Summary
Rheumatoid arthritis (RA) is an autoimmune disease with unknown etiology though both genetic and environmental factors have been suggested to be involved in its pathogenesis. While infections and other environmental factors like smoking have been studies extensively and show some association, a direct link between all the factors has been difficult to prove. With the recent advances in technology, it has become possible to sequence the commensals that are residing in our gut. The gut microbiome may provide the missing link to this puzzle and help solve the mystery of many leaky gut syndromes. The gut commensals are involved in maintaining host immune homeostasis and function suggesting that they might be critical in altering the immune system that leads to autoimmune diseases like RA. Mouse models support the role of the gut microbiota in predisposition to RA. If that is true, the power of gut-derived commensal can be harnessed to our benefit by generating a biomarker profile along with genetic factors to define individuals at risk and by altering the gut microbial composition using various means.
Rheumatoid arthritis (RA) is an autoimmune disease of unknown etiology. It has a world-wide distribution affecting approximately 1% of the population. The main causal factor for RA is the immunologic dysregulation that leads to inflammation. RA is characterized by the presence of autoreactive T and B cells directed to synovial proteins that leads to inflammation of the synovial joints followed by joint damage. Over the decades, significant clinical and scientific work has helped to understand its pathology, yet the factor(s) that cause RA are still unknown. However from the vast literature, it is clear that its pathogenesis requires interaction between genetic and environmental factors [1]. Among genetic factors, several gene variants have been identified. Genome wide association studies (GWAS) and other studies have revealed that the genes located in the Major Histocompatibility complex (MHC) provide the strongest association with susceptibility to develop RA. The presence of certain HLA-class II alleles that share the sequence at position 60-74 in the 3rd hypervariable region 3 (HVR3) with the DRB1*0401 gene, “shared epitope”, were shown to occur more often in RA patients as compared to controls[2]. However, not all individuals carrying those genes develop RA. Recent studies have implicated variants of some genes that could contribute to severity of disease although with much lower impact than HLA genes [3]. Low concordance rate of RA in monozygotic twins indicates involvement of other factors in addition to genetic factors. Interaction between genetic and environmental factors may explain some of the risk for susceptibility to RA. Among the environmental factors, smoking and infections have been associated with development and pathogenesis. Smoking has been studied extensively and has been shown to be associated with seropositive disease in genetically predisposed individuals[1].
An infectious etiology of RA has been proposed for decades although conclusive evidence is lacking. Infectious agents like Proteus mirabilis, specific Escherichia coli, mycoplasma Fermentans, Klebsiella pneumonia and Porphyromonas gingivalis have been linked to inflammatory arthritis in many ethnic populations[4-8]. However, a causal link between infections and onset of RA has been difficult to prove. The concept called “Molecular mimicry” was proposed to explain the role of infectious agents in RA. According to this concept, cross-reactivity between epitopes of the microbial origin with a self-protein that share similarities with infectious agents can cause dysregulation in immune response, crossing a threshold that ensues autoimmunity. Indeed presence of antibodies to certain bacteria has been described in RA patients[9]. For example, sequences ESRRAL found Proteus mirabilis share similarities with sequences at amino acid position 67-74 of the shared epitope[6]. Antibodies to Proteus mirabilis and cell membrane proteins of mycoplasma have been observed in RA patients. Similarly rheumatoid factor positivity has been associated with the presence E. coli and Klebsiella in the gut [5]. As RA is characterized by the presence of autoantibodies, these studies suggest that the inflammatory process may begin many years before the actual onset of disease and environmental factors may contribute as a second hit in genetically predisposed individuals. While the known RA-associated HLA genes and environment may still not explain causation in all individuals, a recent study with humanized mice showing how non-associated HLA molecules can contribute to susceptibility to arthritis supports the interaction between genetic and environmental factors for susceptibility to disease.
Besides bacterial infections, certain viruses like Epstein Barr virus (EBV), cytomegalovirus (CMV) and Parvovirus B19 have also been implicated in RA[10-12].
Microbiome and immune response
Despite enormous efforts, finding a single pathogen that could predict susceptibility to RA has failed. During the last decade our understanding of the interaction between microbes and host has evolved. The major human organs that are exposed to environment are the skin and gut. The human gut harbors vast numbers of bacteria that exceed the number of cells in one’s body. There is conclusive evidence that the gut microflora and its components are essential for maintaining homeostasis in the gut as well as in the systemic immune system [13-15]. Besides maintaining a balanced mucosal immune system, these gut bacteria aid in digestion of certain foods and harvesting energy.
