Abstract
The liver is able to regenerate itself in response to partial hepatectomy or liver injury. This is accomplished by a complex network of different cell types and signals both inside and outside the liver. Bile acids (BAs) are recently identified as liver-specific metabolic signals and promote liver regeneration by activating their receptors: Farnesoid X Receptor (FXR) and G-protein-coupled BA receptor 1 (GPBAR1, or TGR5). FXR is a member of the nuclear hormone receptor superfamily of ligand-activated transcription factors. FXR promotes liver regeneration after 70% partial hepatectomy (PHx) or liver injury. Moreover, activation of FXR is able to alleviate age-related liver regeneration defects. Both liver- and intestine-FXR are activated by BAs after liver resection or injury and promote liver regeneration through distinct mechanism. TGR5 is a membrane-bound BA receptor and it is also activated during liver regeneration. TGR5 regulates BA hydrophobicity and stimulates BA excretion in urine during liver regeneration. BA signaling thus represents a novel metabolic pathway during liver regeneration.
Keywords: liver regeneration, bile acid, FXR, TGR5, FGF15
1. Introduction
Liver is one of the few organs that can powerfully regenerate itself in response to partial ablation or liver injury. Liver regeneration has been widely studied as a paradigm for regenerative organ regrowth since the introduction of a rodent partial hepatectomy (PHx) model in 1931 [1]. Unlike a typically anatomic regeneration, regeneration of the liver is a compensatory hyperplasia of the remaining tissues and is driven by the functional deficit of the organism. Liver regeneration consists of several well orchestrated phases, with rapid induction of proliferating factors activating the quiescent hepatocytes and priming their subsequent progression through the cell cycle, followed by reestablishment of normal liver size and renewed quiescence [2–4]. Growth factors and cytokines are the important early signals to induce the expression of downstream target genes via activation of several key transcription factors [5]. In addition to growth factors and cytokines, metabolic signals are considered as the third major signals during liver regeneration, which is however relatively less studied [6]. Recently, BAs were identified as key metabolic signals during liver regeneration and their roles in promoting liver regeneration have received more and more attention [7, 8]. In this review, the recent advance of BA signaling in liver regeneration will be summarized and discussed.
2. Metabolic signals and liver regeneration
Liver regeneration is an adaptive regrowth response induced by specific stimuli and the subsequently sequential changes in gene expression and morphologic reconstruction. It is generally accepted that the remaining hepatocytes are the major cell types that replicate to regenerate liver in the models of 70% PHx or liver injury. Only in some special injury models, activation and replication of liver progenitors are observed when the hepatocytes fail to replicate normally. In addition to hepatocytes, other cell types are also actively involved in liver regeneration or repair. Recently, several excellent reviews also highlight the roles of liver stellate cells, liver sinusoidal endothelial cells and liver stem/progenitor cells in liver regeneration and repair [9–12].
Liver regeneration includes a highly complex network of signal transductions. The essential circuitry required for this process is defined mainly by three major networks: cytokine, growth factor and metabolic signaling [13]. These three networks subsequently activate specific genes and signaling pathways that are essential for liver regeneration. Compared to the cytokine and growth factor networks, little is known about the roles of metabolic signals in liver regeneration. The identification of several nuclear receptors as receptors for liver metabolites provides novel insight into the roles of metabolic signals in liver regeneration. Among them, the Farnesoid X Receptor (FXR, NR1H4) is identified as a primary BA receptor [14, 15]. FXR belongs to a sub-cluster of metabolic nuclear receptors that also includes Vitamin D Receptor (VDR, NR1I1), Constitutive Androstane Receptor (CAR, NR1I3), Pregnane X Receptor (PXR, NR1I2) and Liver X Receptor alpha and beta (LXRα, NR1H3; LXRβ, NR1H2). Peroxisome proliferator-activated receptors (PPARs) are also important metabolic nuclear receptors. All these receptors bind to DNA either as a monomer or as a heterodimer with a common partner for nuclear receptors, Retinoid X Receptor (RXR, NR2B1) to regulate the expression of various genes involved in BA, lipid, glucose, and drug metabolism [16]. Interestingly, their roles in liver regeneration are also under active investigation. For example, upon PHx, liver regeneration is impaired in mice lacking RXRα in hepatocytes [17]. LXR may suppress liver regeneration after PHx through regulating the cholesterol levels in the liver [18]. CAR activation strongly induces hepatomegaly and may contribute to normal liver regeneration after 70% PHx [7, 19]. Dai et al. indicated that PXR is required for normal progression of liver regeneration by modulating lipid homeostasis and regulating hepatocyte proliferation [20]. In contrast, PPARγ acts as a negative regulator of hepatocyte proliferation and may be responsible for the inhibition of liver growth in the late phase of liver regeneration [21]. There is a detailed summary on nuclear receptors in liver regeneration recently [22].
