Abstract
In Escherichia coli, the ATP-bound form of DnaA (ATP–DnaA) promotes replication initiation. During replication, the bound ATP is hydrolyzed to ADP to yield the ADP-bound form (ADP–DnaA), which is inactive for initiation. The chromosomal site DARS2 facilitates the regeneration of ATP–DnaA by catalyzing nucleotide exchange between free ATP and ADP bound to DnaA. However, the regulatory mechanisms governing this exchange reaction are unclear. Here, using in vitro reconstituted experiments, we show that two nucleoid-associated proteins, IHF and Fis, bind site-specifically to DARS2 to activate coordinately the exchange reaction. The regenerated ATP–DnaA was fully active in replication initiation and underwent DnaA–ATP hydrolysis. ADP–DnaA formed heteromultimeric complexes with IHF and Fis on DARS2, and underwent nucleotide dissociation more efficiently than ATP–DnaA. Consistently, mutant analyses demonstrated that specific binding of IHF and Fis to DARS2 stimulates the formation of ATP–DnaA production, thereby promoting timely initiation. Moreover, we show that IHF–DARS2 binding is temporally regulated during the cell cycle, whereas Fis only binds to DARS2 in exponentially growing cells. These results elucidate the regulation of ATP–DnaA and replication initiation in coordination with the cell cycle and growth phase.
INTRODUCTION
Initiation of chromosomal DNA replication is rigidly regulated to occur only once, at the appropriate time, during each cell cycle. In Escherichia coli, the proteins DnaA and IHF play crucial roles in initiating replication at the chromosomal origin, oriC (1–4) (Figure 1A). DnaA, a member of the AAA+ ATPase family, has an exceptionally high affinity for both ATP and ADP, but only ATP–DnaA is active in initiation. IHF, a member of the nucleoid-associated proteins family, sharply bends DNA at the IHF-binding site (IBS) (5,6). oriC contains an AT-rich duplex unwinding element, a single IBS and at least 12 specific DnaA-binding sites (DnaA boxes) with various affinities (Figure 1A) (3,7,8). Binding of IHF and ATP–DnaA molecules to oriC induces a conformational change in the DNA at the origin, leading to unwinding of the duplex unwinding element (1,3,9). This step is followed by successive loading of DnaB helicase, DnaG primase and DNA polymerase III holoenzyme, which perform DNA synthesis (10). ADP–DnaA can also form multimers on oriC, but these complexes are inactive in initiation.
Figure 1.
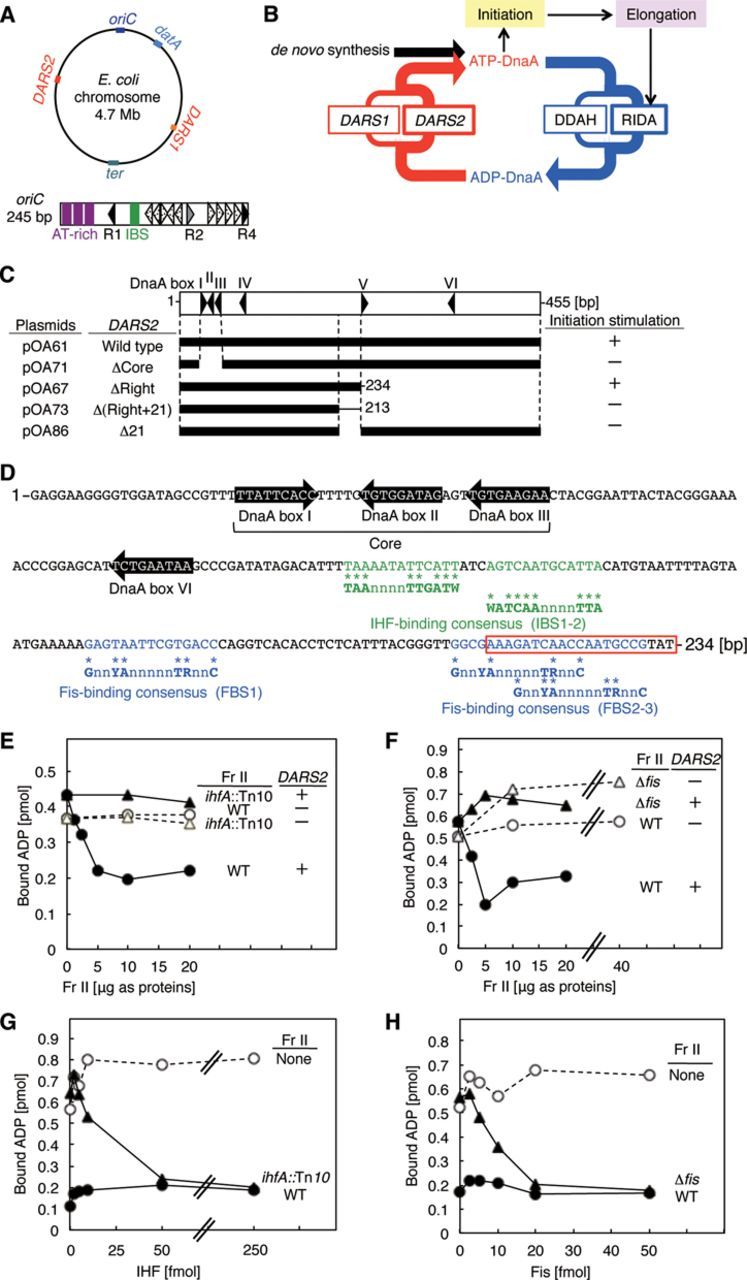
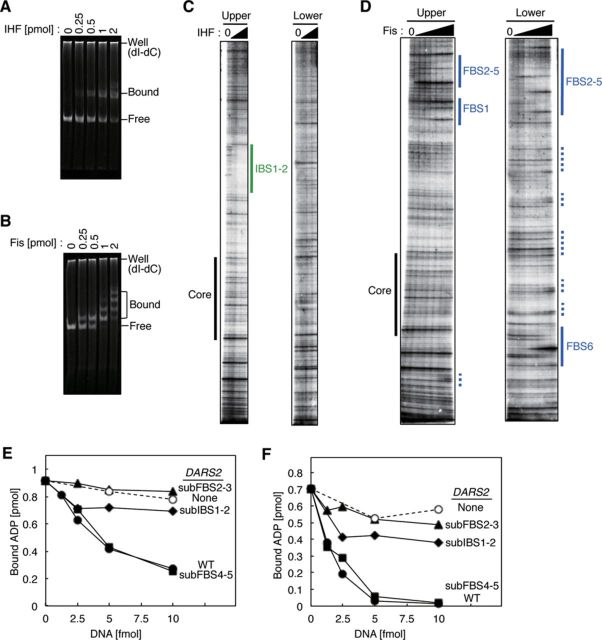
A minimal DARS2 and roles for IHF and Fis. (A) Overall structure of the E. coli chromosome and oriC. The upper figure indicates the relevant loci on the circular chromosome. The lower figure indicates the basic structure of oriC (open rectangle). Filled, gray and dotted arrowheads indicate the high affinity DnaA boxes (R1 and R4), moderate affinity site (R2) and low affinity sites, respectively. The three AT-rich repeats in the duplex unwinding element (AT-rich) and the IBS are also shown by purple bars and a green bar, respectively. (B) Schematic presentation of the regulatory cycle of DnaA. ATP–DnaA initiates replication, resulting in activation of replisomes and RIDA. In addition, DDAH is activated by timely binding of IHF to datA. DARS1 and DARS2 regenerate ATP–DnaA from ADP–DnaA; invivo, DARS2 plays the predominant role in this process. (C) DARS2 deletion analysis. The open rectangle indicates the whole DARS2 region. Filled bars indicate regions carried on vector pACYC177. Cells bearing these plasmids were grown at 37°C in supplemented M9-ampicillin medium, and then subjected to flow cytometry analysis (see Supplementary Figure S1A for data). +, active in stimulation of initiation; -, inactive. (D) Sequence of the minimal DARS2 region on pOA67. Consensus binding sequences of IHF (green letters) and Fis (blue letters) are TAAnnnnTTGATW (where W is A or T) and GnnYAnnnnnTRnnC (Y = C or T; R = A or G; n is any nucleotide) (6,19), respectively. The red box represents the critical 21-mer. (E) DARS2 reaction using IHF-defective crude protein extracts. [3H]ADP–DnaA (2 pmol) and 5 fmol of pOA61 (DARS2, +) or pACYC177 (DARS2, −) were co-incubated at 30°C for 15 min with a crude protein extract (Fr II) of YH014 (WT) or YH014-I (ihfA::Tn10). See also Supplementary Figure S2A. (F) DARS2 reaction using Fis-defective crude protein extracts. Similar experiments were performed using a crude protein extract of MG1655 (WT) or KMG-2 (Δfis). See also Supplementary Figure S2B. (G) In vitro complementation experiments for IHF. Similar experiments were performed using pOA61 and purified IHF with no extract (None) or with a crude extract (Fr II, 5 μg) from YH014 (WT) or YH014-I (ihfA::Tn10).(H) In vitro complementation experiments for Fis. Similar experiments were performed using pOA61 and purified Fis with no extract (None) or crude extract (Fr II, 5 μg) from MG1655 (WT) or KMG-2 (Δfis).
