Abstract
Secretory IgA (SIgA) antibodies in the intestinal tract form the first line of antigen-specific immune defense, preventing access of pathogens as well as commensal microbes to the body proper. SIgA is transported into external secretions by the polymeric immunoglobulin receptor (pIgR). Evidence is reported here that the gut microbiota regulates production of SIgA and pIgR, which act together to regulate the composition and activity of the microbiota. SIgA in the intestinal mucus layer helps to maintain spatial segregation between the microbiota and the epithelial surface without compromising the metabolic activity of the microbes. Products shed by members of the microbial community promote production of SIgA and pIgR by activating pattern recognition receptors on host epithelial and immune cells. Maternal SIgA in breast milk provides protection to newborn mammals until the developing intestinal immune system begins to produce its own SIgA. Disruption of the SIgA-pIgR-microbial triad can increase the risk of infectious, allergic andinflammatory diseases of the intestine.
Keywords: Secretory IgA, polymeric immunoglobulin receptor, gut microbiota, mucus, pattern recognition receptors, breast milk
1. Introduction
The mucosal surfaces of the human body are home to some 100 trillion microorganisms, tenfold more than the total number of host cells throughout the body [1]. The highest concentrations of microbes are found in the intestinal tract, particularly in the colon.The mucosal immune system in the gut faces the challenge of eliminating potential pathogens while maintaining a mutually beneficial relationship with the commensal microbiota. Secretory antibodies of the IgA class (SIgA) represent the first line of antigen-specific immune defense in the gut lumen [2,3].The majority of the IgA antibodies in gut secretions are germline encoded, low affinity and cross-reactive against redundant microbial antigens; however, when challenged, the mucosal B cell system can generate high-affinity, somatically mutated IgA antibodies with unique specificities[4-10]. SIgA antibodies allow beneficial microbes to thrive within the gut lumen while preventing their access to the body proper. When pathogenic microbes threaten to breach the epithelial barrier, SIgA antibodies can cooperate withother elements of the immune system to kill the invading pathogens, albeit at the cost of inflammatory damage to host tissues.
In healthy humans, up to 3 g/day of SIgA is delivered into intestinal secretions[11,12].In the intestine, tight junctions between adjacent epithelial cells maintain a highly selective barrier,which prevents paracellular leakage of luminal contents as well as passive diffusion of antibodies from their site of synthesis by plasma cells in the lamina propria into the gut lumen. Transport of locally synthesized IgA across glandular and mucosal epithelial cells into external secretions is mediated by the polymeric immunoglobulin receptor (pIgR) [13-15] (Fig. 1). Proteolytic cleavage of pIgR at the apical surface of epithelial cells releases a complex of IgA covalently bound to secretory component (SC), the extracellular domain of pIgR. This complex is designated SIgA to distinguish it from IgA devoid of SC, the major form of IgA in the blood circulation. The SC moiety protects SIgA from degradation by host and bacterial proteases in the intestinal tract[16-18], promotes glycan-dependent adherence of SIgA to bacteria [19] and neutralizes inflammatory host factors, such as IL-8 [20,21]. Thus, pIgR-mediated epithelial transcytosis is crucial for the immune and anti-inflammatory functions of SIgA. The discovery that polymorphisms in the PIGR gene locus are linked to increased susceptibility to inflammatory bowel diseases in humans[22,23]highlights the clinical relevance of this pathway.This review will focus on the mechanisms through which epithelial-microbial cross-talk regulates the transport and homeostatic functions of SIgA in the intestine.
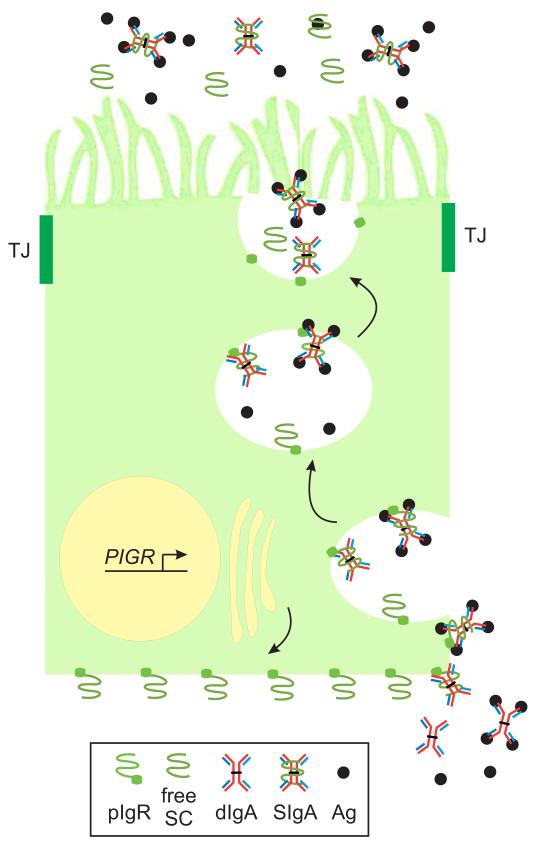
Fig. 1.
Transcytosis of SIgA through a polarized epithelial cell. A polarized columnar epithelial cell is illustrated, with the apical surface at the top and the basolateral surface at the bottom and sides, separated by tight junctions (TJ) with adjacent epithelial cells.Transcription of the PIGR gene is induced by host cytokines and microbial factors. Newly synthesized pIgR is targeted to the basolateral surface, where it binds polymeric Ig (pIg), illustrated here as dimeric (d)IgA, with or without bound antigen (Ag). Following receptor-mediated endocytosis, pIg-bound and unoccupied pIgR molecules are transported through a series of intracellular vesicles to the apical surface. Proteolytic cleavage of pIgR at the extracellular face of the plasma membrane releases free secretory component (SC) and secretory (S)Ig (illustrated here as SIgA).
2. Intestinal SIgA promotes host-microbial mutualism
It has long been appreciated thatcommensal microbes induce IgA responses in the intestine, and more recent evidence demonstrates that SIgA regulates the composition and function of the commensalmicrobiota. The experimental evidence for SIgA-microbial reciprocity will be discussed here (Table 1).Thecellular and molecular mechanisms that mediatethis reciprocal relationship between IgA and the gut microbiota have been discussed in detail in recent reviews [1-3,24-27].
Table 1.
Evidence that intestinal SIgA promotes host-commensal mutualism.
| Evidence | References |
|---|---|
| Colonization of germ-free mice with commensal bacteria induces intestinal IgA responses |
[29-31] |
| Repeated oral administration of commensal bacteria to conventional mice induces specific SIgA responses that prevent bacterial invasion intro draining lymph nodes |
[33] |
| Treatment of humans with prebiotic oligosaccharides changes the composition of the gut microbiota and increases fecal SIgA levels |
[34] |
| Fecal bacteria in humans and mice are coated with SIgA | [32,35,37] |
| IgA-deficient mice exhibit overgrowth of certain types of gut bacteria, which can be reversed by restoration of IgA |
[38] |
| Introduction of bacteria-specific IgA into antibody-deficient mice reduces host inflammatory responses |
[40] |
| Proteobacteria-specific IgA regulates maturation of the intestinal microbiota | [32] |
| pIgR-deficient mice (which lack SIgA in the intestinal lumen) exhibit altered gut microbiota and intestinal epithelial gene expression |
[19,42] |
| SIgA protects the intestinal epithelium by associating with gut microbes in the outer mucus layer |
[48] |
2.1. Commensal microbes induce intestinal IgA responses
Four decades ago,the observation was made that SIgA levels were extremely low in the intestinal contents of germ-free mice (devoid of commensal microorganisms) compared to mice with a normal microbiota [28], suggesting that microbial colonization of the intestine after birth provides the antigenic stimulus for development of IgA responses. Formal proof of this concept was provided by the demonstration that mono-colonization of formerly germ-free mice with various strains of normal gut bacteria resulted in hypertrophy of Peyer’s patches and population of the intestinal lamina propria with IgA-secreting plasma cells [29,30]. More recently, using a model of reversible colonization of germ-free mice with a non-dividing mutant of Escherichia coli, a long-lived SIgA response was observed that was specific for the inducing bacterial strain [31] However, exposure of E. coli-colonized mice to other bacteria limited the duration of the SIgA response against the original colonizer, suggesting that the SIgA response adapts to the dominating members of the microbiota.In another study, antigen-specific fecal SIgA was induced by colonization of adult germ-free mice with the microbiota of infant conventional mice, which was dominated by Proteobacteria of the family Enterobacteriaceae[32]. By contrast, these investigators found no specific increase in fecal SIgA antibodies following colonization of adult germ-free micewith a more diverse microbiota from adult conventional mice. However, other investigators reported that specificSIgA responses could be generated in adult conventional miceafter repeated oral administration of high doses of individual commensal bacterial strains [33]. Although it can be challenging to demonstrate specific effects of the commensal microbiota on IgA responses in humans, a recent study demonstrated that treatment of adult volunteers with prebiotic oligosaccharides altered the composition of the gut microbiota and was correlated with an increase in fecal SIgA levels [34]. Taken together, this evidence suggests that intestinal IgA responses continually adapt to the composition of the resident microbiota, allowing dynamic host-microbial mutualism.
2.2. Intestinal SIgA regulates the composition and activity of the commensal microbiota
If the commensal microbiotais the main driving force for intestinal IgA responses, how then do thesevast quantities of SIgA antibodies affect the composition and activity of the microbiota? A flow cytometric analysis of feces from 22 healthy humans, using antibodies to human IgA, revealed thatthe proportion of gut bacteria coated with IgA ranged from 24-74% (median 45%) [35]. A study of human salivary IgA with specificity fororal bacteria suggested that binding of SIgA to bacterial cell walls does not affect bacterial viability or replication [36], but rather may limit access of bacteria to host tissues. Two recent studies in mice identified specific bacterial taxa in the intestinal contents that were coated with IgA, by first sorting bacteria into IgA-coated and uncoated pools using fluorescence-activated cell sorting, then sequencing the 16S rRNA genes of the IgA-coated bacteria.Cullenderet al.[37], in a study with adult mice, found thatabout 22% of intestinal bacteria were found to be coated with IgA, particularly gram-negative bacteria within the phyla Bacteroidetes and Proteobacteria. Interestingly, when cecal bacteria were analyzed from mice deficient in Toll-like receptor (TLR)5, a pattern recognition receptor broadly reactive to bacterial flagellin, the proportion of IgA-bound Proteobacteria was diminished. The functional significance of reduced IgA coating was demonstrated by the finding that flagellated bacteria breached the mucosal barrier in the intestines of TLR5-deficient mice. In this example of host-microbial mutualism, it appears that flagellated gut bacteria stimulate host IgA responses through TLR5-dependent signaling, and the resultant IgA antibodies limit access of flagellated bacteria to host tissues.In another study, Mirpuri et al.[32] found that the ratio of IgA-bound vs. -unbound Proteobacteria of the family Enterobacteriaceaeincreased significantly in the feces of mice from birth to adulthood, reaching a ratio of about 6:1 by the age of 6 weeks. By contrast, the ratio of IgA-bound vs. -unbound Bacteroides and Firmicutes was much lower (about 1:1), and did not change with age.
