Abstract
BRCA1 is involved in many disparate cellular functions, including DNA damage repair, cell-cycle checkpoint activation, gene transcriptional regulation, DNA replication, centrosome function and others. The majority of evidence strongly favors the maintenance of genomic integrity as a principal tumor suppressor activity of BRCA1. At the same time some functional aspects of BRCA1 are not fully understood. Here, a HAC (human artificial chromosome) module with a regulated centromere was constructed for delivery and expression of the 90 kb genomic copy of the BRCA1 gene into BRCA1-deficient human cells. A battery of functional tests was carried out to demonstrate functionality of the exogenous BRCA1. In separate experiments, we investigated the role of BRCA1 in maintenance of heterochromatin integrity within a human functional kinetochore. We demonstrated that BRCA1 deficiency results in a specific activation of transcription of higher-order alpha-satellite repeats (HORs) assembled into heterochromatin domains flanking the kinetochore. At the same time no detectable elevation of transcription was observed within HORs assembled into centrochromatin domains. Thus, we demonstrated a link between BRCA1 deficiency and kinetochore dysfunction and extended previous observations that BRCA1 is required to silence transcription in heterochromatin in specific genomic loci. This supports the hypothesis that epigenetic alterations of the kinetochore initiated in the absence of BRCA1 may contribute to cellular transformation.
INTRODUCTION
BRCA1 is a well-known tumor suppressor gene, germ line mutations in which predispose women to breast and ovarian cancers. Since the identification of the BRCA1 gene, there have been numerous studies aimed at characterizing the diverse repertoire of its biological functions. BRCA1 is involved in multiple cellular pathways, including DNA damage repair, chromatin remodeling, X-chromosome inactivation, centrosome duplication and cell-cycle regulation (1–7). A recent study has suggested a role in the epigenetic regulation of an oncogenic microRNA (8).BRCA1 associates with constitutive pericentromeric heterochromatin in nuclei (1). Further insight into the role of BRCA1 in pericentromeric heterochromatin and a significant link to maintaining global heterochromatin integrity has been recently gained by Zhu et al. (9). They showed that loss of BRCA1 results in transcriptional de-repression of tandemly repeated satellite DNA in mice and human BRCA1-deficient cells. This impairment of constitutive heterochromatin may lead to de-repression of the normally silenced genes that are located at the tandemly repeated DNA regions, probably through the loss of ubiquitylation of histone H2A. These effects on heterochromatin silencing could potentially account for some aspects of BRCA1 tumor suppression function.
In their experiments, the authors employed a lentivirus vector expressing a BRCA1 cDNA to complement BRCA1-deficiency. Such an approach may not completely recapitulate the physiological expression of the BRCA1 gene for several reasons. These include the lack of a strong copy number control of the transgene, the lack of alternative splice-forms when rescuing function with a cDNA and the absence of the intronic regions of the gene, which may include regulatory elements, and which, when spliced, will increase the efficiency of translation of the resulting mRNA (10–14). We therefore hypothesized that delivery of an entire, single copy of the BRCA1 genomic locus may provide additional information on BRCA1 function.
The use of an alternative HAC-based (human artificial chromosome) vector for gene delivery and expression may potentially overcome some of the limitations associated with the viral-based delivery of the BRCA1 cDNA outlined above. HACs are de novo chromosomes that contain functional centromeres permitting their long-term stable maintenance as single copy mini-chromosomes without integration into the host chromosomes. This minimizes such complications as disruption of endogenous genes (15–18 and references therein). Moreover, HAC vectors have unlimited cloning capacity allowing them to carry entire genomic loci or potentially groups of loci with all regulatory elements that should faithfully mimic the normal pattern of gene expression. At present the carrying capacity is limited to several megabases (Mb) only by technical cloning limitations.A structurally characterized HAC, alphoidtetO-HAC (19–21) with a single gene loading site, has ideal features required for gene function studies. A unique advantage of this HAC is its regulated kinetochore, which provides a unique possibility to compare the phenotypes of the human cell with and without a functional copy of a gene (19). This provides a pure control for phenotypic changes attributed to expression of a HAC-encoded gene by returning the mutant cell line to its original state following loss of the HAC (22). Inactivation of the HAC centromere is accomplished by targeting tet-repressor (tetR) fusion proteins to the alphoid DNA array of the HAC, which contains ∼3000 tetracycline operator (tetO) sequences embedded into each alphoid DNA unit. Certain chromatin-modifying fusion proteins, such as the tTS, inactivate the HAC centromere so that its segregation becomes random and it is gradually lost from growing populations of cells.
In the present study, a 90 kb genomic region spanning the BRCA1 gene, which includes potential regulatory elements in intron regions (23) was inserted into the alphoidtetO-HAC and subsequently used to complement a BRCA1 gene deficiency in human ovarian cancer cell line UWB1.289. The full-length BRCA1 genomic locus was selectively isolated from total genomic DNA by the TAR cloning technique (24,25). Functional expression of BRCA1 in BRCA1-deficient recipient cells was confirmed by a set of specific tests based on the known functions of the BRCA1 protein. In addition, we extended a previous observations of Zhu et al. (9) that BRCA1 is required to silence transcription in heterochromatin in a genomic locus-specific manner. In the separate experiments, using recently released sequence information on higher-order alpha-satellite centromeric repeats (HORs) (26), we used the BRCA1-HAC module to demonstrate a direct link between BRCA1 deficiency and kinetochore dysfunction.
MATERIALS AND METHODS
Cell lines
Hypoxanthine phosphoribosyltransferase (HPRT)-deficient Chinese hamster ovary (CHO) cells (JCRB0218) carrying the alphoidtetO-HAC were maintained in Ham's F-12 nutrient mixture (Invitrogen, USA) plus 10% fetal bovine serum (FBS) with 8 μg/ml Blasticidin S Hydrochloride (Funakoshi, Japan). After insertion of a TAR/BAC/BRCA1 construct and the eGFP gene into alphoidtetO-HAC, CHO cells retaining the tetO-eGFP-BRCA1-HAC were maintained in HAT-containing medium. The BRCA1-deficient human ovarian cancer cell line UWB1.289 carrying a germline BRCA1 mutation within exon 11 and having a deletion of the wild-type allele (27) was obtained from the American Type Culture Collection (ATCC; Manassas, USA). These cells were maintained in 50% RPMI1640 medium containing 50% MEGM (Mammary Epithelial Growth Medium from Clonetics/Lonza) (MEGM Bullet Kit; CC3150) made of Mammary Epithelial Basal Medium (MEBM) basal medium and SingleQuot plus 3% FBS. Swine testis (ST) cells (ATCCRCRL-1746) were obtained from the ATCC. The base medium for ATCCRCRL-1746 cells is ATCC-formulated Eagle's Minimum Essential Medium (Catalog No. 30–2003) plus 10% of FBS.
Retrofitting of YAC/BRCA1 into a BAC
A diagram of the retrofitting of a circular YAC/BRCA1 molecule into a BAC form is shown in Supplementary Figure S2. Retrofitting vector pJBRV1 (22) contains the URA3 yeast selectable marker, F’ factor origin replication, the chloramphenicol acetyltransferase (CmR) gene and a cassette including a 3′ end of the HPRT gene, the eGFP gene, a loxP sequence and two short targeting hooks (∼300 bp each) separated by the unique BamHI site. These targeting hooks flank the ColE1 origin of replication in the pVC604-based TAR cloning vector that was previously used to clone a 90 kb genomic copy of the BRCA1 gene (23). Recombination of the BamHI-linearized pJBRV1 vector with the pVC604 vector carrying the BRCA1 gene in yeast leads to replacement of the ColE1 origin of replication by a cassette containing the F’ factor, the CmR gene, the URA3 gene and 3′-HPRT-loxP-eGFP sequences. For retrofitting, a standard lithium acetate transformation procedure in yeast Saccharomyces cerevisiae strain VL6–48 (MATa,his3-Δ200,trp1-Δ1,ura3–52,lys2,ade2–101,met14) was used. This strain is available under request from the Development Therapy Branch, National Cancer Institute (NCI, NIH) or alternatively from the ATCC (ATCC Number MYA-3666). The YAC/BACs were transferred into Escherichia coli by electroporation. In brief, yeast chromosome-size DNA was prepared in agarose plugs and, after melting and agarase treatment, the DNAs were electroporated into DH10B competent cells (GIBCO/BRL) by using a Bio-Rad Gene Pulser as previously described (22).
Loading of BAC/BRCA1 into the loxP site of the alphoidtetO-HAC in CHO cells
Loading of the BRCA1 gene into HAC was performed using Cre-recombinase as previously described (20). Twenty micrograms of the BAC vector containing the BRCA1 gene and 1 microgram of the Cre expression pCpG-iCre vector were co-transformed into HPRT-deficient CHO cells (105) containing the alphoidtetO-HAC vector by lipofection using either FuGENERHD transfection reagent (Roche) or Lipofectamine 2000 (Invitrogen). HPRT-positive colonies were selected after 3 weeks growth in HAT medium. Insertion of BRCA1 into the alphoidtetO-HAC was confirmed by genomic polymerase chain reaction (PCR) with a specific pair of primers that diagnose reconstitution of the HPRT gene (Supplementary Table S1). Four colonies were confirmed to contain all exons of BRCA1. One clone was chosen for further analysis.