Innate immunity is our first line of defense against pathogens. Toll like receptors (TLRs) present on the surface of cells of innate immunity recognize conserved pathogen associated molecules pattern (PAMPs), activating NFkB pathways leading to activation of T cells and cytokine production [16]. Lipopolysaccharides (LPS) are important components of the outer membrane of all Gram negative bacteria. TLR4 is a natural ligand for LPS and as soon as it binds LPS, all cells expressing TLR4 and pattern recognition receptors (PRR) get activated resulting in secretion of inflammatory cytokines and other defensive mechanisms. Cytokines in turn activate cells of the adaptive immune system that generates antigen-specific response.
Now there is a growing realization that intestinal bacteria may also be involved in adaptive immune response and thus play a significant role in allergies and autoimmune diseases[17].
Microbiome and Rheumatoid arthritis
Predisposition to RA is associated with the presence of certain HLA genes and environmental factors like smoking and infections. Age and sex of a subject can influence development of RA as mean age of patients is around 50 years and it occurs twice as often in women. All the factors that can impact the development of RA including the host genotype, smoking, infections, hormones and aging are connected by one factor, gut microbiome (Fig 1).
Fig 1.
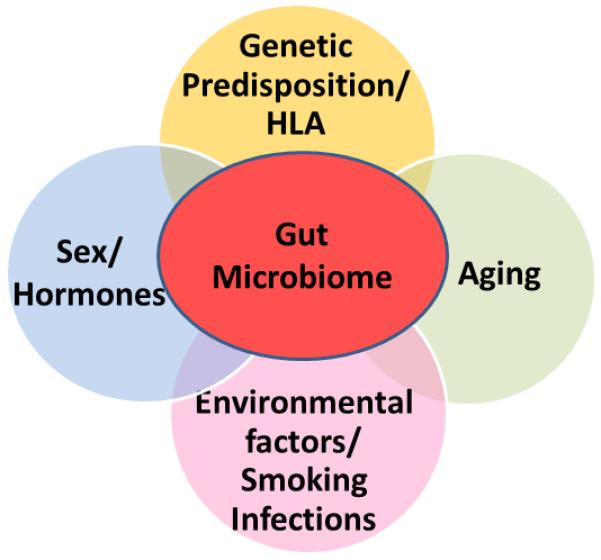
Rheumatoid arthritis is a multifactorial disease that involves both genetic and environmental factors. Among genetic factors, HLA alleles provide the strongest risk while among environmental factors, smoking and infections are involved. A role of hormones and changes in immune system during aging are also associated with pathogenesis of rheumatoid arthritis. All the factors that influence RA also impact the gut microbial composition. Gut microbiome provides a link between all the factors that influence RA. An individual may harbor a core gut microbiome and certain species may contract or expand depending on the exposure to various environmental factors thus influencing the immune system locally in the gut as well as adaptive immune system.
Infection as an etiological factor for RA is supported by the success of antibiotic treatment of some RA patients. While many pathogens have been linked to pathogenesis of RA, Porphyromonas gingivalis, a commensal present in the oral cavity, has been the major focus of recent investigations as it provides a direct link between environmental factors and RA [18]. It is a gram negative bacterium that is present in the gingival crevice of periodontitis patients as well as in healthy individuals, though in lower numbers. Smoking has been strongly correlated to P. gingivalis infection. Epidemiological studies linking periodontal disease and RA were initially suggested on the basis of the presence of antibodies to P. gingivalis. Rheumatoid arthritis is characterized by the presence of antibodies to citrullinated proteins. A high prevalence of periodontitis has been observed in the new onset RA patients even in the absence of smoking [19]. Among the known commensals, P. gingivalis is the only bacterium that has been shown to express the endogenous Peptidyl arginine deiminase (PAD) enzyme required for citrullination of proteins. Thus P. gingivalis could be involved in the autoimmune process by generating citrullinated proteins of self as well as human antigen and the immune response to them[20]. Individuals with a genetic predisposition or other susceptibility factors and P. gingivalis infection are more likely to develop cellular and humoral responses to the citrullinated antigens. Patients with RA have been shown to be positive for antibodies to citrullinated α–enolase peptide 1 (CEP-1) that cross reacts with bacterial enolase. A correlation between the presence of anti-citrullinated antibodies (ACPA) to CEP-1 and positivity for shared epitope further suggests that citrullinated CEP-1 may have a role in pathogenesis of RA [21]. Animal studies have shown that infection with P. gingivalis can exacerbate the disease severity of arthritis[22]. Bacterial PAD enzymes can citrullinate human proteins that may lead to a break in tolerance resulting in an autoimmune response to CEP and other citrullinated synovial proteins. These studies were further confirmed by the presence of DNA of P gingivalis from synovial fluid of RA patients [23]
Gut microbiome in pathogenic response
The notion that gut bacteria are involved in adaptive immunity is not novel. As early as 1908, the Nobel laureate Metchkinov suggested that aging- associated deficiencies came from certain bacterial toxins from the gut and lactic acid producing bacteria can reverse those changes [24]. That was the first inkling into the role of gut commensals in adaptive immunity and its involvement in changes in the systemic immune system. Ever since his discovery, scientists have been trying to determine how gut bacteria affects the overall health of humans. The significance of the intestinal bacteria in rheumatology was realized when presence of certain bacteria were associated with the development of reactive arthritis. Studies showing the presence of certain commensal bacterial DNA in synovial fluids of patients [1, 25, 26] supported the concept that in genetically susceptible individuals exposure to degraded products of the gut bacteria locally in synovium may cause inflammation. Indeed, infections like Yersinia, Shigella, Salmonella and Campylobacter can trigger arthritis, especially in individuals carrying HLA-B27.