The cellular level of ATP–DnaA is tightly regulated by negative and positive regulatory pathways and peaks at the time of replication initiation (11). During replication, DnaA-bound ATP is hydrolyzed by a complex containing Hda and the DNA-loaded clamp subunit of DNA polymerase III holoenzyme, yielding initiation-inactive ADP–DnaA. This replication-coupled negative feedback on DnaA is called RIDA (regulatory inactivation of DnaA; Figure 1A) (2,12). Cells defective in RIDA exhibit constitutively elevated levels of ATP–DnaA and excess initiations, resulting in inhibition of colony formation (12,13). In addition, the chromosomal locus datA, which contains a cluster of DnaA boxes and an IBS, is required to prevent initiation from occurring at inappropriate times (14–16). In a recent study, we showed that ATP–DnaA molecules form oligomers specifically on IHF-bound datA, and that this oligomerization stimulates DnaA–ATP hydrolysis (17). IHF binding to datA is specific to the post-initiation stage of the replication cycle (17). This system for timely inactivation of DnaA, termed DDAH (datA-dependent DnaA–ATP hydrolysis), supports RIDA (Figure 1B). In addition, dnaA gene transcription is autoregulated; it is inhibited more effectively by ATP–DnaA than by ADP–DnaA (16). This inhibition represses de novo ATP–DnaA synthesis around the time of initiation, indirectly assisting RIDA and DDAH.
In contrast to RIDA and DDAH, DARSs (DnaA-reactivating sequences) increase the level of ATP–DnaA by promoting nucleotide exchange on ADP–DnaA (Figure 1B) (18). The E. coli chromosome contains at least two DARSs, DARS1 and DARS2, which are required for timely initiation of replication during the cell cycle. Structurally, these sites share a highly conserved core region containing three DnaA boxes that are necessary for nucleotide exchange (Figure 1C and D). ADP–DnaA oligomers formed on DARS1 promote dissociation of ADP via specific interactions between DnaA molecules, leading to a release of apo-DnaA from the complexes. These apo-DnaA molecules then bind ATP, resulting in the regeneration of ATP–DnaA. In contrast to DARS1, the in vitro activity of DARS2 requires a crude protein extract, suggesting that unidentified proteins regulate DARS2 activity. In addition, DARS2 increases the cellular ATP–DnaA level and stimulates initiation more effectively than DARS1, suggesting that activators for DARS2 play critical roles in regulation of the ATP–DnaA level and initiation (18). However, the nature of the DARS2 activators and how DARS2 function is regulated during the cell cycle or under different growth conditions remains unknown.
In this study, we identified two nucleoid-associated proteins, IHF and Fis, as key DARS2 activators. Fis binds to DNA in a site-specific manner and regulates gene expression, recombination and superhelicity (5). Fis also stimulates replication initiation, but the underlying mechanism remains unknown (19,20,21). We reconstituted DARS2-dependent DnaA reactivation in vitro using purified IHF and Fis. Simultaneous binding of IHF and Fis to specific sites within DARS2 promoted regeneration of ATP–DnaA. Cell-cycle analyses revealed that IHF bound to DARS2 in a temporally regulated manner. By contrast, Fis bound to DARS2 specifically during the exponential growth phase in a cell cycle–independent manner. Based on these observations, we propose that the activation of DARS2 is regulated by two pathways: an IHF pathway that coordinates activation with the cell cycle, and a Fis pathway that coordinates activation with growth phase. The requirement for the simultaneous binding of IHF and Fis to DARS2 for ATP–DnaA regeneration would ensure the proper timing of chromosomal replication.
MATERIALS AND METHODS
Strains, DNA and proteins
Strains, DNA and proteins used in this study are all described in Supplementary Data.
Reconstitution of DARS2 reactions and DnaA cycle in vitro
DARS2–mediated ADP dissociation was performed with a crude protein extract (fraction [Fr] II) and poly (dI-dC) as described previously (18). Reconstituted DARS2 reactions containing IHF and Fis were performed under similar buffer conditions (for details, see Supplementary Data). DARS2–mediated ATP–DnaA regeneration was reconstituted under the same conditions used for ADP dissociation, except that 1.5 μM [α-32P] ATP was added. In the DnaA cycle reconstitution system integrating RIDA or DDAH, regenerated ATP–DnaA was incubated with IHF–datA or DNA–clamp–Hda complexes, respectively, as described previously (17,18). oriC replication coupled with DARS2–mediated ATP–DnaA regeneration was performed using purified replication proteins as described previously (22), except that the reactions contained 80 nM IHF instead of HU, 5 fmol pOA61 or pACYC177, the indicated amounts of Fis and 80 nM DnaG. The details of these reactions are also provided in Supplementary Materials and Methods.
Electrophoretic mobility shift assay
Proteins were incubated at 30°C for 5 min in buffer GS (20 mM Hepes-KOH [pH 7.6], 50 mM potassium glutamate, 10 mM magnesium acetate, 1 mM ethylenediaminetetraacetic acid [EDTA], 8 mM dithiothreitol, 100 μg/ml bovine serum albumin and 5% glycerol) or dissociation buffer (20 mM Tris–HCl [pH 7.5], 100 mM potassium glutamate, 10 mM magnesium acetate, 2 mM ATP, 8 mM dithiothreitol and 100 μg/ml albumin) containing the indicated DNAs and poly (dI-dC), and then subjected to polyacrylamide gel electrophoresis (PAGE) in Tris-borate buffer and stained with GelStar. For details, see Supplementary Materials and Methods.
DNase I footprint analysis
The upper or lower strand of DARS2 was labeled by polymerase chain reaction (PCR) using 32P-labeled primers. The labeled DARS2 DNA (100 fmol) was incubated at 30°C for 10 min in buffer GS (10 μl) containing indicated amounts of IHF or Fis, 120 ng poly (dI-dC), 5 mM calcium acetate and 12.5 miliunits of DNase I (New England Biolabs) and then subjected to 7 M urea-5% PAGE and phosphorimaging as described (7,21).
Pull-down assay and chromatin immunoprecipitaion
Pull-down experiments and chromatin immunoprecipitaion (ChIP) were performed essentially as described previously (17,23). For details, see Supplementary Materials and Methods.
Determination of oriC copy number and the cellular ATP–DnaA level
The experiments were done, as described (18). The details of these experiments are also provided in Supplementary Materials and Methods.
RESULTS
Definition of a minimal DARS2 region
DARS1 and DARS2 are required for timely initiation of replication (18) and contain the core DnaA boxes that promote the formation of DnaA oligomers, a crucial step in mediating ADP dissociation and regenerating ATP–DnaA from ADP–DnaA. The resulting increase of the ATP–DnaA level is a prerequisite for timely initiation in vivo. DARS2 rather than DARS1 plays the dominant factor in mediating ATP–DnaA regeneration in vivo. In addition, unlike DARS1, DARS2 requires trans-acting activators for ADP dissociation (18). It is plausible that the overall conformation of the DARS2-DnaA complex is affected by activators that promote specific inter-DnaA interactions within the DnaA-oligomer, leading to structural changes that reduce the affinity of DnaA for ADP. As previously suggested for DARS1 (18), apo-DnaA may be released from the DARS2-DnaA complex and then bind to ATP, thereby regenerating ATP–DnaA. Also, activators for DARS2 could play a crucial role in the timely regulation of DARS2 function in the cell cycle and during cell growth phase. In this study, we searched for activating factors of DARS2.
Based on the assumption that DARS2 activators directly bind DARS2, we first defined the minimal region of DARS2 necessary for stimulation of initiation, reasoning that this region should contain specific binding sequences for activators. To this end, we ligated three truncated DARS2 derivatives, ΔRight, ΔRight+21 and Δ21, to the moderate–copy number plasmid pACYC177 (Figure 1C and D), and then performed flow cytometry analysis on cells bearing the resultant plasmids. To determine the number of oriC copies, growing cells were incubated in the presence of rifampicin and cephalexin (inhibitors of initiation and cell division, respectively) until the entire chromosome was replicated. After this procedure, the number of chromosomes per cell, deduced by flow cytometry, corresponds to the number of oriC copies per cell (16). In rapidly growing cells, initiation occurs while the previous round of replication is ongoing, resulting in populations of cells containing predominantly two or four (or four or eight) copies of oriC.MG1655 cells bearing pACYC177 predominantly contain two or four copies of oriC (Supplementary Figure S1A). Introduction of a plasmid containing wild-type DARS2 [pOA61, pACYC177-DARS2], but not a mutant lacking the DnaA-box cluster [pOA71, ΔCore], stimulated overinitiation of replication, which was reflected by the increase in oriC copy number (Figure 1B and Supplementary Figure S1A), as previously demonstrated (18). The slightly increased size (mass) of cells bearing pOA61 has been attributed to retardation of cell division by overinitiation (24). Introduction of pOA67 [ΔRight], which lacks the right half of DARS2, also caused overinitiation, whereas introduction of pOA73 [Δ(Right+21)] or pOA86 [Δ21] did not (Figure 1C and Supplementary Figure S1A). Because the latter two plasmids have a 21-bp deletion flanking the right-half region, these results indicate that DARS2 ΔRight contains the minimal DARS2, and that the 21-mer sequence is crucial for DARS2 activation (Figure 1D). Consistent with this, DnaA boxes IV–VI in DARS2 were dispensable for stimulation of initiation under these experimental conditions (Supplementary Figure S1B).
IHF and Fis stimulate DARS2-mediated ADP dissociation
Sequence analysis revealed that the minimal DARS2 region contains two 13-mer IHF-binding consensus sequences (IBS1–2) and three 15-mer Fis-binding consensus sequences (FBS1–3) (Figure 1D). The crucial 21-mer sequence contains FBS2–3. IHF is a heterodimer of α and β subunits, and Fis is a homodimer (5). To investigate the roles of IHF and Fis in DARS2 activation, we assessed ADP dissociation from DnaA using a crude protein extract prepared from wild-type cells or cells defective in ihfA (encoding the IHF α-subunit) or fis (encoding Fis). A wild-type cell extract (fraction [Fr] II) promoted ADP dissociation from DnaA in a DARS2-dependent manner, as reported previously (18), but an extract prepared from either mutant did not (Figure 1E and F). Even low levels of IHF and Fis restored ADP dissociation to the wild-type level in the corresponding mutant extracts (Figure 1G and H). Western blotting analysis indicated that 5 μg of wild-type Fr II contained about 330 fmol IHF and about 35 fmol Fis (Supplementary Figure S2A and B). This means that the level of Fis was the limiting factor determining the activity of wild-type Fr II (Figure 1E and F). In addition, these results suggest that Fis activity in wild-type Fr II (Figure 1F) and purified Fis activity used in the in vitro complementation experiments (Figure 1H) were similar. We previously reported that DARS2 activators are resistant to heat and RNaseA (18), which is consistent with the high stabilities of IHF and Fis (5).