Microcell-mediated chromosome transfer (MMCT)
alphoidtetO-HAC/BRCA1 was transferred from hamster CHO cells to the human UWB1.289 cell line deficient for the BRCA1 gene and ST cells using MMCT protocol described previously (28,29) with minor modifications. First, six Nunc T24 flat flasks of CHO cells containing alphoidtetO-HAC/BRCA1 were grown to 70–80% confluence in 4 ml per flask of F12 media supplemented with 10% FBS. Then the media was changed to fresh media containing 0.1 μg/ml colcemid. The cells were grown with colcemid for ∼72 h. The media was then aspirated and each flask filled up to neck with a solution of 10 μg/ml Cytochalacin B in serum-free Dulbecco's modified Eagle's medium (DMEM) (Cyto-B medium), pre-heated to 37°C, then incubated 30 min at 37°C. Parafilm was used to seal T24 flask. T24 culture flask was stuck to a large centrifuge bottle using laboratory labeling tape with the flat side toward the rotor. The bottles were filled to about 1/3 with pre-warmed 37°C water. Then these six bottles containing the T24 flasks were spun at 7000–7500 revolutions per minute (rpm), 37°C for 60 min in a Sorvall RC 5B. The Cyto-B medium was removed after centrifugation. The Cyto-B medium can be re-used several times after filtration via 0.22 μm filter. Two milliliters of DMEM serum-free medium was added to each flask. Microcells were washed with 5 ml pipette and the suspension of microcells was transferred into 15 ml tube. Then an additional 2 ml was used to wash all tubes to get as much microcells as possible. Microcell suspension was incubated at 4°C for 20 min. The microcell suspension was filtered sequentially through the sterile 8, 5 and 3 μm Isopore Membranes (Millipore, # TETP02500, #TMTP02500 and # TSTP02500) using 6–7 ml suspension for 1 filter set. After filtration, the 15 ml tube containing filtered microcells was centrifuged at 2200 rpm for 10 min. The supernatant was aspirated and the microcells were washed by 1 ml of ice cold Hemagglutinating Virus of Japan (HVJ) buffer [GenomONE-CF EX SeV-E(HVJ-E) 1 Vials Cell Fusion Reagents, Cosmo Bio Co., LTD, # ISK-CF-001-EX], and centrifugated for 5 min at 3000 rpm. The supernatant was removed and the microcells were suspended in 50 μl ice cold HVJ suspension buffer and then kept on ice. During microcell centrifugation step, the recipient cells UWB1.289 cells were split 2 days before MMCT experiment to reach ∼80–90% of confluence by the MMCT day. The cells were trypsinized, washed by complete growth media to inactivate trypsin and resuspended in 1 ml of ice cold HVJ buffer, centrifuged for 1 min at 2000 rpm. The supernatant was aspirated and the cells were resuspended in 50 μl of ice cold HVJ buffer and kept on ice. Suspensions of cells and microcells were mixed together and ice-cooled 10 μl of HVJ viral envelop protein (GenomONE-CF EX SeV-E(HVJ-E) 1 Vials Cell fusion Reagents) was added and mixed by tapping. The mixture was left on ice for 5 min to allow HVJ-E to be adsorbed on the cell/microcell surface, then incubated at 37°C for 15 min when mixing by tapping every 5 min to induce cell/microcell fusion. The complete growth medium was added to the tube containing mixture cells/microcells and transferred to a 10-cm plate for cell proliferation and growth. The alphoidtetO-HAC/BRCA1 carries Bsr genes, thus Blasticidin (BS) was used to select for cells that have taken up the HAC. Blasticidin (2–4 μg/ml) was added in 2 days after MMCT. The blasticidin resistant colonies were isolated in 4–6 weeks after selection. Typically 1–3 BSR colonies were obtained in one MMCT experiment involving HAC transfer into the gene-deficient cells. BSR clones were analyzed by fluorescence in situ hybridization (FISH). Based on FISH analysis, more than 95% of cells in the colony contained a HAC. No co-transfer of CHO chromosomes was detected in our experiments using a sensitive PCR test for rodent SINE elements-hamster B2 repeats (Supplementary Table S1).
FISH analysis
FISH and immuno-FISH analyses were performed as previously described (19,20) with minor modifications. HAC-containing cells were cultured in medium with 0.1 mg/ml of colcemid (Invitrogen, #15212-012) for 3 h at 37°C. Metaphase cells were trypsinized and collected by centrifugation for 2 min at 1000 rpm, treated in hypotonic solution (50 mM KCl) for 20 min at 37°C and washed three times in methanol:acetic acid (3:1) fixative solution. Cells were diluted to the appropriate density with fixative solution, spread onto pre-cleaned slides (Fisher Scientific, # 12-544-7) above of steam (boiling water), check on microscope and when slide is fine allowed to age 2 days at room temperature. Metaphase chromosomes on the slide were denatured by denaturation buffer 70% formamide/2xSSC for 2 min at 72°C. Samples were dehydrated through a 70%, 90% and 100% ethanol series for 4 min each and left to air-dry. Orange 552 dUTP (5-TAMRA-dUTP) (Abbott Molecular) or Green 496 [5-Fluorescein] dUTP (Abbott Molecular) and Red 650 [Cyanine-5E] dUTP (Enzo) labeled probes were denatured in hybridization solution at 78°C for 10 min and left at 37°C for 30 min. The hybridization mix probe (or probes) was applied to the sample and incubated at 37°C overnight. Slides were washed with 0.4× SSC, 0.3% Tween 20 for 2 min at 72°C, briefly rinsed with 2× SSC, 0.1% Tween 20 (5 s–1 min) and air-dried in darkness for 30 min. The samples were counterstained with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Labs). Slides were analyzed by fluorescence microscopy. Images were captured using a DeltaVision microscopy imaging system in the CRC, LRBGE Fluorescence Imaging Facility (NIH) and analyzed using image J software (NIH).
The probes used for FISH analysis
The probes used for FISH were BAC32-2-mer (tetO) DNA containing a 40 kb of alphoid-tetO array cloned into a BAC vector as described previously (19) and cDNA of BRCA1. BAC DNA was labeled using a nick-translation kit with Orange 552 dUTP (5-TAMRA-dUTP) (Abbott Molecular) or Red 650 [Cyanine-5E] dUTP (Enzo) for two colors FISH with cDNA. cDNA was labeled by Green 496 [5-Fluorescein] dUTP (Abbott Molecular).
Genomic DNA preparation
For PCR analysis, genomic DNA from hamster CHO and human UWB1.289 cells containing the alphoidtetO-HAC/BRCA1 was prepared using a DNeasy Blood & Tissue Kit (Qiagen, #69506). Integrity of the BRCA1 gene was confirmed by a set of primers for exonic regions of the gene. These primers are listed in Supplementary Table S1. PCR products were separated by agarose gel electrophoresis, gel extracted and used for sequencing.
Antibodies
The following antibodies were used for immunoblotting, co-immunoprecipitation and immunostaining: anti-BRCA1 (Ab-1) (Catalog#OP92) mouse mAb (MS110) (Calbiochem, EMD Millipore), anti-pol β, anti-phospho-histone H2A.X (Ser139) clone JBW301 (Millipore, # 05–63), Alexa Fluor 555 Goat anti-mouse antibody (Red) (H+L) (Life Technology, #A-21422), anti-Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (14C10) rabbit mAb (Cell Signaling Technology, # 2118). For chromatin immunoprecipitation (ChIP) assay: H3K9me3 (Millipore, #07–523), H3K4me3 (Millipore, # 07–473) and anti-CENP-A. Pol β antibodies were kindly provided by Dr. Samuel H. Wilson (Laboratory of Structural Biology, NIEHS). CENP-A antibodies were previously described (30).
Western analysis
BRCA1 detection using western blotting was performed as following. Briefly, the primary antibodies against BRCA1 (1:50 dilution) and against GAPDH (1:20 000 dilution) were used. GAPDH was used as a loading control. Washes were carried out with Tris-buffered Saline Tween 20 (TBST) buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 30 min with 5 buffer changes. Secondary antibodies used were: Amersham ECL, HRP-linked anti-mouse (1:10 000 dilution) (GE Healthcare, #NA931-1ML) for BRCA1 and Amersham ECL, HRP-linked anti-rabbit (GE Healthcare, #NA934-1ML) (1:10 000 dilution) for GAPDH.
Reverse-transcriptase PCR (RT-PCR)
Transcription of the BRCA1 gene from alphoidtetO-HAC/BRCA1 in CHO, UWB1.289 and ST cell lines was detected by RT-PCR using specific primers listed in Supplementary Table S1. The PCR products were sequenced using a PE-Applied Biosystem 3100 Automated Capillary DNA Sequencer.
alphoidtetO-HAC/BRCA1 elimination by its targeting with chromatin modifiers
Transient expression of the tTS-tet-repressor fusion protein to induce the HAC loss was performed as previously described (20,22). After transfection with the tTS fusion construct or empty vector as control, neomycin-resistant clones were selected in the presence of doxycycline, conditions that support proper HAC segregation and HAC stability (the tTS cannot bind to tetO sites). The HAC elimination assay was then performed by transfer of the stable transfectants to media lacking doxycycline. After 48 h of culturing in the presence of neomycin (800 μg/ml), a fraction of the HAC-less cells was detected by loss of the eGFP signal. The cells that have lost the HAC were selected for the further analysis. Selection of the HAC-less clones takes ∼3 weeks. The absence of the HAC was confirmed by FISH and western blot analyses. HAC-less cells were used as controls for the restoration of the original mutant phenotypes. In separate experiments the cells were sorted using a fluorescence-activated cell sorting (FACS) AriaII cytometer. After cell sorting, two populations of cells, green-containing alphoidtetO-HAC/BRCA1 and non-green (after HAC loss), were cultured independently and used for functional studies.
ChIP analysis
ChIP was carried out according to a previously described method (31). Cultured cells were cross-linked in 1% formaldehyde for 10 min at 37°C. After addition of 1/10 volume of 1.25 M glycine and incubation for 5 min, fixed cells were washed twice with cold phosphate buffered saline (PBS) buffer. Soluble chromatin was prepared by sonication in water bath-based sonicator (Bioruptor sonicator; Cosmo Bio) to an average DNA size of 500 bp and immnnoprecipitated in IP buffer (0.01% sodium dodecyl sulphate (SDS), 1.1% Triton X- 100, 1.2 mM ethylenediaminetetraacetic acid, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl). Protein A agarose/salmon Sperm DNA (Millipore, #16–157) was added, and the antibody-chromatin complex was recovered by centrifugation. The recovery ratio of the immunoprecipitated DNA relative to input DNA was measured by real-time PCR using a 7900HT Fast real-time PCR detection system (Applied Biosystems) and SYBR Green PCR Master Mix (Applied Biosystems, #4309155). Primers for HORs regions of chromosome 21 and chromosome X, tetO-2 mer of the alphoidtetO DNA array, HORs D5Z1 and D5Z2 regions of chromosome 5 were described previously (30,32) (see also Supplementary Table S1).