Liver is a major organ for metabolism. Therefore, there is an immense metabolic demand during liver regeneration. The requirement of metabolic signals for liver regeneration has been known for a long time. However, their direct effect on liver regeneration is still unclear. Among different metabolic signals, BAs are attractive signals for liver regeneration because the levels of BAs are tightly regulated. BAs are intrinsically toxic and cause liver injury if the levels are not controlled properly. As such, liver resection or injury will generate a BA overload in the liver, which is a potential driving force for liver regeneration [7, 8].
3. BA signaling and liver regeneration
BAs are liver-specific metabolites. They are end products from cholesterol catabolism and are important for nutrition absorption in the intestine, which include cholesterol, lipids and fat-soluble vitamins. BAs are synthesized in the liver and stored in the gall bladder. They are secreted into the intestine when a meal is ingested, but 95% BAs are reabsorbed and transported back to the liver through the portal vein. This system is known as enterohepatic circulation. Hepatic BAs comprise less than 5% of the total BA pool and PHx increases bile influx, which rapidly generates a BA overload in the liver. Consistently with this, there is a sharp repression of Cyp7a1 gene expression after 70% PHx or liver injury [23]. Cyp7a1 is the rate-limiting enzyme required for the BA production from cholesterol catabolism. The importance for a stringent control of BA levels is illustrated by a delicate regulation of Cyp7a1 expression. There are many factors and pathways that can regulate the expression of Cyp7a1 gene. A negative feedback loop is identified to regulate BA levels, in which high levels of BA activate FXR to increase the mRNA levels of SHP, which is a negative regulator of Cyp7a1 gene expression. Moreover, additional regulators of Cyp7a1 expression are identified, including cytokines, growth factors and nuclear receptors [24–30]. During liver regeneration, in addition to FXR-SHP axis, hepatocyte growth factor and JNK pathways are also involved in suppressing Cyp7a1 expression during the acute phases of liver regeneration [23].
The strong suppression of BA synthesis during liver regeneration indicates a BA overload stress in the liver. This also suggests that BAs may participate in the liver regeneration. Indeed, interruption of normal enterohepatic biliary circulation has been previously known to inhibit liver regeneration [31, 32]. Moreover, there is also some direct evidence that BAs are able to stimulate hepatocyte proliferation [33–35]. In a study of 70% PHx mouse model, supplementation with a low dose of BAs promotes liver regeneration, while reduction of BA levels by a BA-binding resin delays liver regeneration [7]. Defective BA signaling causes delayed liver regeneration is also demonstrated in other animal models. In the absence of MRP3, a BA transporter, liver regeneration is delayed in mice due to lower BA concentration in the portal blood [36]. Similarly, deletion of Cyp27, an enzyme required for normal BA production and metabolism, results in lower BA pool and defective liver regeneration in mice [37]. In FXR−/− mice, the effect of BAs on promoting liver regeneration is lost [7]. Similarly, in MRP3−/− and Cyp27−/− mice, the delayed liver regeneration is due to impaired FXR activation, suggesting that FXR is the key player to mediate BA effect on liver regeneration. In conclusion, the identification of a novel role of FXR in liver regeneration is key to understand the molecular mechanism by which BAs affect liver regeneration.