In vitro reconstitution of DARS2 reactions by IHF and Fis
We next reconstituted DARS2–mediated ADP dissociation from DnaA in reactions containing only IHF, Fis and the wild-type DARS2 construct pOA61 (Figure 2A–C). The activities of IHF and Fis in this reconstituted reaction were similar to those used for the in vitro complementation of the IHF-defective and Fis-defective crude extracts (Figure 1G and H). By contrast, the activity of HU, a major nucleoid-associated protein and a structural homolog of IHF (5), was low in the ADP dissociation reaction (Supplementary Figure S2C and D), whereas HU is interconvertible with IHF in an in vitro reconstituted system for oriC plasmid replication (25).
Figure 2.
In vitro reconstitution of DARS2 reactions. (A) ADP dissociation from DnaA. [3H]ADP–DnaA (2 pmol), the indicated amounts of IHF and 5 fmol of pOA61 (•,▴) or pACYC177 (○) were co-incubated at 30°C for 15 min without (▴) or with 50 fmol of Fis (•,○). See also Supplementary Figure S2C and D. (B) ADP dissociation from DnaA. Similar experiments were performed using the indicated amounts of Fis without (▴) or with 50 fmol of IHF (•,○). (C) ADP dissociation from DnaA. Similar experiments were performed using the indicated amounts of pOA61 without (○) or with 50 fmol each of Fis (•,▪) and IHF (•,▴). (D) ATP–DnaA generation. ADP–DnaA (2 pmol), 1.5 μM [α-32P]ATP and the indicated amounts of pOA61 (•,▪,▴,♦) or pACYC177 (○,□,Δ,⋄) were co-incubated at 30°C for 15 min with or without (▴,Δ) 50 fmol each of IHF (•,♦,○,⋄) and Fis (•,▪,○,□). apo-DnaA (×) was incubated similarly with [32P]ATP, but without DNA, IHF and Fis. (E) Reconstitution of the DnaA cycle using DARS2 and RIDA. In the 1st stage, ADP–DnaA (•,○) or apo-DnaA (▴,Δ) was incubated as above with or without pOA61 (5 fmol) (DARS2) in buffer containing 1.5 μM [α-32P]ATP and 50 fmol each of IHF and Fis, followed by digestion of pOA61 with PciI. In the 2nd stage, portions of the resultant reactions were incubated at 30°C for 20 min with the DNA-loaded clamp (40 fmol as the clamp) and wild-type Hda (WT) (•,▴) or the Hda Q6A mutant as a negative control (○,Δ). The Hda Q6A mutant is defective in clamp binding and RIDA (26). (F) Reconstitution of the DnaA cycle using DARS2 and DDAH. Similar experiments were performed for the 1st stage. In the 2nd stage, the reactions were incubated at 30°C for 10 min with IHF (0.2 pmol) and 991-bp datA DNA (WT) (•,▴) or datA subDnaAbox2 mutant as a negative control (○,Δ). The datA DnaA box 2 is a crucial DnaA box for DDAH and the datA subDnaAbox2 is a mutant datA bearing a substituted DnaA box 2 sequence that is inactive in DDAH (17). (G) oriC replication coupled with DARS2 ATP–DnaA regeneration. ADP–DnaA (•,▴) or ATP–DnaA (○,Δ) (0.5 pmol [20 nM] each) was incubated at 30°C for 15 min with 5 fmol of pOA61 (•,○) or pACYC177 (▴,Δ) in buffer containing M13KEW101 oriC plasmid (600 pmol nucleotides) and replication proteins, including 2 pmol (80 nM) of IHF and the indicated amounts of Fis.
Incubation of ADP–DnaA with DARS2 and ATP yielded ATP–DnaA (Figure 2D). The ATP–DnaA generated was inactivated by RIDA or DDAH (Figure 2E and F), indicating full reconstitution of ATP–DnaA formation and the DARS2-RIDA/DDAH cycle in vitro (17,18,26). In addition, we successfully reconstituted oriC plasmid replication coupled with DARS2 ATP–DnaA regeneration by IHF and Fis (Figure 2G). In these experiments, we used low concentrations of Fis that did not inhibit oriC replication in previously reported reconstituted systems (27).
Thus, the ATP binding, ATP hydrolysis and oriC initiation activities of DnaA were fully preserved in the reconstituted DARS2–mediated ADP dissociation reaction. Furthermore, these results support the idea that IHF and Fis are crucial activators of DARS2.
Determination of IHF- and Fis-binding sites in DARS2
Next, to analyze the mechanism of DARS2–mediated ADP dissociation from DnaA, we determined the binding sites of IHF and Fis within DARS2 using the electrophoretic mobility shift assay and DNase I footprinting (Figure 3A–D). The results of the electrophoretic mobility shift assay revealed that IHF and Fis directly bind to full-length DARS2 (455 bp) with IHF binding to a predominant single site and Fis binding to at least four sites (Figure 3A and B).
Figure 3.
DARS2 IBS1–2 and FBS2–3 are required for DARS2 activation in vitro. (A, B) Electrophoretic mobility shift assay. IHF (A) or Fis (B) was incubated for 5 min at 30°C in buffer GS containing DARS2 DNA (455 bp, 100 fmol) and poly (dI-dC) (120 ng), followed by 4% PAGE. Well; gel well, Bound; protein-bound DNA, Free; protein-free DNA. Poly (dI-dC) remained in the gel well. (C, D) Footprint analysis. The upper or lower strand of [32P]-DARS2 (100 fmol) was incubated at 30°C for 10 min with DNase I and IHF (0, 1.2, 2.4, or 4.8 pmol) (C) or Fis (0, 0.12, 0.24, 0.48, 0.96, 1.9, 3.8, or 7.6 pmol) (D), followed by urea–PAGE and phosphorimaging. Green lines: IBS1–2. Blue lines: FBS1 FBS2–5, and FBS6. Blue dotted lines: low affinity Fis-binding sites. For detailed sequences, see also Supplementary Figure S3. (E, F) Roles for DARS2 IBS and FBS in ADP dissociation from DnaA. MG1655 Fr II (5 μg) (E) or 50 fmol each of IHF and Fis (F) was co-incubated at 30°C for 15 min with [3H]ADP–DnaA (2 pmol) in the presence of pOA61 (WT) (•), pKX35 (subIBS1–2) (▴), pKX36 (subFBS2–3) (♦), pKX37 (subFBS4–5) (▪) or pACYC177 (None) (○). See also Supplementary Figures S4 and S5.
In the DNase I footprint experiments, we used the upper and lower strands of DARS2 to analyze the left and right halves of DARS2, respectively (Figure 3C and D). IHF protected a single region corresponding to IBS1–2 (Figure 3C and Supplementary Figure S3AB). Two IHF molecules might simultaneously bind to IBS1–2. Alternatively, IHF binding to these two sites might be mutually exclusive, and a mixture comprising DARS2 molecules with IHF bound to IBS1 or to IBS2 might result in protection of the region including IBS1–2. IBS1 and IBS2 are separated by only 3 bp (Figure 1D and Supplementary Figure S3B). The close proximity of these sites may explain why IHF predominantly binds to a single site. DNA-bound IHF covers a 34-bp region, including the 13-bp IBS, and binding is accompanied by sharp DNA bending (6). IHF bound to IBS1 may become a physical obstacle inhibiting binding of IHF to IBS2, and vice versa. In addition, the large structural change induced by IHF binding might affect binding of the second IHF at the flanking site and the DNase I sensitivity of the IBS-flanking regions.
In addition, DNase I footprinting revealed that Fis protected three specific sites, FBS1, FBS2–5 and FBS6 (Figure 3D and Supplementary Figure S3AC). The FBS2–5 region includes four overlapping binding consensuses, but binding occurred predominantly on FBS2–3 (Figure 3D and Supplementary Figure S3C), consistent with our definition of the minimal DARS2 sequence (Figure 1C and D). Several additional binding sites were also detected outside the minimal region when higher concentrations of Fis were present (Figure 3D), consistent with the weak affinity of Fis for AT-rich sites (28).
To confirm the specificity of the identified binding sites, we replaced them with scrambled sequences (Supplementary Figure S4A) and analyzed the resultant DNAs by the electrophoretic mobility shift assay. None of the substituted fragments bound IHF or Fis (Supplementary Figure S4B–F).
IBS1–2 and FBS2–3 are required for ADP dissociation in vitro
To elucidate the functions of IBS and FBS, we assessed ADP dissociation from DnaA in vitro using DARS2 derivatives bearing a substituted IBS or FBS (Supplementary Figure S4A). DARS2 derivatives bearing substituted IBS1–2 and FBS2–3 (subIBS1–2 and subFBS2–3) were inactive for ADP dissociation in the presence of a crude protein extract (Figure 3E), whereas DARS2 variants bearing subFBS1, subFBS4–5 or subFBS6 were fully active (Figure 3E and Supplementary Figure S5A). Similar results were obtained using an in vitro reconstituted system using purified IHF and Fis (Figure 3F and Supplementary Figure S5B). Thus, IBS1–2 and FBS2–3 are required for DARS2 activation in vitro.
Location of IBS1–2 and EBS2–3 is important in DARS2 activation
IHF and Fis bend DNA by ∼180 and ∼60°, respectively (6,34). To assess the importance of DNA bending by IHF and Fis in DARS2 activation, we analyzed DARS2 derivatives in which the location of IBS1–2 or FBS2–3 was changed by 5 or 10 bp (Supplementary Figure S6A). A 5-bp translocation, but not a 10-bp translocation, would result in a change in the direction of the DNA bending. The activities of the DARS2 derivatives were analyzed using the in vitro reconstituted system (Supplementary Figure S6B).