Co-immunoprecipitation of BRCA1 and DNA polymerase pol β
The method was described previously (33) and was applied with minor modifications. Two cell lines were grown to confluence, the UWB1.289 cell line carrying alphoidtetO-HAC /BRCA1 and the isogenic strain that have been induced to lose the HAC. Approximately 15 million cells were harvested from each cell line and individually re-suspended in 1.8 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 25 mM NaF, 0.1 mM sodium orthovanadate, 0.2% Triton X-100, 0.3% Nonidet P-40 with 1× protease inhibitor cocktail) and incubated on ice for 30 min. For each sample, the lysate was centrifuged at 14 000 rpm for 30 min at 4°C and the supernatant fraction was transferred to a new tube. The protein concentration in the extract was determined using the Bio-Rad protein assay with bovine serum albumin (BSA) as standard. For co-immunoprecipitation, 40 μl of affinity purified anti-pol β polyclonal antibody or 10 μl of rabbit non-immune immunoglobulin G (IgG; Upstate, Millipore, #12–370) was added to equal amounts of cell lysate (6 mg of total proteins), and the mixture was incubated with rotation overnight at 4°C. The immunocomplex was adsorbed onto 30 μl suspension of protein A-sepharose (Millipore) beads (pre-washed three times with lysis buffer) by incubating the mixture for 6 h at 4°C. The beads were then washed three times with 600 μl of lysis buffer containing protease inhibitors, with a 2 min, 2000 rpm centrifuge spin between washes. Finally, the beads were re-suspended in 30 μl of SDS sample buffer, heated for 5 min and the soluble proteins were separated by NuPAGE Novex 3–8% Tris-Acetate Protein gel (Life Technology, EA0378BOX). Proteins were transferred onto a nitrocellulose membrane in BioRad transblot apparatus overnight at 5 V and were analyzed by western blot with anti-BRCA1 (Ab-1) (Catalog#OP92), mouse mAb (MS110) antibody or anti-pol β polyclonal antibody.
Micronucleus formation assay (MNi)
UWB1.289 cells were grown to 70–80% confluency, harvested and seeded at 20 000 cells/ml onto chamber slides. After 24 h, cells were washed with 1× PBS, incubated with 0.075 M KCl for 10 min, fixed in ice cold 3:1 v/v methanol:acetic acid and dried overnight under vacuum. The frequency of micronucleated cells was assessed using fluorescence microscopy following DAPI staining.
Quantification of transcripts
Real-time RT-PCR with RNA purified from UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 and from UWB1.289 cells that have lost the HAC was carried out using SYBR Green PCR Master Mix (Applied Biosystems, #4309155) and TaqMan Gene Expression Master Mix (Applied Biosystems, #4369016) according to the manufacturer's protocol. Total RNA was prepared with the RNeasy Mini Kit (Qiagen, #74104) with on column DNase treatment using RNase-Free DNase Set (Qiagen, #79254). First-strand cDNA synthesis was performed using Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV RT) (Invitrogen, #28025-013) according to the manufacturer's protocol. Reverse transcription and PCR were carried out with the primers listed in Supplementary Table S1.
Gamma irradiation
Radiation sensitivity of UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 and the same cells that have lost the HAC was evaluated using the method described previously (34). Exponentially growing cells were suspended to 600–700 cells per 10 ml in sterile 50 ml tube with complete medium in total volume 50 ml and plated in 5–10 cm plates (Nunc) by 10 ml of cell suspension in each plate (600–700 cells per plate). After cells attachment in 12 h, the cells were irradiated at room temperature using a 60Co γ-ray source at a dose rate of 2, 4, 6 and 8 Gy. After irradiation, plates were placed in 10% CO2 incubator at 37°C and removed after 2 weeks for analysis when the colonies were formed. The percentage of survived cells were counted and normalized to the number of colonies counted in unirradiated control cells.
Immunofluorescence
Gamma-H2AX foci accumulation in UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 and in the same cells that have lost the HAC before and after exposure to 2 Gy of gamma-radiation was measured as previously described with minor changes (35). Note that 5 × 104 cells were plated onto sterile glass coverslips (Fisher finest Superslip, round, thickness 1), placed into 12-well plates and grown to ∼80% confluence. The plates were then irradiated using 2 Gy. Following irradiation, cells were placed in the incubator and processed at stated repair times (15 h post-radiation). Cells attached on the coverslip were washed with PBS and then fixed with 2% Paraformaldehyde (PFA) (formaldehyde in PBS, pH 7.4) for 20 min at room temperature. The PFA was then removed and the coverslip was washed three times with PBS over a 15-min period. Pre-chilled (−20°C) 70% ethanol was added and the coverslip was incubated for 20 min at room temperature. Ethanol was removed and the coverslip was washed with PBS three times over a 15-min period. Then the coverslip was incubated in blocking solution 5% BSA in PBS-TT (PBS containing 0.5% Tween 20, 0.1% Triton X-100) for 1 h at room temperature. Gamma-H2AX primary antibody incubation was carried out in 1% BSA in PBS-TT buffer (antibody dilution was 1:500). After 2–3 washes with PBS (5 min each), the coverslips were incubated with Alexa Fluor 555 Goat anti-mouse antibody at a dilution of 1:500 in 1% BSA in PBS-TT for 1 h at room temperature. Then the cells were washed with PBS for 3–5 times, each time for 5 min with slow shaking. Cells were counterstained with DAPI mounting medium. Images were captured using a DeltaVision microscopy imaging system (40× objective) in the CRC, LRBGE Fluorescence Imaging Facility (NIH). Total intensity per nucleus was measured using image J software (NIH).
Paclitaxel treatment
Our experiment protocol was described previously (36). UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 and isogenic cells that have lost the HAC were treated with various doses of the drug (0–1 nM) for 24 h. At the end of the experiment, MTS tetrazolium cell viability assay was done according to the manufacturer's instructions (The CellTiter 96 AQueous One Solution Cell Proliferation Assay, Promega). Experiments were carried out in triplicate.
Transcription timing analysis
UWB1.289 cells containing alphoidtetO-HAC/BRCA1 and the same cells after HAC loss were incubated with 20 μM EdU for 3.5 h. Then cells were fixed, permeabilized and click labeled with modified Alexa Fluor647 azide using Click-iT Plus EdU Flow Cytometry Assay Kits (Molecular Probes, # C10632) according to the manufacturing protocol with minor modifications. These modification include five times dilution of Click-iT fixative (Component D) by PBS. Then, 500 μl of 1× Click-iT saponin-based permeabilization and wash reagent were added to the stained cells and proceed to the next step for staining the cells for DNA content by FxCycle Violet stain (Molecular Probes, # F10347). The staining procedure was performed according to the manufacturing protocol. After staining UWB1.289 cells containing alphoidtetO-HAC/BRCA1 and after HAC loss the cells were sorted on a flow cytometer to separate the cells for G1 and S phases of the cell cycle. The sorting parameters were as follows: 635 nm excitation and a 660/20 nm bandpass emission filter for detection of the EdU Alexa Fluor647 picolyl azide and 405 nm excitation and a 450/40 bandpass emission filter for detection of the FxCycle Violet fluorescence. The sorted cells were used for total RNA purification, cDNA synthesis and analysis of the transcription of centromere regions by qPCR using specific primes (Supplementary Table S1).
Statistical analysis
Statistical analysis was made using Prism (GraphPad Software Inc., La Jolla, CA, USA). An unpaired Student t-test and, Chi square test and two way ANOVA were used. P < 0.05 was considered significant.
RESULTS
Construction of the HAC module carrying the full-length BRCA1 gene
A total of 90 kb of genomic DNA including the BRCA1 gene was previously isolated as a circular YAC by a TAR cloning technique (23) (Figure 1a, Step 1 and Supplementary Figure S1). To prepare this YAC BRCA1-containing molecule for insertion into the alphoidtetO-HAC, the YAC clone was converted into YAC/BAC using a yeast-bacteria-mammalian cell retrofitting vector pJBRV1 (22) (Figure 1a, Step 2). The pJBRV1 vector contains a 3′ HPRT-loxP cassette. This allows gene loading into the unique loxP site of the alphoidtetO-HAC in hamster CHO cells by Cre-loxP mediated recombination. The retrofitting protocol is shown in Supplementary Figure S2.
Figure 1.
