Abstract
We investigate fast iterative image reconstruction methods for fully 3D multispectral bioluminescence tomography for applications in small animal imaging. Our forward model uses a diffusion approximation for optically inhomogeneous tissue, which we solve using a finite element method (FEM). We examine two approaches to incorporating the forward model into the solution of the inverse problem. In a conventional direct calculation approach one computes the full forward model by repeated solution of the FEM problem, once for each potential source location. We describe an alternative on-the-fly approach where one does not explicitly solve for the full forward model. Instead, the solution to the forward problem is included implicitly in the formulation of the inverse problem, and the FEM problem is solved at each iteration for the current image estimate. We evaluate the convergence speeds of several representative iterative algorithms. We compare the computation cost of those two approaches, concluding that the on-the-fly approach can lead to substantial reductions in total cost when combined with a rapidly converging iterative algorithm.
1. Introduction
Bioluminescence tomography is an in vivo imaging technique that localizes and quantifies bioluminescent sources in a small animal. It has recently gained a great deal of attention as a promising means for macroscopic in vivo imaging of gene expression and other molecular and cellular-level processes (Contag and Bachmann 2002, Ntziachristos et al 2005). The objective of bioluminescence tomography is to reconstruct the three-dimensional (3D) spatial distribution of bioluminescent sources inside the animal from images of the light emitted through the animal surface. Collecting measurement data in multiple spectral bands and from more than one view, so that the entire animal surface is imaged, facilitates tomographic reconstruction of the 3D optical source distribution (Chaudhari et al 2005).
We have set up a bioluminescence tomographic imaging system using mirrors to collect four independent views (Chaudhari et al 2005). Multispectral data were acquired to improve localization (Chaudhari et al 2005, Dehghani et al 2006, Cong and Wang 2006, Wang et al 2006b, Kuo et al 2007, Allard et al 2007, Lv et al 2007). We then used an FEM solution of the diffusion equation to construct realistic forward models. We have successfully localized bioluminescent sources with this system, using the TOAST software to compute the forward model (Arridge et al 1993). However, TOAST is primarily intended to solve the diffuse optical tomography problem and is not well adapted for bioluminescence tomography applications; the forward model computation cost is high.
Fully 3D multispectral bioluminescence tomography is a computationally challenging inverse problem because (1) FEM-based forward models, which allow inhomogeneous tissue and realistic geometry, require the inversion of a very large matrix to solve the forward problem, (2) the use of multispectral data, which helps localize deep sources, increases the problem size by the number of spectral bins and (3) the inverse problem is intrinsically ill-posed due to the nature of the photon diffusion process and the limited information contained in data collected only on the animal surface.
In this paper we investigate various numerical techniques in order to minimize the cost of computing inverse solutions. The inverse problem is equivalent to a search for the source distribution which best predicts the measured data while also satisfying an a priori assumption regarding the smoothness of the source distribution. In order to iteratively solve the inverse problem, a forward model which maps a source to the data domain must be computed repeatedly. We explore two different approaches to incorporating the forward solutions into iterative algorithms for the inverse problem: (1) a direct calculation approach where the forward problem is solved in advance for every source location and then a forward solution is computed as a linear combination of these precomputed solutions, and (2) an on-the-fly approach where the FEM matrix inversion problem is solved as needed at each iteration using the current estimate of the source configuration. In this way, the full forward model need never be computed. We evaluate those approaches, combined with several different iterative algorithms, to determine the combination of forward and inverse algorithm that will minimize the computational cost.
Our goal in this paper is to introduce and evaluate methods that can rapidly reconstruct 3D bioluminescent images while still using a realistic FEM forward model. To the best of our knowledge, the on-the-fly approach is novel to this application and can substantially reduce the total reconstruction time, as shown in section 4. The iterative algorithms we consider in this paper include the class of incremental gradient or ordered subset methods, which have been widely explored for applications in nuclear medicine imaging (Hudson and Larkin 1994, Browne and De Pierro 1996, Ahn and Fessler 2003) but have not been investigated in the optical imaging literature. The algebraic reconstruction technique (ART) (Gordon et al 1970), which is a special case of the incremental gradient algorithms, was used for diffuse optical tomography (Arridge and Schweiger 1998). Our comparison of reconstruction times for the different methods described above should help guide the selection of reconstruction and forward modeling algorithms for developers of 3D bioluminescent and fluorescent imaging systems when using FEM-based forward models.
In section 2 we formulate the forward problem based on the diffusion equation using FEM models and describe approaches to incorporating the FEM solvers into iterative algorithms. In section 3 we define the inverse problem using regularized least squares (RLS) and describe various iterative algorithms for computing the inverse solution. Finally, in section 4 we evaluate different reconstruction methods in terms of computation times using in vivo bioluminescent imaging data from a mouse with an implanted brain tumor.
2. Forward problem
In the forward problem one needs to predict the photon flux from the animal surface for a given bioluminescent source distribution using known optical scatter and absorption properties within the animal. Since iterative solutions to the inverse problem require multiple solutions to the forward problem, a computationally efficient forward solution is important.
2.1. Forward model
Photon propagation in turbid media can be described by the radiative transfer or Boltzmann transport equation (Arridge 1999). While this model can be used in bioluminescence tomography (Klose 2007), the high computation cost and detailed knowledge required of the media's optical properties limit its use in practice. In contrast, the diffusion approximation under the assumption of isotropic scattering has been used extensively (Arridge et al 1993, Schweiger et al 1995, Jiang 1998, Arridge 1999, Hielscher et al 1999, Gu et al 2004, Wang et al 2004, Cong et al 2005, Slavine et al 2006, Dehghani et al 2006, Comsa et al 2006, Soloviev 2007, Kuo et al 2007). The diffusion approximation is reasonably accurate in soft tissue in the near-infrared region where scattering dominates absorption (Arridge 1999, Shen et al 2007). We, therefore, focus on the forward model based on the following steady-state diffusion equation (Arridge 1999),
| (1) |
subject to a Robin boundary condition
| (2) |
where q represents the bioluminescent source distribution, ϕ denotes the photon density, denotes the location vector, is the animal volume, ∂Ω is the animal surface, is a unit vector pointed outwardly normal to the surface ∂Ω, λ is the wavelength and , with μa and being the absorption and reduced scattering coefficients, respectively. In the boundary condition (2), G is a parameter modeling internal reflection at the boundary and can be computed as G = (1 + γ)/(1 − γ), where with nint being the refractive index of tissue (Schweiger et al 1995). The measurable photon flux at r ∈ ∂Ω is given by m(r, λ) = ϕ(r, λ)/(2G). The mapping from the source distribution q in the volume to the photon flux m through the surface can be obtained by solving (1) for ϕ given q.
Approaches to solving the diffusion equation (1) include analytical methods that assume simplified geometry and homogeneous tissue (Rice et al 2001) and the more general finite element method (FEM) (Arridge et al 1993, 2000, Schweiger and Arridge 1997, Jiang 1998, Arridge 1999, Roy and Sevick-Muraca 2001, Gu et al 2004, Cong et al 2005, Alexandrakis et al 2005, Chaudhari et al 2005, Lv et al 2006). Although the analytical method is more computationally efficient, simplifying assumptions about geometry and optical properties can produce inaccurate results (Chaudhari et al 2005). Here we focus on the FEM-based forward model.
In the FEM framework, the 3D animal volume Ω is discretized into tetrahedral elements connected at v vertex nodes. The source q and the photon density ϕ are also discretized using finite element basis functions and are represented by and , respectively. Then the problem of solving the diffusion equation reduces to
| (3) |
where . The FEM matrix F is a function of the optical absorption and reduced scattering coefficients and refractive index. The optical properties can be specified using published measurements applied to a segmented MR or CT volume (Wang et al 2004, 2006a, Chaudhari et al 2005, Alexandrakis et al 2005, 2006, Lv et al 2006, Allard et al 2007). A detailed description of how to construct F can be found in Arridge et al (2000). The FEM matrix F is symmetric (Arridge et al (2000), equations (29)–(34)). It is also sparse with on the order of 10 nonzero elements per row.