First, IBS1–2 was translocated by 5- or 10-bp toward FBS2–3, resulting in traIBS-R5 and –R10 (Supplementary Figure S6A). Compared with wild-type DARS2, DARS2 traIBS-R5 was only partially effective in ADP dissociation, whereas DARS2 traIBS-R10 was moderately effective (Supplementary Figure S6B). These results are consistent with the idea that IHF-dependent DNA bending is important for DARS2 activation. The moderate activity of DARS2 traIBS-R10 suggests that the distance between the core DnaA boxes and IBS1–2 is also important for DARS2 function.
Next, the location of IBS1–2 was translocated by 5-bp or 10-bp toward the core DnaA boxes, resulting in traIBS-L5 and –L10 (Supplementary Figure S6A). Both of the DARS2 mutants were only partially effective in ADP dissociation compared with the wild-type DARS2 (Supplementary Figure S6B). This is consistent with the idea that the introduction of a shorter region between the core DnaA boxes and IBS1–2 or a longer region between IBS1–2 and FBS2–3 is inhibitory for DARS2 activation, even if the direction of DNA bending is preserved. Thus, it is conceivable that the location of IBS1–2 and the DNA binding direction would be important for functional assembly of complexes containing IHF, Fis and DnaA bound to DARS2.
In addition, FBS2–3 was translocated by 5 or 10-bp toward IBS1–2, resulting in traFBS-L5 and –L10 (Supplementary Figure S6A). Whereas the DARS2 activity of DARS2 traFBS-L5 was considerably inhibited, the DARS2 activity of traFBS-L10 was only moderately affected (Supplementary Figure S6B).
These results support the idea that DNA bending at specific sites is important for DARS2 activation. However, we cannot exclude the possibility that DnaA bound to the core DnaA boxes interacts with IHF and Fis bound to IBS1–2 and FBS2–3, and for this interaction to occur, the precise locations of the binding sites would be important.
DnaA oligomer formation is crucial for DARS2-mediated ADP dissociation
To elucidate the formation of the DnaA complex on DARS2, we analyzed DnaA mutants using the in vitro reconstituted system described above. DnaA consists of four functional domains: domain I contains specific binding sites for DnaB and self-dimerization; domain II is a flexible linker; domain III is the AAA+ domain; and domain IV is the DnaA box–binding region (1–3). As in the case of DARS1 (18), DnaA domains I–II were dispensable for the DARS2–mediated ADP dissociation, whereas the R399 and T435 residues in domain IV, which are required for DNA binding (29,30), were crucial for dissociation (Figure 4A).
Figure 4.
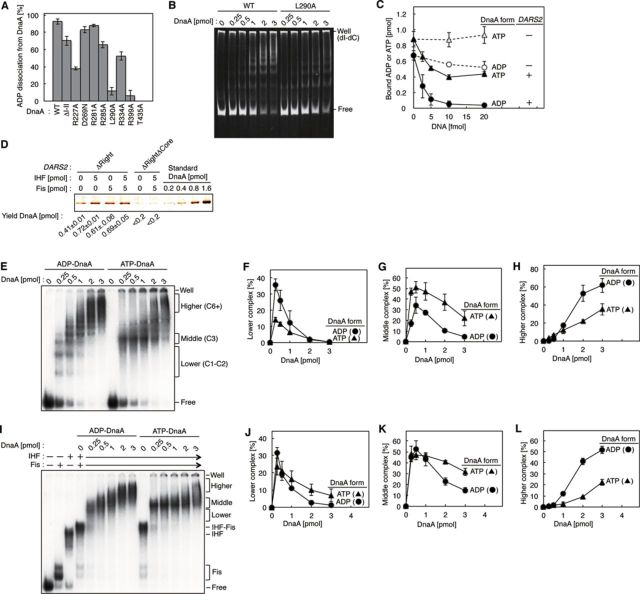
Mechanistic analyses of DnaA oligomer formation on DARS2. (A) Analysis of DnaA mutants. Wild-type and mutant forms of [3H]ADP–DnaA were incubated as described for Figure 2C, with or without 5 fmol of pOA61. Average values and errors of ADP dissociation from DnaA [%] are shown. (B) Electrophoretic mobility shift assay using DnaA AID-2 mutant. Indicated amounts of the ADP forms of wild-type DnaA or the DnaA AID-2 mutant L290A were incubated at 30°C for 5 min in dissociation buffer containing DARS2ΔRight DNA (15 fmol) and poly (dI-dC) (25 ng), followed by 5% PAGE and GelStar staining. Well; gel well, Bound; protein-bound DNA, Free; protein-free DNA. Poly (dI-dC) remained in the gel well. (C) Dissociation of ATP or ADP from DnaA. [α-32P]ATP–DnaA (▴,Δ) or [3H]ADP–DnaA (•,○) (2 pmol) was incubated with pOA61 (•,▴) or pACYC177 (○, Δ) as described above. Two independent experiments were done for each assay, and both data and mean values are shown. (D) Pull-down assay. ADP–DnaA (5 pmol) and 5’-biotinylated DNA (0.5 pmol) of DARS2ΔRight or DARS2ΔRightΔCore were co-incubated with or without IHF and Fis. DNA-bound DnaA was collected using magnetic beads and was quantitatively analyzed by SDS-PAGE and silver staining. DARS2ΔRightΔCore was a DARS2ΔRight derivative bearing ΔCore (Figure 1C). Two independent experiments were done, and both data and the mean values are shown for the recovered DnaA. (E–H) Electrophoretic mobility shift assay using DARS2ΔRight/subFBS1. ADP/ATP–DnaA was incubated at 30°C for 5 min in dissociation buffer containing [32P]DARS2ΔRight/subFBS1 DNA (15 fmol) and poly (dI-dC) (25 ng), followed by 5% PAGE and phosphorimaging (E). Intensities of the signals corresponding to lower (C1–C2) (F), middle (C3) (G) and higher-order multimers (C6+) (H) on the gel image were quantified, and relative levels [% of total] were plotted. Two independent experiments were done, and both data and mean values are shown. (I–L) Electrophoretic mobility shift assay using DARS2ΔRight/subFBS1. Similar experiments were performed in the presence of IHF (0.6 pmol) and Fis (0.3 pmol) (I), followed by analysis of the signals (J–L). Two independent experiments were done, and both data and mean values are shown.
DnaA domain III contains several residues characteristic of the AAA+ family of ATPases: Box VII R281, Arg-finger R285, AID-1 R227 and AID-2 L290. These residues, which are crucial for formation of active initiation complexes, reside on the surface of the domain and are crucial for specific inter-DnaA protomer interactions in initiation complexes (1,7,22). In DARS2–mediated ADP dissociation from DnaA, DnaA R281A was active, and DnaA R285A had moderately reduced activity (Figure 4A), similar to the case for DARS1 (18). By contrast, the R227A and L290A mutants were severely impaired (Figure 4A), suggesting that inter-DnaA interactions via these residues are crucial for DARS2–mediated ADP dissociation. Consistently, DnaA oligomerization on DARS2 was severely inhibited by the L290A mutation (Figure 4B).
In DnaA, the AAA+ sensor I residue D269 and sensor II residue R334 are important for stable nucleotide binding and ATP hydrolysis, respectively (13,31). DnaA D269N was active for DARS2–mediated ADP dissociation (Figure 4A and Supplementary Figure S7A). We found that the addition of poly (dI-dC) to these reactions stimulated ADP dissociation from wild-type DnaA by DARS2, but not significantly by DARS1 (Supplementary Figure S7A and B). Even in the absence of poly (dI-dC), DnaA D269N was basically active in ADP dissociation by DARS2 (Supplementary Figure S7A). ADP dissociation of DnaA D269N by DARS1 was inefficient in the absence of poly (dI-dC), consistent with our previous data (18). In contrast, DnaA R334A was moderately impaired in DARS2-mediated ADP dissociation even in the presence of poly (dI-dC) (Figure 4A), as in the case of DARS1 (18). These results suggest the existence of both common and distinct modes of inter-DnaA interactions on DARS1 and DARS2.
Dissociation of ATP is less efficient than that of ADP
To confirm the specificity of ADP dissociation, we compared the dissociation of ADP and ATP from DnaA by DARS2. In the reconstituted DARS2 reaction, ATP dissociation from DnaA was less efficient than ADP dissociation (Figure 4C), suggesting that ATP–DnaA forms oligomers that impede nucleotide dissociation.
Efficient formation of higher-order complexes of ADP–DnaA on DARS2
To assess the roles of IHF and Fis in DnaA assembly on DARS2, we first performed pull-down assays using ADP–DnaA and biotinylated DARS2ΔRight DNA, the minimal DARS2 sequence (Figure 1C). Yields of biotinylated DNA and DnaA were 0.15–0.20 pmol (i.e. 30–40% of input DNA) and 0.40–0.42 pmol, respectively (Figure 4D). DARS2ΔCore was inactive in DnaA binding (Figure 4D). These results indicate that the core region binds two or three molecules of ADP–DnaA, as predicted from the DARS2 core sequence. Notably, inclusion of IHF, Fis or both proteins increased yields of DnaA to 0.6–0.7 pmol when DARS2 core was present, suggesting that IHF and Fis allow binding of at least three to five ADP–DnaA molecules to a single molecule of DARS2.