Direct evidence for a role of IgA in regulating the gut microbiota was provided by a study of mice deficient in the enzyme activation-induced (cytidine) deaminase (AID), which catalyzes immunoglobulin class switching from IgM to IgA in activated B cells [38]. A persistent expansion of anaerobic bacteria, dominated by segmented filamentous bacteria (SFB), was observed throughout the small intestine of AID-deficient mice, which lack IgA (as well as IgG and IgE). This observation was particularly significant in that SFB are known to adhere tightly to intestinal epithelial cell surfaces and induce robust IgA responses [39]. Restoration of IgA responses in AID-deficient mice by anastomosis with wild-type mice restored a more normal gut microbiota, with dramatic decreases in the numbers of SFB.Another group of investigatorsdeveloped an experimental system in which germ-free, immunodeficient mice (which lacked immunoglobulins of all isotypes) were mono-colonized with Bacteroides thetaiotaomicron, a prominent member of the normal gut microbiota of mice and humans [40]. In the absence of intestinal antibodies,B. thetaiotaomicron induced a robust host inflammatory response. Systemic instillation of a monoclonal IgA antibody specific for this bacterium, which was transported into the intestinal lumen as SIgA, reduced the inflammatory response without altering the numbers of B. thetaiotaomicron in the intestinal lumen. In a more recent study, IgA deficiency in mice was found to result in increased colonization by Proteobacteria and enhanced susceptibility to chemically-induced colitis [32]. The role of SIgA antibodies can be examined more specifically by comparing wild-type mice to pIgR-deficient mice, which have elevated serum IgA but are devoid of SIgA in intestinal secretions [41]. A recent study demonstrated that absence of intestinal SIgA was associated with an altered composition of the gut microbiota and dysregulated gene expression in intestinal epithelial cells[42].Based on these studies, it can be concluded that association of SIgA antibodies with gut bacteria regulates the composition of the commensal microbiota without compromising its beneficial functions. This conclusion, however, relies heavily on studies in experimental animals where genetic and environmental factors can be controlled and monitored. Further studies are needed in humans to explore the reciprocal relationship between intestinal SIgA and the resident microbiota, and its implications for health and disease.
2.3. Association of SIgA and gut bacteria with intestinal mucus protects the epithelium
It has been postulatedthat SIgA mediates immune exclusion at least in part by trapping microbes in the mucus layer overlying the epithelium of mucosal surfaces [20,43]. An in vivostudy in mice demonstrated that N-glycan side chains on theSC moiety anchorSIgA to the mucus gel lining the luminal surface of the respiratory tract[43]. It has been assumed that SIgA carries out a similar role in the intestinal tract. The mucus layer is thicker in the intestinal tract than in other mucosal surfaces, comprising a dense inner layer (increasing in thickness from 15-30 μm in the small intestine to ~ 100 μm in the colon) and a loose outer layer (100-400 μm in the small intestine and ~ 700 μm in the colon) [44]. The inner mucus layer is rich in antimicrobial peptides and is relatively sterile, whereas the outer mucus layer is densely populated by commensal microbes[45] (Fig. 2).
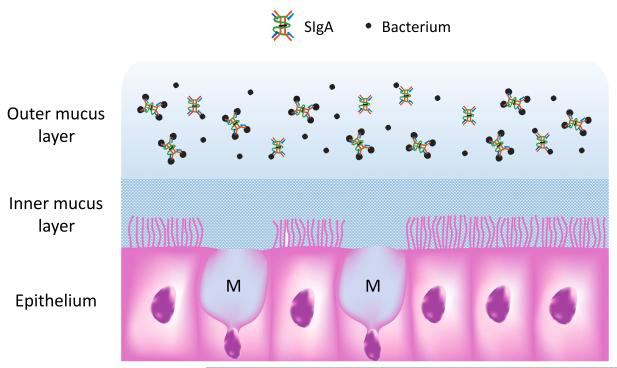
Fig. 2.
SIgA is concentrated in the outer layer of intestinal mucus along with commensal bacteria. Goblet cells extruding newly synthesized mucus (M) are interspersed among columnar epithelial cells in the intestinal ling. Mucus proteins form a gel that adheres to the epithelial surface, comprising a dense inner layer rich in antimicrobial peptides and devoid of bacteria, and a loose outer layer rich in commensal bacteria. SIgA is concentratedin the outer mucus layer, where it coats the surface of resident bacteria.
The major glycoprotein constituent of intestinal mucus is mucin-2 (Muc-2) [46], the product of the MUC2 gene in humans and the Muc2 gene in mice, and is secreted primarily by goblet cells. Mice with a targeted deletion in the Muc2 gene have reduced numbers of goblet cells and are completely devoid of a mucus layer at the luminal surface [47]. To determine whether SIgA is anchored in intestinal mucus via interactions with Muc-2, a recent study from the author’s laboratory analyzed the localization of SIgA in colonic mucus from wild-type and Muc-2-deficient mice[48]. Surprisingly, SIgA was relatively absent from the Muc-2 rich inner mucus layer in wild-type mice, and was instead associated with gut bacteria in the outer mucus layer. Similar localization of SIgA in the outer mucus layer was observed in colonic biopsies from healthy humans. In Muc-2-deficient mice that lacked a mucus layer, SIgA and bacteria were observed in close association with the epithelial surface. In mice deficient in both Muc-2 and pIgR, the epithelial surface was densely populated with bacteria, but SIgA was absent, demonstrating that pIgR is essential for delivering SIgA to the epithelial surface. These findings suggest that anchoring of SIgA to gut mucus is accomplished mainly through interactions of SIgA with bacteria, which could involve antigen-antibody binding via the IgA moiety as well as carbohydrate-dependent binding of the SC moiety to the bacterial surface (Fig. 2). In support of this model, a recent study demonstrated that the N-glycan side chains of SC mediate the association of SIgA with gram-positive commensals [19]. This model does not rule out the possibility of direct interactions between SIgA and mucin proteins, although the affinity of these interactions does not appear to be sufficient to retain SIgA in the inner mucus layer. In the outer mucus layer, weak interactions between SIgA and mucus may synergize with strong interactions between SIgA and gut bacteria, thus impeding migration of bacteria into the inner mucus layer.A decreasing concentration gradient of bacteria from the outer to the inner mucus layer would enhance the effectiveness of antimicrobial peptides in the inner layer. These findings shed new light on the mechanisms through which SIgA mediates immune exclusion in the intestine.
2.4. SIgA protects against intestinal inflammation
Inflammatory bowel diseases (IBD, including Crohn’s disease [CD] and ulcerative colitis [UC])are thought to result from a dysregulated immune response to normal components of the gut microbiota (reviewed in [49]). While many elements of the mucosal immune system are involved in protection against and exacerbation of intestinal inflammation, it is likely that the reciprocal relationship between SIgA and the gut microbiota plays an important role. In a study from the author’s laboratory comparing CD patients with healthy controls, disease severity was found to be correlated with down-regulation of pIgR expression in the colonic mucosa,dysregulated epithelial transcytosis of IgA, accumulation of IgA in the intestinal lamina propria, and increased levels of circulating IgA [50]. We and others have reported that expression of pIgR is down-regulated in colonic epithelial cells in experimental murine colitis induced by oral administration of dextran sulfate sodium to wild-type mice[51,52], or by adoptive transfer of effector T cells into immunodeficient mice [51]. A specific role for SIgA in preventing inflammation was suggested by the finding that pIgR-deficient mice were more susceptible than wild-type mice to chemically-induced colitis [42,53].
If SIgA is important for prevention of intestinal inflammation, it should follow that humans with selective IgA deficiency (IgAD), the most common of the primary antibody deficiencies, would be at increased risk for development of IBD. Indeed, this was the conclusion of a recent population-based matched cohort study in Sweden comparing 2,100 patients with IgAD (defined as having a serum IgA level of <0.07 g/L, with normal IgM and IgG levels) and 18,653 age- and gender-matched healthy controls[54].These investigators found a significantly elevated prevalence of IBD in IgAD patients compared to healthy controls (3.9% vs. 0.81%, prevalence ratio = 5.0), which was more pronounced for CD than for UC. Interestingly, IgAD patients had an even greater prevalence of celiac disease, an antigen-specific autoimmune disease associated with intestinal inflammation (6.7% vs. 0.19% in controls, prevalence ratio = 35). Despite the significantly increased risk, it must be noted that the majority of individuals with IgAD did not develop either IBD or celiac disease, underscoring the multifactorial pathogenesis of these inflammatory diseases of the intestine. Recently, an IBD-associated polymorphism was identified in a non-coding region adjacent to the human PIGR gene locus on chromosome 1 [23]. It will be important to determine whether this polymorphism affects pIgR expression or SIgA levels in mucosal secretions, and whether these changes are associated with increased risk for developing IBD or disease severity. There is also a need to identify other genetic and environmental factors that may interact with reduced SIgA levels to increase disease risk.
Reduction in intestinal SIgA, due to primary deficiency of IgA or pIgR or inflammation-induced down-regulation of pIgR expression, could lead to imbalances in the gut microbiota, further exacerbating inflammation. Molecular characterization of the gut microbiota in IBD patients has indicated a trend toward decreased biodiversity and increased levels of potentially pro-inflammatory bacterial taxa, collectively termed dysbiosis (reviewed in [55]). Whatever the primary cause, disruption of the mutualistic relationship between SIgA and the microbiota could lead to a vicious cycle of decreasing SIgA and increasing dysbiosis over time. There is a clear need to develop novel therapeutic strategies that could restore a more normal balance. Manipulations of the microbiota with antibiotics, probiotics, prebiotics, diet and/or fecal transplants has been suggested [56]. Passive therapy with SIgA antibodies has been successful in some animal models [40], and early exposure to breast milk-derived SIgA may reduce future risk of intestinal inflammation (see Section 3, below).Controlled studies and clinical trials in humans will be needed to determine the efficacy of these approaches. Monitoring of fecal SIgA levels and changes in the gut microbiota in patients with intestinal inflammation could identify those individuals who might profit from therapeutic interventions.
3. Breastmilk-derived maternal SIgA promoteshost-microbiota mutualism
Seminal studies on the structure and function of IgA revealed that SIgA in breast milk provides the first source of antibody-mediated immune protection in the intestinal tract of suckling infants[57-62] (Fig. 3). As the intestinal immune system develops in the offspring, there is a slow transition to endogenous production of SIgA via pIgR-mediated transport of locally synthesized IgA into the gut lumen. In human infants the timing of endogenous SIgA production varies widely, largely depending on environmental factors such as microbial load in the intestine [63]. In developed countries, it may take several years before the concentrations of intestinal SIgA achieve adult levels. In mice, endogenous SIgA is produced only after weaning, at about 3-4 weeks of age [18].While the benefits of breast milk-derived SIgA during infancy are clear, there is some controversy as to whether these beneficial effects persist into later life.