2.2. Forward solution method
There are two standard methods for solving the inversion problem in (3) for φ: (1) Cholesky factorization and substitution (CFS) and (2) preconditioned conjugate gradient (PCG) (Arridge et al 1993, Davies et al 1997, Schweiger and Arridge 1997, Arridge 1999). See Schweiger and Arridge (1997) for an overview of those methods and an analysis of their computational complexities. We chose CFS for our forward problem solver since CFS was faster than PCG in our experiments using a realistic FEM mesh; a comparison can be found in Chaudhari (2006) and Ahn et al (2007a). The reason that CFS was more efficient in our case is that if Cholesky factorization is done once, subsequent forward and back-substitutions can be inexpensively performed for multiple sources, as noted in Schweiger and Arridge (1997). In CFS, F is first decomposed into Cholesky factors as F = U′ U, where U is an upper triangular matrix and ′ represents matrix or vector transpose. Next one solves U′ c = q for by forward substitution and then solves Uφ = c for φ by back-substitution. The Cholesky factorization to calculate U is a one-time computation for a given FEM model F and the forward and back-substitution can be repeatedly computed for different q's using the precomputed Cholesky factor U.
2.3. System matrix
Let be the load vector for a unit source at voxel i and be a matrix of the load vectors. Then the source representation using the load vectors as a basis, and the corresponding source in tessellation nodes have the following relationship:
| (4) |
The data , which denote the photon flux through the animal surface as measured by a CCD camera, are given by
| (5) |
where maps the photon density in the volume to the CCD camera measurements. Typically, p < v and N < v; therefore, W is a thin matrix and D is wide.
Combining (3), (4) and (5), we construct a system matrix that transforms the source x into the measurement data y such that
| (6) |
The matrices D and W are sparse so the cost for multiplying a vector by D or W is negligible. Note that the system matrix A is not sparse whereas F is. To compute F−1q for some q, one needs to solve the forward problem (3), as discussed in section 2.2.
2.4. Forward and back-projector implementation approach
The two important operations involving the system matrix A are `forward projection' and `back-projection,' referred to jointly below as F/B projection. This terminology is borrowed from computed tomography (Herman 1980) although a mapping from a source to measurement data analogous to the system matrix was called the projection operator and the data were called the projection data in the context of optical tomography in Arridge and Schweiger (1998). For forward projection one multiplies a vector by the system matrix, that is, one computes y = Ax for some . For back-projection one computes x = A′ y for some . One needs to compute F/B projection multiple times in iterative algorithms. Since the time required for F/B projection is a major contributor to the total reconstruction time, it is important to implement F/B projection operators efficiently. We investigate two approaches to implementing F/B projectors: a straightforward scheme described below we call the `direct calculation' approach and a new `on-the-fly' method.
2.4.1. Direct calculation approach
In the direct calculation approach, one precomputes the full system matrix A and implements the F/B projector by a matrix-vector multiplication. Each column of the system matrix is computed by taking a unit vector corresponding to each source location as follows:
| (7) |
where is the jth unit vector with only the jth entry being 1. The substitution step in (7) must be repeated for a total of p source locations. However, once A is precomputed, F/B projection can be implemented as direct matrix-vector multiplications, Ax and A′ y.
2.4.2. On-the-fly approach
An alternative that avoids precomputing the full system matrix, is to calculate the F/B projection in an on-the-fly manner. The on-the-fly forward projector can be implemented as follows:
| (8) |
Similarly, in view of the symmetry of F, one can compute the back-projection on the fly as follows:
| (9) |
Every time we compute a forward or back-projection in the on-the-fly approach, we need to perform the substitution step only once.
2.4.3. Comparison
In the direct calculation approach, it is very expensive to precompute the full system matrix because the forward problem must be solved p times where p is the number of source voxels. However, once the matrix has been precomputed, the F/B projection can be computed rapidly as a single matrix vector product. On the other hand, in the on-the-fly approach, whenever one needs to compute the forward or back-projection, one must solve the forward problem in (3), which is more expensive than computing the product of the system matrix and a vector, partly because the FEM matrix is larger than the system matrix (typically, N < v and p < v) despite the sparsity of F. For example, in the multispectral bioluminescence tomography system we consider in section 4, v2 was larger than Np by a factor of >150.
Which of the direct calculation and on-the-fly approaches should be used? This depends on how many images are to be reconstructed for a given forward model, how many iterations per image are required for practical convergence of an iterative algorithm and how many F/B projections per iteration are needed. If one wishes to investigate many different iterative algorithms and regularization schemes for a given subject or if one assumes that the animal's anatomical structure and position do not change much over time in a longitudinal study, the direct calculation approach will be beneficial because once the expensive precomputation is performed, subsequent forward solutions can be rapidly computed by matrix vector multiplication. In contrast, if one is to compute relatively few images with a small number of iterations, the on-the-fly approach should be chosen. Comparisons of image reconstruction times for different approaches using real mouse bioluminescent data are made in section 4.
One of the advantages of the direct calculation approach is that direct access to any column or row in the system matrix is possible when the matrix is precomputed. Column or row access is required by certain iterative algorithms such as coordinate descent and incremental gradient algorithms, as discussed in section 3. Furthermore, the direct calculation approach may be more readily parallelizable than the on-the-fly approach.
Interestingly, the on-the-fly approach is closely connected with the adjoint or reverse differentiation method which is widely used for computing the gradient of a cost function in nonlinear diffuse optical tomography (Davies et al 1997, Hielscher et al 1999), as discussed in the appendix.
2.5. Multispectral system model
In multispectral imaging with K spectral bins, a multispectral system matrix Amul is constructed by concatenating monochromatic system matrices A1(λ1), …, AK(λK) for wavelengths λ1, …, λK (Chaudhari et al 2005):
| (10) |
where with s1, …, sK being known emission spectra of the bioluminescent source, and the multispectral data. It is straightforward to apply the F/B projection methods discussed in section 2.4 by combining individual F/B projections for each wavelength. That is, the forward projection is given by
and the back-projection
Henceforth, we omit the subscript in Amul and ymul for notational simplicity and consider both monochromatic and multispectral systems in a common framework.
3. Inverse problem
The ultimate goal of bioluminescence tomography is to estimate the source x from the photon flux measurement y based on the system model (6) or (10). In this section we formulate the inverse problem and discuss iterative algorithms for the inverse solution.
3.1. Regularized least squares (RLS)
We focus on the following regularized least squares (RLS) estimate:
| (11) |
The nonnegativity constraint, x ≥ 0, is imposed because the source strength is physically nonnegative (Jiang and Wang 2004, Jiang et al 2007). Although the problem size can be reduced by allowing sources only in a permissible region (Cong et al 2004, 2005, Jiang and Wang 2004, Wang et al 2006a, 2006b, Cong and Wang 2006, Jiang et al 2007, Lv et al 2007), we do not consider such constraints here. In (11), L is the data-fit function
| (12) |
β is a regularization parameter, and R is a quadratic regularization function such that
| (13) |
for a nonnegative definite matrix . The regularizer R stabilizes the noise in reconstructed images yet usually introduces bias. In a Bayesian framework, if the source and the additive noise are assumed to be zero-mean Gaussian with covariance of K and σ2I, respectively, then the RLS estimate obtained with choosing β = σ2 and R = K−1 can be viewed as a maximum a posteriori (MAP) estimate, where I is the identity matrix (Tarantola 2005, chapter 3). Other noise models such as the shot-noise model (Ye et al 1999) can also be used; other types of regularization functions include generalized Gaussian Markov random field priors (Bouman and Sauer 1993), non-Gaussian Gibbs priors (Hebert and Leahy 1989), which have an edge-preserving property, and L1-norm regularizers, which are known to produce sparse reconstruction (Cao et al 2007). We focus here on the quadratic cost function using (12) and (13). However, we note that the optimization methods described below are general, and can, with certain restrictions on differentiability and continuity, be used in conjunction with the nonquadratic regularizers referred to above.