Next, we performed the electrophoretic mobility shift assay to investigate further the formation of DnaA oligomers on DARS2. To exclude the effect of FBS1, which is dispensable for ADP dissociation from DnaA (Supplementary Figure S5), we used DARS2ΔRight bearing the subFBS1 mutation. In the absence of IHF and Fis, low levels of ADP–DnaA formed smaller complexes than identical levels of ATP–DnaA (C1–C2; Figure 4E–H). This may be related to the propensity that ATP–DnaA forms homomultimers on oriC more efficiently than ADP–DnaA (2,3,22). However, high levels of ADP–DnaA formed larger complexes more efficiently than identical levels of ATP–DnaA (C3 and greater; Figure 4E–H). Over a wide range of protein concentrations, ATP–DnaA formed complexes of intermediate size that were more abundant than those formed by ADP–DnaA (C3–C4; Figure 4E–H). Notably, even in the presence of IHF and Fis, ADP–DnaA formed larger complexes more efficiently than ATP–DnaA (Figure 4I–L). Also, the fraction of complexes remaining in the wells was considerably reduced by IHF and Fis. These results are consistent with the results of the pull-down assays (Figure 4D) and the idea that IHF and Fis stimulate ordered ADP–DnaA assembly on DARS2.
DARS2-mediated stimulation of initiation in vivo requires IHF and Fis
To investigate the in vivo roles of IHF and Fis in DARS2 activation, we first assessed the sensitivity of cells harboring mutations in ihfA, ihfB or fis (encoding IHF α, IHF β and Fis, respectively) to DARS2 oversupply. Introduction of pKX11 (pBR322 bearing full-length DARS2) severely inhibited colony formation of wild-type cells in an oriC- and dnaA-dependent manner (18). In contrast to wild-type cells, mutant cells bearing pKX11 formed stable colonies at a level similar to those bearing pOA77 [pBR322-DARS2ΔCore] (Figure 5A).
Figure 5.
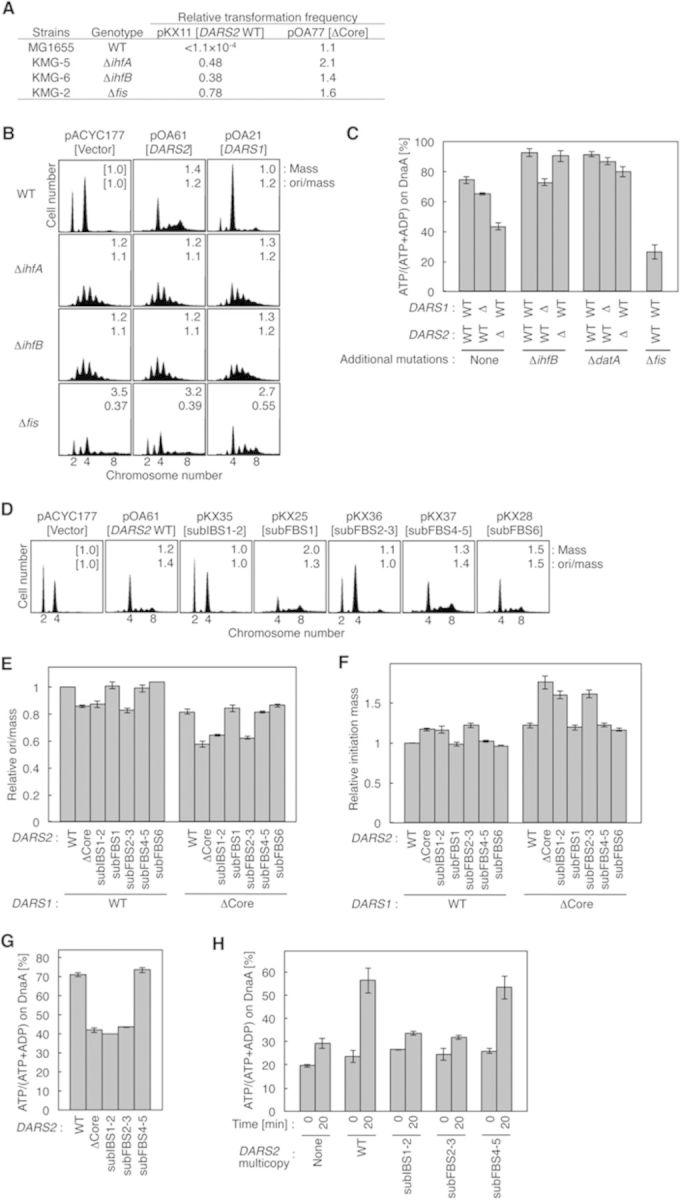
In vivo roles for IHF and Fis in DARS2 activation. (A) Transformation inhibition. Strains MG1655 (WT), KMG-5 (ΔihfA), KMG-6 (ΔihfB) and KMG-2 (Δfis) were transformed with pBR322 (vector), pKX11 (DARS2 WT) or pOA77 (DARS2ΔCore) and incubated at 37°C for 12 h on LB-ampicillin agar plates. Transformation efficiencies of pKX11 and pOA77, relative to those of pBR322, are shown. (B) Flow cytometry analysis using Δihf and Δfis cells. MG1655, KMG-2, KMG-5 and KMG-6 cells bearing pACYC177 (vector), pOA61 (DARS2) or pOA21 (DARS1) were grown at 37°C in supplemented M9-ampicillin medium, and then subjected to flow cytometry analysis as described for Figure 1C. The numbers inserted in the histograms indicate mean cell mass and ori/mass relative to that of MG1655 cells bearing pACYC177. (C) Cellular ATP–DnaA levels. The following strains were grown at 37°C; MK86 (WT), MIT47 (DARS1ΔCore), MIT86 (DARS2ΔCore), KX97 (ΔihfB), KX101 (ΔihfB DARS1ΔCore), KX90 (ΔihfB DARS2ΔCore), KX83 (ΔdatA), KX179 (ΔdatA DARS1ΔCore), KX102 (ΔdatA DARS2ΔCore) and KX29 (Δfis). Error bars represent SD from three independent experiments except for KX29. Two independent experiments were done for KX29, and both data and mean values are shown. (D) Flow cytometry analysis using pOA61 derivatives. MG1655 cells bearing pACYC177 (vector), pOA61 (DARS2 WT) or the indicated pOA61-derivative DARS2 mutant plasmid were analyzed by flow cytometry as described for Figure 1C. Ratios of mean cell mass and ori/mass are shown in the histograms. (E) The ori/mass ratios of the chromosomal DARS2 mutant cells. Cells of the MG1655 (DARS1, WT; DARS2, WT) strain and its derivatives bearing the indicated DARS2 mutations in the presence or absence of the DARS1ΔCore mutation were analyzed by flow cytometry. Ratios of two or four oriC in each strain were calculated. The ori/mass ratios are shown as relative values to those of MG1655, which was used as a standard. (F) The initiation mass of DARS2 mutant cells. The initiation masses relative to that of MG1655 (WT) were deduced from the flow cytometry data obtained above. (G) Cellular ATP–DnaA levels of IBS and FBS mutants. Cells of the KX41 (rnhA::Tn3 ΔoriC Δhda) (WT) strain and its derivatives bearing the indicated DARS2 mutations were analyzed as described for Figure 5C. Two independent experiments were done, and both data and mean values are shown. (H) DARS2-medicated ATP-DnaA regeneration activity in vivo. KA474 (dnaN59) cells bearing pACYC177 (None), pOA61 (WT) or the indicated pOA61-derivative DARS2 mutant plasmid were incubated in TG-ampicillin medium containing [32P]orthophosphate at 28°C until the A660 reached 0.2, transferred to 42°C in the presence of 150 μg/ml chloramphenicol (Time 0) and further incubated for 20 min, followed by immunoprecipitation and thin-layer chromatography. Two independent experiments were done for each assay, and both data and mean values are shown.
Next, we used flow cytometry to analyze the initiation in mutant cells bearing pACYC177-DARS2 (pOA61) or pACYC177-DARS1 (pOA21). Whereas MG1655 cells bearing the parental vector pACYC177 contained two or four origins, those bearing pOA61 predominantly contained four or eight origins, and some cells contained five or seven origins as a result of asynchronous extra initiations (Figure 5B). These cells were larger on average than cells bearing the parental vector, possibly because overinitiation impeded cell division (24). Introduction of pOA21 increased the level of four-origin cells relative to the level in cells bearing the parental vector, indicating that pOA21 moderately stimulated initiation (Figure 5B). These data are consistent with our previous findings (18).ΔihfA or ΔihfB cells bearing pACYC177 initiated replication asynchronously and predominantly contained three to five origins (Figure 5B), in agreement with a previous report (32) and with the idea that IHF plays positive and negative roles in initiation at oriC and datA. Introduction of pOA61 did not change the distribution of origin number in either ihf mutant (Figure 5B). By contrast, mutant cells bearing pOA21 predominantly contained four to six origins (Figure 5B); even the number of cells containing more than six origins was higher in mutant cells bearing pOA21 than in those bearing pACYC177. The reduction in the number of cells containing three origins could have been a consequence of the increase in the number of cells containing four and more origins. These results are consistent with the idea that DARS2 activity, but not DARS1 activity, depends on IHF.Δfis cells bearing pACYC177 also initiated replication asynchronously, and the ratio of origin number to mean cell mass (ori/mass) was reduced to 0.37 relative to the ratio in MG1655, as reported in previous studies (20,21). The ori/mass ratio is used to indicate the relative level of initiation activity (18,33). pOA61 did not affect initiation in Δfis cells, whereas pOA21 stimulated initiation (Figure 5B). These results support the idea that Fis, along with IHF, is specifically required for activation of DARS2 but not for the activation of DARS1. The doubling times of the cells used in these experiments were 32 ± 4 min, which was essentially the same for all strains.
DARS2 requires IHF and Fis for ATP–DnaA regeneration in vivo
Next, we analyzed ATP–DnaA regeneration activity in ΔihfB, ΔdatA and Δfis cells. To eliminate effects of RIDA (Figure 1A), we used strain MK86 [ΔoriC ΔrnhA Δhda] as a common background in these experiments. Δhda inhibits RIDA, resulting in an elevated level of ATP–DnaA and overinitiation at oriC, ultimately inhibiting cell growth (12,34). Introduction of ΔoriC to these cells represses the lethality of the Δhda mutation in the presence of ΔrnhA, a mutation that activates alternative DnaA-independent origins (35). DnaA was isolated by immunoprecipitation (IP) from lysates of 32P-labeled cells, and DnaA-bound nucleotides were eluted and quantitated by thin-layer chromatography, as described previously (11,12,18). In these experiments, nucleotides were labeled and DnaA-bound, labeled nucleotides were analyzed (Supplementary Figure S8). In MK86 cells, the ATP–DnaA level (i.e. the percentage of DnaA in the ATP-bound form) was high (72%), but this level decreased upon deletion of DARS1 or DARS2 (Figure 5C), as we previously demonstrated (11,12,18).