4. FXR and liver regeneration
FXR is highly expressed in the liver, intestine, kidney where the levels of BAs are relatively high [38]. FXR is the primary sensor of BAs and both conjugated and unconjugated bile salts are able to activate FXR at physiological concentrations [39, 40]. FXR regulates BA homeostasis by regulating genes involved in BA synthesis, secretion, transportation, absorption, conjugation, and detoxification [41–45]. As expected, FXR is also the BA receptor to mediate BA’s effect on liver regeneration [7]. FXR is shown to promote liver regeneration after 70% PHx and stimulate liver repair after CCl4-induced liver injury [46]. Interestingly, different from 70% PHx, there is massive cell death in liver injury model. FXR is shown to have a dual role in promoting liver regeneration by both stimulating hepatocyte proliferation and protecting the hepatocyte from death [46, 47].
In addition to metabolic genes, Foxm1b is identified as a FXR direct target gene during liver regeneration [7, 48]. Foxm1b is a key cell cycle regulator essential for G1/S and G2/M progression [49, 50]. Animal studies indicate that Foxm1b is a key transcription factor in liver regeneration. Although liver can fully regenerate itself, aging dramatically reduces this capacity of liver. The delayed and reduced proliferative response has been attributed to the decreased expression of some key transcription factors, such as c-Myc and Foxm1b and to the failure of aging-liver to diminish the age-specific C/EBPα-Brm-HDAC1 complex after PHx [51–53]. The complex suppressed Foxm1b induction after PHx through binding to Foxm1b promoter, which results in age-related proliferation defects upon PHx or liver injury [54]. These studies highlight Foxm1b as one of the key regulators in aging-liver regeneration. Defective activation of FXR occurs in aged regenerating livers [48], which may account for the insufficient Foxm1b induction. Compared with young mice, aging mice did not have altered protein levels of FXR and RXR. Therefore, aging may affect the levels of endogenous FXR ligands such as BAs, which could result in defective activation of FXR during liver regeneration. Interestingly, in pregnant mice, loss of FXR results in reduced liver growth, indicating a similar function of FXR in mediating the liver growth during pregnancy [55].
Sirtuin1 (SIRT1) that can modulate FXR activities also has an effect on liver regeneration through modulating FXR activities during liver regeneration [56]. The transgenic mice that overexpress SIRT1 showed increased mortality, enhanced liver injury and impaired hepatocyte proliferation after PHx. SIRT1 reduces FXR activities through persistent deacetylation and lower FXR expression. In summary, FXR is a key receptor and transcription factor that specifically mediates the effect of BA signaling to promote liver regeneration.
5. Intestine FXR and liver regeneration
FXR is highly expressed in both the liver and intestine. Both hepatic- and intestine-FXR are involved in regulation of BA homeostasis [57]. One critical FXR target gene in the intestine is FGF15. Indeed, several reports suggest that FGF15 secreted from ileum has profound effects on the suppression of Cyp7a1 gene expression and liver metabolism through FGF receptor-mediated signaling pathways in the liver [58–60]. Suppression of Cyp7a1 expression and decreased bile acid synthesis are known to be beneficial for liver regeneration. Therefore, FGF15 induction after liver damage may also contribute to the normal liver regeneration. Indeed, there is significantly delayed liver regeneration and increased liver injury in intestine-specific FXR knockout (ΔIN-FXR) mice compared to FXR Fl/Fl control mice after either 70% PHx or CCl4 injection [61]. During liver regeneration, FXR also activates the expression of FGF15 in the intestine to suppress Cyp7a1 transcription [61]. There results identify an unexpected role of intestine FXR in regulating liver regeneration/repair. First, higher levels of BAs in ΔIN-FXR mice after liver injury may hamper the normal liver regeneration/repair. Second, the metabolic and mitogenic activities of FGF15 may contribute to liver regrowth. Third, the hydrophobic bile acid, deoxycholic acid (DCA) is significantly increased in fecal extracts from ΔIN-FXR mice but not from FXR KO or liver FXR null mice, and DCA may cause hepatocyte apoptosis [62]. This may also be a protective function of intestine FXR during liver regeneration/repair. Finally, exogenous delivery of FGF15 rescued the defect of liver repair in ΔIN-FXR and FXR KO mice [61], suggesting a direct role of FGF15 in promoting liver regeneration.