We use a weighted L2-norm regularizer with a positive-definite diagonal matrix R (Lin et al 2006). If R is the identity matrix, then R(x) becomes a conventional L2-norm regularizer (Cong and Wang 2006, Han et al 2006, Wang et al 2006a, Lv et al 2006, Han and Wang 2007, Soloviev 2007). In this case, as the regularization parameter β increases, the reconstructed image becomes smooth and less noisy. But the reconstructed sources also tend to become superficial, consequently increasing localization errors particularly for deep sources. This occurs because deep sources are highly attenuated so need larger amplitudes to match the surface photon flux from a superficial source; these large amplitudes are in turn penalized by the L2-norm. Quadratic weighting schemes have been proposed to reduce the preference for superficial sources in the related problem of cortical current imaging in EEG and MEG (Lin et al 2006, Baillet et al 2001). Nonquadratic penalties can also have attractive properties in this respect.
We use the following sensitivity-dependent weight matrix, which through column normalization removes the increased penalty for deep sources, effectively removing the bias towards superficial ones:
| (14) |
where diag{·} denotes a diagonal matrix, γj is the sensitivity of the jth source location defined as
| (15) |
and aij are the entries in the system matrix A. One can assume γj > 0 for all j without loss of generality since one can exclude any source xj with γj = 0 from the parameter space. Note that γj's are readily calculated using an on-the-fly back-projection A′1 of uniform data 1 = [1, …, 1]′, without precomputing A.
The cost function in (11) with a positive-definite matrix R is strictly convex and so there exists a unique solution of the nonnegativity-constrained optimization problem in (11) and a local minimum is also the global minimum (Bertsekas 1999, p 685). In the continuous domain, the uniqueness of a solution to the diffusion equation under certain conditions on the source is discussed in Wang et al (2004) for the unregularized case and in Han et al (2006) and Han and Wang (2007) for the regularized one.
Most of the iterative algorithms that we study in section 3.2 use the gradient ∇Φ of the cost function Φ. The gradient is given by
| (16) |
calculation of which requires F/B projections. The Hessian H of the cost function is given by
| (17) |
The diagonal elements of the Hessian are used in certain preconditioners in section 3.2.5.
The quality of the reconstructed image in (11), as reflected in source localization error, quantitation error, resolution and noise characteristics, is determined by the choice of cost function Φ as well as the physical properties and limitations of the imaging system. Inaccuracies in the system model A will introduce a systematic bias into the image. An inaccurate noise model, for which we assume a zero-mean white Gaussian distribution, can lead to suboptimal noise characteristics as well as additional bias. Image quality will also be affected by choice of the regularization function R. However, we note that our goal here is to explore methods for solving the inverse problem once the cost function Φ is specified, rather than to investigate the relative merits of different cost functions. In section 4 we use a single regularization function which produces reasonably good localization results for the data used in our evaluations. We then compare the convergence speeds of different reconstruction algorithms that all converge to the same optimal solution for the chosen cost. Since all algorithms converge to the same solution we do not present comparisons of image quality in this paper.
3.2. Iterative algorithms
We will investigate iterative solutions to the optimization problem (11) since the nonnegativity constraint precludes an analytical solution. We choose four representative iterative algorithms: a gradient projection method (GPM), preconditioned conjugate gradient (PCG), coordinate descent (CD) and an incremental gradient algorithm, OS-SPS. We compare their convergence speeds using real mouse bioluminescent data in section 4. Those algorithms are described in detail below.
A number of different algorithms for bioluminescence tomography have been used in the literature. The expectation maximization (EM) algorithm (Dempster et al 1977) which was originally developed to compute a maximum likelihood solution for linear Poisson inverse problems (Shepp and Vardi 1982) was used in Jiang and Wang (2004), Alexandrakis et al (2005, 2006) and Jiang et al (2007). The EM algorithm maximizes an unregularized Poisson likelihood function and its convergence speed is generally very slow (Meng and van Dyk 1997, Qi and Leahy 2006) so we do not consider it in this paper. A deblurring variant of EM was used in Slavine et al (2006). A regularized Newton's algorithm (Gu et al 2004) and a modified Newton's method with an active set strategy (Cong et al 2004, 2005, Lv et al 2006) have also been used. A major disadvantage of Newton's methods for constrained optimization is that it is computationally costly both to enforce constraints and guarantee convergence to the solution (Bertsekas 1999, p 231); for example, naive orthogonal projection onto the nonnegative constraint set does not necessarily guarantee a decrease in the cost function (Bertsekas 1999, p 245). A constrained Landweber method was used in Jiang and Wang (2004), which requires nontrivial stepsize selection for both global convergence and optimal convergence rates.
The nonnegativity constraint in (11) can be more easily enforced by the four algorithms (GPM, PCG, CD and OS-SPS) considered here than by using active set methods (Cong et al 2004, 2005, Lv et al 2006, 2007). In GPM and PCG, the nonnegativity constraint is enforced by orthogonal projection onto the constraint set and a subsequent bent line search (Mumcuoğlu et al 1994, Qi et al 1998); in CD and OS-SPS, the optimization problem is reduced to a set of one-dimensional constrained problems, which makes it trivial to enforce the constraint. All the algorithms are aimed at minimizing the common RLS cost function in (11) and converge to the optimal point, with the exception of OS-SPS which approaches an approximate solution as stated below. None of these algorithms has any free parameters, such as step size, so that we can directly compare convergence rates for solving the problem in (11).
3.2.1. Gradient projection method (GPM)
A gradient projection method (GPM) is an iterative algorithm for constrained problems analogous to a gradient descent method for unconstrained problems (Bertsekas 1999, p 223). The gradient projection method we focus on here can be summarized as follows: for n = 0, 1, …,
| (18) |
| (19) |
| (20) |
| (21) |
| (22) |
where is the nth iterate, x0 is an initial estimate, is a preconditioner, ∇Φ and H are given in (16) and (17), respectively, and [·]+ denotesthe orthogonal projection onto the nonnegative orthant, that is, q = [t]+ implies qj = min(tj, 0) for all j. We describe preconditioners for this method in section 3.2.5. The above algorithm can be implemented by keeping Ax as a state vector so that GPM requires one F/B projection pair per iteration unless zn leaves the constraint set. Calculating the gradient in (18) and (16) requires one back-projection if Axn is retained as a state vector, and calculating the denominator in (20) needs one forward projection since dn′ Hdn = ∥Adn∥2 + βdn′ Rdn. If [zn]+ ≠ zn, then an additional forward projection is needed in (22) because dn is updated in (21).
3.2.2. Preconditioned conjugate gradient (PCG)
A PCG method is a general purpose optimization algorithm using a conjugate direction as a search direction (Bertsekas 1999, p 138). To enforce the nonnegativity constraint, we use a constrained version of PCG where the search direction is bent when an iterate leaves the constraint set, as in section 3.2.1 (Mumcuoğlu et al 1994, Qi et al 1998). The version we use can be summarized as follows: for n = 0, 1, …,
| (23) |
The PCG method requires one F/B projection pair per iteration with an extra forward projection, depending on whether zn leaves the constraint set.
