Abstract
Human mitochondrial ribosomes are highly divergent from all other known ribosomes and are specialized to exclusively translate membrane proteins. They are linked with hereditary mitochondrial diseases, and are often the unintended targets of various clinically useful antibiotics. Using single-particle electron cryo-microscopy we have determined the structure of its large subunit to 3.4 angstrom resolution, revealing 48 proteins, 21 of which are specific to mitochondria. The structure unveils an adaptation of the exit tunnel for hydrophobic nascent peptides, extensive remodeling of the central protuberance including recruitment of mitochondrial tRNAVal to play an integral structural role, and changes in the tRNA binding sites related to the unusual characteristics of mitochondrial tRNAs.
Introduction
The human mitochondrial (mt) genome encodes 13 essential proteins of the oxidative phosphorylation (OXPHOS) complexes of the inner mitochondrial membrane. These proteins are translated by a dedicated set of ribosomes (mitoribosomes). The mitoribosome has a sedimentation coefficient of 55S and consists of a large (LSU, 39S) and small (SSU, 28S) subunit. These subunits contain a 16S rRNA and 12S rRNA respectively, and no 5S rRNA (1). They differ from cytoplasmic and bacterial ribosomes in having a high protein to RNA ratio. All proteins synthesized by human mitoribosomes are hydrophobic, integral membrane proteins and some require prosthetic groups for folding and functioning. Unlike their cytoplasmic counterparts, human mitoribosomes are permanently tethered to the mitochondrial inner membrane through the LSU (2). Mitochondrial diseases affect >1 in 7500 live births (3) with defects of mitochondrial translation responsible for a subgroup associated with decreased OXPHOS activity (reviewed in (4)). Cancer cells have amplified OXPHOS capacity and elevated mitochondrial protein translation compared with adjacent stromal tissue (5). Specific inhibition of mitoribosomes has successfully induced selective cytotoxicity in leukemia cells (6) establishing mitoribosomes as drug targets for cancer. Furthermore, mitoribosomes are often the unintended targets of various clinically useful antibiotics that target protein synthesis by bacterial ribosomes (7).
The high-resolution structure of the yeast mt-LSU revealed a major remodeling of mitoribosomes compared to bacterial and cytoplasmic ribosomes (8). However, yeast mitoribosomes are themselves very distinct from mammalian mitoribosomes. Structural information for mammalian mitoribosomes is limited to a ~5 Å reconstruction of porcine LSU (9) and a ~7 Å structure of bovine 55S (10) that suggested substantial differences with the yeast mitoribosome. Here we report a stereochemically refined, nearly complete model of human mt-LSU at 3.4 Å resolution, achieved by a combination of rapid biochemical purification in mild conditions (11), and recent developments in data processing (12) and model building (8). We identify 16 more proteins than in the previous porcine model (9) and reveal mt-tRNAVal as a key structural component of the mitoribosome.
Overall structure of human mt-LSU
Intact human mitoribosomes were purified from human embryonic kidney (HEK293) cells within 28 hours of mitochondria disruption (11) and visualized using single-particle electron cryo-microscopy (cryo-EM) (Fig. S1). The data were processed resulting in a reconstruction that extends to 3.8 Å (Fig. S1). The SSU displays considerable conformational heterogeneity with respect to the LSU, so the SSU map could not be interpreted with an atomic model. The use of a soft mask over just the LSU improved its map quality and overall resolution to 3.4 Å (Fig. S1D).