The direct effect of FGF15 on liver regeneration is also examined by directly comparing liver regeneration between WT and FGF15−/− mice [63]. As expected, in FGF15−/− mice, liver regeneration is delayed and there is stronger liver injury in FGF15−/− livers. Furthermore, a recent report indicates that selective activation of intestine FXR or treating mice with FGF19, a human homolog of murine FGF15, could reduce liver necrosis and inflammatory cell infiltration in cholestasis mouse models [64]. Taken together, intestine FXR and its induction of FGF15 may have important roles in liver protection.
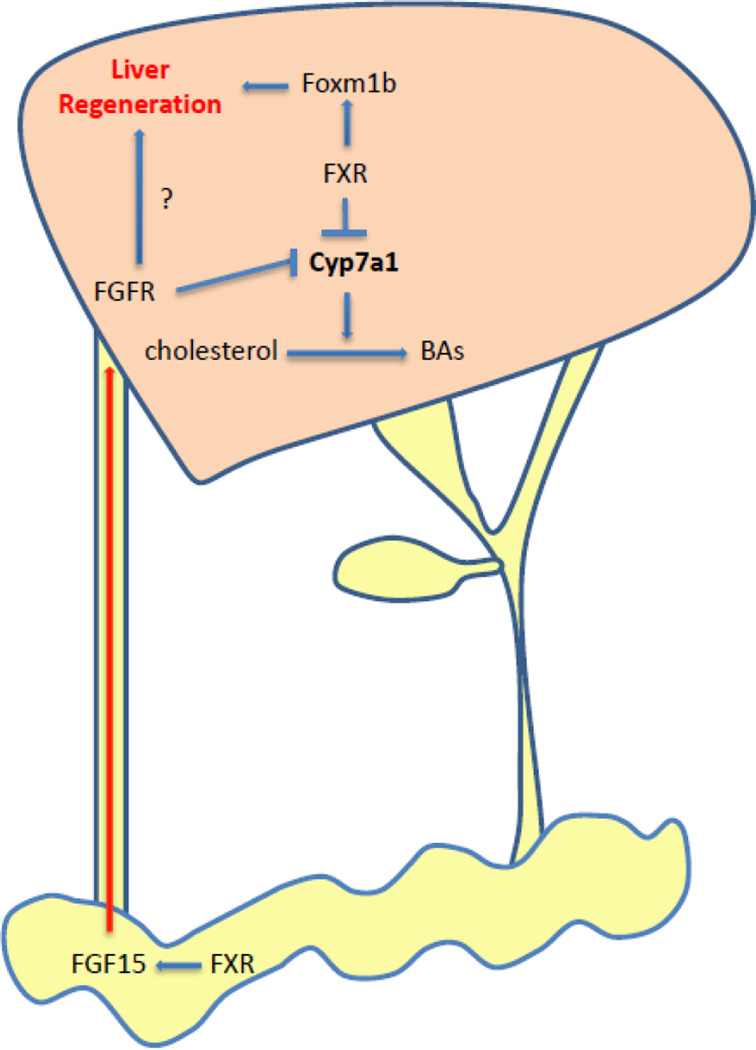
Both liver- and intestine-FXR contribute to liver regeneration. Hepatic FXR directly induces Foxm1b expression and promotes liver regeneration. In contrast, intestine FXR activates FGF15 expression to promote liver regeneration. Therefore, both the cell-autonomous effect of hepatic FXR and the endocrine FGF15 pathway induced by FXR in intestine are required for the liver regeneration (Fig. 1).
Fig. 1. BA signaling activates FXR to promote liver regeneration.
