Abstract
Engineered transcription activator-like effectors, or TALEs, have emerged as a new class of designer DNA-binding proteins. Their DNA recognition sites can be specified with great flexibility. When fused to appropriate transcriptional regulatory domains, they can serve as designer transcription factors, modulating the activity of targeted promoters. We created tet operator (tetO)-specific TALEs (tetTALEs), with an identical DNA-binding site as the Tet repressor (TetR) and the TetR-based transcription factors that are extensively used in eukaryotic transcriptional control systems. Different constellations of tetTALEs and tetO modified chromosomal transcription units were analyzed for their efficacy in mammalian cells. We find that tetTALE-silencers can entirely abrogate expression from the strong human EF1α promoter when binding upstream of the transcriptional control sequence. Remarkably, the DNA-binding domain of tetTALE alone can effectively counteract trans-activation mediated by the potent tettrans-activator and also directly interfere with RNA polymerase II transcription initiation from the strong CMV promoter. Our results demonstrate that TALEs can act as highly versatile tools in genetic engineering, serving as trans-activators, trans-silencers and also competitive repressors.
INTRODUCTION
The strength of mammalian promoters is largely determined by the sequence-specific DNA binding of transcription factors (TFs) in their vicinity (1). Changes in the occupancy of transcription factor binding sites (or response elements, RE) as well as the nature of the binding factors result in different transcription initiation rates. The complexity of early steps of gene expression control in mammals is largely due to the combinatorial action of TFs. Essentially all mammalian promoters are under the control of several different TFs and individual transcription factors control multiple promoters in a context-dependent manner. On the cellular level, this is one of the underlying principles enabling organisms to respond precisely to diverse stimuli and conditions (e.g. to physiological cues or during the execution of developmental programs) (2).
This hallmark of natural, highly adaptive gene regulatory networks substantially complicates the use of endogenous TFs in the engineering of transcriptional control systems. Such artificial expression systems are often employed for gene function analysis, in building artificial genetic control circuits or in biotechnology. In any of these applications, the use of authentic mammalian TFs will result in a lack of monospecificity, making the cell's response pleiotropic (3). This limitation has been partially overcome by designing TFs that are not endogenous to the host cell. Such engineered TFs are often based on prokaryotic or yeast DNA-binding domains and the corresponding REs. Examples are TetR (4) or Gal4 (5) based expression control systems. The application of such engineered TFs has been remarkably successful in the control of transgenes driven by synthetic promoters containing the respective target. However, their utility for direct transcriptional control of endogenous genes is limited. Any such approach requires the engineering of gene regulatory elements in its original chromosomal location. Sophisticated experimental approaches have been realized, but are demanding and restricted to cellular systems amenable to homologous recombination (6).DNA-binding proteins with (almost) freely programmable sequence specificity may overcome this limitation. Pioneering work in designer zinc finger TFs showed the potential of such approaches in controlling transcription of selected target genes (7). However, the design and functional testing of such zinc finger effectors remains demanding. The discovery of a novel class of TFs, so-called transcription activator-like effectors (TALEs), offers new prospects for designer DNA-binding proteins and the way they might be brought to routine application (8–10). TALEs are virulence determinants of plant pathogenic bacteria. Upon secretion in the plant cell, they bind to target gene promoters, thereby manipulating transcription in the host cell (11). They are characterized by tandemly arranged amino acid repeats, which are highly conserved aside from two amino acids at position 12 and 13, the so-called repeat variable diresidues, RVD (12). The modularity of the repeats opens the possibility to create predictable DNA-binding specificities in the custom design of genetic engineering tools. This includes the ability to direct such engineered TALEs to unmodified endogenous promoter sequences and impose transcriptional control via engineered effector domains like trans-activating or trans-repressing domains attached to the TALE (8). Transcriptional repression in eukaryotes can occur via multiple mechanisms (13). In humans, zinc finger proteins with a KRAB repression domain constitute a large family of transcriptional regulators. Their mode of action includes the recruitment of co-repressors and remodeling of chromatin (14).TALENs, TALE-derived site-specific endonucleases, have been widely used as genome engineering tools (15). Custom-engineered TAL effectors that are capable of regulating genes in mammalian cells, thus overriding the endogenous transcription program, have also been reported (8,10,16–19). However, regulation factors were often modest and few of the studies published so far gave conclusive evidence regarding the quantitative aspects of TALE-mediated transcriptional regulation in comparison to other heterologous regulatory systems.
Among the various inducible transcriptional control systems for mammalian cells, those controlled by tetracyclines are most commonly used. Both activating (4,20,21) and repressive systems (22–24) have been established. In consequence, a wide repertoire of extensively pre-characterized tools is available, like plasmid constructs, recombinant viruses and stable cell lines. This makes tetracycline-controlled promoters a preferred target for the analysis of synthetic TFs based on recent advances of freely programmable DNA-binding proteins, which, asides from TALEs, also include guide RNA directed dCas9 transcriptional regulators (25–27).
We chose to design TALEs targeting the 19 base pair prokaryotic tet operator, tetO, that is common to all engineered tetracycline-controlled transcription systems used in eukaryotes. We analyzed the efficacy of such tetTALEs, both in transient and stable transfection experiments. When fused to a trans-acting silencing domain (SD), TALEs can work as efficient long-range repressors. Moreover, the TALE DNA-binding domain alone can repress transcription by efficiently competing with activating TFs sharing the same binding site.
This study demonstrates the favorable DNA-binding properties of engineered TALEs for modulating chromosomal gene activities at will. tetTALEs themselves, lacking inherent inducibility, are only predestined for use in niche applications like the building of artificial transcriptional control circuits. However, they are powerful tools to address the design, optimization and mechanistic characterization of this promising class of programmable DNA-binding proteins for future applications in molecular biology and genetic engineering with unprecedented flexibility.
MATERIALS AND METHODS
Construction of TAL effectors
TAL effectors targeting the 19 bp tet operator sequence (tetO2) referred to as tetTALEs were assembled using the Golden Gate TALEN and TAL Effector kit obtained from Addgene (28). For the recognition of the nucleotides A, T, G and C, the repeats NI, NG, NK and HD were used, respectively. The final assembly construct contained the TAL effector backbone as described in Cermak et al. followed by a nuclear localization signal (NLS) region, a KRAB silencing (SD) or VP64 activation domain (AD), a T2A site and a fluorescent protein marker (see also Figure 1A). Where indicated in the text, a HA-Tag was introduced at the N-terminus for immunoblot detection. The tag had no influence on the transcriptional activity of the tetTALE-AD (data not shown). Two tetTALEs without effector domain were designed, only containing the tetO-specific TALE backbone or in direct fusion with a second fluorescent protein marker. As an unspecific control, a TALE targeting the human FoxP3 promoter (FoxP3TALE) was created. It targets the following sequence: ATGAGAACCCCCCCCCACCCCGTGAT (chrX:49,119,959-49,122,658) and was fused to a KRAB SD.