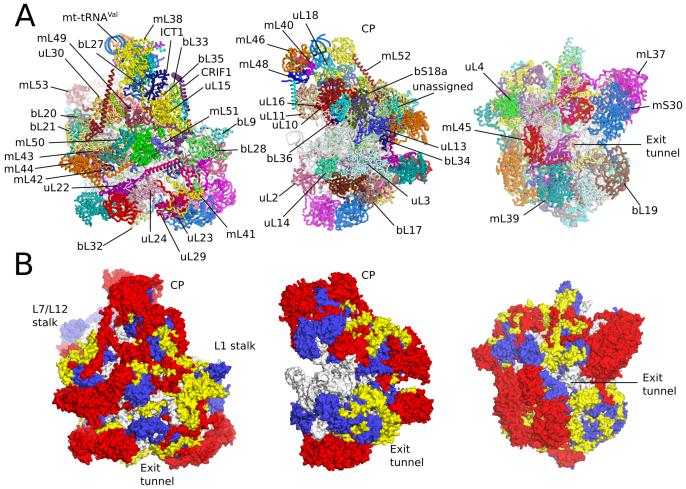
The model of the human mt-LSU contains two structural RNA molecules (16S mt-LSU rRNA and the newly identified mt-tRNAval), as well as a tRNA bound to the E site (Fig. S3], and 48 proteins, of which 21 are specific to mitochondria (Table S2, Fig. 1 and Fig. S4). Five additional short protein elements remain unassigned, but probably correspond to unbuilt protein extensions. The mt-LSU is highly protein-rich with over two-thirds of the total mass of 1.7 MDa consisting of proteins, of which 0.54 MDa can be attributed to mitochondria-specific elements. This gives the human mt-LSU a distinct morphology from both bacterial ribosomes and yeast mitoribosomes (Figs. S5 and S6). The connectivity between proteins has also expanded, with each protein making an average of 4.9 contacts (Fig. S7). Both the average mass of the mitoribosomal proteins and the number of inter-protein contacts exceeds those of the mammalian cytoplasmic ribosome (13).
Fig. 1. Overview of human mt-LSU.
(A] Location of proteins in the human mt-LSU, showing (from left to right) solvent-facing, side and exit tunnel views. (B) Views as in A, proteins conserved with bacteria (blue), extensions of homologous proteins (yellow) and mitochondria-specific proteins (red). rRNA is shown in gray.
While the protein composition has substantially increased, the length of mt-LSU rRNA (1559 nucleotides] has halved compared to bacterial 23S rRNA. Contraction has occurred in all domains (Figs. S8-S10). Using base-pair information extracted from the structure we have constructed a revised secondary structure diagram of mt-LSU rRNA (Fig. S7). In contrast to yeast mitoribosomes where rRNA deletions are minor and primarily occur at the tunnel exit (8), deletions of human mitoribosomal RNA are numerous and evenly distributed. The reduction is frequently a result of shortening surface exposed helices. The extant regions are bridged by 37 short ‘bypass segments’ often of just 2-4 nucleotides (Fig. S9). When internal helices are excised, the location of downstream rRNA elements typically remains unaffected. For example, helices 95-97 (including the sarcin-ricin loop that is essential for GTP-catalyzed steps of translation] have conserved locations despite the absence of connecting stem h94, due to partial stabilization by mitochondria-specific proteins (Fig. S11). Truncation of rRNA and the absence of 5S rRNA have presumably contributed to the loss of uL5, bL25, and bL31 from the mt-LSU compared to bacterial ribosomes (Fig. S8). However, uL6 is absent despite strong conservation of the rRNA to which it binds.
Mitochondria-specific protein elements
Proteins homologous to those in bacteria are, on average, ~60% larger in the human mt-LSU. The extensions are shorter and not conserved with those in the yeast mitoribosome (8) (Fig. S4). Although some of the extensions fill voids left by rRNA deletions, the amount of rRNA replacement by protein extensions is small (Fig. S12). Some extensions protrude into solvent, but predominantly they interact with mitochondria-specific protein elements (Table S3).
Mitochondria-specific proteins are peripherally distributed over the solvent accessible surface of the ribosome (Fig. 1) with clusters at the central protuberance, the L7/L12 stalk and adjacent to the polypeptide exit site. They have generally adopted new positions rather than compensating for lost rRNA (Fig. 1 and Fig. S12), although a large deletion in domain III (h53-h59) is occupied by a ~100 kDa heterodimer of mL37 and mS30 (Fig. S12). Other mitochondria-specific proteins that compensate for lost rRNA (mL41, mL42, mL49, and mL51) are relatively small proteins that help stabilize bypass segments. The effect of not compensating all lost rRNA is an architecture less compact than cytoplasmic ribosomes. Despite the increased porosity, the rRNA accessible to solvent has reduced by 52% compared to bacterial ribosomes. This decrease can only partially be accounted for by reduced rRNA content (41%), with new protein elements contributing by burying 32,500 Å2 of rRNA surface. This agrees with the hypothesis that accretion of mitochondria-specific elements shield the rRNA from reactive oxygen species (14) that are elevated in mitochondria as a by-product of OXPHOS and are a major source of RNA damage.