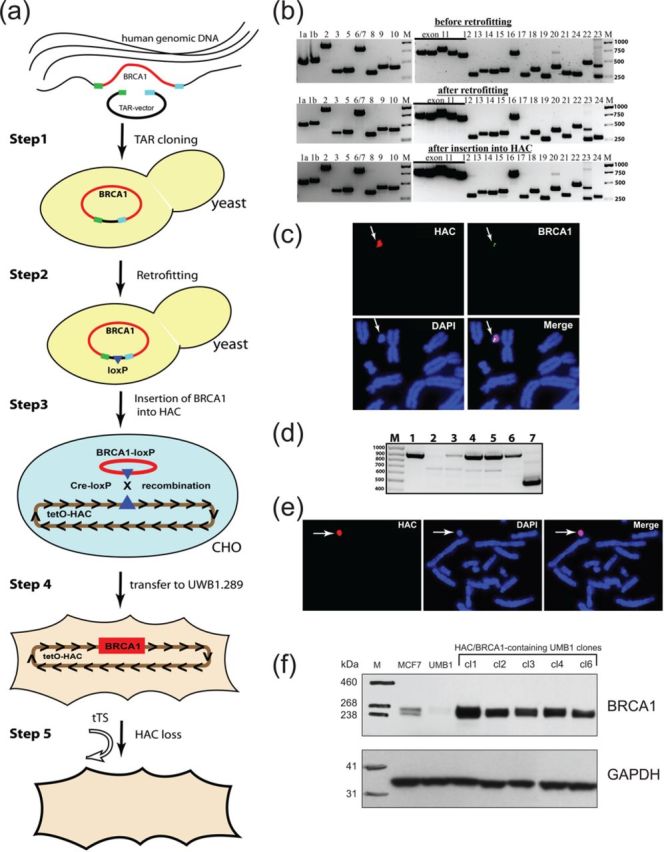
Scheme of consecutive experimental steps from selective BRCA1 gene isolation in yeast S. cerevisiae to its expression in the UWB1.289 BRCA1-deficient human cells. (a) Step 1: A direct TAR isolation of the BRCA1 gene from human genomic DNA. TAR vector contains two gene targeting hooks (green and blue boxes), a yeast centromeric locus (CEN) and a yeast selectable marker HIS3. Recombination between targeting sequences in the TAR vector and the targeted sequences of the genomic DNA fragment leads to the rescue of the BRCA1-containing loci as a circular TAR/YAC molecule. Step 2: Retrofitting of the circular TAR/YAC isolate containing the full-length BRCA1 gene by pJBRV1 vector containing a 3′ HPRT-loxP-eGFP cassette. Recombination of the BamHI-linearized pJBRV1 vector with a TAR/YAC in yeast leads to replacement of the ColE1 origin of replication by the F’′ factor origin of replication that allows enable subsequent propagation in a BAC form. Step 3: BRCA1 gene loading into a unique loxP site of the alphoidtetO-HAC (tetO-HAC) by Cre-loxP recombination system in hamster CHO cells. Step 4: MMCT of alphoidtetO-HAC/BRCA1 from CHO into the human BRCA1-deficient UWB1.289 cells for complementation analyses. Step 5: Elimination of the alphoidtetO-HAC/BRCA1 from UWB1.289 cells by expression of the tTS fusion construct. (b) PCR analysis of the TAR clones containing the full-length BRCA1 gene for the presence of exons before and after retrofitting in yeast and after BRCA1 insertion into a loxP site of the alphoidtetO-HAC in CHO cells. The numbers above correspond to the exon number (from 1 to 24). M, ladder. (c) FISH analysis of the alphoidtetO-HAC/BRCA1 in CHO cells using specific probes for HAC vector (in red) and for cDNA BRCA1 gene sequences (in green). (d) Transcriptional analysis of the human BRCA1 gene in CHO cells. Lane 1 corresponds to a positive control: RT-PCR of RNA purified from human MCF7 cells. Lane 2 corresponds to a negative control: RT-PCR of RNA purified from CHO cells. Lanes from 3 to 6 correspond to RNA purified from five independently obtained alphoidtetO-HAC/BRCA1-containing CHO clones. Lane 7 corresponds to the RT-PCR product of the control ERCCA2 gene. Clones 3, 4, 5 and 6 are BRCA1-positive. All primers designed are presented in Supplementary Table S1. The bands have a predicted size of RT-PCR products. All amplified fragments were gel-purified and sequenced and proved the identity of products to the human BRCA1 transcripts. M, ladder. (e) FISH analysis of the alphoidtetO-HAC/BRCA1 in UWB1.289 cells using specific probes for HAC vector (in red). (f) Western blot analysis of BRCA1-deficient UWB1.289 cells, alphoidtetO-HAC/BRCA1-containing UWB1.289 cells (five independently obtained clones after MMCT transfer from the clone #4 of CHO cells) using human-specific Abs against BRCA1. BRCA1 inserted into the alphoidtetO-HAC produces a protein of the predicted size. The human breast adenocarcinoma MCF7 cell line (hemizygous for the BRCA1 wild type with a reduced BRCA1 expression) was used as a positive control for expression of BRCA1.
To insert the 90 kb genomic copy of the BRCA1 gene into the HAC, the appropriate YAC/BAC construct and a Cre-recombinase expression vector were co-transfected into HPRT-minus CHO cells carrying the alphoidtetO-HAC (Figure 1a, Step 3). A control for quality of BAC DNA is important to efficient gene loading into the HAC. Most of the molecules should be in a covalently closed form (see Supplementary Figure S3). Insertion of the gene into the HAC reconstitutes a functional HPRT gene (Supplementary Figure S4), so HPRT-plus colonies were selected on HAT medium. HPRT reconstitution was confirmed by PCR using a set of specific primers (Supplementary Table S1). Integrity of the BRCA1 gene after insertion into the HAC was confirmed by PCR-amplification of all exon sequences (Figure 1b) using a set of BRCA1-specific primers (Supplementary Table S1). Notably, that only 4 among 30 HPRT-plus colonies selected on HAT medium contained all BRCA1 exons. Such a high rate of deletion of BRCA1 sequences during gene loading into the HAC is presumably due to an abnormal density of Alu repeats representing more than 70% of the BRCA1 gene sequence (37) that can recombine leading to deletions. Note that no deletions were observed after loading of other genes into the HAC (22). Examples of the observed deletions are shown in Supplementary Figure S5. We term the resulting HAC clones containing all BRCA1 exon sequences as alphoidtetO-HAC/BRCA1.A representative FISH image of one alphoidtetO-HAC/BRCA1 clone in CHO cells is shown in Figure 1c. As seen, the HAC is maintained autonomously and the HAC signal co-localizes with the BRCA1 gene signal on metaphase chromosome spreads. Because antibodies against human BRCA1 recognize a band with mobility of hBRCA1 on western blots with the CHO cell lysate, we used RT-PCR analysis to confirm expression of the human BRCA1 gene. RT-PCR products of the predicted size were obtained for four clones containing all exons (Figure 1d), and their identity was confirmed by sequencing (Supplementary Figure S6).
Physiological tests proving functionality of BRCA1 encoded on the HAC
For genetic complementation assays, the BRCA1-containing HAC was transferred from CHO cells to UWB1.289 BRCA1-deficient human cells via MMCT (Figure 1a, Step 4). FISH analysis of five randomly chosen BSR clones showed that alphoidtetO-HAC/BRCA1 propagates autonomously in these cells without detectable integration into chromosomes. A FISH image showing detection of alphoidtetO-HAC/BRCA1 in one representative clone is shown in Figure 1e. Western blot analysis of five HAC positive UWB1.289 clones revealed that they all express BRCA1 (Figure 1f). To confirm that BRCA1 is expressed from the HAC, we targeted the tTS repressor fusion protein to the centromere of the alphoidtetO-HAC/BRCA1 as described previously (19,20,22). The targeting resulted in a high frequency of HAC loss (Figure 1a, Step 5). Based on FISH and western blot analysis, cells that lost the HAC no longer expressed BRCA1 (Supplementary Figure S7). We used five independent tests outlined below to demonstrate the functionality of BRCA1 expressed from the alphoidtetO-HAC/BRCA1 vector.
First test: It is well known that BRCA1 plays a critical role in damage responses to DNA double-strand breaks (DSBs) (4) and within seconds of the generation of DSBs histone H2AX molecules accumulate in foci at sites of DNA damage. These foci then rapidly disappear as the damage is repaired, a process that requires BRCA1 function. To follow the processing of DSBs after irradiation (2 Gy), the total intensity of gamma-H2AX foci was measured at 15 h post-irradiation in UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 and in isogenic cells that been induced to lose the HAC. Figure 2a depicts selected fluorescence microscopy images of non-irradiated (NR) and irradiated cells. When we scored the persistence of the DNA damage foci as defined by gamma-H2AX staining, a statistically significant difference was observed between the two isogenic cell lines (P < 0.05). The number of foci and consequently the total intensity of the gamma-H2AX fluorescence was significantly increased in the cells that have lost the alphoidtetO-HAC/BRCA1 (Figure 2b). This indicates that after 15 h post-irradiation many DSBs are still unrepaired. A similar observation that radiation-induced gamma-H2AX foci are higher in BRCA1-deficient cells compared to isogenic cells expressing BRCA1 has been previously reported (4).
Figure 2.