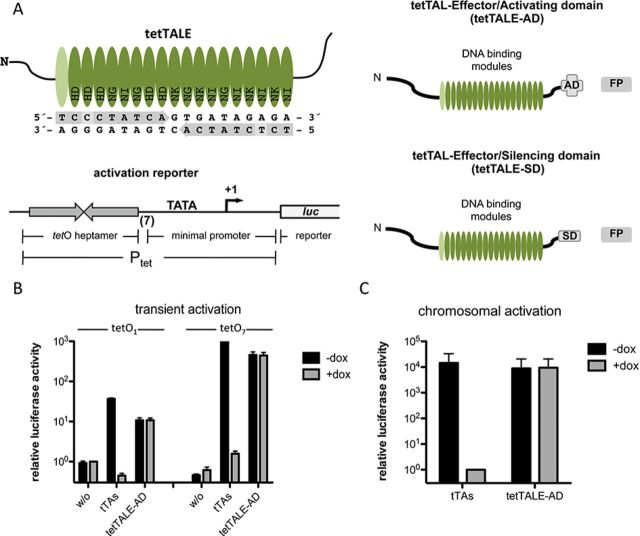
Figure 1.
Activation capacity of tetTALE-AD compared to tet trans-activator (tTAs). (A) Design of the DNA-binding domain of tetTALE with the target sequence and the used RVDs. Light gray boxes mark positions deviating from the operator symmetry (upper left panel). The right panel shows a scheme of tetTALE-AD, tetTALE-SD and the activation reporter construct. tetTALE was constructed to target the 19 bp tetO sequence. The C-terminus encompasses an NLS and a VP64 AD or a KRAB SD. The fluorescence reporter (FP) is linked via a T2A site. The activation reporter consists of a firefly luciferase gene driven by a tet-responsive promotor carrying either one or seven copies of the tet operator (lower left panel). (B) HeLa cells were transfected with an activation reporter construct carrying single or heptameric tetO sequences and either tTAs or tetTALE-AD expression constructs. A Renilla luciferase expression construct was included for internal standardization. Ptet activity in the presence of doxycycline was set to 1. Shown are mean values of three independent experiments with standard deviation. (C) X1/6 cells carrying the reporter with a heptameric tetO sequence stably integrated in the chromosome were transfected with either tTAs or tetTALE-AD expression constructs. A Renilla luciferase expression construct was included for internal standardization. Ptet activity in the presence of doxycycline was set to 1. Shown are mean values of three independent experiments with standard deviation.
For stable cell line generation, TALE expression units were chromosomally integrated via transposition. pT2-based sleeping beauty transposons (29), containing the human elongation factor 1 alpha promoter (EF1α) and a puromycin selection cassette, were used in conjunction with SB100 hyperactive transposase (30). Alternatively, pPB-based piggyBac transposons (31), containing EF1α promoter and a blasticidin selection cassette, were used in conjunction with optimized mPB transposase (32). Details of the TALE expression constructs are available upon request.
Reporter constructs
To test the activation capacity of tetTALE-AD, the pUHC13-3 reporter carrying a tet-responsive promoter (Ptet) upstream of a luciferase gene was used (4). Variants of this reporter with mutations in the tetO sequence have been described (33). Transient repression tests were performed with pUHC13-13, where a cytomegalovirus (CMV) enhancer is located upstream of the tet-responsive promoter of pUHC13-3, rendering it constantly active (22). Expression of the luciferase reporter can be downregulated by binding of tetR-SD or tetTALE-SD to the operator sequences. A tet operator-modified CMV promoter has been isolated from pCDNA4/TO (Invitrogen) and cloned upstream of a transposon-based EGFP reporter. Co-transfections were always performed at a ratio of reporter: tetTALE of 1:1 (w/w) unless stated otherwise.
Cell culture
HAFTL cells (34) used in the stable silencing experiment carry multiple tetO sequences in a TRE context upstream of the EF1α promoter, driving the expression of the destabilized ZsGreen reporter (M. Hofstätter and M. G., unpublished data). They were cultivated in RMPI 1640 medium supplemented with 10% heat inactivated fetal calf serum (FCS) and 50 mM β-mercaptoethanol. Selection of stably transfected pools and clones was achieved by addition of 20 μg/ml blasticidin. Maintenance of HeLa (ATCC:CCL-2) and HeLa-derived stable cell lines was in Dulbecco's Modified Eagle's medium with 10% FCS. HeLa X1/5 cells are stably transfected with pUHC13-3, carrying a tet-responsive promoter upstream of a luciferase reporter as well as an expression cassette for the tet trans-activator tTA (4). Thus, these cells express luciferase in the absence of doxycyline. The expression is abrogated by the addition of doxycycline. X1/6 cells only carry the tet-responsive luciferase reporter, but no functional trans-activator gene any more (21). They were used to determine the activation potential of TAL effectors on chromosomal targets. Both X1/5 and X1/6 cells were cultivated with 50–100 ng/ml doxycycline where indicated. Selection was achieved by addition of 5 μg/ml of blasticidin or 2 μg/ml of puromycin. CHO K1 Tet-ON Advanced and HEK 293 Tet-On Advanced cells from Clontech carry an expression cassette for the reverse tet trans-activator (rtTA) and were cultivated with 1 μg/ml doxycycline where indicated. For all cell lines, clones were obtained by limited dilution. All cells were maintained in medium supplemented with 200 mM of l-glutamine and 100 units/ml of penicillin/streptomycin at 37°C and 5% CO2 in a humidified incubator.
Transfection and transposition
HAFTL cells were transfected by electroporation. In total, 1*107 cells were resuspended in 400 μl of antibiotic free medium and electroporated in 0.4 cm cuvettes with the Gene Pulser Xcell from Biorad with 300V and 950 μF using a single exponentially decaying pulse. Ten micrograms of plasmid DNA plus 30 μg of salmon sperm DNA were used for each transfection. CHO, HEK 293, HeLa and HeLa-derived cells were transfected in six well plates with polyethylenimine (PEI) (35). Four microliter of 7.5 mM PEI solution was mixed with 50 μl of 150 mM NaCl and then added to a total of 1 μg of plasmid DNA in 50 μl of 150 mM NaCl. After vortexing, the mixture was incubated for 10 min at room temperature and then added to 50% confluent cells in a six-well plate. For generation of stable cell lines by transposition, a ratio of transposon to transposase of 4:1 (w/w) was used.