This led to the hypothesis that intestinal bacterial products may be present in circulation and arthritogenic bacteria may cause inflammation in synovial tissues. Evidence for this comes from the presence of bacterial cell wall products in the joints of patients and differences observed between intestinal bacterial composition in RA patients and healthy individuals [27, 28]. Also, there is evidence suggesting that host MHC genes may affect the microbiological milieu of the gut [29, 30]. Increased levels of antibodies directed against antigens of certain gut bacteria in RA patients suggests a pathogenic relationship between these bacteria and rheumatoid arthritis [31]. Animal studies showing susceptibility to develop arthritis when exposed to bacterial cell walls support this contention [32, 33].
Genetics of gut microbiota
There is limited information on the influence of host genotype on the gut microbial composition. Initial studies with twins showed that the composition of fecal and nasal flora was much more similar in identical twins as compared to non-identical twins. Studies carried out with congenic strains of mice showed a strong effect of host genotype on the gut flora [34]. These studies did not show that susceptibility to a disease might be associated with specific gut microbes or dysbiosis. The advent of humanized mice helped resolve this issue. Recent studies with humanized mice expressing RA-associated and non-associated HLA genes show a difference in gut microbial composition between the 2 strains, suggesting that MHC genes may be involved directly or indirectly in determining the intestinal microbial composition[35]. However the mechanism by which MHC molecules can impact gut microbial colonization was unclear. One way MHC molecules impact the gut microbiota could be by binding the proteins on the bacterial cell surface and influencing adherence to the epithelial cells and bacterial colonization.
More recently, studies by Mazmanian and coworkers have shown that polysaccharides (PSA) produced by Bacteroides fragilis may form a complex with MHC II, which is internalized and then presented, leading to activation of CD4 T cells [36]. Introduction of B. fragilis in germ free mice has been shown to correct a defect in the CD4 T cell population observed in germ free mice. These studies suggested that microbes are required for the development of immune system and one can speculate this to be one way for the host genotype to influence microbial colonization.
Hygiene hypothesis
Composition of the gut microbiota is influenced by many factors including diet, geographical location and environmental factors like smoking and infectious agents. The hygiene hypothesis put forth in the 1980s was formulated to explain the role of microbes in shaping immune tolerance [37]. According to the hygiene hypothesis, personal hygiene and smaller family size in the developed world has led to low burden of pathogenic microbes and an increase in autoimmune diseases. This is supported by increased incidence of various autoimmune conditions like type I diabetes, inflammatory bowel disease and rheumatoid arthritis in developed countries. This hypothesis has been modified to assume that the modern life style has altered the gut microbial diversity causing dysbiosis [38, 39]. The low microbial load can cause fixed microflora or change the structure and function of the microbial colonies in the gut which has a strong effect on the immune response by either not generating tolerance or by causing inflammation leading to development of autoimmune diseases. One mechanism used to explain the beneficial effects of commensals is their ability to hinder pathogen adhesion, however it does not necessarily address the potential effects of commensal bacteria on the systemic immune system. The ability of commensals to alter or modulate systemic immune responses may be more germane for modifying systemic autoimmune responses and could provide an experimental framework to explain how the increase in environmental hygiene could result in an increase of autoimmune diseases. It is also clear that bacteria, be they probiotic or commensals, are quite variable in their effects and their general characterization as beneficial or pathogenic organisms do not do justice to the diversity of effects that the different bacteria can induce or prevent.
