Abstract
This study examines metal binding to metallo-β-lactamase VIM-2, demonstrating the first successful preparation of a Co(II)-substituted VIM-2 analogue. Spectroscopic studies of the half- and fully metal loaded enzymes show that both Zn(II) and Co(II) bind cooperatively, where the major species present, regardless of stoichiometry, are apo- and di-Zn (or di-Co) enzymes. We determined the di-Zn VIM-2 structure to a resolution of 1.55 Å, and this structure supports results from spectroscopic studies. Kinetics, both steady-state and pre-steady-state, show that VIM-2 utilizes a mechanism that proceeds through a very short-lived anionic intermediate when chromacef is used as the substrate. Comparison with other B1 enzymes shows that those that bind Zn(II) cooperatively are better poised to protonate the intermediate on its formation, compared to those that bind Zn(II) non-cooperatively, which uniformly build up substantial amounts of the intermediate.
According to the 2013 Antibiotic Resistance Threat Report, more than 2 million people in the U.S. were infected with an antibiotic-resistant bacterial infection, and more than 23,000 of these patients died.1 During the past decade, the incidence of carbapenemase-producing Enterobacteriaceae and Klebsiella pneumoniae has increased, and death rates associated with metallo-β-lactamase (MBL) producers range from 18 to 67%.2 MBL-producing bacteria that exhibited carbapenemase activity were first reported in 1990.3 Since then the emergence of VIM-, IMP-, and NDM-type MBLs has been reported in many countries.2 The activities of these MBLs are not inhibited by clavulanic acid or any other commercially available β-lactamase inhibitors.4 The IMiPenamase (IMP), Verona integrin-encoded (VIM), and New Delhi metallo-β-lactamase (NDM) enzymes are the most clinically significant MBLs because they (and their derivatives) appear in bacterial strains that exhibit high mortality rates.2 All three MBLs belong to the B1 subclass.5 IMP was first identified in the early 1990s in strains of Pseudomonas aeruginosa and Serratia marcescens in Japan,3,6 and currently includes 47 variants isolated in countries across the globe, on every continent except Africa and Antarctica.7,8 So far, 12 variants of NDM MBL have been isolated from 40 countries worldwide except Antarctica.8,9
VIM was first identified in a patient in Italy who was infected with P. aeruginosa.10,11 To date, 39 variants of VIM have been identified,8 and these variants exhibit 74.3–99.6% sequence similarity.12,13 Among these VIM variants, VIM-2 appears to be the one most commonly found in the clinic, and VIM-2-expressing bacterial strains have been found in many countries.14−23 IMP-1 and NDM-1 have been extensively studied biochemically, crystallographically, and spectroscopically.24−27 In contrast, while there are nine crystal structures of VIM variants available, including two of VIM-2 at 1.9 and 2.2 Å resolution,28,29 presently detailed kinetic/mechanistic or spectroscopic studies reported on any variant of VIM are not available. To address this deficit, we have investigated the kinetic mechanism of VIM-2 using stopped-flow kinetics. To further probe the structure of the VIM-2 active site, X-ray crystallographic and X-ray absorption fine structure (EXAFS) spectroscopic studies were performed on the native Zn(II)-containing enzyme. Co(II)-substituted VIM-2 was also prepared and characterized using UV–vis, electron paramagnetic resonance (EPR), and EXAFS spectroscopies. Results of these studies offer insights into metal binding by VIM-2, elucidate emerging patterns in reactivity that track with metal-binding properties, and add to the current efforts in medicinal chemistry to design clinical inhibitors of this class.
Experimental Procedures
Materials
pET24a vector was used to clone blaVIM-2, which was initially isolated from a clinical strain of P. aeruginosa.30Escherichia coli BL21(DE3) cells (Invitrogen) were used for protein overexpression. Lysogeny broth (LB) medium was purchased from Invitrogen (Carlsbad, CA), and minimal medium was prepared according to a previously described procedure.31 Isopropyl β-d-thiogalactoside (IPTG) was purchased from Gold Biotechnology (St. Louis, MO). Q-Sepharose (GE Healthcare) anion-exchange and Sephacryl S-200 gel filtration (Amersham Biosciences AB, Uppasala, Sweden) columns were used to purify recombinant VIM-2. Purified protein solutions were pooled and concentrated with an ultrafiltration cell equipped with YM-10 DIAFLO membranes from Amicon, Inc. (Beverly, MA). Steady-state kinetics and pre-steady-state kinetics studies were conducted using chromacef (Sopharmia, Inc., St. Joseph, MO).26
Overexpression and Purification of VIM-2 in LB and Minimal Media
Plasmid pet24a-VIM-2 was transformed into BL21(DE3) E. coli cells, and the cell mixture was plated on LB-agar plates containing 25 μg/mL kanamycin. A single colony was transferred into 50 mL of LB containing 25 μg/mL kanamycin, and the culture was allowed to shake overnight at 37 °C. Ten milliters of the overnight culture was used to inoculate 4 × 1 L of LB containing 25 μg/mL kanamycin, and the cultures were shaken at 200 rpm and 37 °C to an OD600 nm of 0.6–0.8. Protein production was induced by making the cultures 0.5 mM in IPTG. The resulting cultures were allowed to shake for 3 h at 37 °C, and the cells were harvested by centrifugation for 10 min at 7000 rpm and 4 °C. The cell pellets were resuspended in 30 mL of 30 mM Tris, pH 7.6, containing 500 mM NaCl. The cells were lysed by passing the suspension three times through a French press at a pressure of 1000 psi. The mixture containing the lysed cells was centrifuged for 25 min at 15 000 rpm at 4 °C to remove insoluble components. The cleared solution was dialyzed versus 2 L of 30 mM Tris, pH 7.6, overnight at 4 °C. After centrifugation for 25 min at 15 000 rpm, the cleared solution was loaded onto a Q-Sepharose column (1.5 cm × 20 cm with a 28 mL bed volume). Bound proteins were eluted with a linear gradient of 0–500 mM NaCl in 30 mM Tris, pH 7.6.
Fractions containing VIM-2 were identified using SDS-PAGE, pooled, and then concentrated using an Amicon ultrafiltration concentrator equipped with a YM-10 membrane. The concentrated protein solution (ca. 3 mL) was loaded onto a Sephacryl S-200 gel filtration column (1.5 cm × 40 cm, bed volume of 60 mL), using 30 mM Tris, pH 7.6, containing 100 mM NaCl as the running buffer. The concentration of VIM-2 was determined using an extinction coefficient of 28 500 M–1 cm–1 at 280 nm.32 As-isolated enzyme contained 0.4 equiv of Zn(II). To prepare more fully load Zn(II) analogues of VIM-2, 0.6 or 1.6 equiv of Zn(II) was added to purified VIM-2, and the resulting solutions were dialyzed versus 2 L of Chelex-treated 50 mM Hepes, pH 6.8, for 16 h.
VIM-2 was also overexpressed in minimal medium as previously described.31,33 The protein was purified as described above, except that Chelex-treated 50 mM Hepes, pH 6.8, was used as the buffer during the chromatography steps. One milliliter of 100 mM CoCl2 was added to each 1 L flask at the time of induction.
Preparation of Co(II)-Substituted VIM-2
Metal analyses of purified VIM-2, which was overexpressed in minimal medium, revealed that the protein binds 0.5 equiv of Co(II) and less than 0.1 equiv of Zn(II). Co(II)-containing VIM-2 was concentrated to 1 mM using an Amicon concentrator equipped with a YM-10 membrane. Concentrated Co(II)-containing VIM-2 was centrifuged at 14 000 rpm for 10 min to remove any precipitated protein. CoCl2 solution (0.5 or 1.5 equiv) was added, and the mixtures were incubated on the ice for 20 min. The resulting solutions were dialyzed versus 2 L of Chelex-treated 50 mM Hepes, pH 6.8, for 16 h, and the resulting enzyme was pink and remained pink for several months at 4 °C. UV–vis spectra were obtained on a Hewlett-Packard 5480A UV–vis spectrophotometer, at 25 °C. Difference spectra were generated by subtracting the spectrum of VIM-2 isolated from LB medium from the spectrum of Co(II)-substituted VIM-2.
Metal Analyses
The metal content of the VIM-2 samples was determined using a Perkin-Elmer Optima 7300V inductively coupled plasma spectrometer with optical emission spectroscopy (ICP-OES). Protein samples were diluted to 1–3 μM with 50 mM Hepes, pH 6.8 or 7.6. Calibration curves were generated using serial dilutions of Fisher metal standards (Zn, Co, Fe, and Cu) ranging from 1.0 to 8.0 μM. Emission lines at 202.548, 228.616, 238.196, and 327.394 nm were chosen to ensure the lowest possible detection limits for zinc, cobalt, iron, and copper, respectively.
Steady-State Kinetics
Steady-state kinetic studies were conducted on a Hewlett-Packard 5480A UV–vis spectrophotometer at 25 °C. The hydrolysis of chromacef was monitored by formation of the hydrolyzed product at 442 nm, and absorbance data were converted to concentration data using an extinction coefficient of 18 600 M–1 cm–1.26 The buffer used for the steady-state kinetic studies was 50 mM cacodylate, pH 7.0, and substrate concentrations ranging from 1 to 100 μM were used in these studies. Rate vs substrate concentration curves were fitted to the Michaelis–Menten equation using Igor-Pro to determine KM and Vmax.
Pre-Steady-State Kinetics
Stopped-flow UV–vis studies were conducted using an Applied Photophysics SX 20 stopped-flow spectrophotometer, equipped with a photodiode array detector. Reactions of VIM-2 with chromacef were thermostated at 22 °C, and Chelex-treated, 50 mM cacodylate, pH 7.0, was used as the buffer. Reaction progress curves were generated by converting the absorbance data to concentration data using the following extinction coefficients: substrate ε378 = 22 000 M–1 cm–1, product ε442 = 18 600 M–1 cm–1, and intermediate ε620 = 22 000 M–1 cm–1.26 Simulated progress curves were generated using a previously described MATLAB script,24 derived for the reaction mechanisms shown in Scheme 1. Theoretical kcat and Km values were calculated using the King–Altman method,34 assuming k2 or k3 as the rate-limiting step.26
Scheme 1.
