D) The working model for USP20 regulation of the DDR.
DISCUSSION
Here, we demonstrate a novel role of USP20 in the DDR. USP20 functions by DUB and stabilizing Claspin in response to replication stress, which in turn facilitates the activation of the ATR-Chk1 pathway and triggers cell-cycle checkpoint. On the other hand, USP20 is phosphorylated by ATR, which disrupts the interaction between USP20 and HERC2, resulting in USP20 stabilization. Elevated USP20 further stabilizes Claspin. Thus, our study identified a novel signaling of the ATR-USP20-Claspin axis in checkpoint signal amplification in response to DNA damage (Figure 6D). Furthermore, our xenograft mice study suggested loss of USP20 induces tumorigenesis. Taken together, our results identified the role of HERC2-USP20-Claspin axis in ATR signaling pathway regulation and tumor suppression.USP20, a DUB enzyme, was reported to regulate multiple cellular events through deubiqutinating various target proteins, such as HIF-1, type 2 iodothyronine deiodinase, β2 adrenergic receptor (β2AR) or TRAF6 (43–46). Here, we identified Claspin as USP20 substrates in response to DNA damage. Claspin was reported to be targeted by beta-TRCP and APC/Cdh1 to terminate Chk1 signaling in mitosis and during checkpoint recovery (37–39). These reports suggest that the degradation of Claspin is important for termination and recovery from cell-cycle checkpoint induced by genotoxic stress. The molecular mechanism for Claspin stabilization at the early phase of the DDR is still not clear. We establish here that USP20 is important for initial Claspin stabilization following DNA damage and replication stress (Figure 3C). Interestingly, the levels of USP20 itself are regulated by the DDR. USP20 is upregulated at the early phase of the DDR and then returns to basal levels when cells recover from genotoxic stress. The change in USP20 levels correlates well with the change in Claspin levels, and based on our results is likely an important mechanism for Claspin regulation.A recent report suggested that USP20 regulates Rad17 stability and homologous recombination repair (47). Consistent with this report, we found knockdown of USP20 in cells decreased Rad17 levels. These results suggest USP20 may regulate ATR-dependent DNA damage pathway through targeting multiple substrates. Nevertheless, in USP20 knockdown cells, reconstitution of Claspin was able to rescue phospho-CHK1 levels and significantly suppress tumorigenesis, even if Rad17 levels was decreased (Figures 3E and 6C and Supplementary Figure S2C), suggesting that Claspin is an important target of USP20 in mediating checkpoint activation and tumor suppression.
We demonstrated that an E3 ligase HERC2 is critical for USP20 regulation. In unstressed cells, HERC2 binds USP20 to maintain a low basal level of USP20 in cells. When DNA damage occurs, HERC2 disassociates from USP20, resulting in USP20 upregulation, which in turn stabilizes Claspin and promotes the activation of the DDR. During cell recovery from genotoxic stress, USP20 binds to HERC2 again and gets degraded. This likely contributes to DNA damage signaling termination. Therefore, the E3 ligase and deubiquitinase pair HERC2-USP20 is important for the timely and dynamic regulation of checkpoint activation. Cells deficient in USP20 display defects in the ATR-Chk1 pathway and checkpoint activation. On the other hand, HERC2 deficiency results in increased and sustained activation of the ATR-Chk1 pathway, and delayed cell recovery from genotoxic stress. Therefore, this pathway needs to be tightly regulated.HERC2 was previously reported to promote DNA damage-induced formation of Lys 63-linked ubiquitin chains by facilitating assembly of the ubiquitin-conjugating enzyme Ubc13 with RNF8 (40,48). This function of HERC2, however, is independent of its E3 ligase activity. HERC2 has also been shown to negatively regulate BRCA1 and XPA levels through its E3 ubiquitin ligase activity (41,42). Here we found that HERC2 antagonized USP20 function and negatively regulated the replication stress signaling pathway. These results suggest HERC2 may have different roles in response to various kinds of DNA damage. The HERC2-USP20 pathway is regulated by ATR and ATR is required for USP20 upregulation following the DDR. Mechanistically, the phosphorylation of USP20 by ATR disrupts the interaction between HERC2 and USP20 and is responsible for USP20 upregulation following the DDR. USP20 phosphorylation by ATR does not affect its E3 ligase activity per se, suggesting that USP20 levels, but not catalytic activity, are subjected to ATR regulation. Currently, we do not know how USP20 phosphorylation affects its binding to HERC2. There might be several possibilities. One is that USP20 phosphorylation itself affects the binding between USP20 and HERC2. Consistent with this, we found that HERC2 binds the catalytic domain (aa 145–686) of USP20, which includes all four phosphorylation sites. The other possibility is that USP20 phosphorylation changes USP20 conformation, which in turn prevents HERC2 binding. Further structural studies and conformational analyses of USP20 will be necessary to better understand the precise mechanism.
Tumorigenesis, as a complex pathologic process, is tightly related to genomic instability. Dysfunction of the DDR pathway leads to genomic instability and tumorigenesis. We established USP20 as a tumor suppressor. Both in vitro and in vivo models suggest depletion of USP20 induced cell transformation and tumorigenesis. Cell transformation induced by USP20 depletion was significantly inhibited by ectopic expression of Claspin, suggesting Claspin is a major target for USP20-mediated tumor suppression. In future studies, we will determine the USP20 expression in human cancers and further generate USP20 knockout mice to establish the physiological role of USP20 in tumorigenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr. Larry Karnitz (Mayo Clinic) for providing Claspin constructs. We thank Debra Evans for proofreading the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first three authors should be regarded as Joint First Authors.