Luciferase assay
In transient reporter assays for firefly luciferase activity, cells were lysed in phosphate buffered saline (PBS)/0.25% NP40 48 h after transfection. Ten microliter of lysate was added to 90 μl reaction buffer (25 mM glycylglycine, 15 mM MgSO4). Relative light units were measured with the LB940 Mithras from Berthold 0.5 s after the addition of the substrate buffer (25 mM glycylglycine, 15 mM MgSO4, 5 mM ATP, 200 μM luciferin). Results of transient experiments were normalized to Renilla luciferase expression measured according to standard procedures. Results of double stable experiments were normalized to protein content determined by BCA assay (Pierce).
Flow cytometry
For transient transfection experiments, FACS analysis was performed 48 h post transfection. Adherent cells were trypsinized and resuspended in FACS buffer (PBS, 2% bovine serum albumin, ethylenediaminetetraacetic acid (EDTA)), whereas suspension cells were centrifuged at 300 g for 5 min and then resuspended in the same buffer. FACS analysis was performed on BD Accuri Cytometer for EGFP analysis or on BD LSRII flow cytometer for simultaneous detection of EGFP and mCherry. FlowJo was used for data analysis.
Immunoblotting
Cells were harvested either by trypsinization (adherent cells) or centrifugation (suspension cells) and washed twice with PBS. Afterwards, the cells were directly lysed in 2x loading buffer (100 mM Tris–HCl pH 6.8, 4% sodium dodecyl sulphate (SDS), 20% glycerol, 2% β-mercaptoethanol, 25 mM EDTA, 0.04% bromophenol blue) and sheared with a syringe. All samples were incubated for 10 min at 70°C before loading on a NuPAGE Novex 4–12% Bis-Tris gradient gel (Life Technologies). Mighty Small wet blotting system (Amersham Biosciences) was used to transfer proteins to a nitrocellulose membrane (Millipore). Transfer was for 1 h at 400 mA with constant cooling. After transfer, the blot was blocked for 1 h with Odyssey blocking buffer (Licor). For detection of HA-tagged proteins, a rabbit anti-HA antibody (Sigma) was used. As a reference β-actin was detected with a mouse anti-β-actin antibody (Sigma). Incubation of the blot with the first antibody was done at 4°C overnight in blocking reagent. After three washing steps with TBS-T, the blot was incubated for 1 h at room temperature with an anti-rabbit antibody coupled to the IRDye 800CW as well as an anti-mouse antibody coupled to IRDye 680 (both Licor), followed by two washes in TBS-T and one in TBS. Blot analysis was done on the Odyssey infrared imaging system (Licor) using the manufacturer's software.
RESULTS
Design and validation of tetTALE
We constructed a TAL effector designed to exactly target the 19 bp tetO2 operator sequence of the Tn10 tet operon. This operator sequence is an integral component of all major tetracycline-controlled gene expression systems used in mammalian cells (36), where it is recognized by either TetR itself or by the various TetR-based synthetic trans-activators or -silencers. Our design was facilitated by the presence of a 5’ T in the near-palindromic 19 bp tetO sequence, as functional TALE DNA-binding domains usually start with a N-terminal amino acid repeat recognizing thymidine (37). This tetTALE was either fused to an AD or a SD, linked to a fluorescence protein marker (Figure 1A).
The potency of artificial TF-like TALEs can be most easily addressed in transient trans-activation experiments. To this end, we performed co-transfections of a tet-responsive luciferase reporter containing multimerized tetO sequences (Figure 1A) together with expression vectors encoding either tetTALE-AD or tTAs, the tetracycline-controlled trans-activator (4). As shown in Figure 1B, the tetTALE-AD showed an activation of up to 450-fold, comparable to tTAs-mediated activation. As expected, activation by tetTALE-AD is independent of the Tet system inducer doxycycline. While significantly reduced in the foldness of activation, these principal observations extend to promoters controlled by a tetO monomer (Figure 1B).
The particular advantage of synthetic TALEs is their potential for modifying the expression of chromosomal transcription units. Thus, we analyzed the function of tetTALE in X1/6 cells (21) containing chromosomal copies of the luciferase reporter containing a heptameric tetO. These cells were transiently transfected with tetTALE-AD or tTAs expression vectors. Again, tetTALE-AD was almost as effective in transcriptional activation when compared to tTAs (Figure 1C).
TetTALE-AD was also used to address the specificity of TALE/target sequence interactions by comparing its activation capacity upon binding to wt tetO as shown in Figure 1B compared to analogous 2bp mismatch tet-responsive reporters (33). While such mismatches can almost entirely abrogate TALE binding (Supplementary Figure S1; tetO 6C), the effects are highly context dependent, shown by the rather strong residual activation of tetO 4C constructs.
TAL effectors are able to repress in trans
While most applications of the TALE technology published to date explored its use in activating cellular genes, this study focuses on engineered TALEs in downregulating gene activity, employing various potential modes of action. Initial transient co-transfection experiments demonstrated that silencing efficiency mediated by tetTALE-SD is similar to that of a TetR-based transcriptional silencer and previously described TALEs targeting tet-responsive promoters (25) when tested on a tetO-modified CMV promoter (Supplementary Figure S2) (22). TetTALE-SD was also compared to an analogous TALE construct targeting an unrelated sequence, demonstrating that the observed transcriptional downregulation is target-site dependent (Supplementary Figure S3).
To quantitatively address the silencing capacity of tetTALE-SD on a chromosomal target, we made use of a previously established cell line carrying the green fluorescent protein ZsGreen as a reporter. This gene is driven by a EF1α promoter fused to an upstream tet-responsive element (Figure 2A). This cell line was stably transfected with expression constructs coding for tetTALE-SD or tetTALE without SD, respectively. Both are capable of binding to the tetO target sites upstream of the EF1α promoter. ZsGreen reporter expression and TALE-linked mCherry expression were analyzed by fluorescence microscopy (Figure 2C) and FACS (Figure 2D). For tetTALE-SD expressing cells, pool analysis showed that the vast majority of cells are either positive for ZsGreen or, indicative for tetTALE-SD expression, positive for mCherry. Thus, promoter proximal recruitment of the transcriptional SD of tetTALE-SD completely abrogates the activity of the strong human EF1α promoter. Microscopic and FACS analysis revealed on the single cell level that only in tetTALE-SD+ cells (visualized by the mCherry signal) the ZsGreen signal is reduced (Figure 2C and D). This links directly the presence of tetTALE-SD itself to the observed repression. In tetTALE expressing cell pools, no reduction in ZsGreen signal is detected (Figure 2D), demonstrating that the observed effect is dependent on the SD. Furthermore, clonal analysis of the tetTALE-SD expressing cells shows a remarkable correlation between the tetTALE-SD coupled mCherry signal and the absence of ZsGreen expression. This conclusion was confirmed when directly addressing expression levels of tetTALE-SD by immunoblot analysis (Figure 2B see Supplementary Figure S4 for quantification). Notably, the clonal analysis showed that TALE-mediated silencing can be homogenous. The parental reporter cell line used in these experiments had been established via transposition, with an undetermined copy number of chromosomal integrates. Results obtained by single-copy lentiviral integration of the same reporter were virtually identical (data not shown).