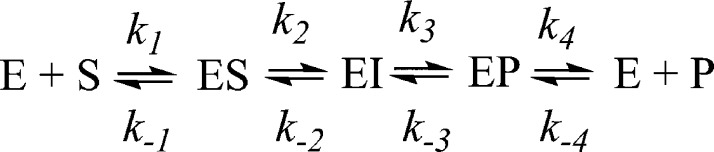
Crystallization and X-ray Diffraction Data Collection
Crystallization trials were prepared in 96-well sitting drop Intelliplate vapor diffusion plates (Art Robbins Instruments, Sunnyvale, CA). The 1.0 μL sitting drops consisted of a 1:1 ratio of 10 mg/mL VIM-2 and reservoir solution. Initial screening was carried out using the sparse matrix crystallization screens MCSG 1–4 (Microlytic, Burlington, MA). Optimization of initial hits from the sparse matrix screens identified a reservoir solution of 0.2 M ammonium acetate, 100 μM zinc chloride, 0.1 M Hepes, pH 7.5, and 25% PEG 3350. Crystals of VIM-2 were cryoprotected by brief transfer through LV CryoOil (MiTeGen) and frozen in liquid nitrogen. X-ray diffraction data were collected at 1.0000 Å for a single VIM-2 crystal on beamline 4.2.2 at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory.
Phases were calculated by molecular replacement using the PHASER35 component of PHENIX36 utilizing the structure of zinc-bound VIM-229 (PDB accession code 1KO3) as the search model. The molecular replacement solution was subjected to iterative rounds of model building in Coot37 and refinement in PHENIX.36 All molecular structure figures were prepared with PyMOL.38 Atomic coordinates and structure factors have been deposited in the PDB (accession code 4NQ2). Stereochemical and geometric analyses of the VIM-2 structure were conducted with MolProbity version 4.02b.39
EXAFS Spectroscopy
Samples for EXAFS (protein concentrations of approximately 1.5 mM) were prepared with 20% (v/v) glycerol as a glassing agent. Samples were loaded in Lucite cuvettes with 6 μm polypropylene windows and frozen rapidly in liquid nitrogen. Data were collected at the National Synchrotron Light Source (NSLS), beamline X3B, equipped with a Si (111) double-crystal monochromator. Harmonic rejection was accomplished using a Ni focusing mirror. Fluorescence excitation spectra for all samples were measured with a 31-element solid-state Ge detector array. Samples were held at approximately 15 K in a Displex cryostat. EXAFS data collection and reduction were performed according to published procedures.40 Data were measured in duplicate, six scans for zinc and eight scans for cobalt each on two samples from independent purifications; fits to the two data sets were equivalent. As both data sets gave similar results, the data were averaged using EXAFSPAK (EXAFSPAK is available free of charge from http://www-ssrl.slac.stanford.edu/exafspak.html); the experimental spectra presented here are the averaged data sets (12 or 16 scans per sample). The data were converted from energy to k-space using E0 = 9680 eV for Zn and E0 = 7730 eV for Co.
Fourier-filtered EXAFS data were fitted using the nonlinear least-squares engine of IFEFFIT, which is distributed with SixPack (SixPack is available free of charge from http://www-ssrl.slac.stanford.edu/∼swebb/sixpack.htm; IFEFFIT is open source software available from http://cars9.uchicago.edu/ifeffit/Ifeffit). Theoretical amplitude and phase functions were calculated with FEFF v. 8.00.41 Zinc–nitrogen single-scattering and zinc–imidazole multiple-scattering were calibrated to the experimental EXAFS of zinc tetrakis-1-methylimidazole Zn(II) perchlorate, [Zn(MeIm)4][ClO4]2. Zinc–sulfur scattering was calibrated to the experimental EXAFS spectrum of tetrabutylammonium zinc tetramesitylthiolate, [Bu4N]2[Zn(Smes)4]. Optimum scale factors (Sc) and ΔE0 were derived from fits to the model data (Sc = 0.78 (Zn–N) or 0.91 (Zn–S); ΔE0 = −21 eV), and they were held fixed at these values for fits to metalloprotein data. The models used for the calibration of cobalt–nitrogen (and cobalt–imidazole) and cobalt–sulfur scattering were hexakis-imidazole cobalt(II) perchlorate, [Co(Im)6][ClO4]2, and tetrabutylammonium cobalt(II) tetramesitylthiolate, [Bu4N]2[Co(Smes)4], respectively. The resulting Sc and ΔE0 values (Sc = 0.79 (Co–N) or 0.85 (Co–S); ΔE0 = −21 eV) were held fixed at these calibrated values in subsequent fits to metalloprotein data. First-shell fits were then obtained for all reasonable coordination numbers, including mixed nitrogen/oxygen/sulfur ligation, while allowing the absorber–scatterer distance, Ras, and the Debye–Waller factor, σas2, to vary. Detailed summaries of the fitting results are presented in Supporting Information. Multiple scattering contributions from histidine ligands were fitted according to published procedures.40 Metal–metal (zinc–zinc and cobalt–cobalt) scattering was modeled with reference to the experimental EXAFS of Zn2 (salpn)2 and Co2 (salpn)2.
EPR Spectroscopy
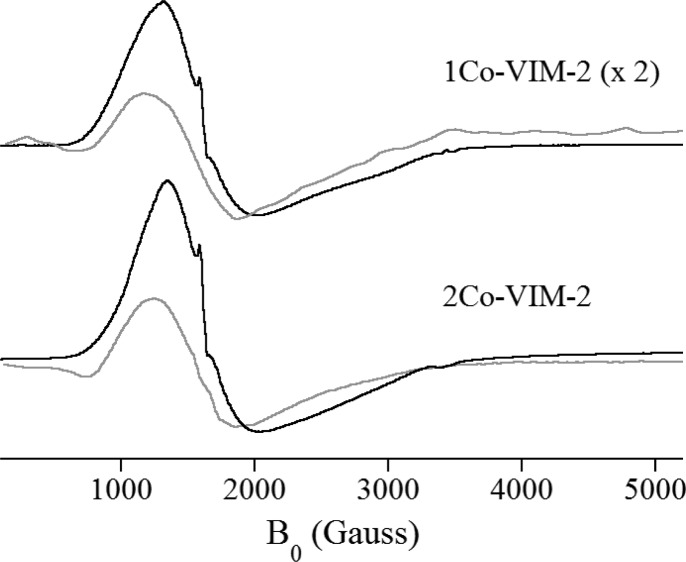
Low-temperature EPR spectra were obtained on a Bruker Elexsys EMX EPR spectrometer equipped with an Oxford Instruments liquid helium flow cryostat. The spectra in Figure 6 were recorded at 9.64 (B0⊥B1) or 9.38 GHz (B0∥B1) using a Bruker DM4116 dual-mode cavity, with 10 G magnetic field modulation (100 kHz). Other parameters: time constant/conversion time = 42 ms, receiver gain = 1 × 105, average of four scans.
Figure 6.
X-band EPR spectra of 1Co- and 2Co-VIM-2, taken with B1∥B0 (gray lines) and B1⊥B0 (black lines). The 1Co-VIM-2 spectra have been scaled by a factor of 2; all other sample and spectrometer conditions were identical.
Results
Overexpression, Purification, and Characterization of VIM-2
Recombinant VIM-2 was overexpressed in LB medium and purified as described above (referred to as “as-isolated (LB)” hereafter). VIM-2 eluted from a Q-Sepharose column between 150 and 250 mM NaCl, and the purity of the protein was shown to be >90% by SDS-PAGE. Gel filtration chromatography further increased the purity to >95%. The overall yield was 10 mg/L of LB growth medium. VIM-2 was also overexpressed in minimal medium in the presence of Co(II) and purified as described above (referred to as “as-isolated (MM)” hereafter). The overall yield of VIM-2 after both columns was 5 mg/L of minimal medium. The purified enzymes were shown to bind 0.4 ± 0.1 equiv of Zn(II) (LB) or 0.5 ± 0.1 Co(II) (minimal medium), and <0.1 equiv of any other metal ion. Metal analyses were not reported for preceding VIM-2 preparations.14,30,32
Direct Addition of Zn(II) or Co(II) to As-Isolated VIM-2
To prepare VIM-2 analogues that contained one or two Zn(II) ions, 0.5 or 1.5 equiv of Zn(II) was added to as-isolated (LB) VIM-2, followed by overnight dialysis against Chelex-treated 30 mM Tris, pH 7.6. ICP-OES studies showed that the resulting enzymes bound 1.0 and 1.9 equiv of Zn(II) (±0.1), respectively (referred to as “1Zn-VIM-2” and “2Zn-VIM-2” hereafter). 1Zn-VIM-2 demostrated kcat = 10 ± 2 s–1 and Km = 9 ± 2 μM, compared to kcat = 22 ± 2 s–1 and Km = 8 ± 2 μM for 2Zn-VIM-2, when using chromacef as substrate (Table 1). Similarly, VIM-2 analogues containing one or two Co(II) ions were prepared by adding 0.5 or 1.5 equiv of Co(II) to as-isolated (MM) VIM-2, followed by overnight dialysis against 2 L of Chelex-treated 50 mM HEPES, pH 6.8 (a slightly acidic pH was seen to help prevent oxidation of Co(II)). The resulting enzymes bound 0.8 and 1.8 equiv of Co(II), respectively (referred to as “1Co-VIM-2” and “2Co-VIM-2” hereafter). 1Co-VIM-2 exhibited kcat = 6 ± 1 s–1 and Km = 5 ± 2 μM, while 2Co-VIM-2 showed kcat = 10 ± 1 s–1 and Km = 6 ± 2 μM (Table 1). The kcat/Km values of the Co(II)-substituted enzymes were roughly equal to those of the Zn(II)-containing analogues, with the 2Zn analogue clearly the most efficient catalyst (Table 1).