FUNDING
National Basic Research Program of China [973 Program, 2013CB530700]; National Natural Science Foundation of China [81222029, 31270806, 81322031 and 31371367]; National Institutes of Health [CA130996, CA189666 and CA148940]. Funding for open access charge: National Basic Research Program of China [973 Program, 2013CB530700].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lukas J., Lukas C., Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell. Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 4.Huen M.S., Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem. Sci. 2010;35:101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downs J.A., Nussenzweig M.C., Nussenzweig A. Chromatin dynamics and the preservation of genetic information. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 6.Bassermann F., Pagano M. Dissecting the role of ubiquitylation in the DNA damage response checkpoint in G2. Cell Death Diff. 2010;17:78–85. doi: 10.1038/cdd.2009.104. [DOI] [PubMed] [Google Scholar]
- 7.Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 8.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 9.Stokes M.P., Rush J., Macneill J., Ren J.M., Sprott K., Nardone J., Yang V., Beausoleil S.A., Gygi S.P., Livingstone M., et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 11.Zeman M.K., Cimprich K.A. Causes and consequences of replication stress. Nat. Cell. Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn R.L., Zou L. ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 2011;36:133–140. doi: 10.1016/j.tibs.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortez D., Guntuku S., Qin J., Elledge S.J. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 14.Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 16.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.Bao S., Lu T., Wang X., Zheng H., Wang L.E., Wei Q., Hittelman W.N., Li L. Disruption of the Rad9/Rad1/Hus1 (9–1–1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- 18.Ellison V., Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou L., Cortez D., Elledge S.J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 21.Mordes D.A., Glick G.G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navadgi-Patil V.M., Burgers P.M. The unstructured C-terminal tail of the 9–1–1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol. Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M. The Rad9-Hus1-Rad1 (9–1–1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J., Dunphy W.G. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol. Biol. Cell. 2010;21:926–935. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S., Shiotani B., Lahiri M., Marechal A., Tse A., Leung C.C., Glover J.N., Yang X.H., Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., Gong Z., Chen J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J. Cell Biol. 2011;193:267–273. doi: 10.1083/jcb.201010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S., Bekker-Jensen S., Mailand N., Lukas C., Bartek J., Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol. Cell. Biol. 2006;26:6056–6064. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X., Zou L., Lu T., Bao S., Hurov K.E., Hittelman W.N., Elledge S.J., Li L. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Kumagai A., Dunphy W.G. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell. Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- 30.Chini C.C., Wood J., Chen J. Chk1 is required to maintain claspin stability. Oncogene. 2006;25:4165–4171. doi: 10.1038/sj.onc.1209447. [DOI] [PubMed] [Google Scholar]
- 31.Jackson S.P., Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 32.Jacq X., Kemp M., Martin N.M., Jackson S.P. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem. Biophys. 2013;67:25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Messick T.E., Greenberg R.A. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bekker-Jensen S., Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 35.Shim H., Dolde C., Lewis B.C., Wu C.S., Dang G., Jungmann R.A., Dalla-Favera R., Dang C.V. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson K.D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 37.Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–267. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Mailand N., Bekker-Jensen S., Bartek J., Lukas J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell. Biol. 2010;12:80–86. doi: 10.1038/ncb2008. supp. 81–12. [DOI] [PubMed] [Google Scholar]
- 41.Wu W., Sato K., Koike A., Nishikawa H., Koizumi H., Venkitaraman A.R., Ohta T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010;70:6384–6392. doi: 10.1158/0008-5472.CAN-10-1304. [DOI] [PubMed] [Google Scholar]
- 42.Kang T.H., Reardon J.T., Sancar A. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res. 2011;39:3176–3187. doi: 10.1093/nar/gkq1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curcio-Morelli C., Zavacki A.M., Christofollete M., Gereben B., de Freitas B.C., Harney J.W., Li Z., Wu G., Bianco A.C. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J. Clin. Investig. 2003;112:189–196. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., Wang D., Messing E.M., Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasunaga J., Lin F.C., Lu X., Jeang K.T. Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell leukemia virus type 1 tax to negatively regulate NF-kappaB signaling. J. Virol. 2011;85:6212–6219. doi: 10.1128/JVI.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berthouze M., Venkataramanan V., Li Y., Shenoy S.K. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009;28:1684–1696. doi: 10.1038/emboj.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanmugam I., Abbas M., Ayoub F., Mirabal S., Bsaili M., Caulder E.K., Weinstock D.M., Tomkinson A.E., Hromas R., Shaheen M. Ubiquitin-specific peptidase 20 regulates Rad17 stability, checkpoint kinase 1 phosphorylation and DNA repair by homologous recombination. J. Biol. Chem. 2014;289:22739–22748. doi: 10.1074/jbc.M114.550459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Danielsen J.R., Povlsen L.K., Villumsen B.H., Streicher W., Nilsson J., Wikstrom M., Bekker-Jensen S., Mailand N. DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 2012;197:179–187. doi: 10.1083/jcb.201106152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.