Figure 2.
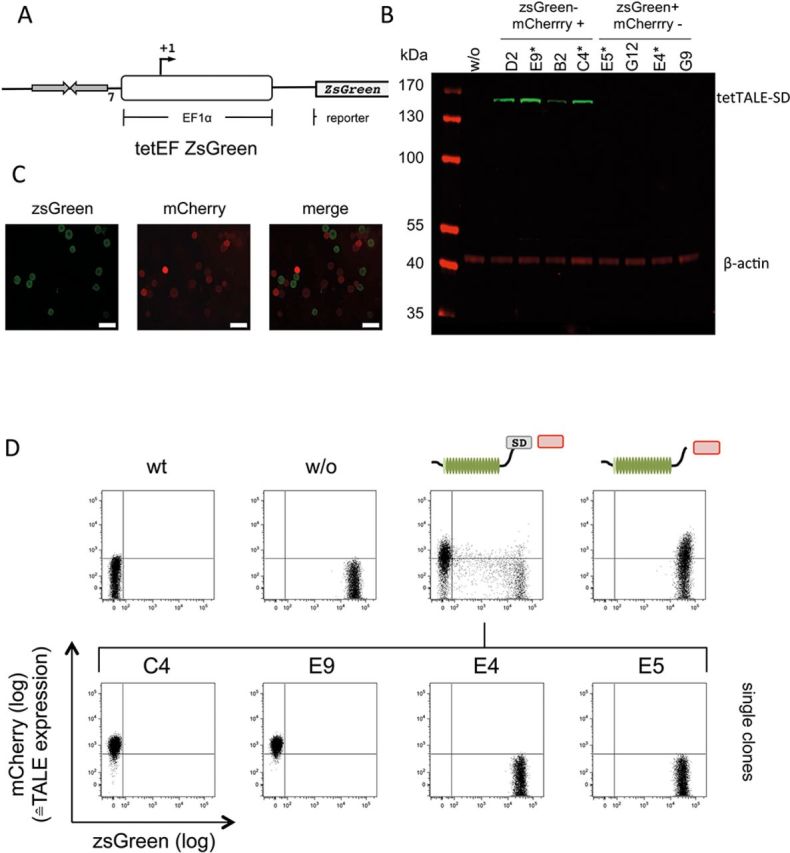
Repression in trans by tetTALE-SD. (A) Schematic picture of the reporter construct stably integrated in HAFTL cells to analyze the trans-repression capacity of tetTALE-SD on chromosomal targets (tetEF ZsGreen). The green fluorescent protein ZsGreen serves as reporter. (B) HAFTL tetEF ZsGreen cells before transfection (w/o) and single clones isolated from the tetTALE-SD+ pool were analyzed by immunoblotting for tetTALE-SD expression levels. β-actin levels served as a loading control. (C) Microscopic picture of HAFTL tetEF ZsGreen cells after stable transfection with tetTALE-SD: ZsGreen (left), mCherry (middle), merge (right). Scale bar: 50 μm. (D) FACS analysis of HAFTL tetEF ZsGreen cells before (w/o) and after stable transfection with tetTALE-SD or tetTALE and selected clones originating from the tetTALE-SD transfected pool.
TALEs are not effective via a roadblock mechanism
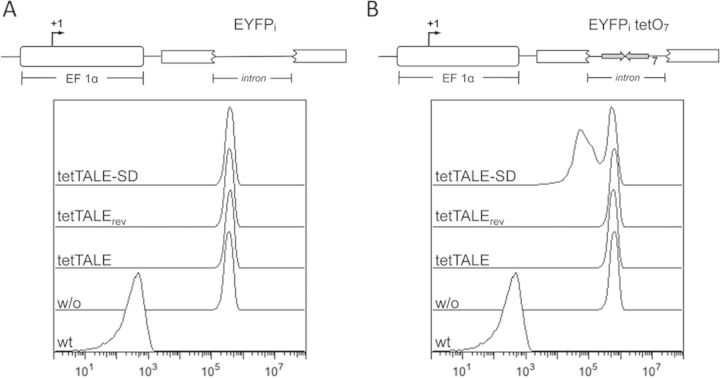
As a second mode of possible TAL effector-mediated silencing, we explored the possibility of DNA-bound TALE protein interfering with transcriptional elongation of mammalian RNA polymerase II (RNAPII) (38). To this end, we utilized a yellow fluorescent protein (EYFP) reporter gene where the original intron-free open reading frame has been interrupted by introduction of a synthetic intron (MG, unpublished), with or without a tetO heptamer sequence (Figure 3A and B). In the former construct, the operator array is located about 1.6 kbp downstream of the transcriptional start site of the EF1α promoter. Cell lines double stable for either of the two reporters and tetTALE were analyzed for downregulation of EYFP expression. As shown in Figure 3B, expression of the tetO containing reporter gene was not affected by tetTALE. Accordingly, in tetTALE+ cells, the presence or absence of the intronic tetO sequences did not make a difference when compared to the control settings (compare Figure 3A and B). This lack of silencing was also observed when we reversed the orientation of DNA-bound TALE protein by synthesizing a tetO-specific TALE DNA-binding domain, tetTALErev, recognizing the anti-parallel tetO2 sequence. However, the expression of tetTALE-SD showed a tetO-dependent reduction of reporter expression, expected for trans-acting SDs, which are functional when placed 3’ to a promoter (39).
Figure 3.
Repression via roadblock. HeLa cells were stably transfected with either of two EF-driven EYFPi constructs, one comprising a synthetic intron alone (EYFPi; A), or an intron with a heptameric tetO sequence (EYFPi tetO7; B). The two resulting cell lines were stably supertransfected with either tetTALE targeting the positive or tetTALrev targeting the negative strand of the tetO sequence. EYFP expression was analyzed by FACS.