Table 1. Steady-State Kinetic Constants and Metal Contents of VIM-2 Samples.
| sample | kcat (s–1) | Km (μM) | kcat/Km (μM–1 s–1) | metal content (equiv)a |
|---|---|---|---|---|
| as-isolated (LB) | 4 ± 1 | 5 ± 2 | 0.8 | 0.4 |
| 1Zn-VIM-2 | 10 ± 2 | 9 ± 2 | 1.1 | 1.0 |
| 2Zn-VIM-2 | 22 ± 2 | 8 ± 2 | 2.8 | 1.9 |
| as-isolated (MM) | 2 ± 1 | 10 ± 1 | 0.2 | 0.5 |
| 1Co-VIM-2 | 6 ± 1 | 5 ± 2 | 1.2 | 0.8 |
| 2Co-VIM-2 | 10 ± 1 | 6 ± 2 | 1.7 | 1.8 |
Uncertainty ±0.1.
Pre-Steady-State Kinetics
To probe the kinetic mechanism of VIM-2, stopped-flow kinetic studies were conducted. The reaction of chromacef with 2Zn-VIM-2 (85 μM, Figure 1A) and 2Co-VIM-2 (125 μM, Figure 1B) was monitored over 500 ms using a stopped-flow mixer and diode-array detection between 300 and 700 nm. Three distinct absorbance bands were observed (378, 442, and 620 nm). The three bands correspond to substrate decay, product formation, and intermediate formation/decay, respectively.26 The resulting progress curves were fitted to the kinetic mechanisms shown in Scheme 1, using the rate constants in Table 2.24 The use of the mechanism in Scheme 1 resulted in good fits to the MATLAB generated progress curves (Figure 1). The shapes of the theoretical progress curves were greatly influenced by the values of k2, k–2, and k3 and not significantly influenced by other microscopic rate constants. We tested other mechanisms with additional intermediates and with no intermediates; however, the use of these mechanisms did not result in improved fits. The “dip” in the substrate decay versus time data has been observed in the past and has been attributed to overlapping absorbances of substrate and product/intermediate.24,26,42−45
Figure 1.
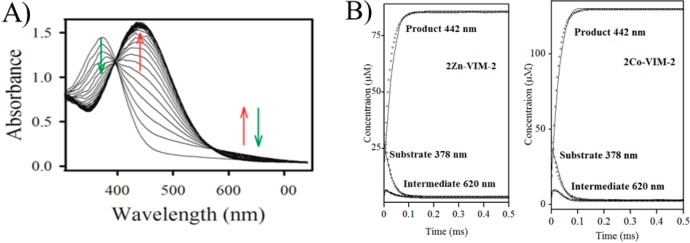
(A) UV–vis spectra monitoring the reaction of 2Zn-VIM-2 (80 μM) and chromacef (80 μM) at pH 7.0 and 22 °C. (B) Progress curves for chromacef hydrolysis by 2Zn-VIM-2 (left) and 2Co-VIM-2 (right). Concentrations of substrate, product, and intermediate were calculated as described in Experimental Procedures. Theoretical progress curves are shown as open symbols, and the experimental progress curves are solid lines. The kinetic mechanism in Scheme 1 and the rate constants in Table 2 were used to generate the theoretical progress curves.
Table 2. Pre-Steady-State Kinetic Parameters of 2Zn-VIM2 and 2Co-VIM2 against Chromacefa.
| 2Zn-VIM-2 | 2Co-VIM-2 | |
|---|---|---|
| k1 (M–1 s–1) | 1 × 108 | 1 × 108 |
| k–1 (s–1) | 500 ± 50 | 200 ± 30 |
| k2 (s–1) | 35 ± 4 | 40 ± 5 |
| k–2 (s–1) | <1 | <1 |
| k3 (s–1) | 170 ± 10 | 300 ± 30 |
| k–3 (s–1) | <1 | <1 |
| k4 (s–1) | 5000 ± 1000 | 5000 ± 1000 |
| k–4 (M–1 s–1) | 1 × 108 | 1 × 108 |
Data were MATLAB theoretical generated curves using the mechanism in Scheme 1. Constants k1 and k–4 as diffusion-controlled limits were fixed during the fitting using the analysis.
To determine whether the kinetic mechanism (Scheme 1) and rate constants in Table 2 correspond to the steady-state experimental data, the King–Altman method was used to determine equations for theoretical kcat and Km values. The theoretical kcat value for 2Zn-VIM-2 was 29 s–1, which nicely matches the experimental kcat = 22 s–1, and the theoretical Km value was 4.4 μM, within a factor of 2 of the experimental Km = 8 μM. The theoretical kcat and Km values for 2Co-VIM-2 were 35 s–1 and 2.1 μM, respectively, within a factor of 3 of the experimental kcat and Km values (10 s–1 and 6 μM).
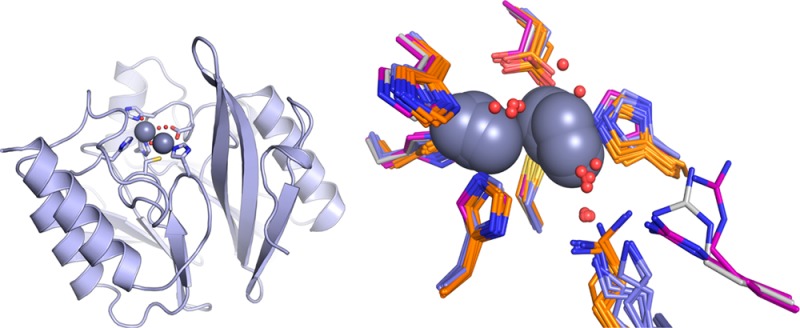
X-ray Crystal Structure of VIM-2
The X-ray crystal structure of 2Zn-VIM-2 was determined to examine the active site structure surrounding the bound Zn(II) atoms (Figure 2). Our structure is the highest resolution (1.55 Å) VIM-2 structure determined to date29 and is of excellent quality as judged by multiple validation parameters, including no Cβ-deviations, a MolProbity clash score of 2.96 (99th percentile, N = 730, 1.550 ± 0.25 Å), and a MolProbity39 score of 1.21 (98th percentile, N = 6779, 1.550 ± 0.25 Å).
Figure 2.
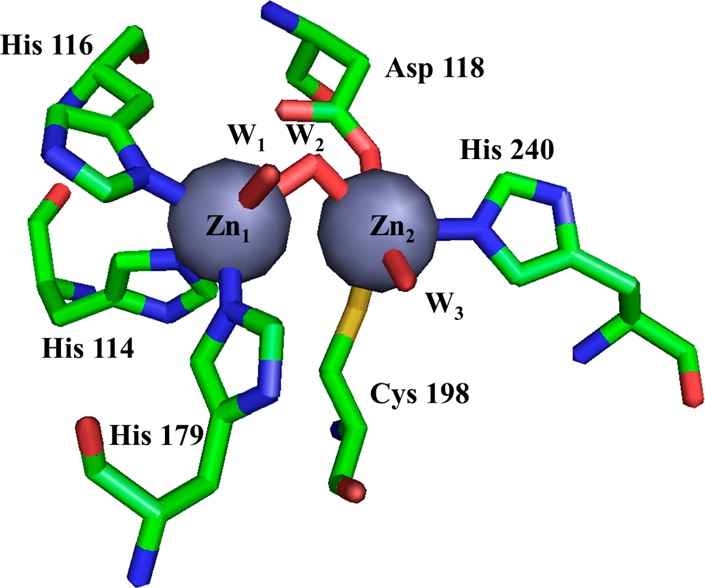
Active site from the VIM-2 crystal structure (PDB ID: 4NQ2) reported here. Zn1 is coordinated by three His and two water molecules, and Zn2 is coordinated by one His, one Cys, one Asp, and two water molecules. Zn1 and Zn2 are connected by a hydroxyl bridge.
The overall structure of our recombinant VIM-2 (PDB ID: 4NQ2) is very similar to those reported by Garcia-Saez et al. (PDB ID: 1KO3, reduced form) and Yamaguchi et al. (PDB ID: 2YZ3, reduced form with bound phenylC3SH inhibitor),28,29 and to those of VIM-7 (PDB ID: 2Y87, reduced form)46 and VIM-31 (PDB ID: 4FR7). In the VIM-4 crystal structure there was a citrate molecule bound in the active site, which results in an unusual six-coordinate Zn2 ion, and the bridging water/hydroxide was replaced by the citrate ion.47
Our structure of VIM-2 showed the Zn(II) in the Zn1 site bound to His114, His116, and His179, and the Zn(II) in the Zn2 site bound to Asp118, Cys198, and His240. In our structure, three water molecules are found at 2.3–2.6 Å from the Zn(II) atoms (Figure 2), while in the previous structures, two water molecules are observed in the VIM-2 structure (PDB ID: 1KO3) and one water molecule is observed in the VIM-2/inhibitor structures (PDB ID: 2YZ3) (Figure 3). The present structure showed a Zn–Zn internuclear distance of 3.46 Å (Table 3), which is similar to the value that we observed by EXAFS (Table 4, below). Saez et al. (PDB ID: 1KO3) and Yamaguchi et al. (PDB ID: 2YZ3) reported different values for the metal–metal (Zn(II)–Zn(II)) distance (4.20 and 3.75 Å, respectively) in VIM-2.28,29 In our structure, a third Zn(II) binding site is found, coordinated by His178 and two acetate ions from the asymmetric unit and His285 from the symmetry mate found at −x + 1/2, −y + 1/2, z + 1/2. Given coordination by residues from two separate VIM-2 molecules, the third Zn(II) binding site is likely a crystallographic artifact. A similar Zn(II) site was reported in the Saez et al. structure.29
Figure 3.