Two of the proteins (mS30 and bS18a) were previously classified as components of the mt-SSU (10,15). The presence of three sequence variants of bS18 had led to suggestions that differential incorporation of bS18 variants generates a heterogeneous population of mitoribosomes (16). However, the identification of bS18a in the mt-LSU suggests that bS18 variants may not promote structural diversity, but represent a duplicated fold incorporated into distinct locations of the mitoribosome.
Mt-tRNAVal is a part of the central protuberance
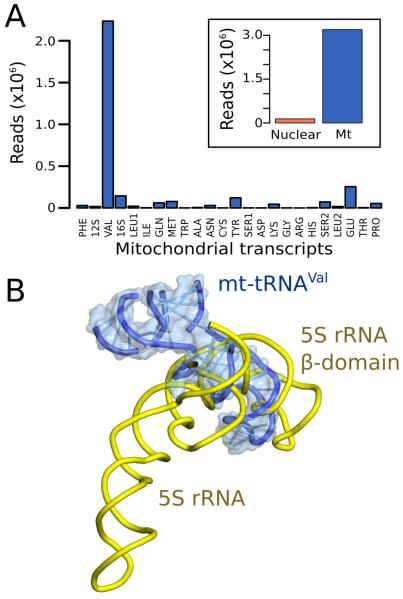
The map of human mt-LSU reveals a density corresponding to an L-shaped RNA molecule located at the top of the central protuberance in a position similar to that occupied by 5S rRNA in cytoplasmic ribosomes (Fig. S13). The RNA component was also reported in the porcine mitoribosome (9). We biochemically extracted the RNA molecule from purified human mt-LSU (Fig. S13A) and using deep RNA-sequencing (11) identified it as mt-tRNAVal (Fig. 2). A structural model of this tRNA agrees well with the density (Fig. S13B-D).
Fig. 2. Mt-tRNAVal is part of the human mitoribosome.
(A) Mapping of rRNA-sequencing reads to total human (inset) and mitochondrial transcripts. (B) The anticodon stem-loop of mt-tRNAVal binds in a similar position to domain β of 5S rRNA.
That mt-tRNAVal becomes incorporated over other tRNAs may result from its location in the mtDNA chromosome (17). The mt-tRNAVal gene is flanked by the two mt-rRNA genes, which are transcribed together as a polycistronic transcript (18) (Fig. S13E). In bacteria, it is the 5S rRNA gene that is located adjacent to the rRNA genes and co-transcribed with them as part of a polycistronic message (19). As with 5S rRNA in bacteria, mt-tRNAVal would be present in stoichiometric amounts after processing, and spatially coincident for incorporation into the LSU. The 12S rRNA-tRNAVal-16S rRNA organization is almost invariant amongst vertebrates (20).