Liver resection or injury increases BA influx, which activates FXR in the liver and intestine. After activation, liver-FXR suppresses Cyp7a1 to reduce BA production and activates Foxm1b to stimulate liver regeneration. Intestine-FXR is also activated to induce FGF15 expression, which circulates with BAs back to liver through enterohepatic circulation. FGF15 binds to its receptor (FGFR) and initiate a signaling pathway to suppress Cyp7a1 expression. At the same time, FGF15 also promotes liver regeneration through unknown mechanism.
6. TGR5 and liver regeneration
TGR5 is a plasma membrane-bound G protein–coupled bile acid (BA) receptor, which displays varied levels of expression in different tissues [65–67]. Hydrophobic BAs, such as lithocholic acid (LCA) and deoxycholic acid (DCA), are potent endogenous ligands of TGR5. TGR5 regulates BA homeostasis, glucose homeostasis, energy metabolism as well as inflammation [68–73].
A role of TGR5 in liver regeneration was recently identified [74]. After 70% PHx, TGR5−/− mice displayed severe hepatocyte necrosis, prolonged cholestasis, exacerbated inflammatory response, and delayed liver regeneration [74]. TGR5 may primarily protect the BA-overloaded remnant liver through control of BA hydrophobicity and suppress inflammatory response. Moreover, TGR5 increases BA efflux in urine through kidney. The regulation of potential BA transporters in kidney needs further investigation. The defective BA clearance in the absence of TGR5 thus lead to exacerbated liver toxicity by BAs. Different from FXR, these results highlight a distinct role of TGR5 during liver regeneration.
Interestingly, a recent report indicates that serotonin also helps increasing BA secretion through urine and reduces liver toxicity, suggesting that BA excretion through kidney may be an important mechanism for liver protection during liver regeneration [75]. Serotonin is not only a neurotransmitter but also a hormone involved in the initiation of liver regeneration [76, 77].
7. Conclusions and perspective
BA signaling is an important metabolic signal during liver regeneration. The novel roles of FXR and TGR5 in promoting liver regeneration are consistent with their defending roles against BA toxicity during liver regeneration. Moreover, there is a close relationship between aberrant liver regeneration and hepatocellular carcinoma (HCC) in FXR null mice [78–80]. Similarly, abnormal BA homeostasis and cell proliferation in SHP null mice also result in HCC development [81, 82]. Therefore, further studies on FXR and TGR5 in this new area will provide novel insights into the complex mechanism of liver regeneration, HCC and other liver diseases. Encouragingly, there is already active research in searching for potent FXR and TGR5 ligands. Thus, both FXR and TGR5 agonist ligands may offer potential approaches to prevent and treat insufficient liver regeneration as well as HCC and other liver diseases.
Highlights.
Bile acids are metabolic signals to promote liver regeneration
Both liver- and intestine-FXR contribute to the promotion of liver regeneration
Membrane-bound BA receptor TGR5 also helps liver regeneration
Acknowledgements
We apologize to colleagues whose work could not be cited due to space limitations. This work is supported by NCI 1R01-CA139158.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Higgins GM, Anderson RM. Experimental pathology of the liver. Arch. Pathol. 1931:186–202. [Google Scholar]
- 2.Fausto N. Liver regeneration. J. Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Taub R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 4.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am. J. Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 6.Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Translational research : the journal of laboratory and clinical medicine. 2014 doi: 10.1016/j.trsl.2014.01.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang WD, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 8.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diehl AM, Chute J. Underlying potential: cellular and molecular determinants of adult liver repair. J Clin Invest. 