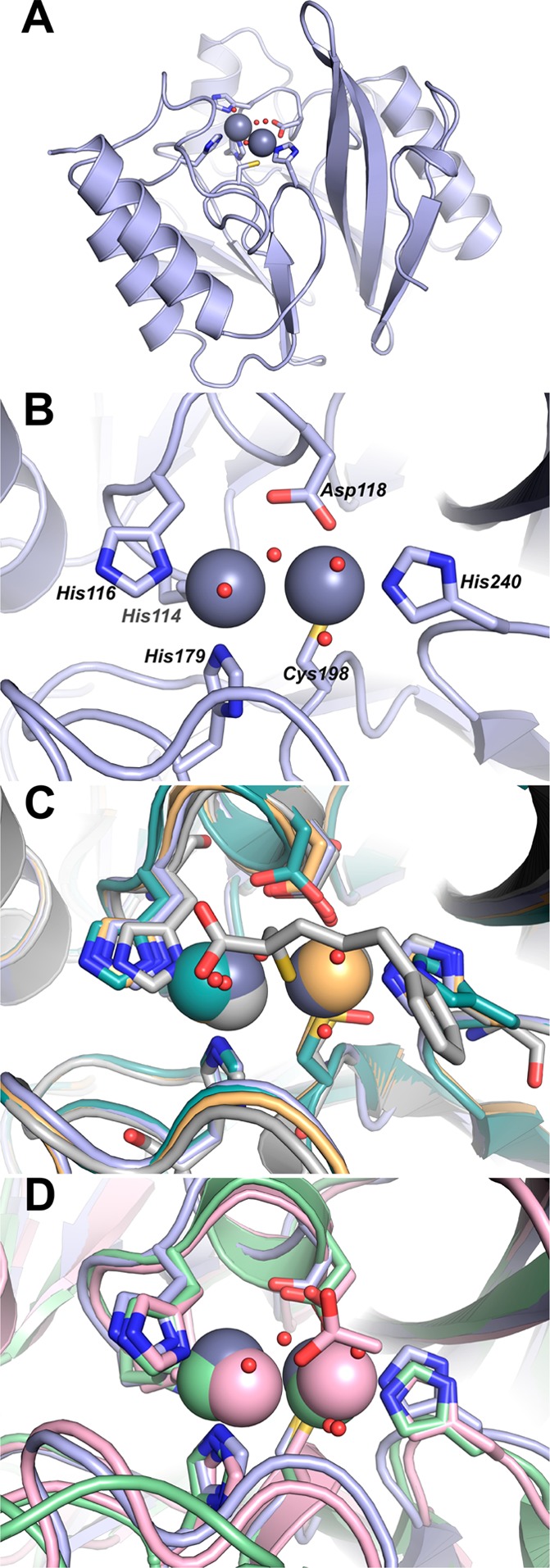
Structure of 2Zn-VIM-2. (A) Cartoon representation of VIM-2 (pale blue). Active site side chains are shown as sticks; Zn(II) ions (gray) and water molecules (red) are shown as spheres. (B) Close-up highlighting coordination of the two Zn(II) ions by residues at the VIM-2 active site. A and B are from this study, PDB ID: 4NQ2. (C) Active site close-up for VIM-2 structures 4NQ2 (pale blue), 1KO2 (light teal), 1KO3 (light orange), and 2YZ3 (gray). Residues coordinating the Zn(II) ions including the 1KO2 cysteinesulfonic acid and the 2YZ3 mercaptocarboxylate inhibitor are shown as sticks; Zn(II) ions (pale blue, light teal, light orange, and gray) and water molecules are shown as spheres. (D) Active site close-up for VIM-2 (4NQ2, pale blue), IMP-1 (1DDK, light pink), and NDM-1 (3PSU, light green). Residues coordinating the Zn(II) ions and the Zn(II)-bound acetate ion from 1DDK are shown as sticks; Zn(II) ions (pale blue, light pink, and light green) and water molecules (red) are shown as spheres.
Table 3. Zinc(II)–Ligand Distances (Å) from Previous Native VIM-2 Enzyme (1KO3) and VIM-2 (4NQ2) from This Study and from EXAFS Experiments.
| Zn(II)–ligand | VIM-2 (1KO3) | VIM-2 (4NQ2) | EXAFS | |
|---|---|---|---|---|
| Zn1 | His114(116/94) | 2.2 | 2.2 | 2.01 |
| His116(118/96) | 2.1 | 2.2 | 2.01 | |
| His179(196/159) | 2.2 | 2.2 | 2.01 | |
| O(W1) | 2.1 | 2.2 | 2.01 | |
| O(W2) | 2.6 | |||
| Zn2 | Asp118(120/98) | 2.3 | 2.3 | 2.01 |
| Cys198(221/178) | 2.3 | 2.3 | 2.31 | |
| His240(263/220) | 2.3 | 2.3 | 2.01 | |
| Cl | 2.9 | |||
| O(W1) | 2.5 | 2.2 | 2.01 | |
| O(W3) | 2.3 | 2.01 | ||
| Zn1 | Zn2 | 4.2 | 3.5 | 3.36 |
Table 4. Best Fits to the EXAFS Spectra of Zn(II)- and Co(II)-Containing VIM-2.
| sample | model | M–Ma | %Ib | fitc |
|---|---|---|---|---|
| as-isolated LB (0.5 Zn) | 4 N/O (2 His) + 0.5 S + Zn–Zn | 3.36 | 51 | S1-4 |
| 1Zn-VIM-2 | 4 N/O (2 His) + 0.5 S + Zn–Zn | 3.37 | 23 | S2-4 |
| 2Zn-VIM-2 | 4 N/O (2 His) + 0.5 S + Zn–Zn | 3.36 | 27 | S3-4 |
| as-isolated MM (0.5 Co) | 4 N/O (2 His) + 0.5 S + Co–Co | 3.51 | 53 | S4-4 |
| 1Co-VIM-2 | 4 N/O (2 His) + 0.5 S + Co–Co | 3.52 | 53 | S5-4 |
| 2Co-VIM-2 | 4 N/O (2 His) + 0.5 S + Co–Co | 3.51 | 37 | S6-4 |
Metal–metal separation in Å.
Percent improvement over a similar fit that lacks a M–M vector.
Fitting results described in Supporting Information, Tables S1–S6.
EXAFS Spectroscopy
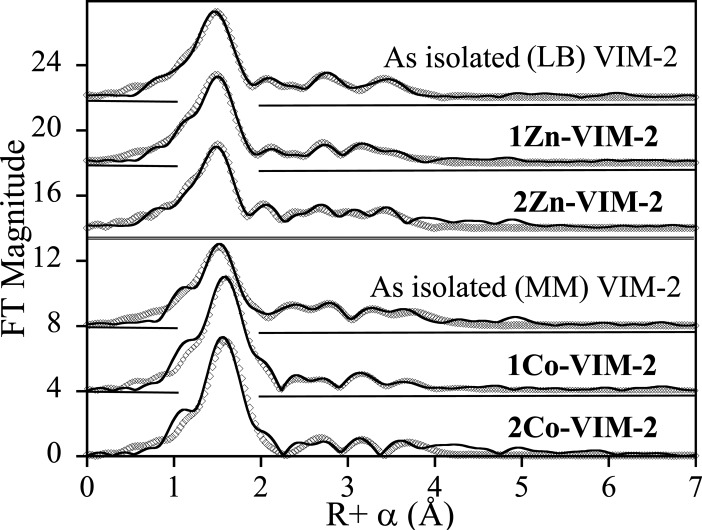
X-ray absorption spectroscopy was used examine the local metal-site structure in the Zn(II)-containing enzymes, for direct comparison with the Co(II)-containing enzymes, in frozen solution. The Fourier-transformed spectra and corresponding best fits are shown in Figure 4, and summarized in Table 4. Detailed fitting results are presented in the Supporting Information.
Figure 4.
EXAFS Fourier-transforms for Zn(II)- and Co(II)-containing VIM-2 (solid lines) and corresponding best fits (open symbols). See Table 4, Tables S1–S6, and Figures S1–S6 for details. From top to bottom: recombinant LB VIM-2 (Zn K-edge), 1Zn-VIM-2 (Zn K-edge), 2Zn-VIM-2 (Zn K-edge), recombinant MM VIM-2 (Co K-edge), 1Co-VIM-2 (Co K-edge), and 2Co-VIM-2 (Co K-edge).
The best fit obtained for as-isolated VIM-2, containing 0.5 equiv of Zn(II), indicates first-shell coordination to 4 N/O at 2.00 Å and 0.5 S scatterers at 2.28 Å (Figure S1 and Table S1, compare fit S1-1 to S1-2). Multiple scattering analyses indicate 2 imidazoles per Zn, while inclusion of a metal–metal interaction at 3.36 Å improved the fit by 51% (Figure S1 and Table S1, compare fit S1-3 to S1-4). Best fit results for 1Zn-VIM-2 are nearly indistinguishable, including a first shell of 4 N/O and 0.5 S scatterers, 2 imidazoles per Zn, and a metal–metal interaction at 3.37 Å. The metal–metal interaction improved the fit by 23% (Figure S2 and Table S2, compare fit S2-3 to S2-4). These results are consistent with VIM-2 containing ≤1 equiv of Zn(II) having both metal binding sites partially occupied. Further, the substantial improvement in fit residual on addition of the metal–metal vector suggests that metal binding is cooperative, with the dominant species present at substoichiometric metal loading being the apo- and dizinc enzymes.
EXAFS of 2Zn-VIM-2 is no different, with a first shell of 4 N/O and 0.5 S, 2 imidazoles per Zn, and a metal–metal interaction at 3.36 Å that improved the fit by nearly 27% (Figure S3 and Table S3, compare fit S3-3 to S3-4). These results are clearly consistent with previous crystallographic studies28,29 and our own structure (Figure 2 and Table 3). The average coordination number of 4.5 and Zn–Zn distance of 3.36 Å are in excellent agreement with our crystallographic studies, which showed an average coordination number of 5 and a Zn–Zn separation of 3.5 Å. The difference in coordination number arises from the second terminal water molecule bound to Zn1 in the crystal structure (Figure 2), which may not be present in the flash frozen EXAFS samples.