3.2.3. Coordinate descent (CD)
A coordinate descent (CD) method is an iterative algorithm that minimizes the cost function along one coordinate direction for each update (Bertsekas 1999, p 267). The method has been successfully applied to image reconstruction applications (Bouman and Sauer 1996, Ye et al 1999). The CD method can be implemented as follows:
| (24) |
| (25) |
where ej is the jth unit vector and A·j is the jth column of A. In (24), the jth diagonal element Hjj of the Hessian H in (17) is given by
| (26) |
where
| (27) |
from (14), (15), and (17). The CD method requires access to each column A·j of the system matrix; therefore, the on-the-fly approach is not suitable for CD.
3.2.4. Incremental gradient methods
Incremental gradient methods (Bertsekas 1999, p 108) or ordered subset (OS) algorithms (Hudson and Larkin 1994) are optimization techniques which use only a subset of the data for each update with the aim of reducing the cost for computing gradients. Various incremental gradient methods have been successfully applied to PET/SPECT image reconstruction to accelerate convergence speeds (Hudson and Larkin 1994, Qi and Leahy 2006). The applicability of an OS algorithm to bioluminescence tomography was mentioned in Jiang and Wang (2004). In this paper we consider an OS version of separable paraboloidal surrogates (OS-SPS) where a line search is not required (Erdoğan and Fessler 1999). We derive the OS-SPS algorithm for our problem by following the derivation of additive updates in Erdoğan and Fessler (1999):
where
and
| (28) |
for m = 1, …, M, with M being the number of subsets Sm or subcost functions Φm. We choose the subsets Sm in (28) by downsampling the data space as Sm = {i : i ≡ m − 1(mod M), i =1, …, KN}.
Generally, OS-SPS does not converge to an optimal solution of the problem in (11) but to a limit cycle of points which are only approximate solutions (Ahn et al 2006). The OS type algorithms can be forced to converge to the optimal solution by using relaxation parameters (Ahn and Fessler 2003), switching to a convergent algorithm at some point (Li et al 2005), or rederiving the algorithm in a framework of incremental optimization transfer (Ahn et al 2006). However, we use the original form of OS-SPS as outlined above for simplicity because it works well enough for our purposes, as shown in section 4. Since OS algorithms, including OS-SPS, require access to each row of the system matrix A to compute the gradient ∇Φm of the subcost function, the on-the-fly approach is not suitable.
3.2.5. Preconditioners
For ill-conditioned problems, gradient based methods suffer from slow convergence but can be accelerated using preconditioning. Newton's method using the inverse of the Hessian as a preconditioner converges very quickly (Bertsekas 1999, p 26). However, calculating the Hessian inverse is impractical for large-scale problems. Here we consider only simple diagonal preconditioners.
A popular choice is a diagonal approximation to Newton's method (Bertsekas 1999, p 27),
| (29) |
with ξj and Rjj defined in (27). Calculating the term requires precomputation of the full system matrix A so the on-the-fly approach is not suitable for this preconditioner. However, one can estimate ξj from where γj ≜ Σi aij since ξj and are usually highly correlated and γj can be readily computed on the fly by back-projecting a vector of ones as discussed in section 3.1. Figure 1 shows an example scatter plot of ξj versus for the multispectral imaging system model from section 4. As shown in the figure, ξj and are strongly correlated (Pearson's correlation coefficient was 0.922). A practical strategy for estimating ξj, which works for the on-the-fly approach, is as follows. First, one computes γj for j = 1, …, p by back-projection A′1. Next, one randomly chooses T source locations j1, …, jT, and calculates projections y(t) = Aejt and subsequently for t = 1, …, T. Then one estimates the slope τ in the scatter plot by fitting a line ξ = τγ2 to the T samples . Finally, one approximates ξj as using the slope estimate for j = 1, …, p. We refer to the resulting preconditioner as the estimated diagonal approximation to Newton's (EN) method,
| (30) |
which can be applied in both direct calculation and on-the-fly approaches. Calculating this preconditioner requires one back-projection for computing 's and T forward projections for estimating τ. In section 4, we choose T = 10 source locations for estimating the slope τ. The coefficient of variation (CV) of due to randomly choosing 10 source locations was about 29%, which is stable enough for our purposes (a very accurate estimate of τ is not necessary).
Figure 1.
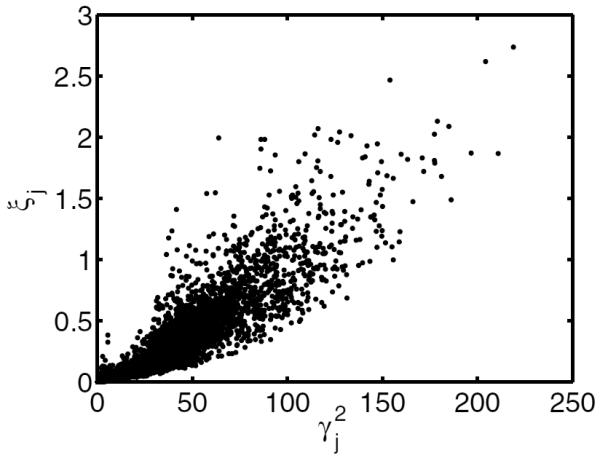
Example scatter plot of versus for the multispectral imaging system used in section 4.
For PCG we also consider an EM-type preconditioner as follows:
| (31) |
where ∊ is a small positive number, e.g., ∊ = 10−3 max(1, maxl xl) > 0. This preconditioner originates from the EM algorithm for maximizing a Poisson likelihood (Shepp and Vardi 1982) and has been used successfully to accelerate PCG algorithms for emission tomography (Kaufman 1987). It has also been used with PCG for bioluminescence tomography (Chaudhari et al 2005).
3.2.6. Comparison
The on-the-fly approach is not suitable for CD and OS-SPS, as discussed above, since they require access to each row or column of the system matrix; neither is it suitable for the diagonally approximated Newton preconditioner PN in (29) since calculating ξj needs the precomputed full system matrix. For GPM and PCG, using PEN in (30) or PEMin (31), both direct calculation and on-the-fly approaches can be applied.
The computation cost for the iterative algorithms is dominated by F/B projections. Therefore, the computational complexity of each algorithm can be represented by the number of F/B projections required per iteration. A forward or back-projection amounts to matrix-vector multiplication for the direct calculation approach, and to one FEM forward solution for the on-the-fly approach. GPM and PCG require one F/B projection pair plus possibly an extra forward projection. The cost per iteration for CD and OS-SPS equals roughly that of one F/B projection pair. Therefore, the computation costs per iteration for those algorithms considered here do not differ substantially once the F/B projection implementation approach is chosen. Other important factors are the cost for precomputation and the convergence speed, that is, how many iterations are required for practical convergence.
Memory requirements for the iterative algorithms are modest. During iterations, one needs a memory space O(p + KN) proportional to the image size p and to the data size KN. Additionally, for the direct calculation approach, one needs to store the KN × p system matrix. On the other hand, for the on-the-fly approach, one must retain the v × v FEM matrix, which is sparse and has about 10v nonzero elements, for each wavelength if PCG is used for the forward problem solver; and one needs to keep the v × v Cholesky factor, which is a sparse upper triangular matrix, for each wavelength if CFS is chosen for the FEM solver.
The cost of all of these methods is dominated by the cost of (possibly partial) forward and back-projections. Since these operations all involve large matrix-vector multiplications for the direct calculation approach, parallelization using multithreading or other methods is relatively straightforward. Among the four methods, the full gradient methods (PCG and GPM) in conjunction with the direct calculation approach benefit most readily since the full forward and back-projection operations can be easily distributed across processors. The CD method which updates one variable at a time will benefit least from parallelization.