Remodeling of the central protuberance
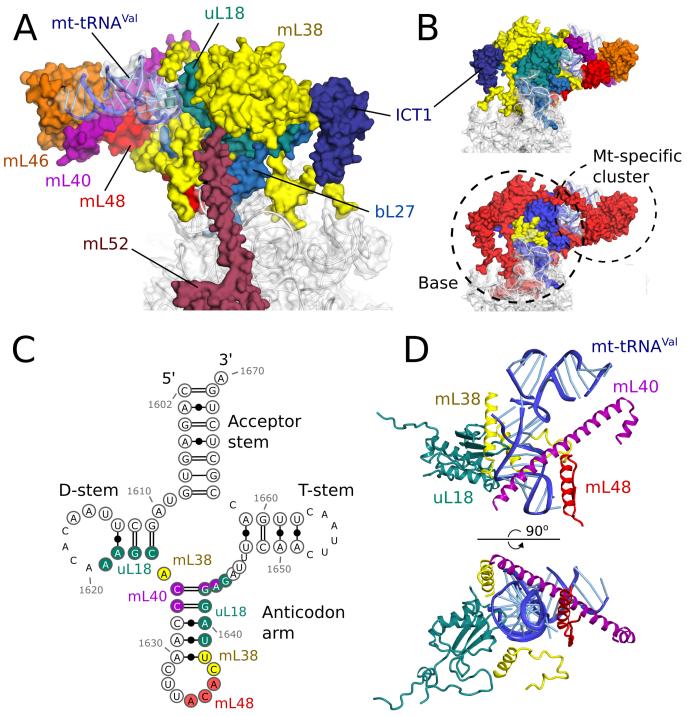
The central protuberance of the human mt-LSU is substantially different from other ribosomes, including the yeast mitoribosome, as a result of the absence of 5S rRNA and its associated proteins (uL5, bL25 and bL31) coupled with the incorporation of mt-tRNAVal and mitochondria-specific proteins (Fig. 3). Despite the remodeling, two functions of the central protuberance are maintained, through entirely mitochondria-specific elements; an interaction with the head of the small subunit, and with tRNAs bound to the ribosome (Fig. S14). This suggests that the interdependence between inter-subunit communication and the fidelity of translation (21) is preserved in mitoribosomes. Due to functional flexibility at the interface, we could not assign the density mediating these functions to specific elements. It is coordinated by an entirely mitochondria-specific cluster (mL40, mL46, mL48) that is bound to the base of the central protuberance (uL18, bL27, mL38, mL52 and ICT1) through mt-tRNAVal (Fig. 3B) and by an unknown mitochondria-specific protein of the small subunit (Fig. S14). This region is likely to undergo structural rearrangement during translation ((reviewed in ref. 22), for which mt-tRNAVal might provide the necessary plasticity. In the yeast mitoribosome this function could be realized through unique rRNA expansion segments (8).
Fig. 3. The central protuberance containing mt-tRNAVal.
(A) Relative locations of proteins and mt-tRNAVal in the central protuberance. (B). View of A rotated by 180°, colored by proteins (top) and conservation (bottom) in accordance with Fig. 1. (C) Secondary structure of mt-tRNAVal. Modeled nucleotides are circled, and those interacting with surrounding proteins are colored. (D) The anticodon arm of mt-tRNAVal (blue) interacts extensively with proteins, whereas the acceptor arm is solvent exposed.
Mt-tRNAVal is located at the top of central protuberance with the acceptor stem exposed to solvent and less well resolved than the anticodon arm, which is ordered due to contact with uL18, mL38, mL40 and mL48 (Fig. 3C-D). mL40 binds through a 70 Å long helix that stretches from the tip of the acceptor arm to the anticodon stem where it makes base-specific interactions in the major groove. uL18, mL38 and mL40 primarily interact with the phosphate backbone. The core architecture of the central protuberance is maintained by mL38, which intertwines with the proteins of the base of the central protuberance and anchors the central protuberance to the rRNA core of the LSU body (Fig. 3), thereby performing a similar role to 5S rRNA in cytoplasmic ribosomes. ICT1 and long helical elements of mL52 and CRIF1 (Fig. 3 and Fig. S15) further connect the central protuberance to the main body. A structurally similar feature of an α-helix bridging the body and central protuberance is seen in the yeast mitoribosome (8).