The EXAFS of all three forms of Co(II)-containing VIM-2 (as-isolated (0.5Co), 1Co, and 2Co) gave similar results, including a first shell of 4 N/O and 0.5 S scatters, 2 histidines per Co(II), and a metal–metal interaction at ∼3.51 Å that improved the fits by 53% (0.5 equiv, Figure S4 and Table S4, compare fit S4-3 to S4-4), 53% (1Co-VIM-2, Figure S5 and Table S5), and 37% 2Co-VIM-2, Figure S6 and Table S6). The same trend was observed in the Zn(II)-containing enzymes, suggesting that Co(II) is also bound cooperatively. The longer Co–Co distance in 2Co-VIM-2 relative to 2Zn-VIM-2 (3.36 Å) is consistent with the slightly larger covalent radius of high-spin Co(II). As Co(II) binding mirrored Zn(II) binding, we examined the Co(II)-substituted enzymes spectroscopically, with confidence the results could be extrapolated back to the native Zn(II)-containing enzymes.
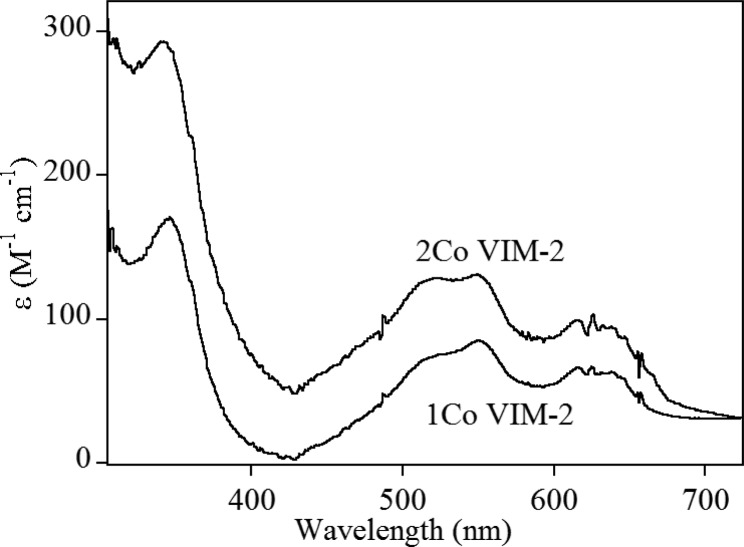
UV–Visible Spectroscopy
The UV–visible spectrum of 1Co-VIM-2 (Figure 5) revealed a broad absorption at 342 nm (ε342 = 170 M–1 cm–1) that is readily assigned to a Cys-S to Co(II) ligand-to-metal charge-transfer transition, requiring Co(II) binding at the Zn2 site.24,48−50 The ligand field transitions between 500 and 650 nm (ε550 = 85 M–1 cm–1) are similar in shape to those for other B1 MBLs,24,51 and they roughly double in intensity for 2Co-VIM-2, as does the charge-transfer transition (ε342 = 293 M–1 cm–1 and ε550 = 131 M–1 cm–1), suggesting binding of Co(II) to both sites in VIM-2 independent of stoichiometry. Similarly low extinction coefficients were observed for Co(II)-substituted IMP-1 (ε342 = 95 M–1 cm–1 and ε550 = 256 M–1 cm–1).24
Figure 5.
Optical spectra of 1Co- and 2Co-VIM-2.
EPR Spectroscopy
The EPR spectra of 1Co- and 2Co-VIM-2, obtained in both parallel and perpendicular modes, are shown in Figure 6. The perpendicular mode spectra (black lines in Figure 6) are typical of Co(II)-substituted metalloproteins,52 with the sharp feature near 1600–1700 G, corresponding to a small fraction of Fe(III) contamination. Comparison of the 1Co- and 2Co-VIM-2 spectra shows that they are nearly indistinguishable, consistent with the EXAFS and optical studies, suggesting that only the apo- and di-Co enzymes are present at measurable concentrations. Neither signal could be simulated using an axial g-tensor, indicating the lack of a unique solution based on the EPR alone.52 Both 1Co- and 2Co-VIM-2 showed strong parallel mode responses (gray lines in Figure 6), nearly one-fourth the normalized intensity of the perpendicular response, with the 2Co enzyme’s signal intensity nearly twice that of the 1Co enzyme. The EPR data indicate that similar metal centers are present in 1Co- and 2Co-VIM-2. This loading-dependent coupling, apparent at substoichiometric levels of metal, supports positive cooperative binding of Co(II) by VIM-2.
Discussion
VIM-2
VIM-2 contains 266 amino acids, with a mass of 29.7 kDa and pI = 5.6, exhibiting 7% structural variation from VIM-1. VIM-2-expressing bacteria have been shown resistant to an array of β-lactam-containing antibiotics, including ureidopenicillins, ticarcillin-clavulanic acid, cefepime, ceftazidime, imipenem, and meropenem, but strains expressing VIM-2 remained susceptible to the monobactam aztreonam.14 VIM-2 shares 32% amino acid identity with BcII from Bacillus cereus, 31% with IMP-1, 27% with CcrA from Bacteroides fragilis, 24% with BlaB from Chryseobacterium meningosepticum, and 24% with IND-1 from Chryseobacterium indologenes. There are significant differences in the steady-state kinetic constants reported for VIM-2 thus far, kcat and Km values ranging from 9.9 to 34 s–1 and from 9 to 10 μM, respectively, with imipenem.14,32 The steady-state kinetic results are difficult to compare, as metal content has not been uniformly reported.
Pre-steady-state kinetic studies have been reported previously for CcrA,49,53 Bla2,54 BcII,55 IMP-1,24 and NDM-126 using either nitrocefin or chromacef as substrate (Scheme 2). For CcrA49,53 and NDM-1,26 the rate-limiting step was reported to be protonation of the anionic, ring-opened intermediate. For Bla2, the mono-Co(II) enzyme quantitively formed the intermediate, suggesting cooperativity in metal binding.54 A similar intermediate was not observed with IMP-124 or BcII.55 In the present study, stopped-flow experiments on 2Zn-VIM-2 showed only small amounts of intermediate, up to ∼10% of the total enzyme concentration (Figure 1A). Interestingly, the chromacef-derived intermediate formed in the reaction with 2Zn-VIM-2 absorbed at 620 nm, a significant shift from the 575 nm λmax observed with NDM-1,26 L1, and CcrA (M. Aitha and M. W. Crowder, unpublished results). The red shift indicates a lower energy intermediate in VIM-2. This is a point under further investigation.
Scheme 2. Comparison of Nitrocefin (Left) and Chromacef (Right) Chromogenic Substrates.
We have previously reported kinetic and spectroscopic studies on Co(II)-substituted analogues of L1,33,43,44,54 ImiS,56 IMP-1,24 CcrA,57 Bla2,54 NDM-1,27 and BcII.58 As discussed in these references, Co(II)-substituted analogues of MBLs have allowed us to probe the reaction mechanisms, inhibitor/substrate binding, and structures of these enzymes even when a crystal structure is/was not available. In this study we have overexpressed, purified, characterized, and structurally probed VIM-2, which is one of the most important clinical MBLs and surprisingly has not been as extensively characterized as other MBLs. The conditions reported above allowed, for the first time, for Co(II)-substituted VIM-2 to be prepared and characterized. Optimization of each preparation required different conditions, with the Zn(II)-containing enzyme obtained from normal culture in LB medium, while our initial efforts to prepare Co(II)-substituted VIM-2 using standard methods, including exposure to metal chelators first to generate apoenzyme followed by direct addition of Co(II), failed. Previous studies have shown that Cys221 is readily oxidized when VIM-2 is exposed to chelators, and the resulting enzyme is unable to bind 2 equiv of metal.29,32 We have successfully used a biological incorporation method to prepare catalytically active Co(II)-substituted MBLs (L133 and CcrA57) previously, and this method resulted in an overexpressed VIM-2 that bound 0.5 equiv of Co(II), and no other detectable metals. VIM-2 analogues containing 0.8 and 1.8 equiv of Co(II) were readily generated from this enzyme by direct addition.
Metal Binding
EXAFS of the Zn(II)-containing analogues clearly show that both metal binding sites are occupied, independent of stoichiometry. The data obtained herein further indicate that they likely reside in a binuclear cluster, based on the improvement in the curve fits on inclusion of a metal–metal interaction around 3.36 Å (see Table 4). The metal–metal distances are consistent with the EXAFS-derived distances in other B1 MBLs24,26,27,54,57,58 and in the present crystal structure (Figure 2 and Table 3). Together with the steady-state kinetics (Table 1), which showed a near-linear dependence in kcat on Zn(II) content, the EXAFS data indicate positive-cooperative binding of Zn(II) to VIM-2, where the major species present, at any stoichiometry, are apo- and di-Zn enzymes.