We have coevolved with our gut commensals. There is considerable evidence now that suggests that the intestinal microflora or its specific components are important for the normal development of the immune system and health of the host and altered microbiota may be associated with disease conditions.
Microbiome and rheumatoid arthritis
Recent studies have suggested that bacterial products can lead to production of pro-inflammatory molecules compromising the intestinal barrier integrity. This can lead to leaky gut syndrome which may explain the presence of gut bacteria in systemic immune system. Altered gut microbial population can affect the overall immune system in the gut with consequent changes in systemic immune system.
The gut harbors around 1013 microbes with a vast majority being anaerobic and difficult to culture. An initial study analyzed fatty acids produced by gastrointestinal bacteria to show that RA patients with erosive disease differed from other subjects[40]. This study was followed by analyzing the fecal microbiome of RA patients that was done by 16S RNA sequencing of microbes in the stool by rtPCR. Patients with RA showed fewer Bifidobacterium, bacteria of the Bacteroides-Porphyromonas-Prevotella group and the Eubacterium rectale–Clostridium coccoides group than the fecal microbiota of patients with non-inflammatory fibromyalgia [27], suggesting dysbiosis as these are the common genera in humans [41-43]. However, the information was very limited due to technical limitations.
Now with the recent advances in technology, it has become possible to sequence 16sRNA of the fecal microbiome of humans. While the fecal microbiome may not reflect the all of the microbiota of the small intestine, it provides a window into the overall gastrointestinal microbiome. Using this approach, Jose Scher and coworkers analyzed stool samples from new onset RA (NORA) patients and controls [44]. They identified an abundance of Prevotella copri with loss of Bacteroides in NORA. Interestingly the relative abundance of P. copri inversely correlated with the presence of shared epitope suggesting a change in the gut microbiota before onset of clinical phenotype. It is known that the inflammatory response in RA begins much before the actual onset of disease. These studies along with humanized mice suggest that MHC genes play a critical role in the colonization of gastrointestinal microbial flora thus indirectly determining the pro-inflammatory conditions in the gut.
In RA patients, smoking has been associated with seropositivity and presence of DR4[21]. Recent studies have suggested that lungs, considered sterile before, harbor microbiota that changes with conditions like COPD[45]. It is possible that mucosal surfaces like lung and gut share microbes as both organs are exposed to environmental factors. In a genetically predisposed individual, the pathogenic microbes generate a pro-inflammatory milieu in both organs and the actual site of initiation of RA might not be the joint.
However, in humans due to variability in genetic complexity, diet, environmental factors and geographical locations, it is impossible to determine causality versus effect. In addition, the likelihood that RA patients are taking drugs before seen in a clinic, that can modulate their gut microbiota, is high.
Gut microbiome in animal models for arthritis
Animal models provide a means to study the pathogen/antigen involved in initiation of disease and its effect on the phenotype. As early as 1977, animal models of arthritis using mycobacteria and streptococcal cell wall components were described [46]. Animals raised in germ free colonies developed severe disease. In contrast, HLA B27 transgenic rats and mice that develop spontaneous arthritis modeling human spondyloarthropathies were protected from spontaneous disease in germ free and specific pathogen free conditions respectively, suggesting a requirement of environment factors [47, 48]. Similarly studies done with spontaneous mouse models of RA, IL-1 receptor antagonist knockout and K/BXN mouse models, have shown a requirement of gut microbiota for arthritis development as mice raised in germ free colony did not develop arthritis [49, 50]. Colonization of germ free mice with a single species could trigger arthritis. The mechanism by which gut microbes trigger extra intestinal systemic disease was analyzed in the K/BXN mouse model of spontaneous arthritis. Introduction of segmented filamentous bacteria in germ free K/BxN mice activates Th17 producing lamina propria cells that migrate out of the gut and produce IL-17, helping differentiation of B cells into antibody producing cells. How gut microbiota exerts its effect locally in joints is remains largely unknown.