Remodeling of tRNA binding sites
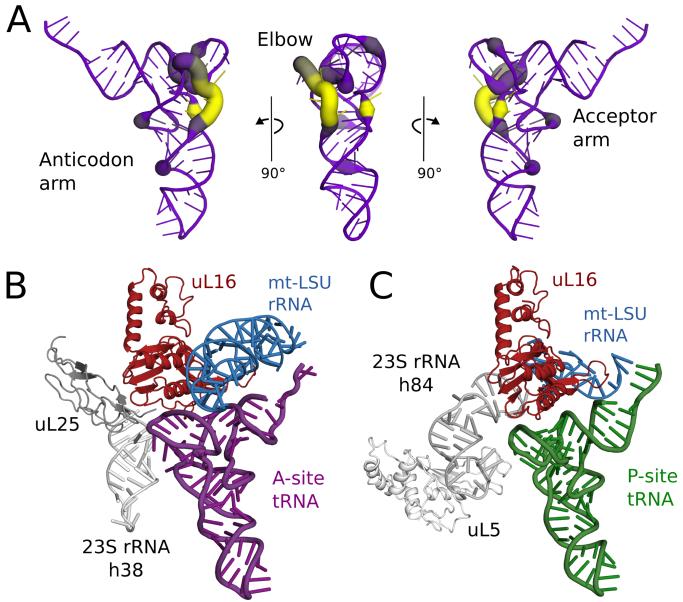
Conventional tRNAs have four-armed cloverleaf secondary structures and L-shaped tertiary structures. However, many human mt-tRNAs have absent or reduced D- and/or T-loops that form the tRNA elbow (Fig. 4A) (23). To accommodate these highly variable loops the tRNA-binding sites have dispensed with elements, common to other ribosomes that interact with the tRNA elbow (Fig. 4B). In the A site, uL25 and the tip of h38 that are responsible for fixing the elbow of A-site tRNA in bacteria (24,25) have been lost. uL25 is also absent in the yeast mitoribosome (8). Similar deletions are observed in the P site, with the loss of the elbow-stabilizing uL5 and h84 (Fig. 4C). The L1 stalk, which controls the dynamics of tRNA ejection (26), also lacks the RNA segments (h76-h77) that bind the elbow of E-site tRNAs (27)(28)(Figs. S8 and S9), although the remodeled stalk is not resolved in our structure. There do not appear to be additional stabilizing interactions at the acceptor stem, suggesting that human mt-tRNAs are less tightly bound to mitoribosomes.
Fig. 4. Co-evolution of mt-tRNAs and their binding sites.
(A) Variability in the elbow region of human mt-tRNAs. The deletion of nucleotides relative to a bacterial tRNA (PDB ID: 2WDI) is shown by line color and thickness, with yellow and thick lines indicating most frequently deleted. (B) Modelling a bacterial A-site tRNA (purple) reveals that uL25 and 23S rRNA h38 (both gray) that stabilize the tRNA elbow region are deleted compared to bacterial ribosomes. (C) Similarly, uL5 and 23S rRNA h84 (both gray) that stabilize the elbow region of P-site tRNA (green), are deleted, but elements that bind the anticodon arm are conserved.
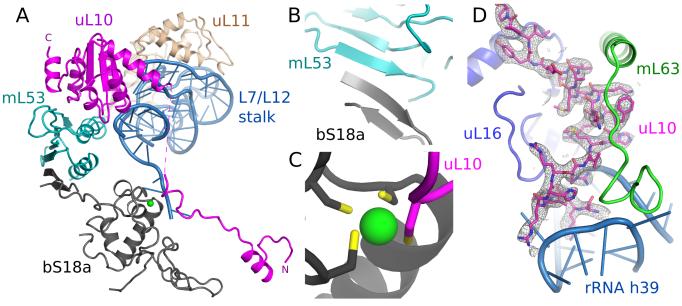
The L7/L12 stalk
The L7/L12 stalk is a large ribosomal protrusion responsible for the recruitment of translation factors, as well as stimulation of factor-dependent GTP hydrolysis. In bacteria it is formed by 23S rRNA (h42-h44), uL10, uL11 and multiple copies of uL12 (29). In the absence of translational factors, the stalk is generally highly flexible and not well resolved in cryo-EM or crystal structures. In our reconstruction, the stalk is partially resolved (Fig. S3B), allowing us to place homology models of uL10 and uL11. The stalk protein-binding platform (h43-h44) is also resolved despite h42, which connects the platform to the main body of the ribosome, being remodeled and flexible.