The present data show that VIM-2 also exhibits positive-cooperative Co(II) binding. The EXAFS of the Co(II)-containing enzymes suggest near-stoichiometric formation of dinuclear enzymes on metal addition, based on even more pronounced fit improvements on inclusion of the M–M interaction (Table 4). EPR studies are clearly consistent with this model, where the perpendicular-mode spectra showed virtually indistinguishable signals whose intensity doubled from 1Co- to 2Co-VIM-2, and strong parallel-mode responses, indicating the presence of relatively strongly spin-coupled dinuclear Co(II) centers in both 1Co- and 2Co-VIM2, as compared to coupling in other MBLs. This may explain the exceptionally poor NMR response of Co(II)-substituted VIM-2 (Figure S7). Finally, the optical spectra are also consistent with distributed, cooperative binding, showing both d–d band structure and charge-transfer (CT) intensity that roughly doubled from 1Co- (ε342 = 170 M–1 cm–1 and ε550 = 85 M–1 cm–1) to 2Co-VIM-2 (ε342 = 293 M–1 cm–1 and ε550 = 131 M–1 cm–1). These extinction coefficients are substantially smaller than those reported for the majority of B1 MBLs, but very similar to that for IMP-1 (ε342 = 256 M–1 cm–1), which also showed highly cooperative Co(II) binding.24
Comparison with Other Dinuclear B1 MBLs
Among the B1 MBLs studied, VIM-2 appears to show the highest level of metal-binding cooperativity, based on the spectroscopic and kinetic properties reported here. Behavior varies widely across the B1 MBL subclass, as summarized in Table 5.24−27,48,49,55,57−60 To support this, we include two reports on CcrA here. One is based on an early study by Benkovic and co-workers that reported an optical titration consistent with distributed Co(II) binding.49 The second is based on our later study, working with the same construct, which suffered from a significant level of irretrievable cysteine oxidation, rendering many of the spectroscopic observables in Table 5 unavailable.57 We have since attempted to repeat these experiments without success. Given that caveat, all six that have been examined both kinetically and spectroscopically show evidence of distributed Co(II) binding, with both sites populated at substoichiometric levels of metal. All, save CcrA, show some level of cooperativity, based on the loading-dependence of kcat and an EXAFS-detected M–M interaction at substoichiometric loading. However, while half of the enzymes bind Zn(II) in the same fashion (BcII, IMP-1, and now VIM-2), with varying levels of cooperativity, the other half (NDM-1, Bla2, and CcrA) bind Zn(II) sequentially, without cooperativity. Without exception, those that bind Zn(II) cooperatively appear less capable of stabilizing the anionic intermediate with these chromogenic substrates, suggesting a shift in the rate-limiting step relative to those that bind Zn(II) sequentially, which uniformly build up a large percentage of the intermediate.
Table 5. Comparison of Metal-Binding, Kinetic, and Spectroscopic Studies of B1 MBLsa.
|
kcate (s–1) |
Co(II) enzymes |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| enzyme | %IVb | %IBc | M | metal-binding | cooperativity | [EI]d | 1 M | 2 M | M–S (Å) | M–M (Å) | ε342 (M–1 cm–1) | β-CH2 (NMR) (ppm) | ∥-EPR | refs |
| VIM-2 | 100 | 34 | Zn | distributed | high | 10 | 10 | 22 | 2.31 | 3.36 | g | |||
| Co | distributed | high | 10 | 6 | 10 | 2.31 | 3.51 | 293 | ND | strong | g | |||
| IMP-1 | 31 | 31 | Zn | distributed | weak | 0 | 18 | 29 | 2.27 | 3.39 | (24) | |||
| Co | distributed | high | 0 | 12 | 25 | 2.33 | 3.45 | 256 | ND | weak | (24) | |||
| BcII | 32 | 100 | Zn | distributed | moderate | 0 | ND | 23 | 2.27 | 3.42 | (55, 58) | |||
| Co | distributed | moderate | ND | ND | ND | 2.31 | 3.55 | 1200 | 170, 175 | weak | (48, 58, 60) | |||
| Bla2 | 34 | 89 | Zn | sequential | none | 0f | 32 | 42 | 2.27 | 3.44 | (59) | |||
| Co | distributed | moderate | 50f | 19 | 47 | 2.27 | ND | 608 | ND | ND | (59) | |||
| NDM-1 | 32 | 27 | Zn | sequential | none | 80 | 4g | 4h | 2.25 | 3.38 | (25−27) | |||
| Co | distributed | moderate | 65 | 2 | 4 | 2.29 | 3.51 | 900 | 84, 170 | weak | (26, 27) | |||
| CcrA | 27 | 30 | Zn | sequential | none | 35 | ND | 221 | 2.33 | 3.44 | (49, 57, 59) | |||
| Co | sequential | none | ND | 98 | 110 | ND | ND | ND | ND | ND | (57) | |||
| distributed | ND | ND | 820 | ND | ND | (49) | ||||||||
ND = not determined; ND = not detected.
Sequence identity with VIM-2.
Sequence identity with BcII.
Percent of enzyme (2 M) concentration that builds up as the anionic intermediate in rapid kinetics using either nitrocefin or chromacef.
kcat vs nitrocefin, except VIM-2 and NDM-1, which used chromacef, for enzymes loaded with 1 equiv (1 M) or 2 equiv (2 M) of the indicated metal.
Doubly loaded Bla2 was not sufficiently stable for rapid kinetics. Studies of the mono-Zn enzyme showed no significant intermediate build up, while studies of the mono-Co enzyme showed near-quantitative intermediate accumulation.
This work.
Steady-state kinetic studies of 2Zn-NDM-1 using nitrocefin show kcat = 15 s–1.25
To gain insight into this apparent distinction, we examined more closely the available structures of the six MBLs in Table 5. Limiting ourselves to doubly zinc-loaded enzymes, and accepting that there are no such structures of Bla2, the 27 available structures (including the present one) of the other 5 MBLs show minimal variability in the position and orientation of metal-coordinating side chains.28,29,61−74 Comparing nearby side chains and structured waters, as illustrated in Figure 7, shows two striking differences between the cooperative and non-cooperative groups. The first is the variability in the position of the Zn2 ion in the cooperative group (Figure 7A), compared to the non-cooperative structures, where placement of the metals is tightly grouped (Figure 7B). Efforts to re-engineer BcII through mutagenesis have shown similar variability in the position of Zn2, with dramatic effects on the reactivity of the resulting enzymes.75
Figure 7.
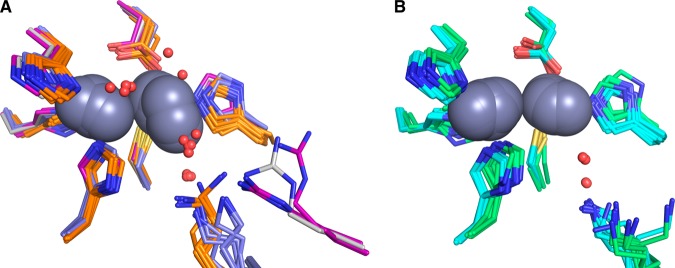
Comparison of available crystal structures for doubly zinc-loaded MBLs. (A) VIM-2 (PDB IDs: 4nq2 chain A, gray; 2yz3 chains A,B, purple; 1ko3 chain A, purple), IMP-1 (PDB IDs: 1dd6 chains A and B, orange; 1ddk chain A, orange; 1vgn chains A and B, orange; 2doo chains A and B, orange; 1jjt chains A and B, orange; and 1jje chains A and B, orange), and BcII (PDB IDs: 2uyx chain A, blue; 2bfx chains A and B, blue; 2bfl chains A and B, blue; and 2bg2 chains A and B, blue). (B) NDM-1 (PDB IDs: 4hl2 chains A and B, cyan; 4eyb chains A and B, cyan; 4exs chains A and B, cyan; 4exy chains A and B, cyan; 4ey2 chains A and B, cyan; 4eyf chains A and B, cyan; 4eyl chains A and B, cyan; 3spu chains B, C, D and E, cyan; and 3q6x chains A and B, cyan) and CcrA (PDB IDs: 1a7t chains A and B, green; 1a8t chains A and B, green; 2bmi chains A and B, green; and 1znb chains A and B, green). All zinc ions are represented as dark gray spheres. Structured waters within the active sites are shown as red spheres.
The second, perhaps more pertinent observation is that the occupancy of the terminally zinc-coordinated water molecule, which has been suggested to serve as the proton source in decay of the intermediate,26,42−44,49,53 is substantially lower in the CcrA and NDM-1 structures (non-cooperative, Figure 7B). While this water molecule occupies a much more variable position in the cooperative group (BcII, IMP-1, and VIM-2, Figure 7A), it is present in a much higher fraction of the structures, suggesting these enzymes are better poised to turn over the intermediate, once it is formed. The terminally coordinated water in VIM-2 structures occupies positions similar to those seen in BcII and IMP-1, despite being held in place by a unique hydrogen bond donor. In all the other five B1 enzymes, the terminal water hydrogen bonds to another water that, in turn, hydrogen bonds to a nearby lysine. VIM-2 lacks the lysine, which is replaced by a tyrosine oriented away from the metal site. The necessary hydrogen bond in VIM-2 is provided by an arginine that is 4 residues away, and oriented as shown in Figure 7A. In the cooperative group (Figure 7A), the hydrogen bond donor appears to sample a larger conformational space than in the non-cooperative group (Figure 7B), which shows little variation in the position of the lysine side chain. It is also worth note that the bridging solvent molecule is readily apparent in most of the cooperative group structures (Figure 7A) and conspicuously absent in the non-cooperative group structures (Figure 7B).
Within the Co(II) enzymes in Table 5, the lines are not as clear. Those that show strong S-to-Co(II) CT bands (BcII and NDM-1) are the only ones that have shown clearly identifiable NMR resonances from the cys β-CH2 protons, with weaker CT bands, presumably associated with substantially reduced hyperfine couplings, and chemical shifts. The strength of the CT band appears loosely correlated to the strength of the magnetic coupling in the di-Co(II) enzymes, based on the strength of the parallel-mode EPR signal, suggesting that more tightly coupled metal ions allow for weaker coupling to the thiolate, which has been shown to aid in stabilization of the anionic intermediate in several B1 MBLs.76 However, this is not a perfect correlation, as among the group that does not stabilize the intermediate, BcII shows the strongest CT band of all six. We are continuing to examine these correlations to better understand the variability in B1 MBLs.