4. Results
We applied iterative reconstruction methods to real bioluminescent data obtained from a mouse with a brain tumor implanted in the right cerebral hemisphere. Our goal is to compare computation cost for different reconstruction methods. All methods described should converge to the RLS image in (11) with the exception of OS-SPS which may enter a limit cycle as described above.
Multispectral bioluminescent data were obtained from top and two side views by the IVIS 200 imaging system for wavelengths 580 nm, 600 nm, 620 nm and 640 nm. Since the tumor was near the dorsal surface of the head, no signal was detected from the bottom view and those three views (top and two sides) had sufficient information for localizing the source (Chaudhari et al 2005).
3D CT scan data were used to segment the anatomical volume into skull and soft tissue and to assign standard optical properties (Cheong et al 1990). The specific values of the absorption and reduced scattering coefficients for the skull and soft tissue we used can be found in Chaudhari et al (2005). A regularization parameter value of β = 0.05 was determined empirically. All image reconstructions were performed on an AMD Opteron 870 2.0 GHz computer. For all the iterative algorithms, a zero uniform image, , was used as an initial estimate x0.
There is a trade-off between the number of FEM mesh nodes and the accuracy of FEM model; 105–106 nodes were recommended for sufficient accuracy in 3D problems (Schweiger and Arridge 1997). Here we used a FEM-based forward model with v = 71256 tessellation nodes, a source space with p = 10814 source locations, and N = 3085 surface measurement nodes for K = 4 spectral bins. We did not attempt to investigate different FEM models in terms of accuracy and computation cost since our purpose is to compare the computation speeds of different reconstruction methods for a given model; however, we achieved reasonably good localization results within a reasonable time, as shown below. See Chaudhari et al (2005) for a study of the effects of the number of surface measurement points on reconstruction results.
First, we precomputed the full system matrix A by using the mldivide function in Matlab (v7.3; Mathworks, MA, USA). We computed 500 columns of the system matrix at a time; that is, we prepared a 71256 × 500 matrix by combining 500 load vectors and solved the forward problem in (3) by putting the matrix on the right-hand side. It took 12 708 s to compute the full system matrix, as shown in table 1. Then we reconstructed the bioluminescence image with the system matrix using the direct calculation approach. The iterative algorithms were implemented in the C language. Table 1 shows, in the columns named iteration cost, the computation time in seconds and the number of iterations, required for each algorithm to achieve the relative error , <5%, and <1%, respectively, where the RLS image in (11) was estimated by 2000 iterations of GPM-N. Note that the relative error is not with respect to the true source distribution which is unknown but with respect to the RLS solution given in (11). Therefore, represents how close an image is to the converged solution and can be used for evaluating the convergence speed of an iterative algorithm. In the table, GPM-U denotes the unpreconditioned GPM; GPM-N and GPM-EN denote GPM with preconditioner PN and PEN given in (29) and (30), respectively; CG, PCG-N, PCG-EN, and PCG-EM denote PCG with no preconditioner and with preconditioners PN, PEN, and PEM, respectively; and OS-SPS-2, OS-SPS-5, and OS-SPS-10 denote OS-SPS with two, five and ten subsets, respectively. In table 1, the total reconstruction time is the sum of the precomputation time and the iteration cost. We did not include the computation cost for constructing the FEM matrix F, which is common in all the methods.
Table 1.
Comparison of computation costs for different reconstruction methods when a single image reconstruction is performed.
|
< 10% |
< 5% |
< 1% |
|||||
|---|---|---|---|---|---|---|---|
| F/B projection approach | Algorithm | Iteration cost (# iters) | Total recon. time | Iteration cost (# iters) | Total recon. time | Iteration cost (# iters) | Total recon. time |
| Direct calculation approach with precomputed system matrix (precomput. time: 12 708 s) | GPM-U | 359 s (216) | 13068 s | 599 s (378) | 13307 s | 1444 s (879) | 14152 s |
| GPM-N | 53 s (32) | 12761 s | 77 s (46) | 12786 s | 129 s (79) | 12838 s | |
| GPM-EN | 54 s (29) | 12762 s | 71 s (42) | 12780 s | 121 s (73) | 12830 s | |
| CG | 563 s (216) | 13272 s | 941 s (378) | 13650 s | 2376 s (878) | 15084 s | |
| PCG-N | 58 s (35) | 12766 s | 88 s (52) | 12796 s | 157 s (93) | 12865 s | |
| PCG-EN | 59 s (33) | 12767 s | 81 s (48) | 12789 s | 146s (86) | 12854 s | |
| PCG-EM | 81 s (45) | 12789 s | 111 s (66) | 12819 s | 268 s (151) | 12976 s | |
| CD | 37 s (20) | 12746 s | 46 s (26) | 12754 s | 64 s (38) | 12773 s | |
| OS-SPS-2 | 48 s (36) | 12756 s | 80 s (61) | 12788 s | 178 s (136) | 12887 s | |
| OS-SPS-5 | 20 s (15) | 12729 s | 35 s (26) | 12743 s | – | – | |
| OS-SPS-10 | 11 s (8) | 12719 s | 20 s (15) | 12728 s | |||
| On-the-fly approach (precomput. time: 110 s) | GPM-U | 4877 s (216) | 4987 s | 8523 s (378) | 8633 s | 16152 s (879) | 16262 s |
| GPM-EN | 688 s (29) | 798 s | 981 s (42) | 1091 s | 1614 s (73) | 1724 s | |
| CG | 6531 s (216) | 6641 s | 11400 s (378) | 11509 s | 21557 s (878) | 21666s | |
| PCG-EN | 735 s (33) | 845 s | 1035 s (48) | 1145 s | 1797 s (86) | 1907 s | |
| PCG-EM | 1033 s (45) | 1143 s | 1480 s (66) | 1590 s | 3291 s (151) | 3401 s | |
Next, we reconstructed images using the on-the-fly approach. For the on-the-fly approach, the precomputation time (110 s) in the table represents the time required for Cholesky factorization. As discussed in section 3, we excluded CD and OS-SPS, and preconditioner PN for this approach. For the on-the-fly approach we implemented the iterative algorithms in Matlab. For the on-the-fly F/B projectors, the one-time computation for Cholesky factorization was performed by the chol function, and repeated substitutions were carried out by the mldivide function in Matlab.
As shown in table 1, for both GPM and PCG, the use of preconditioners PN, PEN and PEM significantly accelerates convergence. In other words, the preconditioned algorithms (GPM-N, GPM-EN and PCG-N, PCG-EN, PCG-EM) require fewer iterations and lower iteration costs in seconds than the unpreconditioned ones (GPM-U and CG). Preconditioners PEN using the estimated diagonal elements of the Hessian and PN using the exact diagonals showed similar convergence speeds. In the direct calculation approach, CD and OS-SPS-10 were much faster than GPM and PCG. In fact, OS-SPS-10 showed the fastest convergence speed; it required only 8 and 15 iterations (11 and 20 s) for < 10% and < 5%, respectively. However, OS-SPS-10 (as well as OS-SPS-5) could not reach a point where ε < 1% because of the limit cycle behavior. This is a typical characteristic of incremental gradient or OS algorithms; as the number of subsets increases the initial convergence speed becomes faster but the limit point moves further from the optimal solution. Therefore, when it comes to the number of subsets for OS algorithms, there is a trade-off between convergence speed and accuracy. For ε < 1%, CD was fastest among the iterative algorithms for the direct calculation approach. On the other hand, for the on-the-fly approach, GPM-EN showed the fastest convergence rate for all three relative error cases, and PCG-EN showed a similar yet little slower convergence rate.