The increased stability of the L7/L12 stalk is the product of an inter-protein network not observed in other ribosomes (Fig. 5A). Firstly, a mitochondria-specific stalk protein, mL53, bridges uL10 on the top of the stalk with bS18a in the body. The interaction between mL53 and bS18a is mediated through a shared β-sheet (Fig. 5B). Secondly, a mitochondria-specific N-terminal extension of uL10 forms a stable interaction with the main body of the mitoribosome, notably through a shared zinc-binding motif with S18a (Fig. 5C), and further coordinated by a C-terminal extension of uL16, mL63 and the loop of h39 (Fig. 5D). A linker region between the N-terminal extension and the conserved part of uL10 is not fully resolved in the maps suggesting some conformational flexibility is maintained. The differences in the L7/L12 stalk may explain why bacterial EF-G is incompatible with mammalian mitoribosomes (30). Additionally, uL6, which interacts with translational factors in bacteria, is not functionally replaced in the human mt-LSU (Fig. S16).
Fig. 5. Remodeling of the L7/L12 stalk.
(A) Overview of new elements at the L7/L12 stalk. (B) bS18a forms a shared β-sheet with mL53 to connect the stalk to the body of the mitoribosome. (C) The novel N-terminal extension of uL10 contributes a cysteine residue to a shared zinc-binding motif with bS18a. (D) Density for the N-terminal extension of uL10 that is highly co-ordinated to the body of the ribosome.
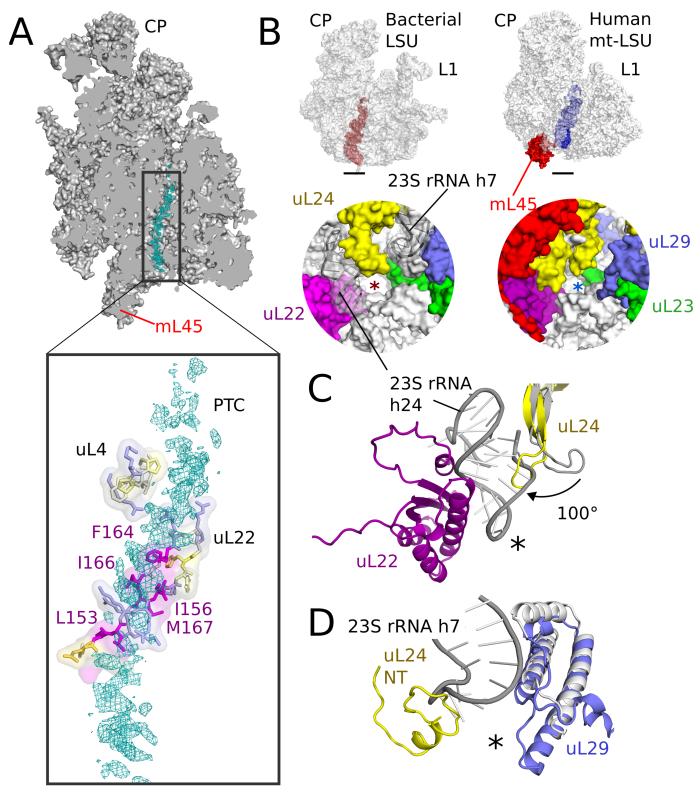
The exit tunnel
The exit tunnel, through which nascent peptides pass before emerging from the ribosome, appears to be adapted for translating hydrophobic membrane proteins. Density for an endogenous polypeptide, or mixture of polypeptides, is seen throughout the exit tunnel and shows clear interactions with hydrophobic residues of the mitoribosomal tunnel wall, mainly from uL22 (Fig. 6A). These residues make the tunnel more hydrophobic than in cytoplasmic ribosomes (13). The hydrophobic nature of both the translated polypeptide and the exit tunnel may explain why a polypeptide remains trapped in the exit tunnel despite the lack of a P-site tRNA to tether it in the ribosome. Although the nascent peptide is better resolved in the upper part of the tunnel, broken density consistent with a helical structure is apparent closer to the exit. Thus helices of mitochondrial OXPHOS proteins may start forming within the mitoribosomal tunnel similar to what has been seen in cytoplasmic ribosomes (13), and the hydrophobic nature of the wall may aid this by mimicking the hydrophobic environment of the membrane that is the eventual site of these proteins. In addition, the increased hydrophobic interactions could act to slow the rate of elongation, allowing more time for transmembrane domains to fold and for assembly of OXPHOS complexes.