The ATP–DnaA levels in ΔihfB or ΔdatA cells were 90–95%, higher than in the parental MK86 cells (Figure 5C), which is consistent with our previous report showing that deletion of ihfB or datA elevates the ATP–DnaA level because of DDAH inactivation (17). Notably, the ATP–DnaA level in ΔihfB cells was not affected by ΔDARS2, but it was decreased to 73% by ΔDARS1 (Figure 5C). These results support the idea that DARS2, but not DARS1, requires IHF for ATP–DnaA regeneration in vivo. Furthermore, the ATP–DnaA level in ΔdatA cells was decreased to 80% by deletion of DARS2 and was decreased slightly by ΔDARS1 (Figure 5C), suggesting that IHF activates DARS2 in a datA-independent manner.
In addition, deletion of fis decreased the ATP–DnaA level to 26% (Figure 5C), supporting the idea that Fis, as well as IHF, is crucial for ATP–DnaA regeneration in vivo (see also Discussion). Growth rates and IP yields were indistinguishable among ΔihfB, ΔdatA and Δfis cells.
IBS1–2 and FBS2–3 are required for ATP–DnaA regeneration in vivo
To investigate the roles of IBS and FBS in DARS2 activation in vivo, we first performed flow cytometry analysis of MG1655 cells bearing pOA61 derivatives. As shown in Figure 5D, initiation was stimulated by pOA61, pKX25 [subFBS1], pKX37 [subFBS4–5] and pKX28 [subFBS6]. By contrast, initiation was not substantially stimulated or only slightly stimulated by pKX35 [subIBS1–2] or pKX36 [subFBS2–3], respectively, compared to initiation stimulated by pOA61. The slight stimulation seen with pKX36 might have been a consequence of the basal, non-specific binding activity of Fis (5,19,28).
Next, we used flow cytometry to analyze the ori/mass ratios in chromosomal DARS2 mutants (Figure 5E). DARS2ΔCore cells exhibited significantly reduced ori/mass ratios. DARS2 subIBS1–2 and subFBS2–3 cells had a reduced ori/mass ratio similar to that of the DARS2ΔCore, whereas the ratio was unaffected by subFBS1, subFBS4–5 or subFBS6. Similar results were obtained using cells with the DARS1ΔCore background (Figure 5E). Furthermore, we used these data to deduce the initiation mass, the cell size per origin at the time of replication initiation (Figure 5F). Initiation mass is an important cell cycle parameter that indicates the potential for replication initiation (36). For instance, a decrease in initiation activity results in an increase in the initiation mass. The data shown in Figure 5 are consistent with the ori/mass ratios (Figure 5E).
In addition, we analyzed ATP–DnaA levels in IBS and FBS mutants with the MK86 background (Figure 5G). Deletion of the DARS2 core in this background decreased the ATP–DnaA level from 72 to 42%, as previously demonstrated (18). Unlike DARS2 subFBS4–5, DARS2 subIBS1–2 or subFBS2–3 decreased the level to 40–45% (Figure 5G). Taken together, these results indicate that specific binding of IHF and Fis to IBS1–2 and FBS2–3 is required for DARS2 activation in vivo, in agreement with the in vitro data.
Based on these results, we investigated ATP–DnaA regeneration in vivo in DARS2 mutants (Figure 5H). We previously demonstrated that ATP–DnaA regeneration is stimulated in a pOA61-dependent manner under conditions where RIDA and de novo DnaA synthesis are inhibited (18). RIDA is a major pathway for the hydrolysis of ATP bound to DnaA. De novo synthesized DnaA will bind ATP, an abundant molecule in the cell, yielding ATP–DnaA. As described in our previous report (18), it is possible to inhibit RIDA and de novo protein synthesis including that of DnaA by introducing a temperature-sensitive mutation (dnaN59) into the dnaN gene encoding the clamp and adding chloramphenicol to the growth medium. The dnaN59 mutant growing at 28°C was transferred to 42°C and chloramphenicol was added to the medium. After incubation for 20 min, IP and thin-layer chromatography analysis indicated a higher ATP–DnaA level in cells bearing pOA61 than that in cells bearing pACYC177 (Figure 5H), in agreement with our previous results (18). A slight increase of the ATP–DnaA level in cells bearing pACYC177 after 20-min incubation might be caused by DARSs carried on the chromosome. Notably, the results obtained using cells bearing pKX35 [DARS2subIBS1–2] or pKX36 [DARS2subFBS2–3] were similar to those obtained using cells bearing pACYC177, and the results obtained using cells bearing pKX37 [DARS2subFBS4–5] were similar to those obtained using cells bearing pOA61. In addition to the data presented in Figure 5C, these results support the idea that specific binding of IHF and Fis to DARS2 promotes ATP–DnaA regeneration in vivo.
Cell cycle-coordinated binding of IHF to DARS2
To investigate whether IHF binding to DARS2 is coordinated with the replication cycle, we performed ChIP or chromatin affinity precipitation (ChAP) assays using the dnaC2 mutant, which is temperature sensitive for helicase loading and replication initiation, but not for the progression of established replisomes at 38°C. When dnaC2 cells grown at 30°C are shifted to 38°C for 90 min, chromosomes are completely replicated, but no new initiations take place. When the cells are returned to 30°C, synchronized initiations occur within 5–10 min after the temperature shift, followed by a second round of initiation at 25–35 min after the shift (11,17,37).
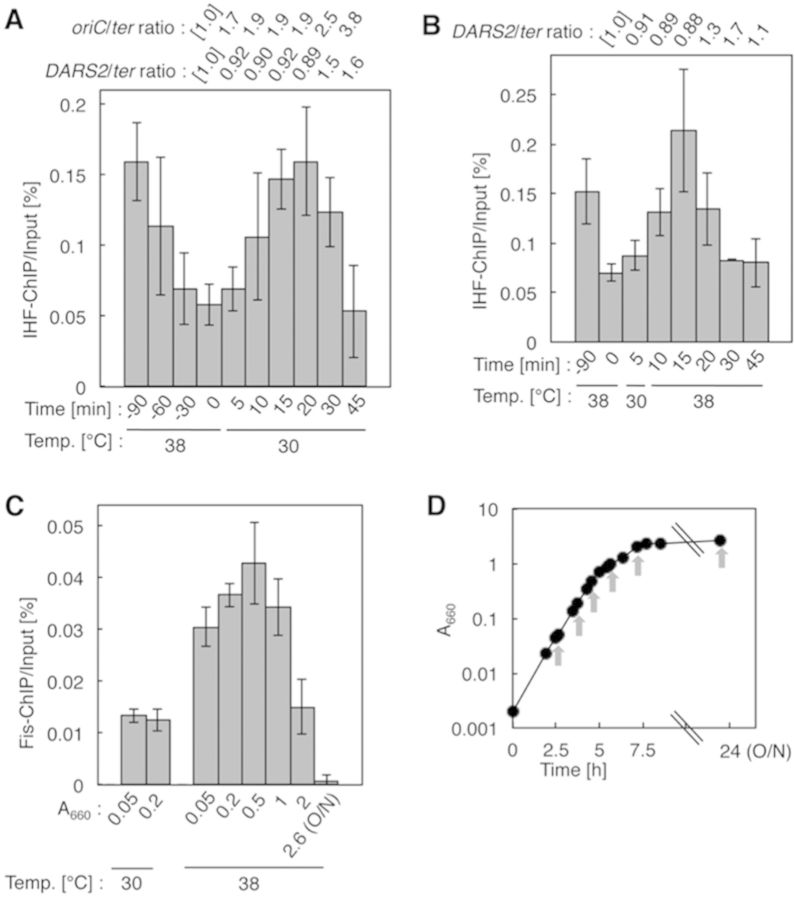
In synchronized cell cultures, oriC/ter (replication terminal locus) ratios suggested that the first and second initiations occurred respectively within 5 and 30–45 min after the shift to 30°C (Figure 6A). Fluctuations in IHF binding to oriC were also consistently observed (Supplementary Figure S9A), as reported previously (17,38). IHF bound to DARS2 was dissociated before the first initiation, bound again to DARS2 between the first and second rounds of initiation and then dissociated again (Figure 6A). ChAP experiments using His-tagged IHF yielded similar results (Supplementary Figure S9B and C). These changes are consistent with the changes in ATP–DnaA levels we observed previously (11): the ATP–DnaA level reached its maximum level before the first round of initiation, decreased to the basal level after initiation and then increased again before the second round of initiation. It is logical to assume that IHF dissociates from DARS2 before the first round of initiation, because the ATP–DnaA level is maximal at this time. IHF binding to DARS2 after the first initiation occurs at an appropriate time to increase the ATP–DnaA level for the second initiation. DARS2/ter ratios, which indicate the time of duplication of DARS2 during replication, suggested that duplication of DARS2 is not required for IHF binding during this period (Figure 6A). The basal level of IHF–DARS2 binding might be a consequence of the non-specific, unstable binding of IHF in the DARS2 region.
Figure 6.