TAL effectors are capable of functionally competing with activators for binding sites
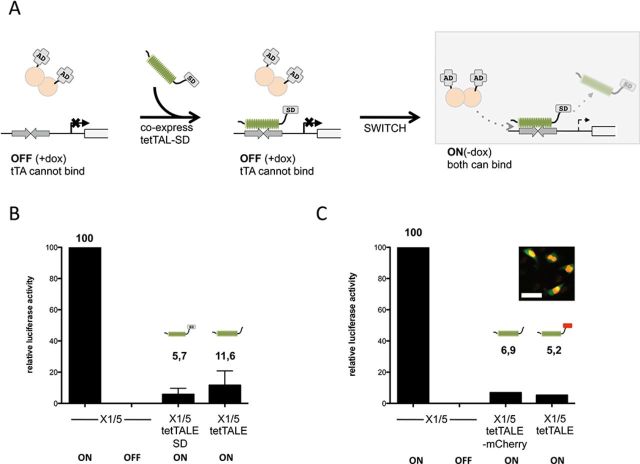
Next we addressed the ability of tetTALE to compete with TFs sharing identical DNA-binding sites, here the tetR-based tTA. Double stable X1/5 cells (4), carrying both chromosomally integrated tet-responsive luciferase reporter and tTA genes, were stably transfected with tetO-specific TALE expression vectors in either ON or OFF state (i.e. absence or presence of doxycycline).
Cells were transfected with tetTALE-SD expression construct in the OFF state of the Tet system, when tTA cannot bind and the reporter is inactive (Figure 4A). Thus, tetTALE-SD was free to bind to the tet operators. Subsequently, the Tet system was switched to ON, addressing if tTA would be able to reestablish itself as the dominant TF (Figure 4A). As shown in Figure 4B, upon expression of tetTALE-SD in induced X1/5 cells (ON) luciferase activity is reduced almost 20fold. However, the efficiency by which tetTALE-SD is able to counter tTA activity could be largely due to the presence of the SD and originate from only a few tetO-bound silencers, rather than by preventing binding of the trans-activator in a more quantitative manner. To this end, we generated X1/5 cells expressing tetTALE, i.e. an effector lacking the SD. As shown in Figure 4B, the absence of the SD only slightly reduced the capacity of the prebound TALE to counter tTA-mediated reporter activation. This slight reduction could be explained by the increased size of the TALE-SD, resulting from the addition of the SD. To test this possibility, we substituted the SD (14 kD) by mCherry (27 kD). As shown in Figure 4C, the more bulky mCherry fusion protein only slightly increased the repressive capacity of tetTALE. Thus, in this experimental setting, the SD is, if at all, only a minor contributor to TALE-mediated repression.
Figure 4.
Competition of tetTALE/tetTALE-SD with tTA for the same binding site. (A) Experimental setup. X1/5 cells containing a Ptet-luciferase reporter and a tTA expression cassette stably integrated were cultured under OFF conditions (+dox). Cells were stably transfected with either a tetTALE or tetTALE-SD expression construct containing a T2A-linked mCherry marker. In the OFF condition only the tetTALE(-SD) can occupy tetO. The Tet system was subsequently switched from OFF to ON. (B) X1/5 cell pools stably transfected with either tetTALE or tetTALE-SD were harvested 7 days after the switch of doxycycline conditions and luciferase activity was analyzed. Ptet-mediated luciferase activation of the TALE-negative parental cell lines with bound tet activator only was set to 100. Shown are mean values of three independent experiments with standard deviation. (C) X1/5 cell pools stably transfected with either tetTALE or tetTALE-mCherry, were harvested 7 days after the switch of dox conditions and luciferase activity was analyzed. Ptet-mediated luciferase activation of the TALE-negative parental cell lines with bound tet activator only was set to 100. Shown is a representative analysis (n = 2) for stably transfected X1/5 cell pools. Insert: microscopic picture of cells stably transfected with tetTALE-mCherry illustrating nuclear localization of TALE. Scale bar: 50 μm.
In the previous experiment, tetTALE was able to prevail on its binding site even upon challenge by tTA. Next we asked if tetTALE repression would also be observed with tetO preoccupied by tTA. Therefore, we stably transfected tetTALE-SD or tetTALE expression constructs in X1/5 cells, which were continuously cultured in the ON state. Thus, tTA was able to bind tetO during the entire selection period (Figure 5A). Analysis of reporter activity showed that both tetTALE and tetTALE-SD effectively reduced tTA-mediated transcriptional activation, arguing for a displacement of this TF by the TALE DNA-binding proteins (Figure 5B). This analysis was extended by clonal analysis of tetTALE-SD+ X1/5 cells generated under ON conditions. When analyzed either by FACS for the T2A-sequence coupled GFP reporter (Figure 5C) or directly by immunoblotting for expression of the TALE itself (Figure 5E), a striking correlation between expression of TALE protein and the reduction in luciferase activity was observed (Figure 5D; see Supplementary Figure S5 for quantification). For example, clone 4 showed the highest GFP signal, a high expression level of the tetTALE-SD and a corresponding low luciferase activity in the ON state of the Tet system. In contrast, clone 1 and 5 only marginally expressed TALE protein. In line with this, the luciferase activity in the ON state is similar to the tetTALE-SD− cells.
Figure 5.