Fig. 6. The exit tunnel.
(A) Slice through the mt-LSU showing nascent chain density (cyan) in the exit tunnel. The nascent polypeptide interacts with a β-hairpin of uL22 enriched with hydrophobic residues. (B) The exit tunnel in bacteria (left, red) and human mitoribosomes (right, blue) showing a view of the polypeptide exit site below. The tunnel exit is marked with an asterisk. The polypeptide exit tunnel in mt-LSU is more proteinaceous than in bacteria as a result of two rRNA deletions. (C) Deletion of h7 in bacteria (gray) is compensated by changes to uL29 and an N-terminal extension of uL24. (D) Deletion of h24 (gray) results in the conserved β-hairpin of uL24 rotating closer to the tunnel exit and exposes uL22 to the nascent polypeptide.
The presence of a nascent polypeptide also unambiguously delineates the overall tunnel path, which is similar to that of bacterial and cytoplasmic ribosomes (Fig. 6B) and different from the yeast mitoribosome (8). The alternative exit tunnel observed in yeast results from deletion of rRNA h16-20 and h24 (Fig. S17). Despite similar deletions in domain I of the human mitoribosome, a short segment that replaces h24 (nucleotides 1806-1813) seals this potential exit and precludes a yeast-like tunnel path being formed in human mitoribosomes (Fig. S17). The peptidyl transfer center and upper tunnel of the human mt-LSU are architecturally similar to bacterial ribosomes, and do not show the constriction observed in yeast (8).
The exit site of the mitoribosomal tunnel, where the nascent chain emerges, has two roles: forming a docking platform for maturation factors and tethering mitoribosomes to the inner mitochondrial membrane. In the human mt-LSU, this region is remodeled, with two rRNA deletions (h7 and h24) compensated by extensions and conformational changes of the conserved proteins that line the tunnel walls (Fig. 6B-D). Deletion of h24 has caused a positional change of a β-hairpin of uL24, which exposes uL22 to the exit site, and h7 is partially replaced by uL29 and an N-terminal extension of uL24. Together these changes result in a more proteinaceous exit site than in other ribosomes and allows binding of mL45 that likely anchors the mitoribosome to the inner mitochondrial membrane in a way that would expose the translated nascent polypeptide to solvent (9), making it accessible to specific maturation factors and chaperones involved in the assembly of OXPHOS complexes.
Mitoribosomal mutations have been linked with hereditary mitochondrial diseases (4). These mutations and their potential effect on the structure are shown in Tables S4 and S5 and Fig. S18. Although all these mutations affect the mitoribosome, their effects are varied, suggesting that in each case they work in conjunction with other mutations to produce the disease. The structural information presented here can be used for the rational design of antibiotics with decreased autotoxicity. In addition, since mitoribosomes are involved in redirection of energy metabolism of tumorogenic cells (31], with many mitoribosomal proteins upregulated in cancer (Table S6), the structure might be useful for the development of novel cancer therapeutics.
Materials and Methods
Isolation of mitochondria
In order to overcome problems related to fast turnover, low abundance and heterogeneity of mammalian mitoribosomes traditionally isolated from animal tissues, we used suspension-adapted and tetracycline resistant human embryonic kidney cells lacking N-acetyl-glucosaminyltransferase I (HEK293S TetR GnTI−) (32) as a source of mitochondria. This cell line stably transfected with a transporter has been found to have a beneficial phenotype for large-scale suspension growth using Wave bioreactors (GE Healthcare). This enabled optimization of growth, harvesting and lysis conditions in a controlled environment, which consequently resulted in an increased and more consistent yield of a more homogenous sample.