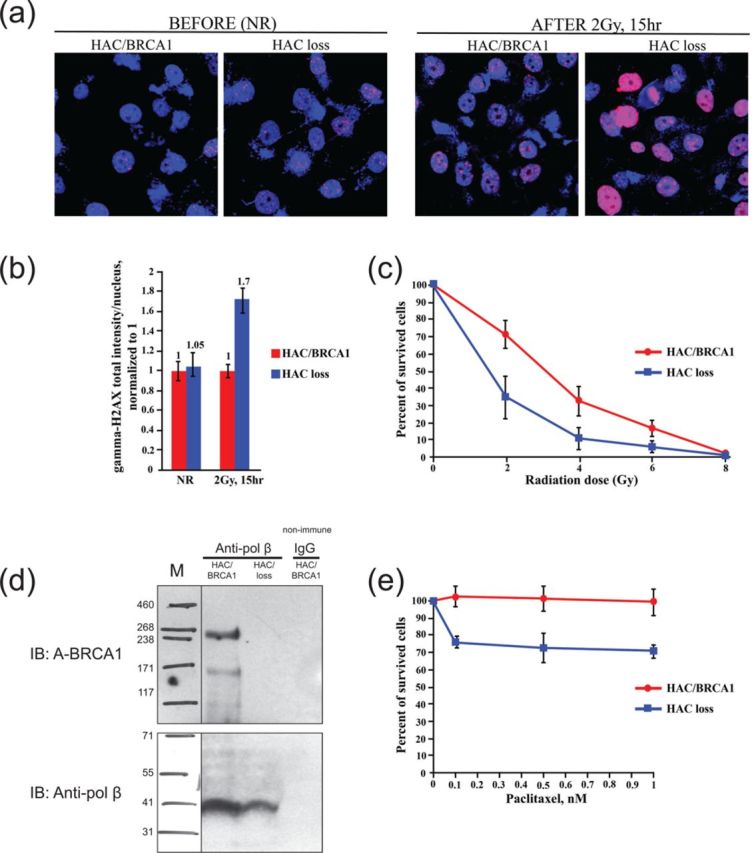
Complementation tests for functionality of BRCA1 in UWB1.289 cells. (a) Accumulation of γ-H2AX in BRCA1-deficient cells (UWB1.289) carrying alphoidtetO-HAC/BRCA1 and in the cells that have lost the HAC after exposure to 2 Gy of γ-rays. Cells were stained 15 h after irradiation with anti-γ-H2AX antibodies (red) and with DAPI (blue). Representative microscopic fluorescence images of γ-H2AX foci accumulation are shown. (b) Quantitative data for γ-H2AX foci processing. Cells were collected after 15 h and the average number of γ-H2AX foci was quantified in BRCA1-positive and BRCA1-negative cells. NR cells have been included as a control. (c) Radiation sensitivity clonogenic survival assay. BRCA1-deficient UWB1.289 cells are radiation sensitive. Cells carrying alphoidtetO-HAC/BRCA1 and after HAC loss were plated in triplicate and irradiated with the indicated doses (from 0 to 8 Gy). Cells were stained with crystal violet and counted visually. Clonogenic survival was calculated as a number of colonies present in each plate normalized to the number of colonies in the NR control plates. Cells carrying alphoidtetO-HAC/BRCA1 (red) are less sensitive than the cells that lost the HAC (blue). Bars, mean of triplicate samples. (d) Co-immunoprecipitation analysis of BRCA1 and pol β proteins interaction. Cell extracts from the cells carrying alphoidtetO-HAC/BRCA1 and from the cells that have lost the HAC were immunoprecipitated with anti-pol β antibody or non-immune IgG. Immunoprecipitated proteins were detected by SDS-polyacrylamide gel electrophoresis and immunoblotting. (e) Viability of UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 (red) and after HAC loss (blue) after treatment with paclitaxel. At 24 h after treatment the cell viability was measured. All experiments were repeated three times with each sample prepared in triplicate.
Second test: Prior studies have shown that UWB1.289 cells are radiation sensitive in clonogenic survival assays and that BRCA1 expression partially corrects this sensitivity (27). Therefore, we compared the colony formation capacity of UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 with that of isogenic cells that had lost the HAC in response to increasing doses of irradiation up to 6 Gy. As expected, UWB1.289 cells carrying alphoidtetO-HAC/BRCA1 cells exhibited greater resistance to irradiation than isogenic cells following induction of HAC loss (Figure 2c) [ANOVA F (1, 4) = 37.27, P = 0.0036].
Third test: Recent studies have demonstrated a partnership between BRCA1 and DNA polymerase β in the base excision repair pathway (33). Therefore, we looked for interactions between the two proteins using co-immunoprecipitation analysis. For this purpose, extracts from UWB1.289 cells expressing BRCA1 (carrying alphoidtetO-HAC/BRCA1) and from isogenic cells that had lost the HAC were prepared and subjected to immunoprecipitation using anti-pol β or anti-BRCA1 antibodies. As expected, BRCA1 co-immunoprecipitated with pol β antibody (IP with anti-pol β) from extracts of cells carrying alphoidtetO-HAC/BRCA1 (e.g. expressing BRCA1), whereas no immunoprecipitation was observed using extracts from cells that have lost the alphoidtetO-HAC/BRCA1 (Figure 2d). As an additional negative control, immunoprecipitation experiments were performed using non-immune IgG. These results confirm that human BRCA1 expressed from alphoidtetO-HAC physically interacts with pol β (33).
Fourth test: Zhou et al. (38) showed that BRCA1 helps to mediate resistance to paclitaxel, the most commonly used chemotherapy drug for patients with ovarian cancer. We therefore measured the viability of UWB1.289 cells (which come from an ovarian carcinoma) either carrying alphoidtetO-HAC/BRCA1 and after loss of the HAC in response to a range of paclitaxel concentrations (0–1 nM) for 24 h. The percentage of viable UWB1.289 cells expressing BRCA1 after exposure to the drug was higher than the percentage of cells that had lost the HAC (Figure 2e) [ANOVA F (1, 4) = 13.43, P = 0.0215]. Thus, the loss of BRCA1 sensitizes UWB1.289 cells to paclitaxel.
Fifth test: To investigate whether the presence of the functional BRCA1 gene is required for accurate chromosome segregation, we performed a micronucleus formation assay (MNi) in UWB1.289 cells either expressing BRCA1 from the HAC or after HAC loss. This assay revealed a significant difference in MNi formation between cells expressing BRCA1 (24%) and BRCA1-deficient cells (38%) (Table 1). Our results therefore support previous reports that BRCA1 is required for accurate chromosome segregation (4,39,40).
Table 1. Micronuclei (MNi) formation in the HAC/BRCA1-containing UWB1.289 cells and in the same cells that have lost the HAC.
| Experiment | No Mni | Mni | Total | %Mni | Mean ± SD (%)** |
|---|---|---|---|---|---|
| BRCA1 positive (A)* | 155 | 53 | 208 | 25.5 | 24.7±1.1 |
| BRCA1 positive (B) | 156 | 49 | 205 | 23.9 | |
| BRCA1 negative (A) | 129 | 81 | 210 | 38.6 | 38.5±0.1 |
| BRCA1 negative (B) | 125 | 78 | 203 | 38.4 |
A, the first experiment; B, the second experiment.
**Number of MN is given as means ± SD in two independent experiments. There is a significant difference between BRCA1 positive and BRCA1 negative cells [χ2 (1) = 18.20, P < 0.0001].
To summarize, our five test experiments demonstrated that the alphoidtetO-HAC/BRCA1 vector expresses a functional BRCA1 protein.
BRCA1 functions to promote maintenance of heterochromatin domains within a functional human kinetochore
Recently Zhu et al. (9) reported that BRCA1 deficiency results in transcriptional de-repression of tandemly repeated satellite DNA in mouse and human, including those specific for centromeric loci. In mouse cells, their results clearly showed that both centromeric and pericentromeric regions are affected by BRCA1 deficiency. Such analysis was possible because these two regions are formed on different types of repeated sequences, i.e. minor and major satellites. In human centromeric regions, most alpha-satellite DNA repeats (∼90%) are organized into chromosome-specific HORs (26) that directly involved in kinetochore assembly. Therefore, two main domains of the human kinetochore, centrochomatin and heterochromatin, are assembled on the same DNA sequences. Such organization complicates the analysis of human centromeric repeats within a functional kinetochore in the human BRCA1-deficient cells. In their work, Zhu et al. used the primers that were not specific to HORs of any individual chromosome (9). Thus, their primers recognized only a minor fraction of diverged alpha-satellite DNA monomers localized outside of the major block of alpha-satellite DNA.
Taking an advantage that a complete sequence representation of alpha-satellite monomers across centromeric regions of the most human chromosomes became available (26) in our study, we focused on well-characterized HORs of several human chromosomes. Initially, we chose single predominant HOR blocks on chromosome 21 (30) and on chromosome X (41), that are required for kinetochore function, for qRT-PCR analysis. Transcription of satellite DNA in these regions was found to be 20- to 30-fold de-repressed in cells that had been ‘cured’ of alphoidtetO-HAC/BRCA1, i.e. in the cells that became BRCA1-deficient relative to the isogenic cells expressing functional BRCA1 (Figure 3a) [chr21a t(4) = 5.7845, P = 0.0044; chr21b t(4) = 21.3600, P < 0.0001; chrX t(4) = 5.2025, P = 0.0065]. Next, we determined the timing of transcription of these regions induced by BRCA1 deficiency. For this purpose, an asynchronous population of human cells was pulse-labeled with EdU. EdU-treated cells were sorted by FACS to two fractions: G1 and S. In each fraction replicating sequences were quantified by qPCR using specific primers (Supplementary Table S1). As seen from Figure 3b, transcription occurs within G1-S phases when the majority of human genes are expressed. We also investigated the effects of BRCA1 loss on transcription of pericentromeric satellite 2 (Sat2), a heterochromatic locus mapped within 10q21, and also observed de-repressed in cells that have lost alphoidtetO-HAC/BRCA1 [SAT2 t(4) = 11.3205, P = 0.0003] (Figure 3c). In separate experiments, we checked the chromatin status of the analyzed regions after loss of BRCA1. Specifically, enrichment in H3K4me3 (a marker for active transcription) and H3K9me3 (a marker for heterochromatin) were evaluated by ChIP analysis. As seen from Supplementary Figure S8, loss of BRCA1 was accompanied by a detectable decrease in levels of the heterochromatin marker at centromere 21 [chr21 t(4) = 9.7901, P = 0.0006]. No significant changes were seen in H3K4me3, but as this mark is typically found proximal to promoters, small local changes may not have been detectible in our analysis (Supplementary Figure S8).
Figure 3.