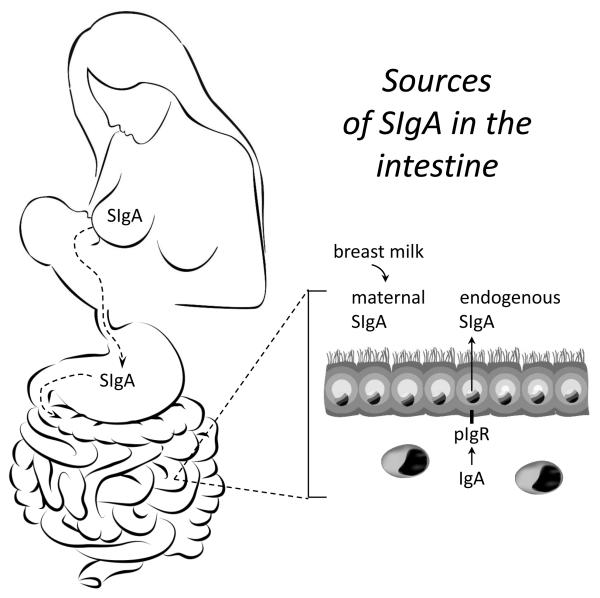
Fig. 3.
Sources of SIgA in the intestine. Polymeric IgA is synthesized by local plasma cells in the lamina propria of lactating mammary glands, and transported across alveolar epithelial cells into milk by pIgR. The pIgR-derived SC moiety protects SIgA from degradation during transit through the GI tract of the suckling infant. Newborn mammals do not produce endogenous SIgA in the intestine, and are reliant on breast milk-derived SIgA for antibody-mediated protection in the intestinal lumen. As the intestinal immune system develops after birth, SIgA can also be supplied by pIgR-mediated epithelial transcytosis of endogenously synthesized polymeric IgA. In mice, intestinal SIgA is derived exclusively from breast milk during the suckling period, and by endogenous transport after weaning. The pace of development of endogenous SIgA production is highly variable in human infants, and make take several years to achieve adult levels.
3.1. Breastfeeding regulates the composition of the gut microbiota in human infants, and provides protection against infectious, allergic and inflammatory diseases
There is considerable evidence from epidemiological studies that the composition of the gut microbiota differs significantly between breast-fed and formula-fed infants [64-71]. However, the specific components in breast milk that regulate the microbiota, including SIgA, have not systematically been investigated. Furthermore, it is controversial whether the composition of the microbiota of older children and adults reflects early breastfeeding experience. These gaps in our knowledge highlight the need for well-controlled longitudinal studies in humans to investigate the long-term effects of breast milk-derived SIgA on the composition of the gut microbiota.
Epidemiological studies and clinical reports suggest that SIgA antibodies in breast milk may reduce the incidence of a wide range of bacterial, viral and parasitic infections in suckling infants [72-93]. In each of these studies, the presence of SIgA antibodies against the infectious agent in question was correlated with protection, implying a specific effect of SIgA in addition to the general benefits of breastfeeding. While the predominant protective mechanism of SIgA in breast milk likely involves immune exclusion of pathogens at mucosal surfaces of the suckling infant, other protective mechanisms could include innate enhancement of the epithelial barrier and promotion of a healthy commensal microbiota.
In addition to preventing infectious diseases, there is evidence that early exposure to SIgA in breast milk may reduce the incidence of allergic diseases later in life [94-101]. However, the benefits of breast milk-derived SIgA remain controversial, largely due to the lack of standardization of epidemiological studies with regard to duration of breastfeeding, concentrations of SIgA and other anti-inflammatory factors in breast milk, effects of other variables such as diet, history of infectious diseases, and the composition of the resident microbiota, and outcomes measured. The PASTURE project (Protection Against Allergy: Study in Rural Environments), a large prospective birth cohort study conducted in Austria, Finland, France, Germany and Switzerland, represents the best effort to date to control for at least some of these variables[102]. In this study, levels of SIgA were analyzed in 610 breast milk samples collected 2 months after delivery. Questionnaires were used to assess duration of breastfeeding, environmental and socioeconomic factors, and the incidence of atopic dermatitis and asthma up to age 6. Atopic status was defined by levels of antigen-specific IgE in blood at the ages of 4 and 6 years. Multivariate logistic regression analysis revealed a significant inverse association between the total amount of SIgA that was ingested via breast milk during the first year of life and the development of atopic dermatitis[101]. However, the incidence of asthma and atopic sensitization were not consistently associated with SIgA consumption. While the results from the PASTURE study represent a step in the right direction for assessing the potential benefits of breast milk-derived SIgA, many questions remain to be answered. In particular, it will be important to assess the relationship between early exposure to SIgA and subsequent allergic disease in other geographic settings (including urban areas) and in other demographic groups. Furthermore, analysis of the gut microbiota at birth and throughout childhood could illuminate the role of host-microbial interactions in allergic diseases.
Given the importance of SIgA in protection against intestinal inflammation (see Section 2.4), the question arises whether early exposure to SIgA in breast milk reduces the incidence of inflammatory diseases in childhood and later in life. A meta-analysis of 4 epidemiological studies involving pediatric and adult IBD patients indicated that breast milk exposure reduced the subsequent risk of developing CD (odds ratio, 0.45; 95% confidence interval (0.26-0.79) and UC (odds ratio 0.56; 95% confidence interval 0.38-0.81) [103].A subsequent meta-analysis of 7 epidemiological studies of pediatric IBD patients indicated that breast milk exposure had a significant protective effect against development of early-onset IBD (odds ratio, 0.69; 95% confidence interval, 0.51-0.94) [104]. However, both groups of investigators noted the generally poor quality of the epidemiological data, and the SIgA content of breast milk was not considered. There is a clear need for well-designed prospective studies to investigate the benefits of breastfeeding in reducing the risk of IBD, and these studies should include an analysis of SIgA in breast milk. These human studies should be complemented with controlled studies in experimental animals that can shed light on the mechanisms through which early exposure to SIgA in breast milk can promote lifelong host-microbial mutualism, and protect against intestinal inflammation.
3.2. Studies on the effects of maternal antibodies in breast milk on the developing intestinal immune system of offspring mice
The relative contributions of passively delivered maternal antibodies vs. endogenously producedantibodies on the developing intestinal immune system have been studied in mice by analyzing offspring of dams with immune deficiencies.The impact of maternal antibodies on intestinal immunity was first studied by crossing wild-type male mice with females with severe combined immunodeficiency (scid/scid), which lack all antibodies in breast milk [105]. Because the scid/+ offspring of these crosses are able to produce their own SIgA (as well as other antibody isotypes), the differential effects of maternal vs. endogenous SIgA could be compared. As an additional approach, scid/+ offspring were cross-fostered to wild-type or scid/scid mothers at birth, thus allowing a direct analysis of the impact of maternal antibodies. Scid/+ pups born to or nursed by scid/scid mothers were observed to undergo accelerated development of endogenous lgA responses, which appeared to be triggered by early colonization of bacteria such as SFB in the intestinal tract. To follow up on this observation, these investigators designed a study involving reciprocal crossings of scid/scid and scid/+ female and male mice that had formerly been germ-free and were mono-colonized with SFB[106].They observed earlier colonization of the intestinal mucosa with SFB in offspring ofscid/scid mothers, regardless of whether the genotype of the pups wasscid/scid or scid/+.Interestingly, scid/+ offspring of scid/scid mothers, which produced endogenous SIgA after weaning, were eventually able to lower their intestinal levels of SFB. By contrast, intestinal levels of SFB remained high until at least 70 days of age in scid/scid offspring of scid/scid mothers.These findings suggest that maternal antibodies in breast milk provide important protection to the newborn intestinal tract during the critical early period when the ability of the newborn to produce its own antibodies is developing. Although the focus of these studies was on SFB, it is likely that the presence or absence of maternal antibodies affected the host response to other members of the commensal microbiota.
Using a variation of this breeding strategy, another group of investigators crossed wild-type male mice with recombination-activating gene-deficient (RAG−/−)females (which lack all B and T cells, and produce no antibodies of any isotype) or JH−/− females (which lack mature B cells and produce no antibodies of any isotype) [107]. Similar to the earlier studies with scid/scid mice, these investigators observed earlier induction of intestinal IgA responses in offspring of immune deficient mothers. They further reported that cross-fostering of JH−/− pups with wild-type mothers resulted in diminished translocation of Enterobacter cloacae to draining mesenteric lymph nodesintragastric challenge, compared to JH−/− pups that were nursed by JH−/− mothers. Using a similar breeding scheme with RAG−/− females and wild-type males, other investigators observed altered expression patterns of selected genes in intestinal epithelial cells of offspring that were deprived of maternal antibodies in breast milk [108].
While these studies with immunodeficient dams suggested that maternal antibodies in breast milk contribute to the development of the intestinal immune system in suckling infants, the specific effects of SIgA were not evaluated. Furthermore, the absence of systemic antibodies (as well as T cells in scid/scid and RAG−/− mice) may have had detrimental effects on the dams that impacted the pre- and post-nataldevelopment of their offspring. Finally, although responses to individual bacterial strains were assessed in these studies, there were no comprehensive analysis of the impact of breast milk antibodies on the composition of the microbiota in the offspring, or the responses of host epithelial cells. To address these issues, a recent study from the author’s laboratory utilized a breeding scheme in which Pigr−/− dams (which lacked SIgA in breast milk) were crossed with Pigr+/− males, and vice versa[18]. Because the offspring of these crosses were evenly distributed between the Pigr+/− (SIgA-sufficient) and Pigr−/− (SIgA-deficient) genotypes, we could study the independent effects of passive (i.e., breast milk-derived) and active (endogenously produced) SIgA. We hypothesized that early exposure to maternal SIgA would foster the development of host-microbial mutualism and enhance epithelial barrier function. In support of this hypothesis, we found that failure to receive SIgA in breast milk resulted in increased translocation of aerobic bacteria from the neonatal gut into draining lymph nodes, regardless of the Pigr genotype of the offspring. A prominent species among the bacteria that breached the epithelial barrier wasOchrobactrum anthropi,which has been identified as an opportunistic pathogen in preterm infants and immunocompromised individuals [109-111]. This finding supports the concept from epidemiological studies in humans that SIgA in breast milk helps to prevent neonatal infections. Enhancement of epithelial barrier function by SIgA could also reduce exposure of the systemic immune system of neonates to environmental and dietary allergens, thus reducing the risk of developing allergic diseases.