2013;123:1858–1860. doi: 10.1172/JCI69966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902–1910. doi: 10.1172/JCI66369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLeve LD. Liver sinusoidal endothelial cells and liver regeneration. J Clin Invest. 2013;123:1861–1866. doi: 10.1172/JCI66025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kordes C, Häussinger D D. Hepatic stem cell niches. J Clin Invest. 2013;123:1874–1880. doi: 10.1172/JCI66027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen WD, Wang YD, Meng Z, Zhang L, Huang W. Nuclear bile acid receptor FXR in the hepatic regeneration. Biochim Biophys Acta. 2011;1812:888–892. doi: 10.1016/j.bbadis.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 15.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 16.Lefebvre P, Benomar Y, Staels B, Retinoid X. receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Imai T, Jiang M, Kastner P, Chambon P, Metzger D. Selective ablation of retinoid X receptor alpha in hepatocytes impairs their lifespan and regenerative capacity. Proc Natl Acad Sci U S A. 2001;98:4581–4586. doi: 10.1073/pnas.071056098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo Sasso G, Celli N, Caboni M, Murzilli S, Salvatore L, Morgano A, Vacca M, Pagliani T, Parini P, Moschetta A. Down-regulation of the LXR transcriptome provides the requisite cholesterol levels to proliferating hepatocytes. Hepatology. 2010;51:1334–1344. doi: 10.1002/hep.23436. [DOI] [PubMed] [Google Scholar]
- 19.Huang W, Zhang J, Washington M, Liu J, Parant JM, Lozano G, Moore DD. Xenobiotic stress induces hepatomegaly and liver tumors via the nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- 20.Dai G, He L, Bu P, Wan YJ. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology. 2008;47:1277–1287. doi: 10.1002/hep.22129. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto Y, Ono T, Dhar DK, Yamanoi A, Tachibana M, Tanaka T, Nagasue N. Role of peroxisome proliferator-activated receptor-gamma (PPARgamma) during liver regeneration in rats. J Gastroenterol Hepatol. 2008;23:930–937. doi: 10.1111/j.1440-1746.2008.05370.x. [DOI] [PubMed] [Google Scholar]
- 22.Vacca M, Degirolamo C, Massafra V, Polimeno L, Mariani-Costantini R, Palasciano G, Moschetta A. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol Cell Endocrinol. 2013;368:108–119. doi: 10.1016/j.mce.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Huang X, Meng Z, Dong B, Shiah S, Moore DD, Huang W. Significance and mechanism of CYP7a1 gene regulation during the acute phase of liver regeneration. Mol Endocrinol. 2009;23:137–145. doi: 10.1210/me.2008-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiao L, Han SI, Fang Y, Park JS, Gupta S, Gilfor D, Amorino G, Valerie K, Sealy L, Engelhardt JF, Grant S, Hylemon PB, Dent P. Bile acid regulation of C/EBPβ, CREB, and c-Jun function, via the extracellular signal-regulated kinase and c-Jun NH2-terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol Cell Biol. 2003;23:3052–3066. doi: 10.1128/MCB.23.9.3052-3066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, Itadani H, Tanaka K. Identification of membrane-type receptor for bile acids (MBAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 26.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G proteincoupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 27.Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors HNF4α and LRH-1 cooperate in regulating Cyp7a1 in vivo. J Biol Chem. 2012;287:41334–41341. doi: 10.1074/jbc.M112.421834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow EC, Magomedova L, Quach HP, Patel RH, Durk MR, Fan J, Maeng HJ, Irondi K, Anakk S, Moore DD, Cummins CL, Pang KS. Vitamin D Receptor Activation Down-Regulates Small Heterodimer Partner and Increases CYP7A1 to Lower Cholesterol. Gastroenterology. 2013;5085:01840–01844. doi: 10.1053/j.gastro.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 30.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda J, Chijiiwa K, Nakano K, Zhao G, Tanaka M. Lack of intestinal bile results in delayed liver regeneration of normal rat liver after hepatectomy accompanied by impaired cyclin E-associated kinase activity. Surgery. 2002;131:564–73. doi: 10.1067/msy.2002.123008. [DOI] [PubMed] [Google Scholar]