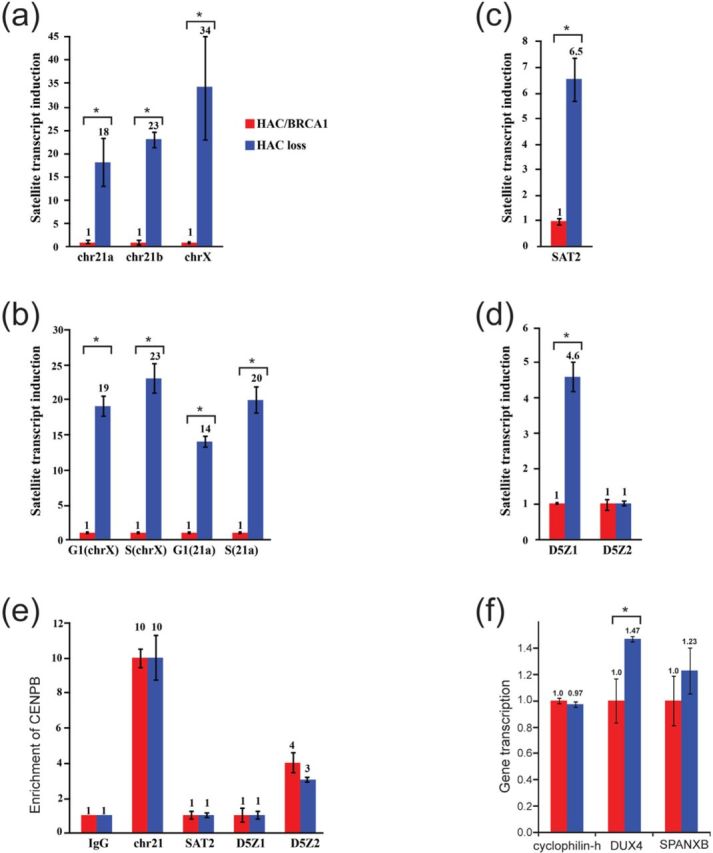
De-repression of alpha-satellite DNA transcription in BRCA1-deficient UWB1.289 cells. (a, c and d) Quantitative RT-PCR experiments showing that the satellite DNA transcripts of HORs regions of the chromosome 21 (chr21a and chr21b), chromosome X (chrX) (a), chromosome 5 (D5Z1 and D5Z2) (d) and DNA transcripts of pericentromeric Sat2 repeats located at 10q21 (c) are significantly repressed in the cells carrying alphoidtetO-HAC/BRCA1 compared to that of the cells that have lost the HAC. Ct values were normalized relative to each other. Error bars indicate SD. (b) Timing of transcription of HORs regions of chromosome X and chromosome 21. (e) ChIP analysis of different centromeric regions using antibodies against CENP-B (centromeric protein B) in BRCA1-deficient UWB1.289 cells containing alphoidtetO -HAC /BRCA1 and in the same cells that have lost the HAC. (f) Quantitative RT-PCR experiments showing the transcripts of the double homeobox 4 (DUX4) gene located in the heterochromatic subtelomeric 4q35 region, the cancer-testis SPANX-B gene located at the Xq27 region and, as a control, a housekeeping gene PPIH, peptidyprolyl isomerase H (cyclophilin H) was used. Asterisk denotes significant difference.
Alpha-satellite or alphoid DNA sequences in HORs are organized into two types of chromatin, heterochromatin (surrounding the kinetochore) and centrochromatin (within the kinetochore domain) corresponding to transcriptionally competent chromatin (41–43). To clarify which compartment of alphoid DNA repeats is predominantly transcribed in BRCA1-deficient cells, we took an advantage of human chromosomes 5 where heterochromatin and centrochromatin are formed on two different HORs: D5Z1 and D5Z2.
These arrays can be distinguished by PCR (32,44,45). Earlier ChIP and PCR analyses revealed that the kinetochore forming CENP-A domain is predominantly localized to D5Z2 while being largely excluded from the pericentromeric D5Z1 domain in a range of human cell lines (32). It is also known that D5Z2 possesses intact CENP-B box sequences (46), a 17 bp motif necessary for binding of the CENP-B protein.
We first checked the CENP-A status on these HORs in BRCA1-deficient UWB1.289 cells as well as in the same cells carrying alphoidtetO-HAC/BRCA1 (Supplementary Figure S9) and confirmed that CENP-A is significantly enriched on D5Z2 compared to D5Z1 [BRCA1 positive cells [t(4) = 4.6093, P = 0.0100; BRCA1 negative cells t(4) = 28.1423, P < 0.0001]. Further, RT-RCR analysis of these regions revealed that transcription is specifically activated within alphoid DNA repeats that are organized into pericentromeric heterochromatin domains, i.e. D5Z1 following the loss of the HAC (Figure 3d) [D5Z1 t(4) = 14.8956, P = 0.0001]. Next, we investigated the CENP-B status on chromosome 5 as well as on chromosome 21 in BRCA1-deficient UWB1.289 cells as well as in the same cells carrying alphoidtetO-HAC/BRCA1. As seen, the level of the CENP-B protein was not significantly affected by the lack of BRCA1 (Figure 3e).
To ask whether the de-repression upon loss of functional BRCA1 described above for certain alpha-satellite loci is indicative of a loss of silencing genome-wide, the transcriptional status of two known human silenced genes, SPANX-B and DUX4 was examined. SPANX-B belongs to the SPANX (sperm protein associated with the nucleus on the X chromosome) gene family of cancer-/testis-specific antigens whose expression is restricted to the normal testis and certain tumors (47–49). These genes are located in segmental duplications at Xq27. The double homeobox 4 (DUX4) gene is located within a 3.3 kb dispersed repeat family that is represented by 10–100 tandem copies on the D4Z4 repeat array in the heterochromatic subtelomeric region 4q35 (50,51). No significant de-repression of transcription of SPANX-B genes was observed in BRCA1-deficient cells (Figure 3e). However, we observe transcriptional de-repression for DUX4 [t(4) = 4.8639, P = 0.0083] in BRCA1-deficient cells in comparison with UWB1.289 cells expressing BRCA1 (Figure 3f).
Thus, our data obtained by a different approach and focused on other chromosome loci are in agreement with those obtained by Zhu et al. (9) and indicate that BRCA1 functions to promote heterochromatin silencing in a genomic locus-specific manner. In addition, we analyzed chromosome-specific HORs. This analysis revealed that BRCA1 deficiency activates transcription of alpha-satellite HORs assembled in heterochromatin domains essential for the kinetochore function. Such activation may explain chromosome instability observed in cells lacking a functional BRCA1.
Expression of the human BRCA1 gene from the HAC-BRCA1 module in porcine cells
The portable HAC module could be potentially used for development of a pig model for breast cancer (52), which offers an alternative to mouse models that do not faithfully mimic the relevant human disease. As the first step toward such a model, we have transferred the BRCA1-containing HAC from CHO cells to the pig's testis fibroblast ST cells using MMCT technique (Figure 4a). FISH analysis of porcine cells containing the alphoidtetO-HAC/BRCA1 in two clones, pl4 and pl5, confirmed an autonomous form of the HAC in the cells without detectable integration into chromosomes (Figure 4b). The BRCA1-HAC module contains the eGFP transgene that is also expressed. Fluorescence image of cells carrying the alphoidtetO -HAC/BRCA1/eGFP is shown in Figure 4c. Expression of human BRCA1 in ST cells bearing alphoidtetO-HAC/BRCA1 was confirmed by RT-PCR (Figure 4e and Supplementary Figure S10) by using primers that specifically amplify the human BRCA1 but not the porcine BRCA1 gene (Supplementary Table S1).
Figure 4.
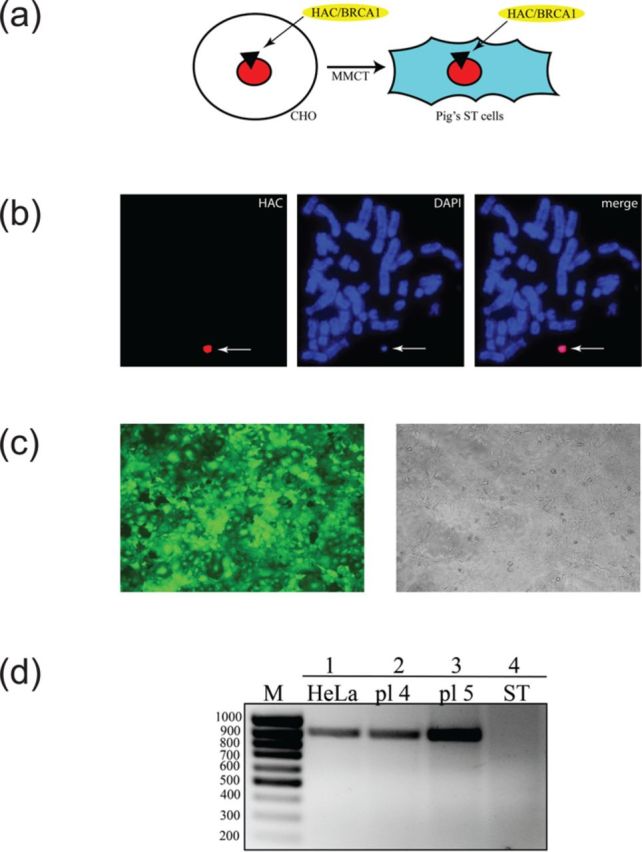
Expression of the human BRCA1 gene in porcine ST cells. (a) From hamster CHO cells exhibiting a high efficiency of microcell formation, the alphoidtetO -HAC/BRCA1 was transferred into the pig's testis fibroblast ST cells using MMCT technique. (b) FISH analysis of porcine cells containing the alphoidtetO-HAC/BRCA1. Chromosomal DNA was counterstained with DAPI (blue). Arrow indicates to the HAC. (c) Fluorescence image of cells carrying the alphoidtetO -HAC/BRCA1 containing the expressed eGFP transgene. (d) Expression of the human BRCA1 gene in porcine cells. RT-PCR analysis of genomic DNA isolated from the original ST cells (lane 4), from two porcine clones (pl4 and pl5) containing the alphoidtetO -HAC/BRCA1 (lanes 2 and 3) and from the human HeLa cells as a positive control (lane 1) using the human BRCA1-specific primers (Supplementary Table S1). The size of the predicted product is 839 bp. M-ladder containing alphoidtetO -HAC/BRCA1
DISCUSSION
In our previous work, we converted the alphoidtetO-HAC, a synthetic artificial human chromosome with a kinetochore that can be inactivated by targeting of chromatin modifiers (19), into a vector able to stably maintain cloned genomic loci over multiple cell generations (20). This HAC contains a unique loxP site that allows selection for gene loading. We previously demonstrated that the alphoidtetO-HAC vector could deliver and stably maintain genomic copies of two functional human genes, VHL and NBS1, and complement the corresponding genetic deficiencies in patient-derived cell lines (22).