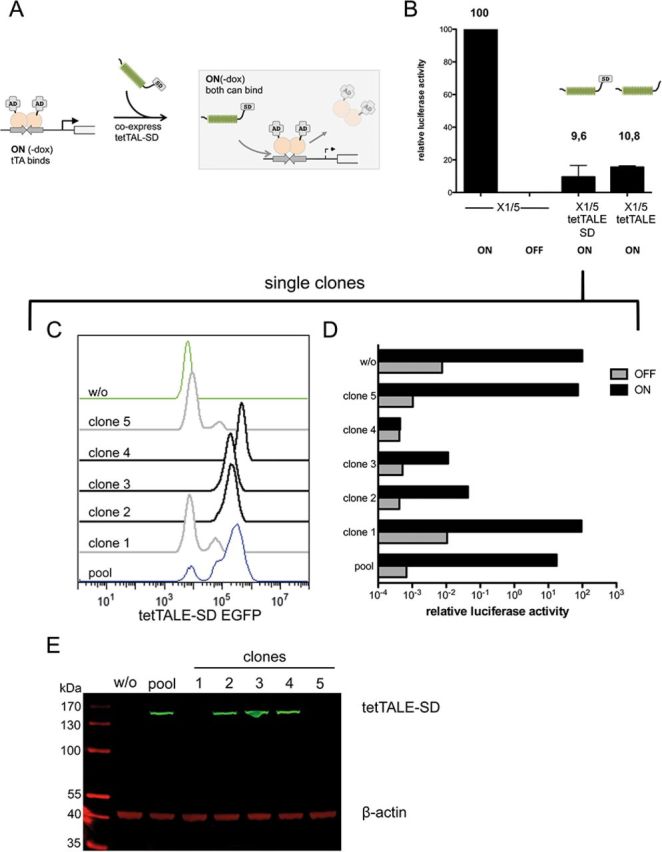
Competition between tTA and tetTALE/tetTALE-SD. (A) Experimental setup. X1/5 cells containing a Ptet-luciferase reporter and a tTA expression cassettes stably integrated were cultured under ON conditions (-dox) where tTA is bound to tetO. Cells were stably transfected with either tetTALE or tetTALE-SD expression construct containing a T2A-linked mCherry marker. (B) Analysis of luciferase activity of X1/5 cell pools (ON) transfected with either tetTALE or tetTALE-SD. Ptet-mediated luciferase activation of the TALE-negative parental cell lines with bound tet activator only was set to 100. Shown are mean values of three independent experiments with standard deviation. (C) Clones isolated from tetTALE-SD-transfected X1/5 cells were analyzed for tetTALE-SD linked GFP expression along with untransfected X1/5 cells (w/o) and the originating pool. (D) Single clones isolated from tetTALE-SD-transfected X1/5 cells grown under ON conditions were analyzed for luciferase activity 7 days after the switch of dox conditions from ON (-dox) to OFF (+dox). Ptet-mediated luciferase activation of the TALE-negative parental cell lines with bound tet activator only was set to 100. (E) Immunoblot analysis of single clones isolated from tetTALE-SD-transfected X1/5 cells grown under ON conditions to monitor tetTALE-SD expression levels. β-actin levels served as loading control.
Largely similar results were obtained in analogous competition experiments with the reverse tTA instead of tTA, using a bidirectional tet-responsive promoter and an additional EGFP reporter. These experiments as shown in Supplementary Figures S6 and S7 were performed in Chinese hamster ovary (CHO) and 293 human embryonic kidney cells.
Taken together, our results support the notion that TALE proteins can prevent binding of and successfully compete with other, prebound TFs sharing the same binding site in an apparently dose-dependent manner.
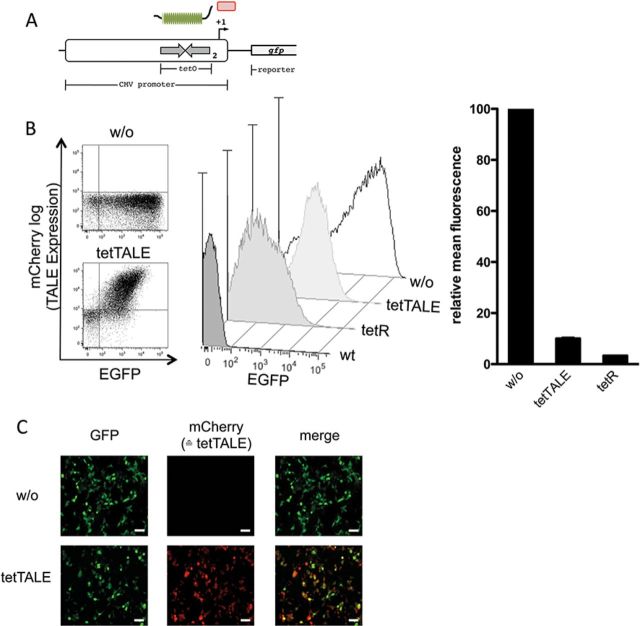
Suppression of CMV promoter activity by TALE binding
Our experiments so far established tetTALE as highly effective in competing with tetR-derived TFs for identical target sites. The resulting downregulation of transcriptional activity was shown to be largely independent of a SD attached to the TALE, most likely by steric hindrance. Next, we asked if this observed repression by TALE would also hold up in a configuration where TALE binding close to a transcriptional control sequence interferes with more complex DNA-binding protein assemblies. Again, we took advantage of preexisting promoter constructs, in this case an engineered full-length CMV promoter designed to respond to tetR-mediated downregulation (‘T-Rex system’) (40). We addressed if binding of tetTALE to the 2 tetO sequences located close to the TATA box would be able to suppress its transcriptional activity as the TALE-binding site partially overlaps with the pre-initiation complex binding region. The principal experimental setting is outlined in Figure 6A. Expression of both tetR and tetTALE efficiently downregulated reporter gene expression driven by a tetO-modified CMV promoter (Figure 6B). These results suggest that the potential of TALEs binding in cis to mediate transcriptional control can be employed as a versatile tool in genetic engineering.
Figure 6.
tetTALE-mediated inhibition of transcription initiation. (A) Scheme of the reporter construct where two copies of tetO are located upstream of the transcriptional start site in the context of a CMV promoter. (B) Left: Analysis of EGFP and mCherry (coupled to tetTALE via T2A) expression in 293Tn cells transiently transfected with the reporter alone (w/o) or co-transfected with tetTALE (1:9 w/w). Middle: Overlay of histograms of 293Tn cells transiently transfected with the reporter alone (w/o), or co-transfected with either tetTALE or tetR for comparison (1:9 w/w). Untransfected cells serve as a control (wt). Right: Quantification of the fluorescence signal showed a 10-fold repression of CMV reporter activity upon tetTALE expression. Signal intensity of the cells transfected with the reporter alone was set to 100. Shown are mean values of three independent experiments with standard deviation. (C) Microscopic picture of 293Tn cells after transient transfection with the reporter alone (w/o) or co-transfected tetTALE: EGFP (left), mCherry (middle), merge (right). Scale bar: 50 μm.
DISCUSSION
Recombinant TALEs enable the targeting of custom-defined nucleotide sequences in the context of mammalian chromosomes, with only few design rules to be considered. Thereby, TALEs provide an easy-to-handle tool for genome engineering and transcriptional control systems. The flexibility in target site selection is a clear advantage of TALE-based DNA-binding proteins when compared to established recombinases (e.g. Cre, Flp), nucleases (e.g. meganucleases) or TFs (e.g. Gal4 or TetR-based), all of which require the target site to be pre-engineered into the genome. However, while TALE DNA-binding proteins in the various functional constellations made a rapid entry into genetic engineering, it remains unclear how such recombinant TALEs compare in quantitative terms with previously established heterologous DNA-binding proteins employed in mammalian cells.