Summary
In summary, this study examines metal binding to the metallo-β-lactamase VIM-2, demonstrating the first successful preparation of a Co(II)-substituted VIM-2 analogue. The spectroscopic studies reveal that Zn(II) and Co(II) bind similarly to VIM-2, with both metal ions showing cooperative binding where the major species present, regardless of stoichiometry, are apo- and di-Zn (or di-Co) enzymes. The crystal structure of the di-Zn enzyme that is presented is the highest resolution VIM-2 structure to date. Kinetic studies strongly suggest that VIM-2 utilizes a mechanism that proceeds through a very short-lived anionic intermediate. Comparison with other B1 enzymes shows that those that bind metal ions cooperatively are better poised to protonate the intermediate on its formation, compared to those that bind Zn(II) non-cooperatively, which uniformly build up substantial amounts of the intermediate. Future studies will address why positive-cooperative metal binding correlates with the abilty to protonate the anionic intermediate.
Glossary
Abbreviations
- MBL
metallo-β-lactamase
- VIM
Verona integrin-encoded metallo-β-lactamase
- NDM
New Delhi metallo-β-lactamase
- EPR
electron paramagnetic resonance
- EXAFS
extended X-ray absorption fine structure spectroscopy
Supporting Information Available
Detailed EXAFS fitting results, crystallization methods table, and NMR spectrum of the 2Co-VIM-2. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by the National Institutes of Health (GM093987 to M.W.C. and D.L.T.; P30-EB-009998 to the Center for Synchrotron Biosciences from the NIBIB, which supports beamline X3B at the NSLS), and the National Science Foundation (CHE-1151658 to M.W.C. and D.L.T.). This work was supported by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Department of Veterans Affairs Merit Review Program 1I01BX001974, the Veterans Integrated Service Network 10 Geriatric Research, Education, and Clinical Center (VISN 10 GRECC), and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers 5R01AI100560-03, R01 AI100560, and R01 AI063517 (to R.A.B.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- CDC (2013) Antibiotic resistance threats in the United States, http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf. [PubMed]
- Nordmann P.; Naas T.; Poirel L. (2011) Global spread of carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 17, 1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M.; Iyobe S.; Inoue M.; Mitsuhashi S. (1991) Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R.; Toleman M. A.; Poirel L.; Nordmann P. (2005) Metallo-β-lactamases: the quiet before the storm?. Clin. Microbiol. Rev. 18, 306–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. (2013) The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 19, 549–559. [DOI] [PubMed] [Google Scholar]
- Osano E.; Arakawa Y.; Wacharotayankun R.; Ohta M.; Horii T.; Ito H.; Yoshimura F.; Kato N. (1994) Molecular characterization of an enterobacterial metallo-b-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P.; Poirel L.; Walsh T. R.; Livermore D. M. (2011) The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. [DOI] [PubMed] [Google Scholar]
- Jacoby G., Bush K., β-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes, http://www.lahey.org/Studies/(accessed July 13, 2014).
- Johnson A. P.; Woodford N. (2013) Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62, 499–513. [DOI] [PubMed] [Google Scholar]
- Lauretti L.; Riccio M. L.; Mazzariol A.; Cornaglia G.; Amicosante G.; Fontana R.; Rossolini G. M. (1999) Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43, 1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Keeney D.; Tang X. J.; Canfield N.; Rasmussen B. A. (1999) Kinetic properties and metal content of the metallo-β-lactamase CcrA harboring selective amino acid substitutions. J. Biol. Chem. 274, 15706–15711. [DOI] [PubMed] [Google Scholar]
- Frasson I.; Biasolo M. A.; Bartolini A.; Cavallaro A.; Richter S. N.; Palu G. (2013) Rapid detection of blaVIM-1-37 and blaKPC1/2-12 alleles from clinical samples by multiplex PCR-based assays. Int. J. Antimicrob. Agents 42, 68–71. [DOI] [PubMed] [Google Scholar]
- Cornaglia G.; Giamarellou H.; Rossolini G. M. (2011) Metallo-β-lactamases: a last frontier for β-lactams?. Lancet Infect. Dis. 11, 381–393. [DOI] [PubMed] [Google Scholar]
- Poirel L.; Naas T.; Nicolas D.; Collet L.; Bellais S.; Cavallo J. D.; Nordmann P. (2000) Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44, 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J. J.; Hsueh P. R.; Ko W. C.; Luh K. T.; Tsai S. H.; Wu H. M.; Wu J. J. (2001) Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45, 2224–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavroidi A.; Tsakris A.; Tzelepi E.; Pournaras S.; Loukova V.; Tzouvelekis L. S. (2000) Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46, 1041–1042. [DOI] [PubMed] [Google Scholar]
- Pallecchi L.; Riccio M. L.; Docquier J. D.; Fontana R.; Rossolini G. M. (2001) Molecular heterogeneity of bla(VIM-2)-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195, 145–150. [DOI] [PubMed] [Google Scholar]
- Lolans K.; Queenan A. M.; Bush K.; Sahud A.; Quinn J. P. (2005) First nosocomial outbreak of Pseudomonas aeruginosa producing an integron-borne metallo-β-lactamase (VIM-2) in the United States. Antimicrob. Agents Chemother. 49, 3538–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. S.; Qu T. T.; Zhou J. Y.; Wang J.; Li H. Y.; Walsh T. R. (2006) Integrons containing the VIM-2 metallo-β-lactamase gene among imipenem-resistant Pseudomonas aeruginosa strains from different Chinese hospitals. J. Clin. Microbiol. 44, 4242–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duljasz W.; Gniadkowski M.; Sitter S.; Wojna A.; Jebelean C. (2009) First organisms with acquired metallo-β-lactamases (IMP-13, IMP-22, and VIM-2) reported in Austria. Antimicrob. Agents Chemother. 53, 2221–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras S.; Maniati M.; Petinaki E.; Tzouvelekis L. S.; Tsakris A.; Legakis N. J.; Maniatis A. N. (2003) Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51, 1409–1414. [DOI] [PubMed] [Google Scholar]
- Cardoso O.; Alves A. F.; Leitao R. (2008) Metallo-β-lactamase VIM-2 in Pseudomonas aeruginosa isolates from a cystic fibrosis patient. Int. J. Antimicrob. Agents 31, 375–379. [DOI] [PubMed] [Google Scholar]
- Castanheira M.; Bell J. M.; Turnidge J. D.; Mathai D.; Jones R. N. (2009) Carbapenem resistance among Pseudomonas aeruginosa strains from India: evidence for nationwide endemicity of multiple metallo-β-lactamase clones (VIM-2, -5, -6, and -11 and the newly characterized VIM-18). Antimicrob. Agents Chemother. 53, 1225–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. H.; Richmond T. K.; Sanchez C.; M?ller A. J.; Breece R. M.; Tierney D. L.; Bennett B.; Crowder M. W. (2011) Structural and kinetic studies on metallo-β-lactamase IMP-1. Biochemistry 50, 9125–9134. [DOI] [PubMed] [Google Scholar]
- Thomas P. W.; Zheng M.; Wu S.; Guo H.; Liu D.; Xu D.; Fast W. (2011) Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 50, 10102–10113. [DOI] [PubMed] [Google Scholar]
- Yang H.; Aitha M.; Hetrick A. M.; Richmond T. K.; Tierney D. L.; Crowder M. W. (2012) Mechanistic and spectroscopic studies of metallo-β-lactamase NDM-1. Biochemistry 51, 3839–3847. [DOI] [PubMed] [Google Scholar]
- Yang H.; Aitha M.; Marts A. R.; Hetrick A.; Bennett B.; Crowder M. W.; Tierney D. L. (2014) Spectroscopic and mechanistic studies of heterodimetallic forms of metallo-β-lactamase NDM-1. J. Am. Chem. Soc. 136, 7273–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y.; Jin W.; Matsunaga K.; Ikemizu S.; Yamagata Y.; Wachino J. I.; Shibata N.; Arakawa Y.; Kurosaki H. (2007) Crystallographic investigation of the inhibition mode of a VIM-2 metallo-β-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor. J. Med. Chem. 50, 6647–6653. [DOI] [PubMed] [Google Scholar]
- Garcia-Saez I.; Docquier J. D.; Rossolini G. M.; Dideberg O. (2008) The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375, 604–611. [DOI] [PubMed] [Google Scholar]
- Borgianni L.; Vandenameele J.; Matagne A.; Bini L.; Bonomo R. A.; Frere J. M.; Rossolini G. M.; Docquier J. D. (2010) Mutational analysis of VIM-2 reveals an essential determinant for metallo-β-lactamase stability and folding. Antimicrob. Agents Chemother. 54, 3197–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan P. T. R.; Grimme S.; Pei D. H. (2000) Characterization of cobalt(II)-substituted peptide deformylase: Function of the metal ion and the catalytic residue Glu-133. Biochemistry 39, 779–790. [DOI] [PubMed] [Google Scholar]
- Docquier J. D.; Lamotte-Brasseur J.; Galleni M.; Amicosante G.; Frere J. M.; Rossolini G. M. (2003) On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chem. 51, 257–266. [DOI] [PubMed] [Google Scholar]
- Hu Z. X.; Periyannan G. R.; Crowder M. W. (2008) Folding strategy to prepare Co(II)-substituted metallo-β-lactamase L1. Anal. Biochem. 378, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel I. H. (1993) Enzyme Kinetics, John Wiley and Sons, Inc., New York. [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Cryst. D 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.6.0.0 (2013), Schrödinger, LLC.