Using transgenic mice expressing RA-associated DRB1*0401 and resistant DRB1*0402 genes, we have developed a collagen-induced arthritis (CIA) model that shares similarities with human disease in sex-bias, autoantibody profile and phenotype [51-53]. We sequenced fecal flora of *0401 and *0402 mice to identify compositional differences in the intestinal microbiota between the 2 strains. Our data showed a significant shift in the ratio of Bacteroidetes/Firmicutes with *0401 mice showing even abundance of both phyla while in *0402 mice Bacteroidetes occurred with twice the abundance of Firmicutes suggesting, 1) HLA genes may have a role in gut microbial colonization and 2) a dysbiosis may be present in genetically susceptible subjects indicating a crucial role of gut flora in the autoimmune process [35]. This was supported by the observations in this study since *0401 mice did not show dynamic changes in fecal microbiome with age although *0402 mice, showed a sex and age dependent microbiome. Moreover, gut permeability was much higher in *0401 mice compared to *0402 mice further providing a mechanism by which the products of gut commensals may be migrating in extra intestinal organs. Arthritis-susceptible mice had an abundance of bacteria with similarities to Clostridials gp. while resistant mice had abundance of Bifidobacteria. Presence of Bifidobacteria was negatively correlated to Th17 expression in the gut suggesting that commensals in this group may have a dominant role in protection. Bacterial rRNA from naturally occurring commensals like clostridium have been isolated from synovial tissue of RA but not non-RA patients [26]. These studies suggest that the intestine may be a critical organ in triggering disease through altered immune homeostasis and a leaky gut with proinflammatory conditions may be an event that begins before the actual onset of clinical phenotype.
This suggests that the gut microbiome may dictate a pro or anti-inflammatory environment that can have a significant impact on adaptive immune response away from gut. We propose that the gut microbiome, sex and genetic factors may be able to predict susceptibility to develop autoimmunity like arthritis. Treatment of arthritis-susceptible mice with a potential anti-inflammatory commensal might help in stopping progression of disease and reducing severity by changing gut permeability and the immune environment.
The only probiotic tried as a treatment for RA is the lactobacillus. Their therapeutic application did not significantly improve the American College of Rheumatology (ACR) Scores in one study, but did show benefit in another [54, 55]. In mouse models of arthritis L. casei potentiates antigen-specific oral tolerance and suppresses Th1-type immune responses [56, 57]. A normal intestinal microbiota can positively influence immune responses and protect against the development of inflammatory diseases in various inflammatory models [58-60]. Molecules produced by intestinal bacteria like B. fragilis can ameliorate inflammatory disease [59, 61, 62]. Despite these tantalizing reports, there is a paucity of data on the composition of gut microbial population and its use as therapeutic agents in inflammatory autoimmune diseases, particularly RA.
Current treatment of RA involves non-Steroid anti-inflammatory drugs, known to increase gut permeability, as well as immunosuppressive and biologic drugs. Although biologics like TNFα inhibitors and CD20 depletion have been used successfully in RA patients, not all patients respond to these therapies and those that do respond may develop side effects. Pharmacological treatments for arthritis target the inflammatory process by suppressing the host reaction whereas commensals may regulate rather than suppress the host response. If this proves to be successful, it would open the possibility of inducing immune tolerance in other autoimmune diseases with unknown etiology.
Conclusions
Recent technology has made it possible to culture the gut microflora and understand their involvement in homeostasis of immune system in the gut and in various diseases. From the published observations, one can speculate that autoimmunity is the result of many factors including genetic factors and environmental factors like smoking, infections as well diet that can influence the gut microbiota (Fig 2). Thus a healthy gut maintains homeostasis that maintains integrity of the gut and help in various functions. On the other hand, in genetically predisposed individuals, environmental factors can influence dysbiosis of the gut microbiome causing a change in the local and adaptive immune system the gut that compromises its integrity resulting in various disease conditions. We propose that the characterization of the composition of the intestinal microflora of Rheumatoid arthritis (RA) patients and their healthy family members could potentially define specific microbial species or ecological properties such as biodiversity that may be responsible for dysbiosis in patients. This will help in improving predictive and diagnostic protocols for individuals at high risk of developing RA and future treatment strategies. There is still a lot that needs to be answered. Observations in mouse models and RA patients suggest that the gut may be an important contributor to onset or severity of disease. The next step will be to find a way to modulate disease by generating symbiosis in the gut microbial composition by using diet, prebiotics or probiotics.
Fig 2.