We performed a comparative analysis between synthetic TALE-based TFs and heterologous TFs of the Tet system. For direct comparison, we designed a tetTALE binding to tetO2. This operator sequence is naturally recognized by the tetR DNA-binding domain, that is common to all engineered tet TFs. When comparing these two DNA-binding proteins fused to trans-activating domains, similar activation levels were observed, both in double transient assays and for chromosomal reporters. This is in line with previous findings from Li et al. for TALEs partially covering tetO sequences. In that study, TALEs mediated a modest activation when targeted to a chromosomally integrated tet-responsive promoter (25). While some of the current limitations in the effectiveness of individual TALEs might be overcome by improved design rules including the use of improved effector domains (26), the fact that our tetO-specific TALE constructs stand comparison to one of the strongest TFs known for mammalian cells and can compete with the transcription initiation machinery itself already suggests that custom TALE TFs can function as a powerful tool in regulating endogenous genes.
The modular structure of many eukaryotic TFs is since long established (41,42) and has been capitalized on in the design of numerous heterologous transcriptional control systems. This observation extends to TALE TFs, where the activating domains could be successfully exchanged for a KRAB SD causing repression (10,25,43). Using tetTALE fused to a SD or, alternatively, the tetTALE DNA-binding domain itself, we could discriminate between possible modes of repression.
Firstly, our experiments demonstrate the ability of TALE silencers to repress expression from the strong human EF1α promoter to background level. Furthermore, we could affirm that this effect was attributed to the presence of the KRAB SD. Results obtained so far with this new technology often attest rather modest repression of endogenous genes using TALE silencers (10,44). Our findings suggest that powerful repression of endogenous promoters is feasible, up to total abrogation of expression with the correct loci targeted, in this case 5’ of the EF1α promoter site. Nevertheless, it is important to keep in mind that a heptameric target was used in these experiments. Prior studies show a synergistic effect of multiple TALEs binding in one region (17,45,46). In line with these observations, we also observed a synergistic effect for tetTALE activators when comparing 1 versus 7 tetO-binding sites fused to a minimal reporter construct. Quantitative limitations in the silencing of target promoters might also be overcome using more potent SDs, like the human SID domain (10). The outcome of more systematic investigations of alternative SDs might augment repression of even stronger endogenous promoters and clarify a potential context dependency of these effector domains.
Secondly, we asked if TALEs could act by a roadblock mechanism. For prokaryotic cells, it was shown that TALEs could decrease gene expression when bound intragenically by stalling the RNA polymerase. However, the measured repression was rather moderate (47). Our data suggest that this principle is not applicable in eukaryotic cells, as we observe no reduction in target gene expression when the tetTALE is bound to multiple binding sites within the transcribed region of a reporter gene. Conceptually similar experiments using gRNA-based recruitment of catalytically inactive Cas9 to intragenic regions resulted in a moderately efficient repression of gene activity (48). The observed effects could be improved by fusing the dCas protein with a SD (27). In this setting, additional repressive effects are most likely mediated in trans, as would be concluded by our roadblock experiments using tetTALE-SD versus tetTALE (Figure 3). In addition, the results by Gilbert et al. suggest that the distance of effector binding sites to the transcriptional start site may be an important parameter, which has not been addressed in our experiments (27). Previous studies showed that prokaryotic repressors can act as roadblocks for elongating RNAPII (38), depending on the composition of the polymerase complex (49). Contrary to the previously investigated DNA-bound prokaryotic repressors, TALEs are actually wrapped around the DNA, with the TALE repeats arranged in a helical structure around the DNA (50). Still, even such a compact form of protein–DNA interaction is, according to our analysis, no obstacle for a mammalian RNA polymerase.
Thirdly, and most strikingly, we found that DNA binding of tetTALE is potent enough, as to efficiently counter the activity of a tet trans-activator, tTA. Our analysis was performed upon stable co-expression of the competing TFs in an attempt to avoid distortion of the readout due to imbalanced effector protein levels that might result from transient transfections. Taking advantage of the inducible nature of the tTA/tetO interaction, we were able to show that the TALE can either prevent tTA binding or effectively compete with tTA pre-bound to its recognition site. The associated decrease of reporter activity was only marginally dependent on the SD fused to tetTALE. Thus, we conclude that TALEs can successfully compete with TFs for identical or overlapping binding sites, a finding validated in several different cell lines also extending to competition with the reverse tet trans-activator, rtTA. This substantiates previous observations where upon transient overexpression of a shorter tetO-binding TALE prevented transcriptional activation by stably expressed rtTA (25). In this context, it is worth noting that the dissociation constant of TetR to the tetO2 operator site is in the high picomolar range (51). By contrast, only few studies analyzed the affinity of TALEs toward their target sites, with one study employing a backbone similar to that used by us showing a KD in the low nanomolar range (52). In conclusion, designer TALEs have the capacity to act efficiently on target sequences preoccupied by other DNA-binding proteins. It remains to be determined to which extent the efficacy of TALEs in interfering with endogenous mammalian TFs depends on the type or identity of the endogenous DNA-binding proteins. However, our experiments demonstrating tetTALEs ability to substantially hinder hCMV-driven transcription when binding to the core promoter region argue for a broader validity of our claim. Here, tetTALE binding is directed to the assembly site of the transcription pre-initiation complex (53). Thus, the previously reported repression of an exceptionally potent TALE-SD targeted to sequences close to the transcriptional start site of an unmodified CMV promoter might at least be in part due to successful binding competition with the pre-initiation complex and independent of the SD used in that study (43). Examples like those from proteins of the E2F family show that antagonism of TFs from mutually exclusive DNA binding is not an artificial setting, but a principle employed in natural transcriptional control circuits (54). Comparative analyses between TALEs and Cas/CRISPR based TFs binding to proximal sites in the promoter regions of endogenous genes suggest that TALEs are the more potent heterologous TFs (55,56). While our results demonstrate their efficacy in sterically competing with other DNA-binding proteins, it has been discussed that dCas/CRISPR efficacy may be compromised if binding sites are preoccupied by endogenous DNA-binding proteins (55).
Further studies, in particular with conditional TALEs (46), are required to probe the reversibility and long-term effects of repression, either in trans silencing or by direct competition. In the latter case, it is expected that repression by tetTALE without SD is totally reversible. Whether the observed KRAB-mediated repression via tetTALE-SD is fully reversible needs to be addressed as it might manifest itself epigenetically (57). The detailed understanding of these different aspects contributing to TALE efficacy is expected to improve their utility as an alternative tool controlling gene activities of mammalian cells at their authentic chromosomal loci.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
R. Lauster, A. Lendlein and A. Leutz are acknowledged for their support, and in particular A. Leutz for critically reading this manuscript. We thank the BCRT Flow Cytometry Lab for assistance. Past and present members in our research group are acknowledged for continuous technical support.