- 32.Otao R, Beppu T, Isiko T, Mima K, Okabe H, Hayashi H, Masuda T, Chikamoto A, Takamori H, Baba H. External biliary drainage and liver regeneration after major hepatectomy. Br J Surg. 2012;99:1569–1574. doi: 10.1002/bjs.8906. [DOI] [PubMed] [Google Scholar]
- 33.Barone M, Francavilla A, Polimeno L, Ierardi E, Romanelli D, Berloco P, Di Leo A, Panella C. Modulation of rat hepatocyte proliferation by bile salts: in vitro and in vivo studies. Hepatology. 1996;23:1159–1166. doi: 10.1053/jhep.1996.v23.pm0008621149. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]
- 35.Columbano A, Ledda-Columbano GM. Mitogenesis by ligands of nuclear receptors: an attractive model for the study of the molecular mechanisms implicated in liver growth. Cell Death Differ. 2003;1:S19–S21. doi: 10.1038/sj.cdd.4401113. [DOI] [PubMed] [Google Scholar]
- 36.Fernández-Barrena MG, Monte MJ, Latasa MU, Uriarte I, Vicente E, Chang HC, Rodriguez-Ortigosa CM, Elferink RO, Berasain C, Marin JJ, Prieto J, Ávila MA. Lack of Abcc3 expression impairs bile-acid induced liver growth and delays hepatic regeneration after partial hepatectomy in mice. J Hepatol. 2012;56:367–373. doi: 10.1016/j.jhep.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Meng Z, Liu N, Fu X, Wang X, Wang YD, Chen WD, Zhang L, Forman BM, Huang W. Insufficient bile acid signaling impairs liver repair in CYP27(−/−) mice. J Hepatol. 2011;55:885–895. doi: 10.1016/j.jhep.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 38.Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 39.Wang YD, Chen WD, Moore DD, Huang W. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18:1087–1095. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- 40.Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020–2030. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 41.Cariou B, Staels B. FXR: a promising target for the metabolic syndrome? Trends Pharmacol Sci. 2007;28:236–243. doi: 10.1016/j.tips.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 43.Edwards PA, Kast HR, Anisfeld AM. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- 44.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Pircher PC, Schulman IG, Westin SK. Regulation of complement C3 expression by the bile acid receptor FXR. J Biol Chem. 2005;280:7427–7434. doi: 10.1074/jbc.M411473200. [DOI] [PubMed] [Google Scholar]
- 46.Meng Z, Wang Y, Wang L, Jin W, Liu N, Pan H, Liu L, Wagman L, Forman BM, Huang W. FXR regulates liver repair after CCl4-induced toxic injury. Mol Endocrinol. 24:886–897. doi: 10.1210/me.2009-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang YD, Yang F, Chen WD, Huang X, Lai L, Forman BM, Huang W. Farnesoid X receptor protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo. Mol Endocrinol. 2008;22:1622–1632. doi: 10.1210/me.2007-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen WD, Wang YD, Zhang L, Shiah S, Wang M, Yang F, Yu D, Forman BM, Huang W. Farnesoid X receptor alleviates age-related proliferation defects in regenerating mouse livers by activating forkhead box m1b transcription. Hepatology. 2010;51:953–962. doi: 10.1002/hep.23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa RH. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7:108–110. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 50.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Krupczak-Hollis K, Tan Y, Dennewitz MB, Adami GR, Costa RH. Increased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J Biol Chem. 2002;277:44310–44316. doi: 10.1074/jbc.M207510200. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent agerelated proliferation defects in regenerating liver. Proc Natl Acad Sci U S A. 2001;98:11468–11473. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krupczak-Hollis K, Wang X, Dennewitz MB, Costa RH. Growth hormone stimulates proliferation of old-aged regenerating liver through forkhead box m1b. Hepatology. 2003;38:1552–1562. doi: 10.1016/j.hep.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 55.Milona A, Owen BM, Van Mil S, Dormann D, Mataki C, Boudjelal M, Cairns W, Schoonjans K, Milligan S, Parker M, White R, Williamson C. The normal mechanisms of pregnancy-induced liver growth are not maintained in mice lacking the bile acid sensor FXR. Am J Physiol Gastrointest Liver Physiol. 2010;298:G151–G158. doi: 10.1152/ajpgi.00336.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Rodríguez JL, Barbier-Torres L, Fernández-Álvarez S, Juan VG, Monte MJ, Halilbasic E, Herranz D, Alvarez L, Aspichueta P, Marín JJ, Trauner M, Mato JM, Serrano M, Beraza N, Martínez-Chantar ML. SIRT1 controls liver regeneration by regulating BA metabolism through FXR and mTOR signaling. Hepatology. 2013 in press. [Google Scholar]