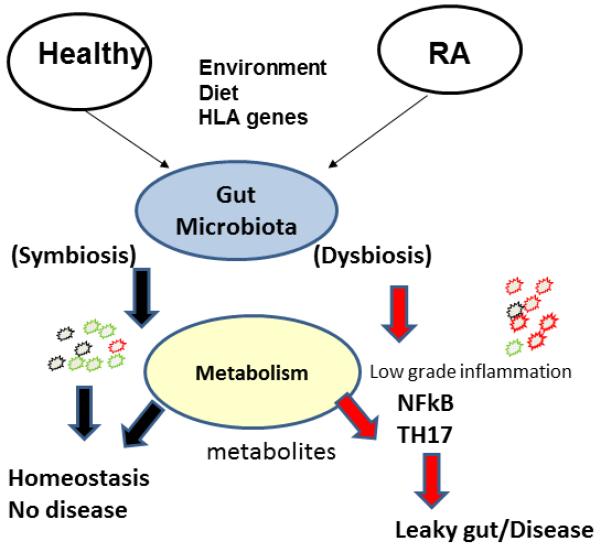
In genetically predisposed individual, environmental factors like diet, infections and smoking can cause dysbiosis in the gut microbiota resulting in expansion of contraction of certain species of a genus. This dysbiosis may be related to production of some metabolites as well as NFkB activation and Th17 cytokine production, compromising the integrity of the gut epithelial layer and causing a leaky gut and disease conditions. On the other hand a healthy individual will maintain homeostasis in the gut so that the local and systemic immune systems stay healthy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: Genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nature clinical practice. 2006;2:425–433. doi: 10.1038/ncprheum0249. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis and rheumatism. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 3.Alauzet C, Mory F, Carlier JP, Marchandin H, Jumas-Bilak E, Lozniewski A. Prevotella nanceiensis sp. nov., isolated from human clinical samples. International journal of systematic and evolutionary microbiology. 2007;57:2216–2220. doi: 10.1099/ijs.0.65173-0. [DOI] [PubMed] [Google Scholar]
- 4.de Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. The Journal of rheumatology. 2008;35:70–76. [PubMed] [Google Scholar]
- 5.Newkirk MM, Zbar A, Baron M, Manges AR. Distinct bacterial colonization patterns of Escherichia coli subtypes associate with rheumatoid factor status in early inflammatory arthritis. Rheumatology (Oxford) 2010;49:1311–1316. doi: 10.1093/rheumatology/keq088. doi: 10.1093/rheumatology/keq088. [DOI] [PubMed] [Google Scholar]
- 6.Ebringer A, Rashid T, Wilson C. Rheumatoid arthritis, Proteus, anti-CCP antibodies and Karl Popper. Autoimmun Rev. 2010;9:216–223. doi: 10.1016/j.autrev.2009.10.006. doi: 10.1016/j.autrev.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Sato N, Oizumi T, Kinbara M, Sato T, Funayama H, Sato S, Matsuda K, Takada H, Sugawara S, Endo Y. Promotion of arthritis and allergy in mice by aminoglycoglycerophospholipid, a membrane antigen specific to Mycoplasma fermentans. FEMS Immunol Med Microbiol. 2010;59:33–41. doi: 10.1111/j.1574-695X.2010.00657.x. doi: 10.1111/j.1574-695X.2010.00657.x. [DOI] [PubMed] [Google Scholar]
- 8.Hitchon CA, El-Gabalawy HS. Infection and rheumatoid arthritis: still an open question. Curr Opin Rheumatol. 2011;23:352–357. doi: 10.1097/BOR.0b013e3283477b7b. doi: 10.1097/BOR.0b013e3283477b7b. [DOI] [PubMed] [Google Scholar]
- 9.Rashid T, Jayakumar KS, Binder A, Ellis S, Cunningham P, Ebringer A. Rheumatoid arthritis patients have elevated antibodies to cross-reactive and non cross-reactive antigens from Proteus microbes. Clinical and experimental rheumatology. 2007;25:259–267. [PubMed] [Google Scholar]
- 10.Meron MK, Amital H, Shepshelovich D, Barzilai O, Ram M, Anaya JM, Gerli R, Nicola B, Shoenfeld Y. Infectious aspects and the etiopathogenesis of rheumatoid arthritis. Clin Rev Allergy Immunol. 2010;38:287–291. doi: 10.1007/s12016-009-8158-6. doi: 10.1007/s12016-009-8158-6. [DOI] [PubMed] [Google Scholar]
- 11.Magnusson M, Brisslert M, Zendjanchi K, Lindh M, Bokarewa MI. Epstein-Barr virus in bone marrow of rheumatoid arthritis patients predicts response to rituximab treatment. Rheumatology (Oxford) 2010;49:1911–1919. doi: 10.1093/rheumatology/keq159. doi: 10.1093/rheumatology/keq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colmegna I, Alberts-Grill N. Parvovirus B19: its role in chronic arthritis. Rheum Dis Clin North Am. 2009;35:95–110. doi: 10.1016/j.rdc.2009.03.004. doi: 10.1016/j.rdc.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Current opinion in immunology. 2010;22:455–460. doi: 10.1016/j.coi.2010.06.008. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 14.Round JL, O’Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. Journal of autoimmunity. 34:J220–225. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Advances in immunology. 107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen VL, Kasper DL. Interactions between the intestinal microbiota and innate lymphoid cells. Gut microbes. 2013:5. doi: 10.4161/gmic.27289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, Kasper LH. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. Journal of immunology. 2009;183:6041–6050. doi: 10.4049/jimmunol.0900747. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 18.Rosenstein ED, Weissmann G, Greenwald RA. Porphyromonas gingivalis, periodontitis and rheumatoid arthritis. Med Hypotheses. 