- Chen V. B.; Arendall W. B.; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. W.; Stone E. M.; Costello A. L.; Tierney D. L.; Fast W. (2005) The quorum-quenching lactonase from Bacillus thuringiensis is a metalloprotein. Biochemistry 44, 7559–7565. [DOI] [PubMed] [Google Scholar]
- Ankudinov A. L.; Ravel B.; Rehr J. J.; Conradson S. D. (1998) Real-space multiple-scattering calculation and interpretation of X-ray-absorption near-edge structure. Phys. Rev. B 58, 7565–7576. [Google Scholar]
- McManus-Munoz S.; Crowder M. W. (1999) Kinetic mechanism of metallo-β-lactamase L1 from Stenotrophomonas maltophilia. Biochemistry 38, 1547–1553. [DOI] [PubMed] [Google Scholar]
- Garrity J. D.; Bennett B.; Crowder M. W. (2005) Direct evidence that the reaction intermediate of metallo-β-lactamase L1 is metal bound. Biochemistry 44, 1078–1087. [DOI] [PubMed] [Google Scholar]
- Hu Z.; Periyannan G.; Bennett B.; Crowder M. W. (2008) Role of the Zn1 and Zn2 sites in metallo-β-lactamase L1. J. Am. Chem. Soc. 130, 14207–14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.; Spadafora L. J.; Hajdin C. E.; Bennett B.; Crowder M. W. (2009) Structure and mechanism of copper- and nickel-substituted analogues of metallo-β-lactamase L1. Biochemistry 48, 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra P. S.; Leiros H. K. S.; Ahmad R.; Spencer J.; Leiros I.; Walsh T. R.; Sundsfjord A.; Samuelsen O. (2011) Structural and computational investigations of VIM-7: insights into the substrate specificity of vim metallo-β-lactamases. J. Mol. Biol. 411, 174–189. [DOI] [PubMed] [Google Scholar]
- Lassaux P.; Hamel M.; Gulea M.; Delbruck H.; Mercuri P. S.; Horsfall L.; Dehareng D.; Kupper M.; Frere J. M.; Hoffmann K.; Galleni M.; Bebrone C. (2010) Mercaptophosphonate compounds as broad-spectrum inhibitors of the metallo-β-lactamases. J. Med. Chem. 53, 4862–4876. [DOI] [PubMed] [Google Scholar]
- Orellano E. G.; Girardini J. E.; Cricco J. A.; Ceccarelli E. A.; Vila A. J. (1998) Spectroscopic characterization of a binuclear metal site in Bacillus cereus β-lactamase II. Biochemistry 37, 10173–10180. [DOI] [PubMed] [Google Scholar]
- Wang Z. G.; Benkovic S. J. (1998) Purification, characterization, and kinetic studies of a soluble Bacteroides fragilis metallo-β-lactamase that provides multiple antibiotic resistance. J. Biol. Chem. 273, 22402–22408. [DOI] [PubMed] [Google Scholar]
- Crowder M. W.; Yang K. W.; Carenbauer A. L.; Periyannan G.; Seifert M. E.; Rude N. E.; Walsh T. R. (2001) The problem of a solvent exposable disulfide when preparing Co(II)-substituted metallo-β-lactamase L1 from Stenotrophomonas maltophilia. J. Biol. Chem. 6, 91–99. [DOI] [PubMed] [Google Scholar]
- Garmer D. R.; Krauss M. (1993) Ab initio quantum chemical study of the cobalt d-d spectroscopy of several substituted zinc enzymes. J. Am. Chem. Soc. 115, 10247–10257. [Google Scholar]
- Bennett B. (2010) in EPR of cobalt-substituted zinc enzymes, Metals in Biology (Hanson G., Berliner L., Eds.) pp 345–370, Springer, Berlin. [Google Scholar]
- Wang Z. G.; Fast W.; Benkovic S. J. (1998) Direct observation of an enzyme-bound intermediate in the catalytic cycle of the metallo-β-lactamase from Bacteroides fragilis. J. Am. Chem. Soc. 120, 10788–10789. [Google Scholar]
- Hawk M. J.; Breece R. M.; Hajdin C. E.; Bender K. M.; Hu Z. X.; Costello A. L.; Bennett B.; Tierney D. L.; Crowder M. W. (2009) Differential binding of Co(II) and Zn(II) to metallo-β-lactamase Bla2 from Bacillus anthracis. J. Am. Chem. Soc. 131, 10753–10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasia R. M.; Vila A. J. (2003) Mechanistic study of the hydrolysis of nitrocefin mediated by B. cereus metallo-β-lactamase. ARKIVOC 3, 507–516. [Google Scholar]
- Sharma N. P.; Hajdin C.; Chandrasekar S.; Bennett B.; Yang K. W.; Crowder M. W. (2006) Mechanistic studies on the mononuclear ZnII-containing metallo-β-lactamase ImiS from Aeromonas sobria. Biochemistry 45, 10729–10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyannan G. R.; Costello A. L.; Tierney D. L.; Yang K. W.; Bennett B.; Crowder M. W. (2006) Sequential binding of cobalt(II) to metallo-β-lactamase CcrA. Biochemistry 45, 1313–1320. [DOI] [PubMed] [Google Scholar]
- Breece R. M.; Llarrull L. I.; Tioni M. F.; Vila A. J.; Tierney D. L. (2012) X-ray absorption spectroscopy of metal site speciation in the metallo-β-lactamase BcII from Bacillus cereus. J. Inorg. Biochem. 111, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk M. J.; Breece R. M.; Hajdin C. E.; Bender K. M.; Hu Z.; Costello A. L.; Bennett B.; Tierney D. L.; Crowder M. W. (2009) Differential binding of Co(II) and Zn(II) to metallo-β-lactamase Bla2 from Bacillus anthracis. J. Am. Chem. Soc. 131, 10753–10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llarrull L. I.; Tioni M. F.; Vila A. J. (2008) Metal content and localization during turnover in B. cereus metallo-β-lactamase. J. Am. Chem. Soc. 130, 15842–15851. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y.; Kuroki T.; Yasuzawa H.; Higashi T.; Jin W.; Kawanami A.; Yamagata Y.; Arakawa Y.; Goto M.; Kurosaki H. (2005) Probing the role of Asp-120(81) of metallo-β-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography. J. Biol. Chem. 280, 20824–20832. [DOI] [PubMed] [Google Scholar]
- Concha N. O.; Janson C. A.; Rowling P.; Pearson S.; Cheever C. A.; Clarke B. P.; Lewis C.; Galleni M.; Frere J. M.; Payne D. J.; Bateson J. H.; Abdel-Meguid S. S. (2000) Crystal structure of the IMP-1 metallo-β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39, 4288–4298. [DOI] [PubMed] [Google Scholar]
- Kurosaki H.; Yamaguchi Y.; Higashi T.; Soga K.; Matsueda S.; Yumoto H.; Misumi S.; Yamagata Y.; Arakawa Y.; Goto M. (2005) Irreversible inhibition of metallo-β-lactamase (IMP-1) by 3-(3-mercaptopropionylsulfanyl)propionic acid pentafluorophenyl ester. Angew. Chem., Int. Ed. 44, 3861–3864. [DOI] [PubMed] [Google Scholar]
- Kurosaki H.; Yamaguchi Y.; Yasuzawa H.; Jin W.; Yamagata Y.; Arakawa Y. (2006) Probing, inhibition, and crystallographic characterization of metallo-β-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups. ChemMedChem 1, 969–972. [DOI] [PubMed] [Google Scholar]
- Toney J. H.; Hammond G. G.; Fitzgerald P. M.; Sharma N.; Balkovec J. M.; Rouen G. P.; Olson S. H.; Hammond M. L.; Greenlee M. L.; Gao Y. D. (2001) Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-β-lactamase. J. Biol. Chem. 276, 31913–31918. [DOI] [PubMed] [Google Scholar]
- Llarrull L. I.; Fabiane S. M.; Kowalski J. M.; Bennett B.; Sutton B. J.; Vila A. J. (2007) Asp-120 locates Zn2 for optimal metallo-β-lactamase activity. J. Biol. Chem. 282, 18276–18285. [DOI] [PubMed] [Google Scholar]
- Davies A. M.; Rasia R. M.; Vila A. J.; Sutton B. J.; Fabiane S. M. (2005) Effect of pH on the active site of an Arg121Cys mutant of the metallo-β-lactamase from Bacillus cereus: implications for the enzyme mechanism. Biochemistry 44, 4841–4849. [DOI] [PubMed] [Google Scholar]
- King D. T.; Worrall L. J.; Gruninger R.; Strynadka N. C. (2012) New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J. Am. Chem. Soc. 134, 11362–11365. [DOI] [PubMed] [Google Scholar]
- King D.; Strynadka N. (2011) Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 20, 1484–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Hao Q. (2011) Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J. 25, 2574–2582. [DOI] [PubMed] [Google Scholar]
- Toney J. H.; Fitzgerald P. M.; Grover-Sharma N.; Olson S. H.; May W. J.; Sundelof J. G.; Vanderwall D. E.; Cleary K. A.; Grant S. K.; Wu J. K.; Kozarich J. W.; Pompliano D. L.; Hammond G. G. (1998) Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-β-lactamase. Chem. Biol. 5, 185–196. [DOI] [PubMed] [Google Scholar]
- Carfi A.; Duee E.; Paul-Soto R.; Galleni M.; Frere J. M.; Dideberg O. (1998) X-ray structure of the ZnII β-lactamase from Bacteroides fragilis in an orthorhombic crystal form. Acta Crystallogr. D 54, 45–57. [DOI] [PubMed] [Google Scholar]
- Fitzgerald P. M.; Wu J. K.; Toney J. H. (1998) Unanticipated inhibition of the metallo-β-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-Å resolution. Biochemistry 37, 6791–6800. [DOI] [PubMed] [Google Scholar]
- Concha N. O.; Rasmussen B. A.; Bush K.; Herzberg O. (1996) Crystal structure of the wide-spectrum binuclear zinc β-lactamase from Bacteroides fragilis. Structure 4, 823–836. [DOI] [PubMed] [Google Scholar]
- Gonzalez J. M.; Medrano Martin F. J.; Costello A. L.; Tierney D. L.; Vila A. J. (2007) The Zn2 position in metallo-β-lactamases is critical for activity: a study on chimeric metal sites on a conserved protein scaffold. J. Mol. Biol. 373, 1141–1156. [DOI] [PubMed] [Google Scholar]
- Gonzalez J. M.; Meini M. R.; Tomatis P. E.; Medrano Martin F. J.; Cricco J. A.; Vila A. J. (2012) Metallo-β-lactamases withstand low Zn(II) conditions by tuning metal-ligand interactions. Nat. Chem. Biol. 8, 698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.