FUNDING
Helmholtz President's Initiative and Networking Fund. Funding for open access charge: Institutional Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 2.Spitz F., Furlong E.E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 3.Yarranton G.T. Inducible vectors for expression in mammalian cells. Curr. Opin. Biotechnol. 1992;3:506–511. doi: 10.1016/0958-1669(92)90078-w. [DOI] [PubMed] [Google Scholar]
- 4.Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. U.S.A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Gossen M., Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 7.Pollock R., Giel M., Linher K., Clackson T. Regulation of endogenous gene expression with a small-molecule dimerizer. Nat. Biotechnol. 2002;20:729–733. doi: 10.1038/nbt0702-729. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F., Cong L., Lodato S., Kosuri S., Church G.M., Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao X., Yang J., Tsang J.C., Ooi J., Wu D., Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Reports. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong L., Zhou R., Kuo Y.C., Cunniff M., Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boch J., Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 12.Boch J. TALEs of genome targeting. Nat. Biotechnol. 2011;29:135–136. doi: 10.1038/nbt.1767. [DOI] [PubMed] [Google Scholar]
- 13.Gaston K., Jayaraman P.S. Transcriptional repression in eukaryotes: repressors and repression mechanisms. Cell. Mol. Life Sci. 2003;60:721–741. doi: 10.1007/s00018-003-2260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupo A., Cesaro E., Montano G., Zurlo D., Izzo P., Costanzo P. KRAB-Zinc finger proteins: a repressor family displaying multiple biological functions. Curr. Genomics. 2013;14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun N., Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 2013;110:1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 16.Geissler R., Scholze H., Hahn S., Streubel J., Bonas U., Behrens S.E., Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeder M.L., Linder S.J., Reyon D., Angstman J.F., Fu Y., Sander J.D., Joung J.K. Robust, synergistic regulation of human gene expression using TALE activators. Nat. Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J., et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 19.Bultmann S., Morbitzer R., Schmidt C.S., Thanisch K., Spada F., Elsaesser J., Lahaye T., Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron U., Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/s0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 21.Baron U., Gossen M., Bujard H. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freundlieb S., Schirra-Müller C., Bujard H. A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J. Gene Med. 1999;1:4–12. doi: 10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Deuschle U., Meyer W.K., Thiesen H.J. Tetracycline-reversible silencing of eukaryotic promoters. Mol. Cell. Biol. 1995;15:1907–1914. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao F., Eriksson E. A novel tetracycline-inducible viral replication switch. Hum. Gene Ther. 1999;10:419–427. doi: 10.1089/10430349950018869. [DOI] [PubMed] [Google Scholar]
- 25.Li Y., Moore R., Guinn M., Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci. Rep. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Z., Geurts A.M., Liu G., Kaufman C.D., Hackett P.B. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 30.Mates L., Chuah M.K., Belay E., Jerchow B., Manoj N., Acosta-Sanchez A., Grzela D.P., Schmitt A., Becker K., Matrai J., et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 31.Yusa K., Rad R., Takeda J., Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadinanos J., Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron U., Schnappinger D., Helbl V., Gossen M., Hillen W., Bujard H. Generation of conditional mutants in higher eukaryotes by switching between the expression of two genes. Proc. Natl Acad. Sci. U.S.A. 1999;96:1013–1018. doi: 10.1073/pnas.96.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes K.L., Pierce J.H., Davidson W.F., Morse H.C., III Murine hematopoietic cells with pre-B or pre-B/myeloid characteristics are generated by in vitro transformation with retroviruses containing fes, ras, abl, and src oncogenes. J. Exp. Med. 1986;164:443–457. doi: 10.1084/jem.164.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed S.E., Staley E.M., Mayginnes J.P., Pintel D.J., Tullis G.E. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Berens C., Hillen W. Gene regulation by tetracyclines. Genet. Eng. 2004;26:255–277. doi: 10.1007/978-0-306-48573-2_13. [DOI] [PubMed] [Google Scholar]
- 37.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 38.Deuschle U., Hipskind R.A., Bujard H. RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science. 1990;248:480–483. doi: 10.1126/science.2158670. [DOI] [PubMed] [Google Scholar]
- 39.Moosmann P., Georgiev O., Thiesen H.J., Hagmann M., Schaffner W. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Kruppel-type zinc finger factor. Biol. Chem. 1997;378:669–677. doi: 10.1515/bchm.1997.378.7.669. [DOI] [PubMed] [Google Scholar]
- 40.Yao F., Svensjo T., Winkler T., Lu M., Eriksson C., Eriksson E. Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum. Gene Ther. 1998;9:1939–1950. doi: 10.1089/hum.1998.9.13-1939. [DOI] [PubMed] [Google Scholar]
- 41.Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 42.Frankel A.D., Kim P.S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 43.Garg A., Lohmueller J.J., Silver P.A., Armel T.Z. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rennoll S.A., Scott S.A., Yochum G.S. Targeted repression of AXIN2 and MYC gene expression using designer TALEs. Biochem. Biophys. Res. Commun. 2014;446:1120–1125. doi: 10.1016/j.bbrc.2014.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Pinera P., Ousterout D.G., Brunger J.M., Farin A.M., Glass K.A., Guilak F., Crawford G.E., Hartemink A.J., Gersbach C.A. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer A.C., Gaj T., Sirk S.J., Lamb B.M., Barbas C.F., III Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth. Biol. 2013;3:723–730. doi: 10.1021/sb400114p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Politz M.C., Copeland M.F., Pfleger B.F. Artificial repressors for controlling gene expression in bacteria. Chem. Commun. 2013;49:4325–4327. doi: 10.1039/c2cc37107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reines D., Mote J., Jr Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl Acad. Sci. U.S.A. 1993;90:1917–1921. doi: 10.1073/pnas.90.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.K., Shi Y., Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kamionka A., Bogdanska-Urbaniak J., Scholz O., Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meckler J.F., Bhakta M.S., Kim M.S., Ovadia R., Habrian C.H., Zykovich A., Yu A., Lockwood S.H., Morbitzer R., Elsaesser J., et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Res. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenhard B., Sandelin A., Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–245. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 54.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 55.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szulc J., Wiznerowicz M., Sauvain M.O., Trono D., Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat. Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.