A comprehensive analysis of the fecal microbiota in Pigr+/−offspring of Pigr+/−and Pigr−/−mothers highlighted the importance of breast milk-derived SIgA in regulating the composition of the gut microbiota[18].By the age of weaning, mice that received maternal SIgA in breast milk had a significantly different gut microbiota from mice that did not receive SIgA. Importantly,the differences in the gut microbiota were magnified when the offspring of Pigr+/− vs.Pigr−/−mothers reached adulthood, despite the fact that the endogenous production of SIgA was comparable in all the offspring. This finding suggests that the microbiota that develops in the presence of maternal SIgA antibodiessets up a long-lasting pattern of SIgA-microbial reciprocity.Certain parallels were evident between the altered microbiota of mice that did not receive passive SIgA in breast milk and the microbiota of patients with inflammatory bowel disease (IBD). For example, increased abundance of bacterial taxa in the families Pasteurellaceae (phylum Proteobacteria) and Lachnospiraceae (phylum Firmicutes) has been observed in the gut microbiota of pediatric IBD patients [112], and increased abundance oftaxa in the family Comamonadaceae (phylum Proteobacteria) has been observed in the microbiota of adult IBD patients with chronic pouchitis [113]. Although caution must be exercised in translating results from controlled animal experiments to the complex human condition, our findings are consistent with the concept that early exposure to SIgA in breast milk promotes the development of a healthy microbiota that may provide protection against inflammatory diseases later in life. There is a clearly a need for more longitudinal studies to evaluate changes in the composition and function of the microbiota from infancy through childhood and adulthood, controlling for the presence and duration of breastfeeding and the concentration of SIgA in breast milk.
Having observed that the effects of breast milk-derived SIgA included promotion of epithelial barrier function and establishment of a distinctive microbiota, we hypothesized that the expression profile of epithelial cells could be altered as well. In support of this hypothesis, we found that the pattern of global gene expression in intestinal epithelial cells of adult mice was affected by early exposureto SIgA in breast milk as well as continuing production of endogenous SIgA [18]. Furthermore, this altered pattern of gene expression was correlated with increased susceptibility to colonic inflammation caused by the epithelial-disrupting agent dextran sulfate sodium. Interestingly, some of the genes whose expression wasregulatedby SIgA were orthologs of genes that have been associated with IBD and other intestinal inflammatory diseases in humans, including FUT2, IRF1, PLA2G2A, SLC26A3, VDR and ZMIZ1.In this context it is significant that down-regulation of pIgR expression by colonic epithelial cells has been correlated with disease severity in IBD patients [50], and that polymorphisms in the non-coding regions of the human PIGR gene locus have been associated with increased risk for IBD [23] (see Section 2.4, above). It will be important to determine whether PIGR gene polymorphisms affect the level of pIgR expression in the intestinal epithelium, and its potential consequences on global gene expression. It will also be important to study the effects of PIGR gene polymorphisms on SIgA levels in the milk of breastfeeding women, which could impact the development of host-microbial mutualism in the intestinal tract of the suckling infant.
4. Epithelial transcytosis of SIgA is regulated by the gut microbiota
The evidence described above supports a reciprocal relationship in which the gut microbiota induces host SIgA responses, which in turn regulate the composition and activity of the microbiota. Intestinal epithelial cells are important players in this host-microbial mutualism, by receiving signals from microbes and their products, transmitting these signals to immune cells, producing mucins and a wide range of antimicrobial peptides, and transporting IgA from its site of synthesis in the lamina propria into gut secretions[114]. Although it was initially proposed that intestinal epithelial cells were “ignorant” or “tolerant” of microbial antigens, it is now appreciated that the epithelium actively senses microbes through pattern recognition receptors (PRRs) that respond to redundant microbe-associated molecular patterns (MAMPs) [115]. We now know that the PIGR gene is one of the many targets of PRR signaling in intestinal epithelial cells [13-15]. Because one molecule of pIgR is consumed for every molecule of IgA that is transported across intestinal epithelial cells (Fig. 1), the rate of SIgA delivery into gut secretions is limited by the number of pIgR molecules at the epithelial surface.By inducing pIgR expression, the commensal microbiota can increase the concentration of SIgA in the intestinal lumen, thus adding an additional layer to its reciprocal relationship with SIgA.
4.1. Evidence that the commensal microbiota induces pIgR expression
The first demonstration that gut bacteria may modulate SIgA transport came with the report that butyrate, a bacterial fermentation product and important energy source in the colon, up-regulated expression of pIgR in the human colonic epithelial cell-line HT-29 [116]. A direct role for commensal bacteria in stimulating pIgR expression was demonstrated by the finding that mono-colonization of formerly germ-free mice with the commensal bacterium B. thetaiotaomicron restored intestinal expression of pIgR to levels comparable to those in mice with a normal microbiota[117]. In the intestinal tract of humans and mice,the level of expression of pIgR is directly correlated with the density of luminal bacteria, being substantially higher in the colon than in the small intestine [50,118].In vitroexperiments with HT-29 cells demonstrated that expression of pIgR was induced by co-culturewith E. coli or Salmonella typhimurium, gram-negative bacteria of the family Enterobacteriaceae [119].
Theoretical models for the regulation of gene expressionin colonic epithelial cells must be reconciled with the observation that in the colon, most commensal bacteria are spatially segregated from the epithelial surface, residing in the outer layer of mucus (Fig. 2) (see section 2.3, above). The finding that live and heat-killed bacteria induced similar levels of pIgR expression in HT-29 cells suggested that signaling pathways can be induced by shed bacterial products in addition to direct adherence of bacteria to the epithelial cell surface. Indeed,stimulation of HT-29 cells with purified lipopolysaccharide (LPS) from S. typhimuriumor E. coli caused a robust increase in pIgR expression [118-120]. It has been conjectured that shed products from dead bacteria can diffuse through the dense inner layer of mucus in the colon, thus gaining access to PRRs on the mucosal surface of epithelial cells [121]. The fact that low concentrations of LPS (1-200 pg/ml) can be detected in the circulation of healthy humans and experimental animals in the absence of bacteremia [122-124] suggests that LPS shed by gut bacteria may diffuse through the mucus layer and breach the epithelial barrier. A study using freshly isolated rat colons mounted in diffusion chambers demonstrated that luminally-administered LPS could migrate from the mucosal to the serosal surface, and that this process was enhanced by prior exposure of the rats to LPS by the oral route [125]. Importantly, these investigators detected expression of the LPS receptor TLR4 and its co-receptor CD14 at the mucosal surface of freshly isolated rat colons. Although it is difficult to ascertain the actual amounts of LPS that diffuse through the mucus to reach the mucosal surface, it is reasonable to posit that the large numbers of commensal bacteria in the colon would provide an abundant supply of LPS and other by-products that could stimulate epithelial PRRs. Much remains to be determined regarding the mechanisms through which bacterial by-products cross the mucus layer and gain access to the epithelial surface, and how changes in the composition of the gut microbiota may alter the concentration and composition of PRR ligands.
In addition to direct stimulation of epithelial cells, microbial products mayregulate pIgR expression indirectly stimulating the release of cytokines by host innate and adaptive immune cells, including interferon (IFN)-γ, interleukin (IL)-1, IL-4, IL-17, tumor necrosis factor (TNF)and lymphotoxin (LT)-β(reviewed in [13,15]).Most of these studies involved direct stimulation of culturedintestinal epithelial cell-lines with recombinant cytokines. The in vivosignificance of these findings was confirmed in a recent study demonstrating that mice deficient in T cells (TCR-βxδ−/−) mice exhibited reduced expression of pIgR and secretion of SIgA in the intestinal tract [126]. Mice deficient in the IL-17 receptor (IL17R−/−) also had reduced levels of pIgR and SIgA, suggesting an important role for IL-17 signaling. Adoptive transfer of IL-17 secreting Th17 cells specific for an immunodominant antigen of the commensal microbiota (CBir1 flagellin) caused a significant increase in pIgR levels, as well as total SIgA and CBir1-specific IgA. These findings highlight the importance of microbial antigens in stimulating pIgR expression via host cytokines, and demonstrate the need for additional research on the role of the gut microbiota in regulating host cytokine production.
4.2. Signaling pathways that regulate pIgR expression
Molecular cloning of the human PIGR and mouse Pigr genes revealed the presence of regulatory elements that are activated by TLR and cytokine receptor signaling pathways [120,127-130] (Fig. 4). Ligation of TLR4 by bacterial LPS transduces signals through the cytoplasmic adaptor protein myeloid differentiation primary response protein 88 (MyD88), which initiatesthe “classical” pathway of nuclear factor (NF)-κB activation. Phosphorylation of IκBα by IKKβ and its subsequent degradation results in nuclear translocation of an NF-κB dimer comprising p65/RelA and p50 subunits, which bind to a cognate element in intron 1 of the PIGR gene.Knockdown of RelA expression using small inhibitory RNA(siRNA) or mutation of the intron 1 NF-κB site was shown to block the induction of PIGR gene expression by LPS in HT-29 cells [119,120]. Knockdown of MyD88 expressionalso blocked LPS-inducedPIGR gene transcription in HT-29 cells, and mice with a targeted deletion of the Myd88 gene in intestinal epithelial cells had reduced expression of pIgR and transport of SIgA compared to wild-type littermates [131]. A model is proposed in which continuous stimulation of intestinal epithelial cells by MAMPs (including, but not limited to LPS) maintains robust expression of pIgR by MyD88- and NF-κB-dependent signaling (Fig. 4). It is now clear that intestinal epithelial cells express a wide range of cell surface and intracellular PRRs [132]. Future studies on the signaling pathways used by these receptors should reveal additional mechanisms through which commensal and pathogenic microbes regulate pIgR expression in intestinal epithelial cells.
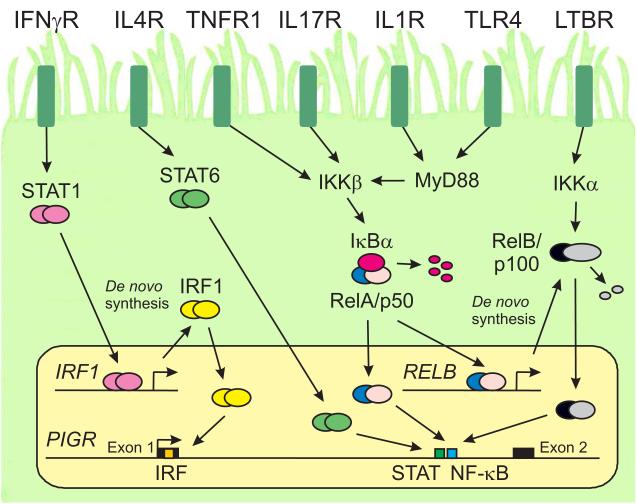
Fig. 4.