The precomputation time (12 708 s) for the direct calculation approach is much larger than that (110 s) for the on-the-fly approach whereas the cost per iteration for the direct approach is smaller than that for the on-the-fly approach. If few images are to be reconstructed for a given system, then the on-the-fly approach can substantially reduce the total computation cost. For example, for the relative error <10%, GPM-EN, the fastest among the on-the-fly algorithms in total reconstruction time, is faster than OS-SPS-10, the fastest of the direct calculation algorithms, by a factor of 12719/798 ≈ 15.9; similarly, for < 5%, by a factor of 12728/1091 ≈ 11.7. For ε < 1%, GPM-EN in the on-the-fly approach is faster than CD in the direct calculation approach by a factor of 12773/1724 ≈ 7.4. As the number of required iterations increases, the factor of computational saving of the on-the-fly approach compared to the direct approach decreases.
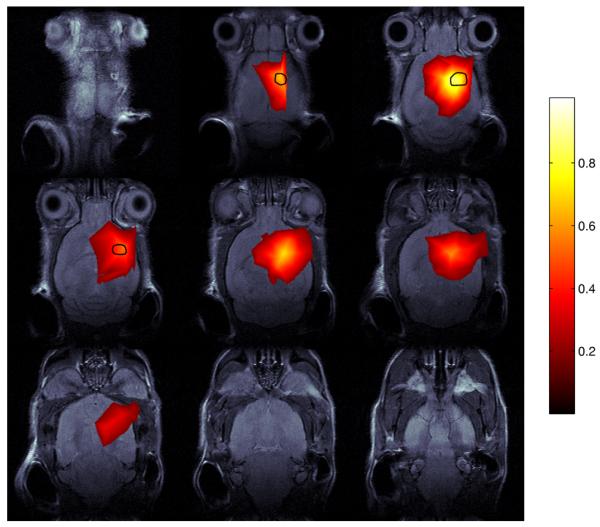
Figure 2 shows the RLS image and figure 3 shows an image reconstructed by 29 iterations, taking 798 s in total reconstruction time, of GPM-EN, which is the fastest among the on-the-fly algorithms. Although the relative error between the images is about 10%, they look similar and the localization is reasonably accurate.
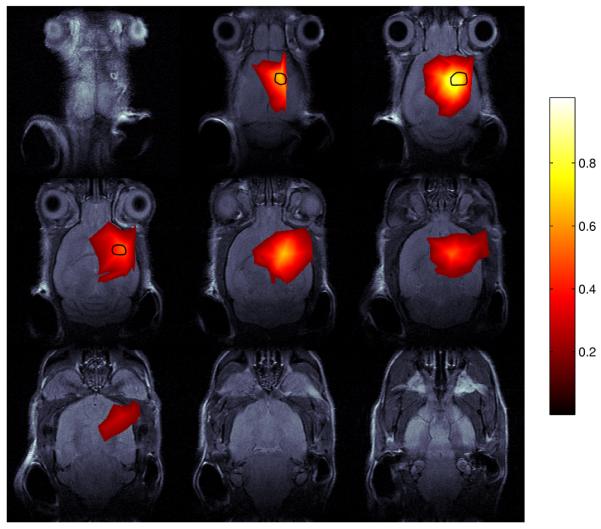
Figure 2.
The RLS image , which minimizes the cost function Φ given in (11), shown on nine horizontal planes. The reconstructed bioluminescence image was obtained by 2000 iterations of GPM-N and is overlaid on co-registered MR slices. Reconstructed values greater than or equal to 10% of the maximum value are displayed. The black contour denotes the boundary of the tumor obtained from the MR image. The color scale on the right shows relative intensity. These results are not quantitatively calibrated.
Figure 3.
The same as figure 2 except that this image was reconstructed using 29 iterations of GPM-EN using the on-the-fly approach. The relative error is about 10%.
5. Conclusion
We have explored computationally efficient methods for fully 3D multispectral bioluminescence image reconstruction. When it comes to incorporating F/B projectors into iterative algorithms, we investigated the straightforward direct calculation approach and an on-the-fly approach where one does not have to precompute the system matrix. We evaluated those approaches combined with various iterative algorithms by using real mouse bioluminescence data. We achieved a substantial speed-up using the proposed on-the-fly approach by a factor of up to 15.9 when a single image reconstruction is required. However, if multiple image reconstructions are to be performed for a given model, the speed-up factor decreases and in this case the direct calculation approach may be more efficient. Using proper preconditioners accelerates convergence speeds significantly; the estimated diagonal approximation to Newton's preconditioner (EN), which applies to both direct calculation and on-the-fly approaches, reduced the iteration cost by a factor of >7 for both GPM and PCG. In the on-the-fly approach, GPM-EN yielded the fastest convergence speed and PCG-EN showed a little slower convergence rates. For the direct calculation approach, OS-SPS, an incremental gradient algorithm, was the fastest one although it converged to an approximated RLS solution; CD was the second fastest, which converges to the RLS solution.
We note that the emphasis in this paper is on fast methods for computing inverse solutions. With the exceptions described above, all methods implemented should converge to the same solution. For this reason we have not explored measures of image quality such as bias, resolution and variance. The methods presented here represent the simplest (quadratic) error models and regularization functions. However, most of the ideas discussed can be extended to incorporate more sophisticated noise models and nonquadratic regularization functions. We also restricted our attention here to bioluminescence imaging. However, the methods can be extended to the related problem of reconstructing images in 3D fluorescence optical tomography (Ahn et al 2007b). In this case, the models must be extended to also include the propagation of the excitation light through the animal. Since this process can also be approximated using the diffusion equation, similar fast FEM and on-the-fly methods can be developed.
Acknowledgment
This work was supported by the National Cancer Institute under Grant 1R01CA121783.
Appendix. Relationship between adjoint or reverse differentiation and F/B projection approaches
Suppose that F = F (θ) and q = q(θ) where F and q are defined in (3) and is an unknown parameter vector. In diffuse optical tomography, θ represents the optical absorption and reduced scattering coefficients and q is a known constant in θ. In bioluminescence tomography, θ is the source distribution and F is a known constant. From (3), one can show
| (A.1) |
for s = 1, …, S. Let a data-fit function be defined as
| (A.2) |
where D is defined as in (5). Here our goal is to compute the gradient ∇L(θ) efficiently.
Using (A.1), one obtains
| (A.3) |
where 〈·,·〉 denotes the real inner product. If one is to use the above expression to compute ∇L(θ). one needs to solve the forward problem in (3) to compute ϕ and additionally `S' forward problems to calculate F−1{∂q/∂θs – ∂F/∂θs)ϕ} for s = 1, …, S by solving Fu = ∂q/∂θs – ∂F/∂θs)ϕ. Now one can rewrite (A.3) as
| (A.4) |
where (F−1)* is the adjoint of F−1 and (A.4) is due to the fact that F is a real symmetric matrix, in other words, a self-adjoint operator. Computing ∇L(θ) by (A.4) requires only two forward solutions, one for computing ϕ and the other for computing the vector F−1D′ (y – Dϕ) by solving Fu = D′ (y – D ϕ) for u, hence a computational saving by a factor of (S + 1)/2 is achieved if the cost for forward solutions is dominant in the gradient computation. Using (A.4) for computing the gradient is often referred to as adjoint or reverse differentiation in diffuse optical tomography (Davies et al 1997, Hielscher et al 1999). A similar idea is also used in Arridge and Schweiger (1995, 1998).
Now we consider the special case of bioluminescence tomography, where F is a known constant and q = Wθ as in (4). Then (A.3) becomes
where es represents the sth unit vector. Calculating the vector DF−1Wes in the above equation for s = 1, …, S is equivalent to computing the full system matrix A = DF−1W as in the direct calculation approach described in section 2.4.1. On the other hand, (A.4) for the adjoint differentiation method reduces to
where calculating A′(y−Dϕ) by a forward solution method without precomputing the system matrix A corresponds to the on-the-fly implementation of F/B projectors.