2009;73:457–458. doi: 10.1016/j.mehy.2009.04.008. doi: 10.1016/j.mehy.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, Lipuma L, Attur M, Pillinger MH, Weissmann G, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis and rheumatism. 2012;64:3083–3094. doi: 10.1002/art.34539. doi: 10.1002/art.34539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, Kinloch A, Culshaw S, Potempa J, Venables PJ. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis and rheumatism. 2010;62:2662–2672. doi: 10.1002/art.27552. doi: 10.1002/art.27552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegner N, Lundberg K, Kinloch A, Fisher B, Malmstrom V, Feldmann M, Venables PJ. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunological reviews. 2010;233:34–54. doi: 10.1111/j.0105-2896.2009.00850.x. doi: 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- 22.Marchesan JT, Gerow EA, Schaff R, Taut AD, Shin SY, Sugai J, Brand D, Burberry A, Jorns J, Lundy SK, et al. Porphyromonas gingivalis oral infection exacerbates the development and severity of collagen-induced arthritis. Arthritis Res Ther. 2013;15:R186. doi: 10.1186/ar4376. doi: 10.1186/ar4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichert S, Haffner M, Keysser G, Schafer C, Stein JM, Schaller HG, Wienke A, Strauss H, Heide S, Schulz S. Detection of oral bacterial DNA in synovial fluid. J Clin Periodontol. 2013;40:591–598. doi: 10.1111/jcpe.12102. doi: 10.1111/jcpe.12102. [DOI] [PubMed] [Google Scholar]
- 24.Podolsky S. Cultural divergence: Elie Metchnikoff’s Bacillus bulgaricus therapy and his underlying concept of health. Bull Hist Med. 1998;72:1–27. doi: 10.1353/bhm.1998.0056. [DOI] [PubMed] [Google Scholar]
- 25.Moen K, Brun JG, Valen M, Skartveit L, Eribe EK, Olsen I, Jonsson R. Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clinical and experimental rheumatology. 2006;24:656–663. [PubMed] [Google Scholar]
- 26.Kempsell KE, Cox CJ, Hurle M, Wong A, Wilkie S, Zanders ED, Gaston JS, Crowe JS. Reverse transcriptase-PCR analysis of bacterial rRNA for detection and characterization of bacterial species in arthritis synovial tissue. Infection and immunity. 2000;68:6012–6026. doi: 10.1128/iai.68.10.6012-6026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. The Journal of rheumatology. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 28.Gerard HC, Wang Z, Wang GF, El-Gabalawy H, Goldbach-Mansky R, Li Y, Majeed W, Zhang H, Ngai N, Hudson AP, et al. Chromosomal DNA from a variety of bacterial species is present in synovial tissue from patients with various forms of arthritis. Arthritis and rheumatism. 2001;44:1689–1697. doi: 10.1002/1529-0131(200107)44:7<1689::AID-ART293>3.0.CO;2-K. doi: 10.1002/1529-0131(200107)44:7<1689::AID-ART293>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Vaahtovuo J, Toivanen P, Eerola E. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS microbiology ecology. 2003;44:131–136. doi: 10.1016/S0168-6496(02)00460-9. [DOI] [PubMed] [Google Scholar]
- 30.De Palma G, Capilla A, Nadal I, Nova E, Pozo T, Varea V, Polanco I, Castillejo G, Lopez A, Garrote JA, et al. Interplay Between Human Leukocyte Antigen Genes and the Microbial Colonization Process of the Newborn Intestine. Current issues in molecular biology. 2009;12:1–10. [PubMed] [Google Scholar]
- 31.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nature reviews. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Broek MF, van den Berg WB, van de Putte LB, Severijnen AJ. Streptococcal cell wall-induced arthritis and flare-up reaction in mice induced by homologous or heterologous cell walls. Am J Pathol. 1988;133:139–149. [PMC free article] [PubMed] [Google Scholar]
- 33.Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 2003;5:775–780. doi: 10.1016/s1286-4579(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 34.Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infection and immunity. 2001;69:2372–2377. doi: 10.1128/IAI.69.4.2372-2377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, White BA, Taneja V. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PloS one. 2012;7:e36095. doi: 10.1371/journal.pone.0036095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- 37.Strachan DP. Family size, infection and atopy: the first decade of the “hygiene hypothesis”. Thorax. 2000;55(Suppl 1):S2–10. doi: 10.1136/thorax.55.suppl_1.s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Makela MJ, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eerola E, Mottonen T, Hannonen P, Luukkainen R, Kantola I, Vuori K, Tuominen J, Toivanen P. Intestinal flora in early rheumatoid arthritis. British journal of rheumatology. 1994;33:1030–1038. doi: 10.1093/rheumatology/33.11.1030. [DOI] [PubMed] [Google Scholar]