Regulation of PIGR gene transcription by cytokines and microbial factors. A mucosal epithelial cell expressing cytokine receptors and Toll-like receptors (TLRs) is shown. These receptors are depicted on one surface of the cell for purposes of illustration; however, it should not be implied that these receptors are localized preferentially to the apical and/or basolateral surface of a polarized epithelial cell. It should further be noted that epithelial cells express a wide range of cytokine and pattern recognition receptors in addition to those illustrated here.Ligation of cytokine receptors and TLRs results in translocation of activated transcription factors to the nucleus and de novo synthesis of additional transcription factors. The cytoplasmic adapter protein MyD88 transduces signals from cytokine receptors in the IL1R superfamily as well as many TLRs. The PIGR gene contains binding sites for members of the Interferon Regulatory Factor (IRF), Signal Transducer and Activator of Transcription (STAT) and Nuclear Factor-κB (NF-κB) families of transcription factors. The signaling pathways that regulate PIGR gene transcription are described in the text.
Multiple intracellular signaling pathways are involved in the regulation of pIgR expression by host cytokines, including Janus kinase-signal transduction and activator of transcription (JAK-STAT), NF-κB, mitogen-activated protein kinase (MAPK) and likely others(reviewed in [13-15]). Ligation of cell surface IFN-γresults in translocation of activated STAT1 dimers to the nucleus, where they bind to a cognate element in the gene encoding Interferon Regulatory Factor (IRF)1 (Fig. 4). Newly synthesizedIRF1then induces transcription of the PIGR gene by binding to a cognate element in exon 1. IL-4 signaling causes nuclear translocation of activated STAT6 dimers, which bind to a cognate element in intron 1 of the PIGR gene, adjacent to the NF-κB site.Ligation of IL-1 receptors activates the MyD88 signaling pathway, and may synergize with bacterial MAMPs to activate the intron 1 NF-κB site in the PIGR gene. By contrast, ligation of receptors for the pro-inflammatory cytokines TNF and IL-17initiates the classicalNF-κB activation pathwaythrough MyD88-independent signaling. TNF was shown to activate PIGR gene transcription through the same site in intron 1 that is activated by LPS [119,120,130]. Inhibition of the classical NF-κB activation pathway was found to block the induction of pIgR expression by IL-17 in HT-29 cells[126], suggesting that IL-17 signaling activates the PIGR gene through the same intron 1 NF-κB element that is activated by LPS, IL-1 and TNF.The induction of pIgR expression by IL-17 is particularly important in light of the role that that intestinal microbes, especially SFB, play in inducing Th17 responses [133], as well as the production of intestinal IgA [106].
PIGR gene transcription has also been shown to be induced by ligation of cell surface receptors for lymphotoxin-β (LTBR), which activates the IKKα-dependent “alternative” activation pathway of NF-κB and leads to nuclear translocation of RelB/p52 dimers [119]. Experiments with siRNA-mediated knockdown of individual NF-κB subunits suggested that both RelA/p50 and RelB/p52 dimers can activate the NF-κB element in intron 1 of the PIGR gene. The involvement of multiple signaling pathways and transcription factors in regulation of PIGR gene transcription suggests that the gut microbiota promotes robust expression of pIgR in intestinal epithelial cells through direct binding of microbial products to cell surface receptors as well as by stimulating secretion of cytokines by host innate and adaptive immune cells.Future studies are likely to identify additional cytokines that regulate pIgR expression, and it will be important to examine synergy and/or antagonism among these cytokines in the complex milieu of the gut lamina propria.
4.3. Microbial stimulation may enhance the rate of pIgR-mediated transcytosis of SIgA
Epithelial transcytosis of SIgA is unidirectional, initiated by binding of polymeric IgA to pIgR at the basolateral surface, and culminating with proteolytic degradation of pIgR and release of SIgA at the apical surface (Fig. 1). Intracellular targeting of the pIgR is similar to other integral membrane proteins destined for the apical plasma membrane in polarized epithelial cells, which generally follow an indirect route involving initial targeting the basolateral surface and subsequent transcytosis to the apical surface[134].While constitutive transcytosis of pIgR and release of free SC can occur in the absence of pIgA, binding of pIgA to pIgR at the basolateral surface increases the transcytotic rate of the pIgR-pIgA complex by initiating a signaling cascade that involves the tyrosine kinase p62YES. This kinase phosphorylates several substrates including phospholipase Cγ1, leading an increased rate of exocytosis of pIgR-pIgA complexes from apical recycling endosomes[135].It was subsequently demonstrated that p62YES could also phosphorylate the epidermal growth factor receptor in an IgA-dependent manner, suggesting that other extracellular signals might also affect transcytosis[136]. Enhanced transcytosis of pIgR depended on phosphorylation and activation of extracellular regulated kinase (ERK), which in turn was found to phosphorylate the Rab11 effector Rab11-FIP5. Thus stimulation of IgA production by the commensal microbiota could promote secretion of SIgA by enhancing the rate of pIgR-mediated epithelial transcytosis.
While the ability of IgA to stimulate its own transcytosis makes teleological sense, there is evidence that bacterial MAMPs can directly stimulate transcytosis of pIgR. Stimulation of HT-29 cells with live or dead E. coli or with LPS was found to enhance both pIgR expression and the rate of pIgA transport across polarized epithelial monolayers[137]. Because transcytosis was significantly enhanced prior to the increase in pIgR expression, the authors concluded that LPS could also directly stimulate transcytosis. Although the signal transduction pathway was not investigated in this study, it is possible that enhancement of pIgR transcytosis by LPS andpIgA stimulation converged at the point of ERK phosphorylation. Additional stimulation may have been mediated by other components of whole bacteria, such as formyl peptides, which have been shown to activate ERK in intestinal epithelial cells [138].
It should be noted that almost all the studies of epithelial transcytosis of IgA to date have utilized polarized monolayers of cultured epithelial cell-lines. While these models have been extremely useful for identifying the transcytotic pathway of SIgA and its regulation by intracellular signal transduction, there is a critical need for studying SIgA transcytosis in more physiological in vivoand ex vivo models. A recently published paper described method for the culture of epithelial spheroids from mouse colons, which allowed the expansion of intestinal epithelial stem/progenitor cells as a source of primary epithelial cells [139]. When grown on Transwell culture membranes, these colonic epithelial cells differentiated into monolayers that displayed high transepithelial electrical resistance, an index of cellular polarization and tight junction formation. Interestingly, transcytosis of IgA bythese epithelial monolayers was found to require induction of pIgR expression by treatment with LPS. This finding is consistent with the concept that robust pIgR expression is maintained in vivothrough continuous microbial stimulation, likely supplied by diffusion of LPS and other MAMPs through the mucosal barrier to the epithelial surface. These investigators confirmed earlier studies with epithelial cell-lines by demonstrating that TNF, IL-1β, IL-17 and heat-killed microbes stimulated pIgR expressionand SIgA transcytosis. A major advantage of this ex vivo system is that cells can be harvested from mice with a wide range of genetic mutations, allowing precise identification of key signaling molecules and transcriptional regulatory motifs. For example, this system could be used to explore the impact on pIgR expression and SIgA transcytosis of single-nucleotide polymorphisms that have been linked to IBD risk, including polymorphisms in the PIGR gene. It will also be important to develop in vivoand ex vivo experimentalmodels in whichSIgA transcytosis can be studied in the presence of an intact mucus layer and a diverse microbial community, the ultimate goal being to study cooperativity amongSIgA, pIgR and the gut microbiota.
5. Concluding remarks
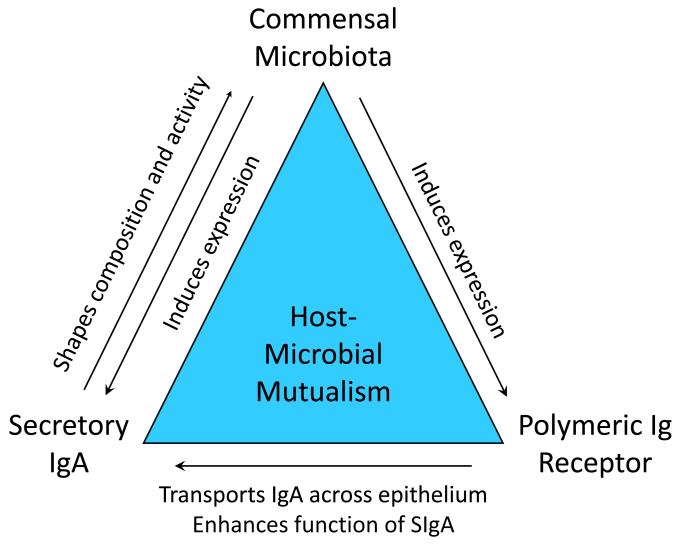
At mucosal surfaces, cooperativitybetween the commensal microbiota, secretory IgA and the polymeric immunoglobulin receptorpromotes a mutually beneficial relationship between the vertebrate host and some 100 trillion microbes (Fig. 5). The commensal microbiota sends signals to host epithelial and immune cells, leading to increased expression of pIgR. In turn, pIgR mediates epithelial transcytosis of SIgA and donates the SC moiety, which enhances immune functions of SIgA. The microbiota also sends signals to B cells and T helper cells, which promote IgA class switching and production of polymeric IgA by plasma cells in the intestinal lamina propria. SIgA plays its part by regulating the composition and activity of the microbiota in ways that are beneficial to the host. By anchoring bacteria in the outer mucus layer of the intestine, SIgA helps to maintain spatial segregation between the microbiota and the epithelial surface. In newborn mammals, breast milk-derived maternal SIgA performs these functions until the developing intestinal immune system of the newborn can take over.
Fig. 5.
Reciprocal interactions between the commensal microbiota, SIgA and pIgR promote host-microbial mutualism.
In the ideal situation, early exposure to SIgA in breast milk and robust production of endogenous SIgA throughout life promotes a mutually beneficialrelationship between the host and its microbiota and provides protection against infectious, allergic and inflammatory diseases. Achieving a better understanding of how this process works in healthy individuals offers the potential to develop novel and improved strategies for prevention and treatment of intestinal diseases. Where possible, breastfeeding should be encouraged, and further research should be devoted to finding ways to enhance SIgA levels in breast milk. Purified SIgA from breast milk could be explored as a potential supplement for infant formula, especially for premature and other high-risk infants. Purified SIgA may also offer promise as a therapeutic modality for allergic and inflammatory diseases. Manipulation of the gut microbiota though probiotics, prebiotics, and/or fecal transplants, could provide another avenue for optimizing SIgA production and function, although implementation of these strategies will require more knowledge about what constitutes a “healthy” microbiota at various stages of life. Finally, there is a need for a holistic approach in which SIgA status and the composition/function of the gut microbiota is studied in the broader context of a healthy lifestyle, including diet, weight management, exercise, vaccinations and other approaches to promote intestinal homeostasis and overall wellness.
Highlights.
Secretory IgA (SIgA) is transported across epithelial cells into intestinal secretions by the polymeric immunoglobulin receptor (pIgR).
The gut microbiota regulates production of SIgA and pIgR, which in turn regulate the composition and activity of the microbiota.
SIgA in the intestinal mucus layer helps to maintain spatial segregation between the microbiota and the epithelial surface without compromising the metabolic activity of the microbes.
Maternal SIgA in breast milk provides protection to newborn mammals until the developing intestinal immune system begins to produce its own SIgA.