- 57.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez A, Ratliff EP, Andres AM, Huang X, McKeehan WL, Davis RA. Bile acids decrease hepatic paraoxonase 1 expression and plasma high-density lipoprotein levels via FXR-mediated signaling of FGFR4. Arterioscler. Thromb. Vasc. Biol. 2006;26:301–306. doi: 10.1161/01.ATV.0000195793.73118.b4. [DOI] [PubMed] [Google Scholar]
- 60.Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu H, Kemper JK. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;23:986–996. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Wang YD, Chen WD, Wang X, Lou G, Liu N, Lin M, Forman BM, Huang W. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology. 2012;56:2336–2343. doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HC, Carotti S, Vespasiani-Gentilucci U, Morini S, Vicente E, Concepcion AR, Medina JF, Marin JJ, Berasain C, Prieto J, Avila MA. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. [DOI] [PubMed] [Google Scholar]
- 64.Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, Di Tullio G, Palasciano G, Moustafa T, Halilbasic E, Trauner M, Moschetta A. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. Gastroenterology. 2012;142:355–365. doi: 10.1053/j.gastro.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 65.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 66.Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2003;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 67.Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J. 2006;398:423–430. doi: 10.1042/BJ20060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 69.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191:197–205. doi: 10.1677/joe.1.06546. [DOI] [PubMed] [Google Scholar]
- 71.Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 72.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-Protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Péan N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, Schoonjans K, Rainteau D, Tordjmann T. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58:1451–1460. doi: 10.1002/hep.26463. [DOI] [PubMed] [Google Scholar]
- 75.Jang JH, Rickenbacher A, Humar B, Weber A, Raptis DA, Lehmann K, Stieger B, Moritz W, Soll C, Georgiev P, Fischer D, Laczko E, Graf R, Clavien PA. Serotonin protects mouse liver from cholestatic injury by decreasing bile salt pool after bile duct ligation. Hepatology. 2012;56:209–218. doi: 10.1002/hep.25626. [DOI] [PubMed] [Google Scholar]
- 76.Lesurtel M, Graf R, Aleil B, Walther DJ, Tian Y, Jochum W, Gachet C, Bader M, Clavien PA. Platelet-derived serotonin mediates liver regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 77.Matondo RB, Punt C, Homberg J, Toussaint MJ, Kisjes R, Korporaal SJ, Akkerman JW, Cuppen E, de Bruin A. Deletion of the serotonin transporter in rats disturbs serotonin homeostasis without impairing liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2009;296:G963–G968. doi: 10.1152/ajpgi.90709.2008. [DOI] [PubMed] [Google Scholar]
- 78.Liu N, Meng Z, Lou G, Zhou W, Wang X, Zhang Y, Zhang L, Liu X, Yen Y, Lai L, Forman BM, Xu Z, Xu R, Huang W. Hepatocarcinogenesis in FXR−/− mice mimics human HCC progression that operates through HNF1α regulation of FXR expression. Mol Endocrinol. 2012;26:775–785. doi: 10.1210/me.2011-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan H, Fu X, Huang W. Molecular mechanism of liver cancer. Anticancer Agents Med Chem. 2011;11:493–499. doi: 10.2174/187152011796011073. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Fu X, Van Ness C, Meng Z, Ma X, Huang W. Bile Acid Receptors and Liver Cancer. Curr Pathobiol Rep. 2013;1:29–35. doi: 10.1007/s40139-012-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song KH, Ellis E, Strom S, Chiang JY. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 2007;46:1993–2002. doi: 10.1002/hep.21878. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–298. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]