In the present study, the alphoidtetO-HAC was used for delivery and expression of a 90-kb genomic region encoding the functional BRCA1 gene. We demonstrated that the BRCA1 gene isolated by TAR cloning (24,25) can be introduced into the alphoidtetO-HAC, resulting in indefinite propagation of the artificial chromosome in hamster CHO cells. Because CHO cells exhibit a high efficiency of microcell formation, this enables transfer of the HAC to other cell types via MMCT (28,29). The alphoidtetO-HAC/BRCA1 was accordingly transferred into the ovarian carcinoma cell line UWB1.289, derived from a patient with a BRCA1 deficiency. Functional expression of BRCA1 in recipient cells was proven by a set of five independent tests, based on the known functions of the protein. Importantly, this system allows corresponding controls to be conducted in isogenic cells, following elimination of the HAC from the cells after targeted inactivation of its kinetochore.
An important property of the alphoidtetO-HAC is its ability to support long-term expression of cloned genes without transgene-induced silencing. Thus, no significant changes in the level of BRCA1 expression were detected after more than 6 months of propagation of alphoidtetO-HAC/BRCA1 in the patient-derived cell line that is in agreement with our results obtained with other genes loaded in the HAC (22).
In this work, we examined the recently proposed role of BRCA1 in maintenance of heterochromatin integrity within pericentromeric and centromeric satellite repeats. A link between BRCA1 foci and pericentromeric chromatin was observed several years ago (1). More recently, Zhu et al. (9) identified histone H2A as a specific substrate of BRCA1 within pericentromeric satellite DNA in both mouse and human. Their study reported that depletion of BRCA1 results in de-repression of satellite DNA transcription and abnormal heterochromatin structure. Although the mechanism is not known, it is likely to involve decreased levels of the monoubiquitylated histone H2A (UbH2A).
For human cells, three types of human satellite repeats, satellite III (localized on the short arms of acrocentric chromosomes and in the heterochromatin of the long arms of chromosome 1, 9 and Y), gamma-satellite (localized on the chromosome X) (53) and alphoid DNA were analyzed by RT-PCR in the original study (9). However, the alphoid DNA families amplified by chosen primers were not distinguished in the prior study and the genomic positions of the transcribed sequences of these repeats cannot be mapped unambiguously. Also, it is difficult to conclude what fraction of the repeats is transcribed.
In our work, using the alphoidtetO-HAC carrying the full-length BRCA1 gene we carried out a more detailed analysis of chromatin status of chromosome-specific alpha-satellite repeats after loss of BRCA1. For this purpose, we chose well-characterized alpha-satellite regions corresponding to megabase-size HORs of human chromosomes 21 and X. For each HOR, specific diagnostic primers were used, allowing us to cover ∼90% of the alpha-satellite DNA monomers in these chromosomal alphoid arrays (26). This analysis revealed that a loss of the BRCA1 gene is accompanied by 20- to 30-fold increase of alphoid DNA transcripts within HORs of these two chromosomes. Transcription was also activated within pericentromeric satellite 2 repeats located at 10q21, a region that is commonly used as a control to define pericentromeric heterochromatin (19,30). In the same experiments, a detectable increase in the level of transcription was observed at one region enriched in heterochromatin, i.e. for the peritelomeric region 4q35 (51), and no increase for a genomic region carrying cancer-testis specific genes at Xq27 (47). Thus, these data agree with those obtained by Zhu et al. (9) and indicate that BRCA1 functions to promote transcriptional silencing of heterochromatin in a genomic locus-specific manner.
To clarify which fraction of alphoid DNA repeats, organized into heterochromatin or centrochromatin, is predominantly transcribed in BRCA1-deficient cells, we analyzed HOR arrays of the human chromosome 5, which consists of two different blocks of alphoid DNA repeats. This analysis clearly demonstrated that transcription is specifically activated within alphoid DNA repeats that are organized into heterochromatin domains that are distal to the kinetochore forming alpha-satellite arrays.
It is well documented that loss of BRCA1 is accompanied by chromosome instability (4,39,40). One accepted explanation for defects in chromosome transmission is due to misregulation of centrosome copy number (54). However, elevated transcription of centromeric repeats can also lead to kinetochore disassembly (42). Based on this observation and our data on analysis of HORs transcription, we propose that impairment of kinetochore function due to elevated transcription may also contribute chromosome instability in BRCA1-deficient cells and may contribute to cellular transformation.
Large heterochromatic blocks of satellite DNA are typically clustered in the nucleus and could provide anchor points for genomic organization. De-regulated transcription in these regions in BRCA1-deficient cells would be expected to lead to epigenetic alterations that could impact on regulation of nearby gene(s) and nuclear structural interactions (55). It is worth noting that there are multiple lines of evidence to support the idea that overexpression of satellite DNA and other repetitive elements may be widely prevalent in many types of cancer (56,57).
An increasing set of evidence strongly indicates that a principal tumor suppressor activity of BRCA1 is a repair of DNA damages (58,59). Clarification of a role of chromosome instability in cell transformation will require additional experiments of which functional complementation like that reported here may contribute important information. The portable HAC module carrying the full-length BRCA1 gene should be useful for analysis of the tumor suppressor function of BRCA1. As mentioned above, the HAC/BRCA1 module assembled in hamster CHO cells can be transferred into any recipient cells, including transgenic animals. For example, the module could potentially be used for development of a pig model for breast cancer (52), which offers an alternative to mouse models that do not faithfully mimic the relevant human disease. As the first step toward such a model, we have demonstrated the stable propagation of the alphoidtetO-HAC/BRCA1 and expression of hBRCA1 in porcine cells.
To summarize, we have here constructed a portable BRCA1-HAC module and demonstrated its utility for delivery of the full-length BRCA1 gene and correction of genetic deficiencies in human and porcine cells. Using this module, in which the full-length transcript, including any splice variants and intronic signals, can be expressed in cell lines or removed from them without altering the genomic DNA complement, we have extended an earlier study (9) suggesting that BRCA1 is required for the normal silencing of certain DNA repeats. More importantly, we demonstrated a clear link between specific disruption of centromeric heterochromatin essential for kinetochore and loss of BRCA1 function. A BRCA1-HAC module is available under request from Developmental Therapeutics Branch, NCI, NIH.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors would like to thank the CRC, LRBGE Fluorescence Imaging Facility (NIH) and personally Drs. Karpova and Dr. McNally for instructions, consultations and help with the usage of a DeltaVision microscopy imaging system.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, USA [to V.L. and N.K]; Wellcome Trust Principal Research Fellowship [073915 to W.C.E.]; Grand-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology of Japan [23247030, 23114008 to H.M.]; Kazusa DNA Research Institute Foundation [to H.M.]; Indiana Genomics Initiative (INGEN) [to B.R.G.]. INGEN is supported in part by the Lilly Endowment. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pageau G.J., Lawrence J.B. BRCA1 foci in normal S-phase nuclei are linked to interphase centromeres and replication of pericentric heterochromatin. J. Cell Biol. 2006;175:693–701. doi: 10.1083/jcb.200602055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pageau G.J., Hall L.L., Ganesan S., Livingston D.M., Lawrence J.B. The disappearing Barr body in breast and ovarian cancers. Nat. Rev. Cancer. 2007;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- 3.Moynahan M.E., Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell. Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hair J.M., Terzoudi G.I., Hatzi V.I., Lehockey K.A., Srivastava D., Wang W.X., Pantelias G.E., Georgakilas A.G. BRCA1 role in the mitigation of radiotoxicity and chromosomal instability through repair of clustered DNA lesions. Chem. Biol. Interact. 2010;188:350–358. doi: 10.1016/j.cbi.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 5.Huen M.S., Sy S.M., Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell. Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joosse S.A. BRCA1 and BRCA2: a common pathway of genome protection but different breast cancer subtypes. Nat. Rev. Cancer. 2012;12:372. doi: 10.1038/nrc3181-c2. [DOI] [PubMed] [Google Scholar]
- 8.Chang S., Sharan S.K. The role of epigenetic transcriptional regulation in BRCA1-mediated tumor suppression. Transcription. 2013;4:24–28. doi: 10.4161/trns.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Q., Pao G.M., Huynh A.M., Suh H., Tonnu N., Nederlof P.M., Gage F.H., Verma I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:U179–U176. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lufino M.M., Edser P.A., Wade-Martins R. Advances in high-capacity extrachromosomal vector technology: episomal maintenance, vector delivery, and transgene expression. Mol. Ther. 2008;16:1525–1538. doi: 10.1038/mt.2008.156. [DOI] [PubMed] [Google Scholar]
- 11.Matrai J., Chuah M.K., Vanden Driessche T. Recent advances in lentiviral vector development and applications. Mol. Ther. 2010;18:477–490. doi: 10.1038/mt.2009.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchholz C.J., Muhlebach M.D., Cichutek K. Lentiviral vectors with measles virus glycoproteins - dream team for gene transfer. Trends Biotechnol. 2009;27:259–265. doi: 10.1016/j.tibtech.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Banasik M.B., McCray P.B., Jr Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- 14.Wanisch K., Yanez-Munoz R.J. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Therapy. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouprina N., Earnshaw W.C., Masumoto H., Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell. Mol. Life Sci. 2013;70:1135–1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouprina N., Tomilin A.N., Masumoto H., Earnshaw W.C., Larionov V. Human artificial chromosome-based gene delivery vectors for biomedicine and biotechnology. Expert Opin. Drug Deliv. 2014;11:517–535. doi: 10.1517/17425247.2014.882314. [DOI] [PubMed] [Google Scholar]