Disruption of the SIgA-pIgR-microbial triad can increase the risk of infectious, allergic and inflammatory diseases of the intestine.
Acknowledgments
This work was supported by grant AI069027 from the National Institutes of Health of the United States of America (and an associated American Recovery and Reinvestment Act supplement) and a Senior Research Award from the Crohn’s and Colitis Foundation of America.
Abbreviations
- AID
activation-induced (cytidine) deaminase
- Ag
antigen
- ERK
extracellular receptor kinase
- IgA
immunoglobulin A
- IBD
inflammatory bowel disease
- IFN
interferon
- IRF
interferon regulatory factor
- IL
interleukin
- JAK
Janus kinase
- LPS
lipopolysaccharide
- LT
lymphotoxin
- MAMPs
microbe-associated molecular patterns
- MAPK
mitogen-activated protein kinase
- NF
nuclear factor-kappaB-κB
- PRR
pattern recognition receptor
- pIgA
polymeric IgA
- pIgR
polymeric immunoglobulin receptor
- RAG
recombination activating gene
- SC
secretory component
- SIgA
secretory IgA
- SFB
segmented filamentous bacteria
- scid
severe combined immunodeficiency
- STAT
signal transducer and activator of transcription
- siRNA
small inhibitory RNA
- TCR
T cell receptor
- TJ
tight junction
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The author declares that there are no conflicts of interest regarding the publication of this article.
References
- [1].Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012;245:132–46. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- [3].Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Berg RD, Savage DC. Immunological responses and microorganisms indigenous to the gastrointestinal tract. Am J Clin Nutr. 1972;25:1364–71. doi: 10.1093/ajcn/25.12.1364. [DOI] [PubMed] [Google Scholar]
- [5].Berg RD, Savage DC. Immune responses of specific pathogen-free and gnotobiotic mice to antigens of indigenous and nonindigenous microorganisms. Infect Immun. 1975;11:320–9. doi: 10.1128/iai.11.2.320-329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Arnold RR, Mestecky J, McGhee JR. Naturally occurring secretory immunoglobulin A antibodies to Streptococcus mutans in human colostrum and saliva. Infect Immun. 1976;14:355–62. doi: 10.1128/iai.14.2.355-362.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tlaskalova-Hogenova H, Sterzl J, Stepankova R, Dlabac V, Veticka V, Rossmann P, et al. Development of immunological capacity under germfree and conventional conditions. Ann N Y Acad Sci. 1983;409:96–113. 96–113. doi: 10.1111/j.1749-6632.1983.tb26862.x. [DOI] [PubMed] [Google Scholar]
- [8].Mellander L, Carlsson B, Jalil F, Soderstrom T, Hanson LA. Secretory IgA antibody response against Escherichia coli antigens in infants in relation to exposure. J Pediatr. 1985;107:430–3. doi: 10.1016/s0022-3476(85)80528-x. [DOI] [PubMed] [Google Scholar]
- [9].Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol. 1998;6:13–8. doi: 10.1155/1998/68382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–51S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- [11].Mestecky J, Russell MW, Jackson S, Brown TA. The human IgA system: a reassessment. Clin Immunol Immunopath. 1986;40:105–14. doi: 10.1016/0090-1229(86)90073-5. [DOI] [PubMed] [Google Scholar]
- [12].Conley ME, Delacroix DL. Intravascular and mucosal immunoglobulin A: two separate but related systems of immune defense? Ann Intern Med. 1987;106:892–9. doi: 10.7326/0003-4819-106-6-892. [DOI] [PubMed] [Google Scholar]
- [13].Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011;4:598–602. doi: 10.1038/mi.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kaetzel CS. The polymeric immunoglobulin receptor. eLS. 2013 DOI: 10.1002/9780470015902.a0024237. [Google Scholar]
- [15].Baker K, Blumberg RS, Kaetzel CS. Immunoglobulin transport and immunoglobulin receptors. In: Mestecky J, Strober W, Cheroutre H, Kelsall B, Lambrecht BN, Russell MW, editors. Mucosal Immunology. 4th ed. Academic Press/Elsevier; Waltham, MA: 2014. in press. [Google Scholar]
- [16].Chintalacharuvu KR, Morrison SL. Production of secretory immunoglobulin A by a single mammalian cell. Proc Natl Acad Sci USA. 1997;94:6364–8. doi: 10.1073/pnas.94.12.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Crottet P, Corthesy B. Secretory component delays the conversion of secretory IgA into antigen-binding competent F(ab’)2: a possible implication for mucosal defense. J Immunol. 1998;161:5445–53. [PubMed] [Google Scholar]
- [18].Rogier EW, Frantz AL, Bruno MEC, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA. 2014;111:3074–9. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mathias A, Corthésy B. N-Glycans on secretory component: mediators of the interaction between secretory IgA and gram-positive commensals sustaining intestinal homeostasis. Gut Microbes. 2011;2:287–93. doi: 10.4161/gmic.2.5.18269. [DOI] [PubMed] [Google Scholar]
- [20].Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–11. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Corthésy B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmun Rev. 2012;12:661–5. doi: 10.1016/j.autrev.2012.10.012. [DOI] [PubMed] [Google Scholar]
- [22].Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Macpherson AJ, Geuking MB, McCoy KD. Homeland security: IgA immunity at the frontiers of the body. Trends Immunol. 2012;33:160–7. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- [25].Macpherson AJ, Geuking MB, McCoy KD. Innate and adaptive immunity in host-microbiota mutualism. Front Biosci. 2012;4:685–98. 685–98. doi: 10.2741/s293. [DOI] [PubMed] [Google Scholar]
- [26].Macpherson AJ, McCoy KD. Stratification and compartmentalisation of immunoglobulin responses to commensal intestinal microbes. Semin Immunol. 2013;25:358–63. doi: 10.1016/j.smim.2013.09.004. [DOI] [PubMed] [Google Scholar]
- [27].Kato LM, Kawamoto S, Maruya M, Fagarasan S. Gut TFH and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunol Cell Biol. 2014;92:49–56. doi: 10.1038/icb.2013.54. [DOI] [PubMed] [Google Scholar]
- [28].Benveniste J, Lespinats G, Salomon J. Serum and secretory IgA in axenic and holoxenic mice. J Immunol. 1971;107:1656–62. [PubMed] [Google Scholar]
- [29].Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–9. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shroff KE, Meslin K, Cebra JJ. Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun. 1995;63:3904–13. doi: 10.1128/iai.63.10.3904-3913.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2013;5 doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–5. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- [34].Vulevic J, Juric A, Tzortzis G, Gibson GR. A mixture of trans-galactooligosaccharides reduces markers of metabolic syndrome and modulates the fecal microbiota and immune function of overweight adults. J Nutr. 2013;143:324–31. doi: 10.3945/jn.112.166132. [DOI] [PubMed] [Google Scholar]
- [35].van der Waaij LA, Limburg PC, Mesander G, van der WD. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–54. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Brandtzaeg P, Fjellanger I, Gjeruldsen ST. Adsorption of immunolgobulin A onto oral bacteria in vivo. J Bacteriol. 1968;96:242–9. doi: 10.1128/jb.96.1.242-249.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–81. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci USA. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Suzuki K, Ha SA, Tsuji M, Fagarasan S. Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Semin Immunol. 2007;19:127–35. doi: 10.1016/j.smim.2006.10.001. [DOI] [PubMed] [Google Scholar]
- [40].Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- [41].Johansen FE, Pekna M, Norderhaug IN, Haneberg B, Hietala MA, Krajci P, et al. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J Exp Med. 1999;190:915–22. doi: 10.1084/jem.190.7.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Reikvam DH, Derrien M, Islam R, Erofeev A, Grcic V, Sandvik A, et al. Epithelial-microbial cross-talk in polymeric Ig receptor deficient mice. Eur J Immunol. 2012;42:2959–70. doi: 10.1002/eji.201242543. [DOI] [PubMed] [Google Scholar]
- [43].Phalipon A, Cardona A, Kraehenbuhl JP, Edelman L, Sansonetti PJ, Corthésy B. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity. 2002;17:107–15. doi: 10.1016/s1074-7613(02)00341-2. [DOI] [PubMed] [Google Scholar]
- [44].McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–78. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- [45].Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Perez-Vilar J. Mucin granule intraluminal organization. Am J Respir Cell Mol Biol. 2007;36:183–90. doi: 10.1165/rcmb.2006-0291TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Velcich A, Yang W, Heyer J, Fragale A, Nicholas C, Viani S, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726–9. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- [48].Rogier EW, Frantz AL, Bruno MEC, Kaetzel CS. Secretory IgA is concentrated in the outer layer of intestinal mucus along with gut bacteria. Pathogens. 2014;3:390–403. doi: 10.3390/pathogens3020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Strober W. Impact of the gut microbiome on mucosal inflammation. Trends Immunol. 2013;34:423–30. doi: 10.1016/j.it.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Arsenescu R, Bruno MEC, Rogier EW, Stefka AT, McMahan AE, Wright TB, et al. Signature biomarkers in Crohn’s disease: toward a molecular classification. Mucosal Immunol. 2008;1:399–411. doi: 10.1038/mi.2008.32. [DOI] [PubMed] [Google Scholar]
- [51].Frantz AL, Bruno ME, Rogier EW, Tuna H, Cohen DA, Bondada S, et al. Multifactorial patterns of gene expression in colonic epithelial cells predict disease phenotypes in experimental colitis. Inflamm Bowel Dis. 2012;18:2138–48. doi: 10.1002/ibd.22923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Takiguchi H, Endo S, Omagari D, Okabayashi K, Watanabe T, Asano M, et al. Reduced production of polymeric immunoglobulin receptor in murine dextran sodium sulfate-induced colitis. J Oral Sci. 2012;54:23–32. doi: 10.2334/josnusd.54.23. [DOI] [PubMed] [Google Scholar]