- 41.Finegold SMSV, Mathisen GE. Academic Press; New York: 1983. pp. 3–31. [Google Scholar]; Hentges DJ, editor. Human intestinal microflora in health and disease. Academic Press; New York: 1983. Normal indigenous intestinal flora; pp. 3–31. [Google Scholar]
- 42.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Applied and environmental microbiology. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura M, Turroni F, Canchaya C, Vaughan EE, O’Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci. 2009;14:3214–3221. doi: 10.2741/3445. [DOI] [PubMed] [Google Scholar]
- 44.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, Young VB, Toews GB, Curtis JL, Sundaram B, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PloS one. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginsburg I. Can chronic and self-perpetuating arthritis in the human be caused by arthrotropic undegraded microbial cell wall constituants? A working hypothesis. Rheumatol Rehabil. 1977;16:141–149. doi: 10.1093/rheumatology/16.3.141. [DOI] [PubMed] [Google Scholar]
- 47.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, Balish E, Hammer RE. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. The Journal of experimental medicine. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khare SD, Luthra HS, David CS. Spontaneous inflammatory arthritis in HLA-B27 transgenic mice lacking beta 2-microglobulin: a model of human spondyloarthropathies. The Journal of experimental medicine. 1995;182:1153–1158. doi: 10.1084/jem.182.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, Heuvelmans-Jacobs M, Akira S, Nicklin MJ, Ribeiro-Dias F, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. The Journal of clinical investigation. 2008;118:205–216. doi: 10.1172/JCI32639. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behrens M, Trejo T, Luthra H, Griffiths M, David CS, Taneja V. Mechanism by which HLA-DR4 regulates sex-bias of arthritis in humanized mice. Journal of autoimmunity. doi: 10.1016/j.jaut.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA-DR4-transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis and rheumatism. 2007;56:69–78. doi: 10.1002/art.22213. [DOI] [PubMed] [Google Scholar]
- 53.Taneja V, Krco CJ, Behrens MD, Luthra HS, Griffiths MM, David CS. B cells are important as antigen presenting cells for induction of MHC-restricted arthritis in transgenic mice. Molecular immunology. 2007;44:2988–2996. doi: 10.1016/j.molimm.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hatakka K, Martio J, Korpela M, Herranen M, Poussa T, Laasanen T, Saxelin M, Vapaatalo H, Moilanen E, Korpela R. Effects of probiotic therapy on the activity and activation of mild rheumatoid arthritis--a pilot study. Scandinavian journal of rheumatology. 2003;32:211–215. doi: 10.1080/03009740310003695. [DOI] [PubMed] [Google Scholar]
- 55.Nenonen MT, Helve TA, Rauma AL, Hanninen OO. Uncooked, lactobacilli-rich, vegan food and rheumatoid arthritis. British journal of rheumatology. 1998;37:274–281. doi: 10.1093/rheumatology/37.3.274. [DOI] [PubMed] [Google Scholar]
- 56.So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, Hwang KC, Jeon YH, Im SH. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Molecular immunology. 2008;45:2690–2699. doi: 10.1016/j.molimm.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 57.So JS, Lee CG, Kwon HK, Yi HJ, Chae CS, Park JA, Hwang KC, Im SH. Lactobacillus casei potentiates induction of oral tolerance in experimental arthritis. Molecular immunology. 2008;46:172–180. doi: 10.1016/j.molimm.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 58.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 60.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal immunology. 2010;3:487–495. doi: 10.1038/mi.2010.29. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 62.Ochoa-Reparaz J, Mielcarz DW, Haque-Begum S, Kasper LH. Induction of a regulatory B cell population in experimental allergic encephalomyelitis by alteration of the gut commensal microflora. Gut microbes. 2010;1:103–108. doi: 10.4161/gmic.1.2.11515. doi: 10.4161/gmic.1.2.11515. [DOI] [PMC free article] [PubMed] [Google Scholar]