References
- Allard H, Cote D, Davidson L, Dazai J, Henkelman RM. Combined magnetic resonance and bioluminescence imaging of live mice. J. Biomed. Opt. 2007;12:034018. doi: 10.1117/1.2745298. [DOI] [PubMed] [Google Scholar]
- Ahn S, Chaudhari AJ, Darvas F, Bouman CA, Leahy RM. Fast image reconstruction methods for fully 3D multispectral optical bioluminescence tomography. Proc. IEEE Intl. Symp. Biomed. Imaging. 2007a:229–32. [Google Scholar]
- Ahn S, Dutta J, Chaudhari AJ, Leahy RM. Computationally efficient image reconstruction methods for multispectral optical fluorescence tomography using FEM-based forward models. Abstract Book Joint Molecular Imaging Conf. 2007b:329–30. [Google Scholar]
- Ahn S, Fessler JA. Globally convergent image reconstruction for emission tomography using relaxed ordered subsets algorithms. IEEE Trans. Med. Imaging. 2003;22:613–26. doi: 10.1109/TMI.2003.812251. [DOI] [PubMed] [Google Scholar]
- Ahn S, Fessler JA, Blatt D, Hero AO. Convergent incremental optimization transfer algorithms: Application to tomography. IEEE Trans. Med. Imaging. 2006;25:283–96. doi: 10.1109/TMI.2005.862740. [DOI] [PubMed] [Google Scholar]
- Alexandrakis G, Rannou FR, Chatziioannou AF. Tomographic bioluminescence imaging by use of a combined optical-PET (OPET) system: a computer simulation feasibility study. Phys. Med. Biol. 2005;50:4225–41. doi: 10.1088/0031-9155/50/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrakis G, Rannou FR, Chatziioannou AF. Effect of optical property estimation accuracy on tomographic bioluminescence imaging: simulation of a combined optical-PET (OPET) system 2006. Phys. Med. Biol. 2006;51:2045–53. doi: 10.1088/0031-9155/51/8/006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arridge SR. Optical tomography in medical imaging. Inverse Problems. 1999;15:41–93. [Google Scholar]
- Arridge SR, Dehghani H, Schweiger M, Okada E. The finite element model for the propagation of light in scattering media: a direct method for domains with nonscattering regions. Med. Phys. 2000;27:252–64. doi: 10.1118/1.598868. [DOI] [PubMed] [Google Scholar]
- Arridge SR, Schweiger M. Photon-measurement density functions: part 2. Finite-element-method calculations. Appl. Opt. 1995;34:8026–37. doi: 10.1364/AO.34.008026. [DOI] [PubMed] [Google Scholar]
- Arridge SR, Schweiger M. A gradient-based optimisation scheme for optical tomography. Opt. Express. 1998;2:213–26. doi: 10.1364/oe.2.000213. [DOI] [PubMed] [Google Scholar]
- Arridge SR, Schweiger M, Hiraoka M, Delpy DT. A finite element approach for modeling photon transport in tissue. Med. Phys. 1993;20:299–309. doi: 10.1118/1.597069. [DOI] [PubMed] [Google Scholar]
- Baillet S, Mosher JC, Leahy RM. Electromagnetic brain mapping. IEEE Signal Process. Mag. 2001;18:14–30. [Google Scholar]
- Bertsekas DP. Nonlinear Programming. 2nd edn Athena Scientific; Belmont, MA: 1999. [Google Scholar]
- Bouman C, Sauer K. A generalized Gaussian image model for edge-preserving MAP estimation. IEEE Trans. Image Process. 1993;2:296–310. doi: 10.1109/83.236536. [DOI] [PubMed] [Google Scholar]
- Bouman CA, Sauer K. A unified approach to statistical tomography using coordinate descent optimization. IEEE Trans. Image Process. 1996;5:480–92. doi: 10.1109/83.491321. [DOI] [PubMed] [Google Scholar]
- Browne JA, De Pierro AR. A row-action alternative to the EM algorithms for maximizing likelihoods in emission tomography. IEEE Trans. Med. Imaging. 1996;15:687–99. doi: 10.1109/42.538946. [DOI] [PubMed] [Google Scholar]
- Cao N, Nehorai A, Jacob M. Image reconstruction for diffuse optical tomography using sparsity regularization and expectation-maximization algorithm. Opt. Express. 2007;15:13695–708. doi: 10.1364/oe.15.013695. [DOI] [PubMed] [Google Scholar]
- Chaudhari AJ. PhD Thesis. University of Southern California; Los Angeles, CA: 2006. Hyperspectral and multispectral optical bioluminescence and fluorescence tomography in small animal imaging. [Google Scholar]
- Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, Cherry SR, Leahy RM. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Phys. Med. Biol. 2005;50:5421–41. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- Cheong WF, Prahl SA, Welch AJ. A review of the optical properties of biological tissues. IEEE J. Quantum Electron. 1990;26:2166–85. [Google Scholar]
- Comsa DC, Farrell TJ, Patterson MS. Quantification of bioluminescence images of point source objects using diffusion theory models. Phys. Med. Biol. 2006;51:3733–46. doi: 10.1088/0031-9155/51/15/009. [DOI] [PubMed] [Google Scholar]
- Cong AX, Wang G. Multispectral bioluminescence tomography: methodology and simulation. Int. J. Biomed. Imaging. 2006;2006:57614. doi: 10.1155/IJBI/2006/57614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong W, Kumar D, Liu Y, Cong A, Wang G. A practical method to determine the light source distribution in bioluminescent imaging. Proc. SPIE 5535: Developments in X-Ray Tomography IV. 2004:679–86. [Google Scholar]
- Cong W, et al. Practical reconstruction method for bioluminescence tomography. Opt. Express. 2005;13:6756–71. doi: 10.1364/opex.13.006756. [DOI] [PubMed] [Google Scholar]
- Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 2002;4:235–60. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- Davies AJ, Christianson DB, Dixon LCW, Roy R, van der Zee P. Reverse differentiation and the inverse diffusion problem. Adv. Eng. Softw. 1997;28:217–21. [Google Scholar]
- Dehghani H, Davis SC, Jiang S, Pogue BW, Paulsen KD, Patterson MS. Spectrally resolved bioluminescence optical tomography. Opt. Lett. 2006;31:365–7. doi: 10.1364/ol.31.000365. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B. 1977;39:1–38. [Google Scholar]
- Erdoğan H, Fessler JA. Ordered subsets algorithms for transmission tomography. Phys. Med. Biol. 1999;44:2835–51. doi: 10.1088/0031-9155/44/11/311. [DOI] [PubMed] [Google Scholar]
- Gordon R, Bender R, Herman GT. Algebraic reconstruction techniques (ART) for the three-dimentional electron microscopy and X-ray photography. J. Theor. Biol. 1970;29:471–81. doi: 10.1016/0022-5193(70)90109-8. [DOI] [PubMed] [Google Scholar]
- Gu X, Zhang Q, Larcom L, Jiang H. Three-dimensional bioluminescence tomography with model-based reconstruction. Opt. Express. 2004;12:3996–4000. doi: 10.1364/opex.12.003996. [DOI] [PubMed] [Google Scholar]
- Han W, Cong W, Wang G. Mathematical theory and numerical analysis of bioluminescence tomography. Inverse Problems. 2006;22:1659–75. [Google Scholar]
- Han W, Wang G. Theoretical and numerical analysis on multispectral bioluminescence tomography. IMA J. Appl. Math. 2007;72:67–85. [Google Scholar]