- [53].Murthy AK, Dubose CN, Banas JA, Coalson JJ, Arulanandam BP. Contribution of polymeric immunoglobulin receptor to regulation of intestinal inflammation in dextran sulfate sodium-induced colitis. J Gastroenterol Hepatol. 2006;21:1372–80. doi: 10.1111/j.1440-1746.2006.04312.x. [DOI] [PubMed] [Google Scholar]
- [54].Ludvigsson JF, Neovius M, Hammarstrom L. Association Between IgA Deficiency & Other Autoimmune Conditions: A Population-Based Matched Cohort Study. J Clin Immunol. 2014 doi: 10.1007/s10875-014-0009-4. [DOI] [PubMed] [Google Scholar]
- [55].Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. doi: 10.1097/QCO.0b013e32832a8a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].D’Haens GR, Sartor RB, Silverberg MS, Petersson J, Rutgeerts P. Future directions in inflammatory bowel disease management. J Crohns Colitis. 2014;10 doi: 10.1016/j.crohns.2014.02.025. [DOI] [PubMed] [Google Scholar]
- [57].Axelsson H, Johansson BG, Rymo L. Isolation of immunoglobulin A (IgA) from human colostrum. Acta Chem Scand. 1966;20:2339–48. doi: 10.3891/acta.chem.scand.20-2339. [DOI] [PubMed] [Google Scholar]
- [58].Vierucci A, Panero C, Morgese G. The role of IgA in the antibacterial defense of the newborn infant: immuno-chemical analysis of anti-E. coli antibodies in colostrum, milk and blood. Riv Clin Pediatr. 1967;80:354–62. [PubMed] [Google Scholar]
- [59].Halpern MS, Koshland ME. Noval subunit in secretory IgA. Nature. 1970;228:1276–8. doi: 10.1038/2281276a0. [DOI] [PubMed] [Google Scholar]
- [60].Porter P, Noakes DE, Allen WD. Secretory IgA and antibodies to Escherichia coli in porcine colostrum and milk and their significance in the alimentary tract of the young pig. Immunology. 1970;18:245–57. [PMC free article] [PubMed] [Google Scholar]
- [61].Eddie DS, Schulkind ML, Robbins JB. The isolation and biologic activities of purified secretory IgA and IgG anti-Salmonella typhimurium “O” antibodies from rabbit intestinal fluid and colostrum. J Immunol. 1971;106:181–90. [PubMed] [Google Scholar]
- [62].Kraehenbuhl JP, Racine L, Galardy RE. Localization of secretory IgA, secretory component, and alpha chain in the mammary gland of lactating rabbits by immunoelectron microscopy. Ann N Y Acad Sci. 1975;254:190–202. doi: 10.1111/j.1749-6632.1975.tb29169.x. [DOI] [PubMed] [Google Scholar]
- [63].Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2012;12:9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- [64].Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology. 2010;156:3329–41. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- [65].Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–92. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- [66].Tsuji H, Oozeer R, Matsuda K, Matsuki T, Ohta T, Nomoto K, et al. Molecular monitoring of the development of intestinal microbiota in Japanese infants. Benef Microbes. 2012;3:113–25. doi: 10.3920/BM2011.0038. [DOI] [PubMed] [Google Scholar]
- [67].Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–94. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fan W, Huo G, Li X, Yang L, Duan C, Wang T, et al. Diversity of the intestinal microbiota in different patterns of feeding infants by Illumina high-throughput sequencing. World J Microbiol Biotechnol. 2013;29:2365–72. doi: 10.1007/s11274-013-1404-3. [DOI] [PubMed] [Google Scholar]
- [70].Gomez-Llorente C, Plaza-Diaz J, Aguilera M, Munoz-Quezada S, Bermudez-Brito M, Peso-Echarri P, et al. Three main factors define changes in fecal microbiota associated with feeding modality in infants. J Pediatr Gastroenterol Nutr. 2013;57:461–6. doi: 10.1097/MPG.0b013e31829d519a. [DOI] [PubMed] [Google Scholar]
- [71].Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. 2014;80:2889–900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Eibl MM, Wolf HM, Furnkranz H, Rosenkranz A. Prevention of necrotizing enterocolitis in low-birth-weight infants by IgA-IgG feeding. N Engl J Med. 1988;319:1–7. doi: 10.1056/NEJM198807073190101. [DOI] [PubMed] [Google Scholar]
- [73].Ali HM, Scott R, Toms GL. The effect of foster feeding and bottle feeding expressed breast-milk on the susceptibility of guinea-pig infants to influenza virus. Br J Exp Pathol. 1989;70:183–91. [PMC free article] [PubMed] [Google Scholar]
- [74].Merhav HJ, Wright HI, Mieles LA, Van Thiel DH. Treatment of IgA deficiency in liver transplant recipients with human breast milk. Transpl Int. 1995;8:327–9. doi: 10.1007/BF00346889. [DOI] [PubMed] [Google Scholar]
- [75].Dickinson EC, Gorga JC, Garrett M, Tuncer R, Boyle P, Watkins SC, et al. Immunoglobulin A supplementation abrogates bacterial translocation and preserves the architecture of the intestinal epithelium. Surgery. 1998;124:284–90. [PubMed] [Google Scholar]
- [76].Prentice A, Watkinson M, Prentice AM, Cole TJ, Whitehead RG. Breast-milk antimicrobial factors of rural Gambian mothers. II. Influence of season and prevalence of infection. Acta Paediatr Scand. 1984;73:803–9. doi: 10.1111/j.1651-2227.1984.tb17779.x. [DOI] [PubMed] [Google Scholar]
- [77].Cruz JR, Gil L, Cano F, Caceres P, Pareja G. Breast milk anti-Escherichia coli heat-labile toxin IgA antibodies protect against toxin-induced infantile diarrhea. Acta Paediatr Scand. 1988;77:658–62. doi: 10.1111/j.1651-2227.1988.tb10726.x. [DOI] [PubMed] [Google Scholar]
- [78].Jayashree S, Bhan MK, Kumar R, Bhandari N, Sazawal S. Protection against neonatal rotavirus infection by breast milk antibodies and trypsin inhibitors. J Med Virol. 1988;26:333–8. doi: 10.1002/jmv.1890260313. [DOI] [PubMed] [Google Scholar]
- [79].Achi R, Dac CP, Forsum U, Karlsson K, Saenz P, Mata L, et al. Titres of class-specific antibodies against Shigella and Salmonella lipopolysaccharide antigens in colostrum and breast milk of Costa Rican, Swedish and Vietnamese mothers. J Infect. 1992;25:89–105. doi: 10.1016/0163-4453(92)93657-c. [DOI] [PubMed] [Google Scholar]
- [80].Nachamkin I, Fischer SH, Yang XH, Benitez O, Cravioto A. Immunoglobulin A antibodies directed against Campylobacter jejuni flagellin present in breast-milk. Epidemiol Infect. 1994;112:359–65. doi: 10.1017/s0950268800057769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Espinoza F, Paniagua M, Hallander H, Svensson L, Strannegard O. Rotavirus infections in young Nicaraguan children. Pediatr Infect Dis J. 1997;16:564–71. doi: 10.1097/00006454-199706000-00006. [DOI] [PubMed] [Google Scholar]
- [82].Lehmann D, Pomat WS, Riley ID, Alpers MP. Studies of maternal immunisation with pneumococcal polysaccharide vaccine in Papua New Guinea. Vaccine. 2003;21:3446–50. doi: 10.1016/s0264-410x(03)00348-7. [DOI] [PubMed] [Google Scholar]
- [83].Deubzer HE, Obaro SK, Newman VO, Adegbola RA, Greenwood BM, Henderson DC. Colostrum obtained from women vaccinated with pneumococcal vaccine during pregnancy inhibits epithelial adhesion of Streptococcus pneumoniae. J Infect Dis. 2004;190:1758–61. doi: 10.1086/424597. [DOI] [PubMed] [Google Scholar]
- [84].Obaro SK, Deubzer HE, Newman VO, Adegbola RA, Greenwood BM, Henderson DC. Serotype-specific pneumococcal antibodies in breast milk of Gambian women immunized with a pneumococcal polysaccharide vaccine during pregnancy. Pediatr Infect Dis J. 2004;23:1023–9. doi: 10.1097/01.inf.0000143651.54880.09. [DOI] [PubMed] [Google Scholar]
- [85].Bouhlal H, Latry V, Requena M, Aubry S, Kaveri SV, Kazatchkine MD, et al. Natural antibodies to CCR5 from breast milk block infection of macrophages and dendritic cells with primary R5-tropic HIV-1. J Immunol. 2005;174:7202–9. doi: 10.4049/jimmunol.174.11.7202. [DOI] [PubMed] [Google Scholar]
- [86].Tanriverdi HA, Acun C, Ustundag G, Barut A, Tekin IO, Ustundag Y. Investigation of human colostrum Helicobacter pylori IgA content in lactating women. Eur J Obstet Gynecol Reprod Biol. 2006;124:58–60. doi: 10.1016/j.ejogrb.2005.02.027. [DOI] [PubMed] [Google Scholar]
- [87].Carrero JC, Cervantes-Rebolledo C, Aguilar-Diaz H, Diaz-Gallardo MY, Laclette JP, Morales-Montor J. The role of the secretory immune response in the infection by Entamoeba histolytica. Parasite Immunol. 2007;29:331–8. doi: 10.1111/j.1365-3024.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- [88].Castelletti E, Lo CS, Kuhn L, Borelli M, Gajardo J, Sinkala M, et al. The mucosae-associated epithelial chemokine (MEC/CCL28) modulates immunity in HIV infection. PLoS ONE. 2007;2:e969. doi: 10.1371/journal.pone.0000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Harrison LM, Morris JA, Lauder RM, Telford DR. The effect of Staphylococcus aureus carriage in late pregnancy on antibody levels to staphylococcal toxins in cord blood and breast milk. FEMS Immunol Med Microbiol. 2008;54:137–43. doi: 10.1111/j.1574-695X.2008.00463.x. [DOI] [PubMed] [Google Scholar]
- [90].Mota-Ferreira DM, Goncalves-Pires MR, Junior AF, Sopelete MC, Abdallah VO, Costa-Cruz JM. Specific IgA and IgG antibodies in paired serum and breast milk samples in human strongyloidiasis. Acta Trop. 2009;109:103–7. doi: 10.1016/j.actatropica.2008.09.023. [DOI] [PubMed] [Google Scholar]
- [91].Bhuiyan TR, Saha A, Lundgren A, Qadri F, Svennerholm AM. Immune responses to Helicobacter pylori infection in Bangladeshi children during their first two years of life and the association between maternal antibodies and onset of infection. J Infect Dis. 2010;202:1676–84. doi: 10.1086/657085. [DOI] [PubMed] [Google Scholar]
- [92].Kadooka Y, Tominari K, Sakai F, Yasui H. Prevention of rotavirus-induced diarrhea by preferential secretion of IgA in breast milk via maternal administration of Lactobacillus gasseri SBT2055. J Pediatr Gastroenterol Nutr. 2012;55:66–71. doi: 10.1097/MPG.0b013e3182533a2b. [DOI] [PubMed] [Google Scholar]
- [93].Mabuka J, Nduati R, Odem-Davis K, Peterson D, Overbaugh J. HIV-specific antibodies capable of ADCC are common in breastmilk and are associated with reduced risk of transmission in women with high viral loads. PLoS Pathog. 2012;8:e1002739. doi: 10.1371/journal.ppat.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Calbi M, Giacchetti L. Low breast milk IgA and high blood eosinophil count in breast-fed newborns determine higher risk for developing atopic eczema after an 18-month follow-up. J Investig Allergol Clin Immunol. 1998;8:161–4. [PubMed] [Google Scholar]