Cell cycle- and growth phase-coordinated regulation of DARS2. (A, B) IHF-ChIP of DARS2. KYA018 (dnaC2) cells growing at 30°C were transferred to 38°C and incubated for 90 min. The cells were then transferred to 30°C (Time 0) and incubated for 5 min, followed by further incubation at 30°C (A) or 38°C (B). The relative DARS2 levels before (Input) and after (IHF-ChIP) IP using anti-IHF antiserum were determined using real-time qPCR, yielding the ChIP/Input ratio [expressed as%]. Error bars represent SD from at least three independent experiments. In addition, cellular levels of oriC, DARS2 and ter in the Input samples were quantified, and the relative ratios of oriC/ter and DARS2/ter are expressed relative to the ratio at 0 min (defined as 1). (C, D) Fis-ChIP of DARS2. MG1655 cells were incubated at 30 or 38°C in supplemented M9 medium. At the indicated A660, samples were withdrawn for determination of the Fis-ChIP/Input ratio [expressed as%] (C). Error bars represent SD from at least three independent experiments. O/N, overnight incubation. Growth curve at 38°C (D). Arrows: sampling times.
Similar results were obtained in synchronized cultures in which the second round of initiation was inhibited by a temperature up-shift (Figure 6B) or addition of rifampicin (Supplementary Figure S9D). These results indicate that IHF binding to and dissociation from DARS2 is independent of replication initiation as well as transcription, and are consistent with the idea that the IHF–DARS2 interaction is coordinated with the cell cycle. These features of IHF interaction at DARS2 are similar to those observed at datA (17) (see Discussion).
Growth phase-dependent binding of Fis to DARS2
We performed similar cell-cycle analyses of Fis binding. Unlike IHF-DARS2 binding, Fis-DARS2 binding was essentially sustained throughout the replication cycle (Supplementary Figure S9E), although a slight loss in binding might have occurred temporarily after initiation (i.e. at 45 min of Supplementary Figure S9E). When the cultures were incubated at 30°C for a limited time, Fis–DARS2 binding temporarily decreased. These changes cannot be interpreted as being linked to the replication cycle, however, because Fis–DARS2 binding was considerably lower in cells incubated at 30°C (Figure 6C). ChIP experiments, which used 3 M urea-denaturing conditions after the crosslinking step, yielded consistent results (Supplementary Figure S9F).
Expression of Fis depends on growth rate and growth phase: it is abundant (10 000–50 000 molecules/cell) in early exponential phase, but the levels decrease to <100 molecules/cell from late exponential phase to stationary phase (5,39). Fis–DARS2 binding showed similar kinetics: binding was high in exponential phase and significantly lower in late exponential phase to stationary phase (Figure 6C and D). These results support the idea that Fis is the determinant of growth phase-coordinated activation of DARS2. By contrast, IHF is abundant throughout all growth phases (5), and we observed that its binding to DARS2 was basically independent of growth phase (Supplementary Figure S9G).
Evolutionary conservation of IBS1–2 and FBS2–3
Whereas IHF is broadly conserved across all eubacterial species, Fis is conserved specifically among γ-proteobacterial species related to E. coli (40), as is the DARS2 core sequence (18). The IBS1–2 and FBS2–3 sequences are highly conserved in representative bacterial species in which Fis and IHF homologs are conserved and in which the DARS2 core has a genomic position similar to that in E. coli (Supplementary Figure S10A). These observations suggest that the regulatory mechanism for DARS2 is shared among these species, including some pathogens.
DISCUSSION
The primary regulatory system responsible for the temporal increase in the ATP–DnaA level during the cell cycle has remained elusive. However, in this study, we revealed that simultaneous binding of IHF and Fis to DARS2 activates ATP–DnaA production (Figure 7A). In contrast to initiation at oriC, HU cannot substitute for IHF in DARS2 activation, indicating a crucial role for IHF in this regulation. Cell-cycle and growth-phase analyses revealed that IHF binds to specific sites within DARS2 in a cell cycle-coordinated manner that is independent of DNA replication, whereas Fis binds specifically in the exponential phase. In addition, DARS2–mediated ADP dissociation from DnaA was more efficient than ATP dissociation, which is consistent with the more efficient formation of higher-order complexes by ADP–DnaA and DARS2. Taken together, these results demonstrate that activation of DARS2 is regulated in coordination with the cell cycle and growth phase via temporal changes in the binding of IHF and Fis.
Figure 7.
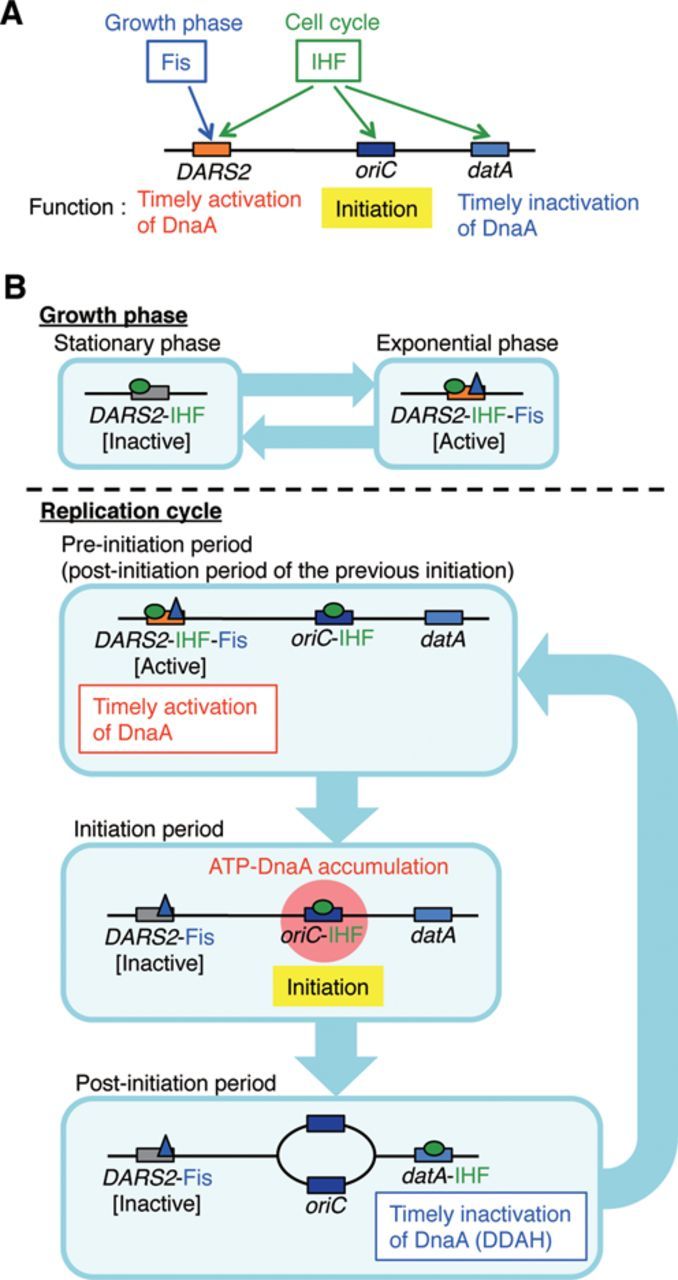
Regulated activation of DARS2 by IHF and Fis in the DnaA cycle. (A) A schematic view of the roles for IHF and Fis in regulatory systems for DARS2, oriC and datA. See text for details. (B) A model of the timely regulation of DARS2 by IHF and Fis. In exponential phase cells, Fis binds to DARS2. In the pre-initiation stage, the time when the ADP–DnaA level is high, IHF binds to oriC and DARS2 in a cell cycle-coordinated manner, and the DARS2–IHF–Fis complex increases the level of ATP–DnaA. Next, the elevated ATP–DnaA level allows the IHF-bound oriC to initiate replication, at which point IHF dissociates from DARS2. In the post-initiation stage, IHF binds to datA, activating DDAH. IHF then dissociates from datA in a cell cycle-coordinated manner, and re-binds to DARS2 and oriC for the next round of initiation. For simplicity, only single molecules of IHF and Fis are shown to bind to DARS2 in this figure. In growing cells, the pre-initiation stage overlaps the post-initiation stage of the previous round of initiation.
IHF plays multiple crucial roles in the regulation of initiation. First, IHF binds to oriC at the pre-initiation stage (Figure 7A and B), stimulating DnaA assembly on oriC and unwinding of the duplex unwinding element (8,9,25,38). The requirement for IHF in replication was first demonstrated using the plasmids pSC101 and R6K, whose origins bear IBS (41,42). Second, IHF binds to datA, and this interaction is required for activation of DDAH (Figure 7A and B). In replication cycle-synchronized cultures, IHF–datA binding occurs only for a short time at a post-initiation stage: levels of binding increase immediately after initiation and peak 15 min later, followed by rapid dissociation (17). Third, as demonstrated in this study, IHF binds to DARS2, activating ATP–DnaA regeneration (Figure 7A and B). At 30°C, IHF–DARS2 binding began to increase 5 min after initiation, peaked within 20 min and was sustained for an additional 10 min, followed by dissociation; binding levels returned to the basal level at 45 min. This fluctuation is consistent with the observation that under the same conditions, the second initiation occurs 30–45 min after the first, as well as with the fact that the cellular ATP–DnaA level peaks around the time of replication initiation. Thus, IHF binding to DARS2, oriC and datA is regulated differently, resulting in cell cycle-coordinated regulation of initiation (Figure 7A and B). In other words, IHF plays both positive and negative roles in initiation that are regulated temporally and by the locations of DNA binding during the cell cycle.
The dynamic interactions of IHF–DARS2 are independent of replication initiation: these events occur in a timely manner even under conditions in which the second round of initiation is inhibited (Figure 6B and Supplementary Figure S9D). This observation supports the idea that a mechanism regulating dynamic IHF–DARS2 interactions is coupled with a cell cycle-regulatory pathway that is epistatic to replication regulation. Similarly, temporal regulation of the IHF–datA interaction also would be regulated in a cell cycle-coupled manner (17). A recent study, which applied genome conformation capture analysis to the E. coli genome, suggested that a DARS2 region spatially interacts with oriC only in growing cells (43), consistent with the existence of a possible mechanistic link in the timely delivery of IHF and ATP–DnaA between oriC and DARS2. In addition, a specific factor that dissociates IHF–DARS2 complexes, if present, might be crucial for cell cycle-coordinated replication regulation. Recently, we found a crude protein extract that contains IHF–DARS2 dissociating activity, but not Fis–DARS2 dissociating activity.