- Hebert T, Leahy RM. A generalized EM algorithm for 3-D Bayesian reconstruction from Poisson data using Gibbs priors. IEEE Trans. Med. Imaging. 1989;8:194–202. doi: 10.1109/42.24868. [DOI] [PubMed] [Google Scholar]
- Hebert T, Leahy RM, Singh M. Three-dimensional maximum-likelihood reconstruction for an electronically collimated single-photon-emission imaging system. J. Opt. Soc. Am. A. 1990;7:1305–13. doi: 10.1364/josaa.7.001305. [DOI] [PubMed] [Google Scholar]
- Herman GT. Image Reconstruction from Projections: The Fundamentals of Computerized Tomography. Academic; New York: 1980. [Google Scholar]
- Hielscher AH, Klose AD, Hanson KM. Gradient-based iterative image reconstruction scheme for time-resolved optical tomography. IEEE Trans. Med. Imaging. 1999;18:262–71. doi: 10.1109/42.764902. [DOI] [PubMed] [Google Scholar]
- Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans. Med. Imaging. 1994;13:601–9. doi: 10.1109/42.363108. [DOI] [PubMed] [Google Scholar]
- Jiang H. Frequency-domain fluorescent diffusion tomography: a finite-element-based algorithm and simulations. Appl. Opt. 1998;37:5337–43. doi: 10.1364/ao.37.005337. [DOI] [PubMed] [Google Scholar]
- Jiang M, Wang G. Image reconstruction for bioluminescence tomography. Proc. SPIE 5535: Developments in X-Ray Tomography IV. 2004:335–51. [Google Scholar]
- Jiang M, Zhou T, Cheng J, Cong W, Wang G. Image reconstruction for bioluminescence tomography from partial measurement. Opt. Express. 2007;15:11095–116. doi: 10.1364/oe.15.011095. [DOI] [PubMed] [Google Scholar]
- Kaufman L. Implementing and accelerating the EM algorithm for positron emission tomography. IEEE Trans. Med. Imaging. 1987;6:37–51. doi: 10.1109/TMI.1987.4307796. [DOI] [PubMed] [Google Scholar]
- Klose AD. Transport-theory-based stochastic image reconstruction of bioluminescent sources. J. Opt. Soc. Am. A. 2007;24:1601–8. doi: 10.1364/josaa.24.001601. [DOI] [PubMed] [Google Scholar]
- Kuo C, Coquoz O, Troy TL, Xu H, Rice BW. Three-dimensional reconstruction of in vivo bioluminescent sources based on multispectral imaging. J. Biomed. Opt. 2007;12:024007. doi: 10.1117/1.2717898. [DOI] [PubMed] [Google Scholar]
- Li Q, Ahn S, Leahy RM. Fast hybrid algorithms for PET image reconstruction. Proc. IEEE Nucl. Sci. Symp. Medical Imaging Conf. 2005;4:1851–5. [Google Scholar]
- Lin FH, Witzel T, Ahlfors SP, Stufflebeam SM, Belliveau JW, Hamalainen MS. Assessing and improving the spatial accuracy in MEG source localization by depth-weighted minimum-norm estimates. NeuroImage. 2006;31:160–71. doi: 10.1016/j.neuroimage.2005.11.054. [DOI] [PubMed] [Google Scholar]
- Lv Y, Tian J, Cong W, Wang G, Luo J, Yang W, Li H. A multilevel adaptive finite element algorithm for bioluminescence tomography. Opt. Express. 2006;14:8211–23. doi: 10.1364/oe.14.008211. [DOI] [PubMed] [Google Scholar]
- Lv Y, Tian J, Cong W, Wang G, Yang W, Qin C, Xu M. Spectrally resolved bioluminescence tomography with adaptive finite element analysis: methodology and simulation. Phys. Med. Biol. 2007;52:4497–512. doi: 10.1088/0031-9155/52/15/009. [DOI] [PubMed] [Google Scholar]
- Mumcuoğlu EÜ, Leahy R, Cherry SR, Zhou Z. Fast gradient-based methods for Bayesian reconstruction of transmission and emission PET images. IEEE Trans. Med. Imaging. 1994;13:687–701. doi: 10.1109/42.363099. [DOI] [PubMed] [Google Scholar]
- Meng XL, van Dyk D. The EM algorithm - An old folk song sung to a fast new tune. J. R. Stat. Soc. Ser. B. 1997;59:511–67. [Google Scholar]
- Ntziachristos V, Ripoll J, Wang LV, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat. Biotechnol. 2005;23:313–20. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- Qi J, Leahy RM. Iterative reconstruction techniques in emission computed tomography. Phys. Med. Biol. 2006;51:541–78. doi: 10.1088/0031-9155/51/15/R01. [DOI] [PubMed] [Google Scholar]
- Qi J, Leahy RM, Hsu C, Farquhar TH, Cherry SR. Fully 3D Bayesian image reconstruction for the ECAT EXACT HR+ IEEE Trans. Nucl. Sci. 1998;45:1096–103. [Google Scholar]
- Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J. Biomed. Opt. 2001;6:432–40. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- Roy R, Sevick-Muraca EM. Three-dimensional unconstrained and constrained image-reconstruction techniques applied to fluorescence, frequency-domain photon migration. Appl. Opt. 2001;40:2206–15. doi: 10.1364/ao.40.002206. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Arridge SR. The finite-element method for the propagation of light in scattering media: frequency domain case. Med. Phys. 1997;24:895–902. doi: 10.1118/1.598008. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Arridge SR, Hiraoka M, Delpy DT. The finite element method for the propagation of light in scattering media: boundary and source conditions. Med. Phys. 1995;22:1779–92. doi: 10.1118/1.597634. [DOI] [PubMed] [Google Scholar]
- Shen H, Cong W, Qian X, Durairaj K, Wang G. Numerical study on the validity of the diffusion approximation for computational optical biopsy. J. Opt. Soc. Am. A. 2007;24:423–29. doi: 10.1364/josaa.24.000423. [DOI] [PubMed] [Google Scholar]
- Shepp LA, Vardi Y. Maximum likelihood reconstruction for emission tomography. IEEE Trans. Med. Imaging. 1982;1:113–22. doi: 10.1109/TMI.1982.4307558. [DOI] [PubMed] [Google Scholar]
- Slavine NV, Lewis MA, Richer E, Antich PP. Iterative reconstruction method for light emitting sources based on the diffusion equation. Med. Phys. 2006;33:61–8. doi: 10.1118/1.2138007. [DOI] [PubMed] [Google Scholar]
- Soloviev VY. Tomographic bioluminescence imaging with varying boundary conditions. Appl. Opt. 2007;46:2778–884. doi: 10.1364/ao.46.002778. [DOI] [PubMed] [Google Scholar]
- Tarantola A. Inverse Problem Theory and Methods for Model Parameter Estimation. SIAM; Philadelphia, PA: 2005. [Google Scholar]
- Wang G, Cong W, Durairaj K, Qian X, Shen H, Sinn P, Hoffman E, McLennan G, Henry M. In vivo mouse studies with bioluminescence tomography. Opt. Express. 2006a;14:7801–9. doi: 10.1364/oe.14.007801. [DOI] [PubMed] [Google Scholar]
- Wang G, Shen H, Durairaj K, Qian X, Cong W. The first bioluminescence tomography system for simultaneous acquisition of multiview and multispectral data. Int. J. Biomed. Imaging. 2006b;2006:58601. doi: 10.1155/IJBI/2006/58601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Li Y, Jiang M. Uniqueness theorems in bioluminescence tomography. Med. Phys. 2004;31:2289–99. doi: 10.1118/1.1766420. [DOI] [PubMed] [Google Scholar]
- Ye JC, Webb KJ, Bouman CA, Millane RP. Optical diffusion tomography by iterative-coordinate-descent optimization in a Bayesian framework. J. Opt. Soc. Am. A. 1999;16:2400–12. [Google Scholar]