- [95].Bottcher MF, Haggstrom P, Bjorksten B, Jenmalm MC. Total and allergen-specific immunoglobulin A levels in saliva in relation to the development of allergy in infants up to 2 years of age. Clin Exp Allergy. 2002;32:1293–8. doi: 10.1046/j.1365-2222.2002.01470.x. [DOI] [PubMed] [Google Scholar]
- [96].Mosconi E, Rekima A, Seitz-Polski B, Kanda A, Fleury S, Tissandie E, et al. Breast milk immune complexes are potent inducers of oral tolerance in neonates and prevent asthma development. Mucosal Immunol. 2010;3:461–74. doi: 10.1038/mi.2010.23. [DOI] [PubMed] [Google Scholar]
- [97].Tomicic S, Johansson G, Voor T, Bjorksten B, Bottcher MF, Jenmalm MC. Breast milk cytokine and IgA composition differ in Estonian and Swedish mothers-relationship to microbial pressure and infant allergy. Pediatr Res. 2010;68:330–4. doi: 10.1203/PDR.0b013e3181ee049d. [DOI] [PubMed] [Google Scholar]
- [98].Ismail IH, Licciardi PV, Oppedisano F, Boyle RJ, Tang ML. Relationship between breast milk sCD14, TGF-beta1 and total IgA in the first month and development of eczema during infancy. Pediatr Allergy Immunol. 2013;24:352–60. doi: 10.1111/pai.12075. [DOI] [PubMed] [Google Scholar]
- [99].Bernard H, Ah-Leung S, Drumare MF, Feraudet-Tarisse C, Verhasselt V, Wal JM, et al. Peanut allergens are rapidly transferred in human breast milk and can prevent sensitization in mice. Allergy. 2014;10 doi: 10.1111/all.12411. [DOI] [PubMed] [Google Scholar]
- [100].Jarvinen KM, Westfall JE, Seppo MS, James AK, Tsuang AJ, Feustel PJ, et al. Role of maternal elimination diets and human milk IgA in the development of cow’s milk allergy in the infants. Clin Exp Allergy. 2014;44:69–78. doi: 10.1111/cea.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Orivuori L, Loss G, Roduit C, Dalphin JC, Depner M, Genuneit J, et al. Soluble immunoglobulin A in breast milk is inversely associated with atopic dermatitis at early age: the PASTURE cohort study. Clin Exp Allergy. 2014;44:102–12. doi: 10.1111/cea.12199. [DOI] [PubMed] [Google Scholar]
- [102].von ME, Schmid S. The PASTURE project: EU support for the improvement of knowledge about risk factors and preventive factors for atopy in Europe. Allergy. 2006;61:407–13. doi: 10.1111/j.1398-9995.2006.01009.x. [DOI] [PubMed] [Google Scholar]
- [103].Klement E, Cohen RV, Boxman J, Joseph A, Reif S. Breastfeeding and risk of inflammatory bowel disease: a systematic review with meta-analysis. Am J Clin Nutr. 2004;80:1342–52. doi: 10.1093/ajcn/80.5.1342. [DOI] [PubMed] [Google Scholar]
- [104].Barclay AR, Russell RK, Wilson ML, Gilmour WH, Satsangi J, Wilson DC. Systematic review: the role of breastfeeding in the development of pediatric inflammatory bowel disease. J Pediatr. 2009;155:421–6. doi: 10.1016/j.jpeds.2009.03.017. [DOI] [PubMed] [Google Scholar]
- [105].Kramer DR, Cebra JJ. Early appearance of “natural” mucosal IgA responses and germinal centers in suckling mice developing in the absence of maternal antibodies. J Immunol. 1995;154:2051–62. [PubMed] [Google Scholar]
- [106].Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. 2001;69:3611–7. doi: 10.1128/IAI.69.6.3611-3617.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, et al. Mechanisms of neonatal mucosal antibody protection. J Immunol. 2006;177:6256–62. doi: 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- [108].Jenkins SL, Wang J, Vazir M, Vela J, Sahagun O, Gabbay P, et al. Role of passive and adaptive immunity in influencing enterocyte-specific gene expression. Am J Physiol Gastrointest Liver Physiol. 2003;285:G714–G725. doi: 10.1152/ajpgi.00130.2003. [DOI] [PubMed] [Google Scholar]
- [109].Stiakaki E, Galanakis E, Samonis G, Christidou A, Maraka S, Tselentis Y, et al. Ochrobactrum anthropi bacteremia in pediatric oncology patients. Pediatr Infect Dis J. 2002;21:72–4. doi: 10.1097/00006454-200201000-00018. [DOI] [PubMed] [Google Scholar]
- [110].Duran R, Vatansever U, Acunas B, Basaran UN. Ochrobactrum anthropi bacteremia in a preterm infant with meconium peritonitis. Int J Infect Dis. 2009;13:e61–e63. doi: 10.1016/j.ijid.2008.06.027. [DOI] [PubMed] [Google Scholar]
- [111].Naik C, Kulkarni H, Darabi A, Bhanot N. Ochrobactrum anthropi: a rare cause of pneumonia. J Infect Chemother. 2013;19:162–5. doi: 10.1007/s10156-012-0436-1. [DOI] [PubMed] [Google Scholar]
- [112].Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–91. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Tannock GW, Lawley B, Munro K, Lay C, Taylor C, Daynes C, et al. Comprehensive analysis of the bacterial content of stool from patients with chronic pouchitis, normal pouches, or familial adenomatous polyposis pouches. Inflamm Bowel Dis. 2012;18:925–34. doi: 10.1002/ibd.21936. [DOI] [PubMed] [Google Scholar]
- [114].Duerr CU, Hornef MW. The mammalian intestinal epithelium as integral player in the establishment and maintenance of host-microbial homeostasis. Semin Immunol. 2012;24:25–35. doi: 10.1016/j.smim.2011.11.002. [DOI] [PubMed] [Google Scholar]
- [115].Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–44. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- [116].Kvale D, Brandtzaeg P. Constitutive and cytokine induced expression of HLA molecules, secretory component, and intercellular adhesion molecule-1 is modulated by butyrate in the colonic epithelial cell line HT-29. Gut. 1995;36:737–42. doi: 10.1136/gut.36.5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–4. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- [118].Bruno MEC, Rogier EW, Frantz AL, Stefka AT, Thompson SN, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor in intestinal epithelial cells by Enterobacteriaceae: Implications for mucosal homeostasis. Immunol Invest. 2010;39:356–82. doi: 10.3109/08820131003622809. [DOI] [PubMed] [Google Scholar]
- [119].Bruno ME, Frantz AL, Rogier EW, Johansen FE, Kaetzel CS. Regulation of the polymeric immunoglobulin receptor by the classical and alternative NF-κB pathways in intestinal epithelial cells. Mucosal Immunol. 2011;4:468–78. doi: 10.1038/mi.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Schneeman TA, Bruno MEC, Schjerven H, Johansen FE, Chady L, Kaetzel CS. Regulation of the polymeric Ig receptor by signaling through Toll-like receptors 3 and 4: linking innate and adaptive immune responses. J Immunol. 2005;175:376–84. doi: 10.4049/jimmunol.175.1.376. [DOI] [PubMed] [Google Scholar]
- [121].Pearson JP, Brownlee IA. The interaction of large bowel microflora with the colonic mucus barrier. Int J Inflam. 2010;2010:321426. doi: 10.4061/2010/321426. doi: 10.4061/2010/321426.:321426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am J Clin Nutr. 2007;86:1286–92. doi: 10.1093/ajcn/86.5.1286. [DOI] [PubMed] [Google Scholar]
- [123].Ghoshal S, Witta J, Zhong J, de VW, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res. 2009;50:90–7. doi: 10.1194/jlr.M800156-JLR200. [DOI] [PubMed] [Google Scholar]
- [124].Laugerette F, Vors C, Geloen A, Chauvin MA, Soulage C. Lambert-Porcheron S et al. Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation. J Nutr Biochem. 2011;22:53–9. doi: 10.1016/j.jnutbio.2009.11.011. [DOI] [PubMed] [Google Scholar]
- [125].Tomita M, Ohkubo R, Hayashi M. Lipopolysaccharide transport system across colonic epithelial cells in normal and infective rat. Drug Metab Pharmacokinet. 2004;19:33–40. doi: 10.2133/dmpk.19.33. [DOI] [PubMed] [Google Scholar]
- [126].Cao AT, Yao S, Gong B, Elson CO, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–73. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Piskurich JF, Youngman KR, Phillips KM, Hempen PM, Blanchard MH, France JA, et al. Transcriptional regulation of the human polymeric immunoglobulin receptor gene by interferon-γ. Mol Immunol. 1997;34:75–91. doi: 10.1016/s0161-5890(96)00079-x. [DOI] [PubMed] [Google Scholar]
- [128].Blanch VJ, Piskurich JF, Kaetzel CS. Cutting edge: coordinate regulation of IFN regulatory factor-1 and the polymeric Ig receptor by proinflammatory cytokines. J Immunol. 1999;162:1232–5. [PubMed] [Google Scholar]
- [129].Schjerven H, Brandtzaeg P, Johansen FE. Mechanism of IL-4-mediated up-regulation of the polymeric Ig receptor: Role of STAT6 in cell type-specific delayed transcriptional response. J Immunol. 2000;165:3898–906. doi: 10.4049/jimmunol.165.7.3898. [DOI] [PubMed] [Google Scholar]
- [130].Schjerven H, Brandtzaeg P, Johansen FE. A novel NF-κB/Rel site in intron 1 cooperates with proximal promoter elements to mediate TNF-α-induced transcription of the human polymeric Ig receptor. J Immunol. 2001;167:6412–20. doi: 10.4049/jimmunol.167.11.6412. [DOI] [PubMed] [Google Scholar]
- [131].Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–12. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Fukata M, Arditi M. The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunol. 2013;6:451–63. doi: 10.1038/mi.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ivanov II, Littman DR. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3:209–12. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Mostov KE, Verges M, Altschuler Y. Membrane traffic in polarized epithelial cells. Curr Opin Cell Biol. 2000;12:483–90. doi: 10.1016/s0955-0674(00)00120-4. [DOI] [PubMed] [Google Scholar]
- [135].Rojas R, Apodaca G. Immunoglobulin transport across polarized epithelial cells. Nat Rev Mol Cell Biol. 2002;3:944–55. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- [136].Su T, Bryant DM, Luton F, Verges M, Ulrich SM, Hansen KC, et al. A kinase cascade leading to Rab11-FIP5 controls transcytosis of the polymeric immunoglobulin receptor. Nat Cell Biol. 2010;12:1143–53. doi: 10.1038/ncb2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Diebel LN, Liberati DM. Disparate effects of bacteria and toll-like receptor-dependant bacterial ligand stimulation on immunoglobulin a transcytosis. J Trauma. 2011;70:691–700. doi: 10.1097/TA.0b013e31820c780e. [DOI] [PubMed] [Google Scholar]
- [138].Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS. Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol. 2010;177:2782–90. doi: 10.2353/ajpath.2010.100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Moon C, Vandussen KL, Miyoshi H, Stappenbeck TS. Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 2013;10 doi: 10.1038/mi.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]