The molecular mechanism responsible for initiation stimulation by Fis has remained obscure (20,21). A previous report suggests that Fis binding to oriC, even if it occurs, does not affect replication initiation in vivo (44). The present study provides a reasonable solution to the long standing question of how Fis contributes to initiation; the present solution is that Fis actives DARS2 in the presence of IHF, stimulating ATP–DnaA production and initiation. Fis binds to DARS2 during exponential phase, but not late stationary phase (Figure 6C), indicating that Fis determines the growth-phase specificity of DARS2 activity. This might be related to the dramatic fluctuations in the cellular level of Fis, which peaks early in exponential phase and disappears in late stationary phase. Also, depending on the growth rate, the cellular level of Fis might regulate DARS2 activity. Deletion of fis causes a severe reduction in the cellular ATP–DnaA level (Figure 5C). In addition to providing a mechanism for Fis-dependent stimulation of initiation, these data suggest the existence of a Fis-dependent third DARS, which is yet to be unidentified.
Specific residues for inter-DnaA interactions, as well as those for DNA binding, are crucial for DARS2–mediated ADP dissociation. The basic AID-1 (R227) and hydrophobic AID-2 (L290) residues are important for ADP dissociation (Figure 4A). As in the case of initiation complexes (22), these residues might stabilize inter-DnaA interactions, leading to construction of functional higher-order complexes. In support to this idea, AID-2 stimulated DnaA oligomerization on DARS2 (Figure 4B). The requirements for other DnaA AAA+ residues for DARS2 are similar to those for DARS1, suggesting that the mechanistic principles are conserved between the two loci.
In addition, the requirement for a DNA region that binds to IHF and Fis during DARS2 activation is consistent with the fact that in DARS1, a specific DNA region flanking the core DnaA boxes stimulates ADP dissociation from DnaA (18). In the case for DARS2, the DNA conformational change induced by binding of IHF and Fis might be crucial for the interaction of this region with the DnaA complex formed at core DnaA boxes (Supplementary Figure S10B). Thus, it is plausible that, as in the case of DARS1 complexes (18), IHF-Fis-bound DARS2 causes changes in inter-DnaA interactions in the DnaA complex that lead to the conformational changes in ADP–DnaA protomers that induce ADP dissociation. The resultant apo-DnaA would bind ATP after its release from DARS2, yielding initiation-competent ATP–DnaA (Figure 2). ATP–DnaA is less active for higher complex formation on DARS2 than ADP–DnaA (Figure 4), which might allow preferential binding of ADP–DnaA to DARS2, enhancing cyclic reactivation of DnaA (Supplementary Figure S10B).
In eukaryotes, ORC (origin recognition complex) is regulated by its nucleotide-bound state, and its major subunits exhibit structural similarity to the DnaA AAA+ domain (45). Eukaryotic HMG (high mobility group) proteins are thought to play roles similar to those of the bacterial nucleoid-associated proteins, i.e. to promote DNA conformational changes (5). Yeast NHP6-A, a member of the HMG family, complements defects in nucleoid compaction and in λ-excision caused by the lack of nucleoid-associated proteins in E. coli (46). Although sequences homologous to DARS have not been reported in eukaryotic cells, this does not exclude the possibly that nucleotide-bound states of ORC or eukaryotic AAA+ proteins are regulated by HMG proteins and DARS2-like sequences in these cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Yuki Ohyama, Dr Mika Yoshimura and NBRP (NIG, Japan) for participating in an initial search for DARS2 activating factors, for generous gifts of pSTV28-derived plasmids and for providing E. coli strains, respectively.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as Joint First Authors.
Present address: Kazuyuki Fujimitsu, Cell Cycle Control Group, UCL Cancer Institute, University College London, London WC1E 6BT, UK.
FUNDING
JSPS and MEXT, Japan [KAKENHI, 26291004, 22370064, 19770150, 11J03114]. Funding for open access charge: Ministry of Education, Culture, Sports, Science and Technology, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaguni J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011;15:606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katayama T., Ozaki S., Keyamura K., Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 3.Leonard A.C., Grimwade J.E. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillon S.C., Dorman C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 6.Aeling K.A., Opel M.L., Steffen N.R., Tretyachenko-Ladokhina V., Hatfield G.W., Lathrop R.H., Senear D.F. Indirect recognition in sequence-specific DNA binding by Escherichia coli integration host factor: the role of DNA deformation energy. J. Biol. Chem. 2006;281:39236–39248. doi: 10.1074/jbc.M606363200. [DOI] [PubMed] [Google Scholar]
- 7.Kawakami H., Keyamura K., Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 8.McGarry K.C., Ryan V.T., Grimwade J.E., Leonard A.C. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl Acad. Sci. U.S.A. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki S., Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40:1648–1665. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell M., Langston L., Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013;5:a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato J., Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida S., Fujimitsu K., Sekimizu K., Ohmura T., Ueda T., Katayama T. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J. Biol. Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- 14.Nozaki S., Yamada Y., Ogawa T. Initiator titration complex formed at datA with the aid of IHF regulates replication timing in Escherichia coli. Genes Cells. 2009;14:329–341. doi: 10.1111/j.1365-2443.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 15.Kitagawa R., Ozaki T., Moriya S., Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skarstad K., Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb. Perspect. Biol. 2013;5:a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasho K., Katayama T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl Acad. Sci. U.S.A. 2013;110:936–941. doi: 10.1073/pnas.1212070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujimitsu K., Senriuchi T., Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23:1221–1233. doi: 10.1101/gad.1775809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stella S., Cascio D., Johnson R.C. The shape of the DNA minor groove directs binding by the DNA-bending protein Fis. Genes Dev. 2010;24:814–826. doi: 10.1101/gad.1900610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filutowicz M., Ross W., Wild J., Gourse R.L. Involvement of Fis protein in replication of the Escherichia coli chromosome. J. Bacteriol. 1992;174:398–407. doi: 10.1128/jb.174.2.398-407.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flåtten I., Skarstad K. The Fis protein has a stimulating role in initiation of replication in Escherichia coli in vivo. PLoS One. 2013;8:e83562. doi: 10.1371/journal.pone.0083562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozaki S., Noguchi Y., Hayashi Y., Katayama T. Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 2012;287:37458–37471. doi: 10.1074/jbc.M112.372052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009;284:25038–25050. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama T., Takata M., Sekimizu K. CedA is a novel Escherichia coli protein that activates the cell division inhibited by chromosomal DNA over-replication. Mol. Microbiol. 1997;26:687–697. doi: 10.1046/j.1365-2958.1997.5941967.x. [DOI] [PubMed] [Google Scholar]
- 25.Hwang D.S., Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 26.Nakamura K., Katayama T. Novel essential residues of Hda for interaction with DnaA in the regulatory inactivation of DnaA: unique roles for Hda AAA Box VI and VII motifs. Mol. Microbiol. 2010;76:302–317. doi: 10.1111/j.1365-2958.2010.07074.x. [DOI] [PubMed] [Google Scholar]
- 27.Margulies C., Kaguni J.M. The FIS protein fails to block the binding of DnaA protein to oriC, the Escherichia coli chromosomal origin. Nucleic Acids Res. 1998;26:5170–5175. doi: 10.1093/nar/26.22.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho B., Knight E.M., Barrett C.L., Palsson B.Ø. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Shigeyuki Y. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton M.D., Kaguni J.M. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 1997;272:23017–23024. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami H., Ozaki S., Suzuki S., Nakamura K., Senriuchi T., Su'etsugu M., Fujimitsu K., Katayama T. The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 2006;62:1310–1324. doi: 10.1111/j.1365-2958.2006.05450.x. [DOI] [PubMed] [Google Scholar]
- 32.Von Freiesleben U., Rasmussen K.V., Atlung T., Hansen F.G. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 2000;37:1087–1093. doi: 10.1046/j.1365-2958.2000.02060.x. [DOI] [PubMed] [Google Scholar]
- 33.Skarstad K., Løbner-olesen A., Atlung T., von Meyenburg K., Boye E. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol. Gen. Genet. 1989;218:50–56. doi: 10.1007/BF00330564. [DOI] [PubMed] [Google Scholar]
- 34.Fujimitsu K., Su'etsugu M., Yamaguchi Y., Mazda K., Fu N., Kawakami H., Katayama T. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J. Bacteriol. 2008;190:5368–5381. doi: 10.1128/JB.00044-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wold S., Skarstad K., Steen B., Stokke T., Boye E. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 1994;13:2097–2102. doi: 10.1002/j.1460-2075.1994.tb06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Withers H.L., Bernander R. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 1998;180:1624–1631. doi: 10.1128/jb.180.7.1624-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cassler M.R., Grimwade J.E., Leonard A.C. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14:5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J., Yoshimura S.H., Hizume K., Ohniwa R.L., Ishihama A., Takeyasu K. Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res. 2004;32:1982–1992. doi: 10.1093/nar/gkh512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filutowicz M., Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K γ origin and is essential for replication. Nucleic Acids Res. 1988;16:3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamas P., Burger A., Churchward G., Caro L., Galas D., Chandler M. Replication of pSC101: effects of mutations in the E. coli DNA binding protein IHF. Mol. Gen. Genet. 1986;204:85–89. doi: 10.1007/BF00330192. [DOI] [PubMed] [Google Scholar]
- 43.Cagliero C., Grand R.S., Jones M.B., Jin D.J., O'Sullivan J.M. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 2013;41:6058–6071. doi: 10.1093/nar/gkt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weigel C., Messer W., Preiss S., Welzeck M., Morigen, Boye E. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001;40:498–507. doi: 10.1046/j.1365-2958.2001.02409.x. [DOI] [PubMed] [Google Scholar]
- 45.Kawakami H., Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem. Cell Biol. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- 46.Paull T.T., Johnson R.C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.