- 17.Kazuki Y., Oshimura M. Human artificial chromosomes for gene delivery and the development of animal models. Mol. Ther. 2011;19:1591–1601. doi: 10.1038/mt.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshimura M., Kazuki Y., Iida Y., Uno N. eLS. Chichester: John Wiley & Sons Ltd.; 2013. [Google Scholar]
- 19.Nakano M., Cardinale S., Noskov V.N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W.C., Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iida Y., Kim J.H., Kazuki Y., Hoshiya H., Takiguchi M., Hayashi M., Erliandri I., Lee H.S., Samoshkin A., Masumoto H., et al. Human artificial chromosome with a conditional centromere for gene delivery and gene expression. DNA Res. 2010;17:293–301. doi: 10.1093/dnares/dsq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kouprina N., Samoshkin A., Erliandri I., Nakano M., Lee H.S., Fu H.G., Iida Y., Aladjem M., Oshimura M., Masumoto H., et al. Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth. Biol. 2012;1:590–601. doi: 10.1021/sb3000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J.H., Kononenko A., Erliandri I., Kim T.A., Nakano M., Iida Y., Barrett J.C., Oshimura M., Masumoto H., Earnshaw W.C., et al. Human artificial chromosome (HAC) vector with a conditional centromere for correction of genetic deficiencies in human cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20048–20053. doi: 10.1073/pnas.1114483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Annab L.A., Kouprina N., Solomon G., Cable P.L., Hill D.E., Barrett J.C., Larionov V., Afshari C.A. Isolation of a functional copy of the human BRCA1 gene by transformation-associated recombination in yeast. Gene. 2000;250:201–208. doi: 10.1016/s0378-1119(00)00180-3. [DOI] [PubMed] [Google Scholar]
- 24.Kouprina N., Larionov V. Innovation - TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 2006;7:805–812. doi: 10.1038/nrg1943. [DOI] [PubMed] [Google Scholar]
- 25.Kouprina N., Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 26.Miga K.H., Newton Y., Jain M., Altemose N., Willard H.F., Kent W.J. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DelloRusso C., Welcsh P.L., Wang W., Garcia R.L., King M.C., Swisher E.M. Functional characterization of a novel BRCA1-null ovarian cancer cell line in response to ionizing radiation. Mol. Cancer Res. 2007;5:35–45. doi: 10.1158/1541-7786.MCR-06-0234. [DOI] [PubMed] [Google Scholar]
- 28.Fournier R.E.K., Ruddle F.H. Microcell-mediated transfer of murine chromosomes into mouse, chinese-hamster, and human somatic-cells. Proc. Natl. Acad. Sci. U.S.A. 1977;74:319–323. doi: 10.1073/pnas.74.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koi M., Shimizu M., Morita H., Yamada H., Oshimura M. Construction of mouse A9 clones containing a single human-chromosome tagged with neomycin-resistance gene via microcell fusion. Jpn. J. Cancer Res. 1989;80:413–418. doi: 10.1111/j.1349-7006.1989.tb02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohzeki J., Bergmann J.H., Kouprina N., Noskov V.N., Nakano M., Kimura H., Earnshaw W.C., Larionov V., Masumoto H. Breaking the HAC barrier: Histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kononenko A.V., Lee N.C., Earnshaw W.C., Kouprina N., Larionov V. Re-engineering an alphoid(tetO)-HAC-based vector to enable high-throughput analyses of gene function. Nucleic Acids Res. 2013;41:e107. doi: 10.1093/nar/gkt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slee R.B., Steiner C.M., Herbert B.S., Vance G.H., Hickey R.J., Schwarz T., Christan S., Radovich M., Schneider B.P., Schindelhauer D., et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31:3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- 33.Masaoka A., Gassman N.R., Horton J.K., Kedar P.S., Witt K.L., Hobbs C.A., Kissling G.E., Tano K., Asagoshi K., Wilson S.H. Interaction between DNA polymerase beta and BRCA1. Plos ONE. 2013;8:e66801. doi: 10.1371/journal.pone.0066801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scully R., Ganesan S., Vlasakova K., Chen J., Socolovsky M., Livingston D.M. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol. Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 35.Francisco D.C., Peddi P., Hair J.M., Flood B.A., Cecil A.M., Kalogerinis P.T., Sigounas G., Georgakilas A.G. Induction and processing of complex DNA damage in human breast cancer cells MCF-7 and nonmalignant MCF-10A cells. Free Rad. Biol. Med. 2008;44:558–569. doi: 10.1016/j.freeradbiomed.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 36.Lee H.S., Lee N.C.O., Grimes B.R., Samoshkin A., Kononenko A.V., Bansal R., Masumoto H., Earnshaw W.C., Kouprina N., Larionov V. A new assay for measuring chromosome instability (CIN) and identification of drugs that elevate CIN in cancer cells. BMC Cancer. 2013;13:252. doi: 10.1186/1471-2407-13-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T.M., Lee M.K., Szabo C.I., Jerome N., McEuen M., Taylor M., Hood L., King M.C. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- 38.Zhou C.Y., Smith J.L., Liu J.S. Role of BRCA1 in cellular resistance to paclitaxel and ionizing radiation in an ovarian cancer cell line carrying a defective BRCA1. Oncogene. 2003;22:2396–2404. doi: 10.1038/sj.onc.1206319. [DOI] [PubMed] [Google Scholar]
- 39.Stolz A., Ertych N., Bastians H. Loss of the tumour-suppressor genes CHK2 and BRCA1 results in chromosomal instability. Biochem. Soc. Trans. 2010;38:1704–1708. doi: 10.1042/BST0381704. [DOI] [PubMed] [Google Scholar]
- 40.Kwei K.A., Kung Y., Salari K., Holcomb I.N., Pollack J.R. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol. Oncol. 2010;4:255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mravinac B., Sullivan L.L., Reeves J.W., Yan C.M., Kopf K.S., Farr C.J., Schueler M.G., Sullivan B.A. Histone modifications within the human X centromere region. Plos ONE. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergmann J.H., Jakubsche J.N., Martins N.M., Kagansky A., Nakano M., Kimura H., Kelly D.A., Turner B.M., Masumoto H., Larionov V., et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 2012;125:411–421. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergmann J.H., Martins N.M.C., Larionov V., Masumoto H., Earnshaw W.C. HACking the centromere chromatin code: insights from human artificial chromosomes. Chromosome Res. 2012;20:505–519. doi: 10.1007/s10577-012-9293-0. [DOI] [PubMed] [Google Scholar]
- 44.Finelli P., Antonacci R., Marzella R., Lonoce A., Archidiacono N., Rocchi M. Structural organization of multiple alphoid subsets coexisting on human chromosomes 1, 4, 5, 7, 9, 15, 18, and 19. Genomics. 1996;38:325–330. doi: 10.1006/geno.1996.0635. [DOI] [PubMed] [Google Scholar]
- 45.Wevrick R., Willard H.F. Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res. 1991;19:2295–2301. doi: 10.1093/nar/19.9.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosandic M., Paar V., Basar I., Gluncic M., Pavin N., Pilas I. CENP-B box and pJalpha sequence distribution in human alpha satellite higher-order repeats (HOR) Chromosome Res. 2006;14:735–753. doi: 10.1007/s10577-006-1078-x. [DOI] [PubMed] [Google Scholar]
- 47.Kouprina N., Mullokandov M., Rogozin I.B., Collins N.K., Solomon G., Otstot J., Risinger J.I., Koonin E.V., Barrett J.C., Larionov V. The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3077–3082. doi: 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kouprina N., Noskov V.N., Pavlicek A., Collins N.K., Bortz P.D.S., Ottolenghi C., Loukinov D., Goldsmith P., Risinger J.I., Kim J.H., et al. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. Plos ONE. 2007;2:e359. doi: 10.1371/journal.pone.0000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kouprina N., Pavlicek A., Noskov V.N., Solomon G., Otstot J., Isaacs W., Carpten J.D., Trent J.M., Schleutker J., Barrett J.C., et al. Dynamic structure of the SPANX gene cluster mapped to the prostate cancer susceptibility locus HPCX at Xq27. Genome Res. 2005;15:1477–1486. doi: 10.1101/gr.4212705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexiadis V., Ballestas M.E., Sanchez C., Winokur S., Vedanarayanan V., Warren M., Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim. Biophys. Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 52.Luo Y., Bolund L., Sorensen C.B. Pig gene knockout by rAAV-mediated homologous recombination: comparison of BRCA1 gene knockout efficiency in Yucatan and Gottingen fibroblasts with slightly different target sequences. Transgenic Res. 2012;21:671–676. doi: 10.1007/s11248-011-9563-1. [DOI] [PubMed] [Google Scholar]
- 53.Spence J.M., Critcher R., Ebersole T.A., Valdivia M.M., Earnshaw W.C., Fukagawa T., Farr C.J. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J. 2002;21:5269–5280. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kais Z., Chiba N., Ishioka C., Parvin J.D. Functional differences among BRCA1 missense mutations in the control of centrosome duplication. Oncogene. 2012;31:799–804. doi: 10.1038/onc.2011.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carone D.M., Lawrence J.B. Heterochromatin instability in cancer: from the Barr body to satellites and the nuclear periphery. Semin. Cancer Biol. 2013;23:99–108. doi: 10.1016/j.semcancer.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eymery A., Horard B., El Atifi-Borel M., Fourel G., Berger F., Vitte A.L., Van den Broeck A., Brambilla E., Fournier A., Callanan M., et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009;37:6340–6354. doi: 10.1093/nar/gkp639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ting D.T., Lipson D., Paul S., Brannigan B.W., Akhavanfard S., Coffman E.J., Contino G., Deshpande V., Iafrate A.J., Letovsky S., et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S.J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 59.Francisco D.C., Peddi P., Hair J.M., Flood B.A., Cecil A.M., Kalogerinis P.T., Sigounas G., Georgakilas A.G. Induction and processing of complex DNA damage in human breast cancer cells MCF-7 and nonmalignant MCF-10A cells. Free Radic. Biol. Med. 2008;44:558–569. doi: 10.1016/j.freeradbiomed.2007.10.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