HEK293 cells were grown in FreeStyle media (Gibco, Life technologies) supplemented with 5% fetal bovine serum (Gibco, Life technologies). The cells were pelleted by centrifugation at 1,000 × g for 7 min and washed with phosphate-buffered saline (PBS) buffer. The pellet was resuspended in homogenization buffer (50 mM Hepes-KOH pH 7.45, 10 mM KCl, 1.5 mM MgOAc, 70 mM sucrose, 210 mM mannitol, 1 mM EDTA, 1 mM EGTA) and lysed with 120 strokes in a 400 ml glass homogenizer. Cell debris and nuclei were separated at 800 × g for 15 min. Crude mitochondria were collected at 10,000 × g for 15 min, treated with DNase and further purified on a sucrose gradient in SEM buffer (250 mM sucrose, 20 mM Hepes-KOH pH 7.45, 1 mM EDTA). Mitochondria were pooled and stored at −80°C.
Purification of Mitoribosomes
To avoid loss of proteins and potential degradation, we adopted an approach of rapid and gentle purification, keeping low salt and high magnesium concentrations throughout, as described below. Intact mitoribosomes were purified within 28 hours and used immediately for grid preparation. We avoided biochemical separation of mitoribosomal subunits as it requires harsher conditions and results in preferential orientation of particles on the grid, thereby impairing the quality of the map.
To purify the mitoribosomes, 4 volumes of Lysis buffer (25 mM Hepes-KOH pH 7.45, 100 mM KCl, 25 mM MgOAc, 1.7% Triton X-100, 2 mM DTT) were added to purified mitochondria and incubated for 15 min at 4°C. The membranes were then separated by centrifugation at 30,000 × g for 20 min. The supernatant was loaded on a 1 M sucrose cushion in buffer (20 mM Hepes-KOH pH 7.45, 100 mM KCl, 20 mM MgOAc, 1% Triton X-100, 2 mM DTT). The resuspended pellet was then loaded onto a 10%-25% sucrose gradient in the same buffer without Triton X-100 and run for 16 h at 85,000 × g. Fractions corresponding to mitoribosomes were collected and sucrose removed by buffer exchange, also in the same buffer.
Identification of mt8tRNAVal
The mitochondrial subunits were separated using sucrose gradients and total rRNA was extracted from the mt-LSU using phenol-chloroform and precipitated using ethanol. The small RNA was separated from LSU rRNA on a 12% denaturating polyacrylamide gel, excised and eluted overnight at 4°C. A library was prepared using the TruSeq small RNA sample preparation kit (Illumina, Ca) and sequenced with single-end reads on the Illumina MiSeq system (Edinburgh Genomics, UK). The sequenced data are available on a webserver: http://genepool.bio.ed.ac.uk/seqdata/2014037_Ramakrishnan_Venki/. The adaptor sequence was trimmed using the FASTX-Toolkit (33) and reads shorter than 25 nucleotides were discarded. A total of 4,374,074 reads were of sufficient length for further processing. To detect the genomic source of the rRNA, the reads were mapped to human transcripts from the Ensembl database (release 67) (34) using bowtie aligner v. 0.12.7 (35). Overall, 3,361,202 reads were successfully aligned, with the majority corresponding to tRNAVal encoded by the mitochondrial chromosome (Fig. 2). However, this preliminary analysis revealed that in some cases tRNA genes annotated by Ensembl are not complete, including mt-tRNAVal. Thus, to further confirm the identity of the RNA molecule, the reads were mapped to the human mitochondrial genome tRNA database (36). Since most of the reads ended with CCA, this sequence was also added to the target database. Out of 3,022,966 aligned reads, 2,235,105 were found to correspond to mt-tRNAVal (Fig. 2).
Electron microscopy
Aliquots of 3 μl of purified mitochondrial ribosomes at a concentration of ~80 nM (0.23 mg/ml) were incubated for 30 s on glow-discharged holey carbon grids (Quantifoil R2/2), onto which a home-made continuous carbon film (estimated to be ~50 Å thick) had previously been deposited. Grids were blotted for 2.5 s in 100% ambient humidity and flash frozen in liquid ethane using an FEI Vitrobot. Grids were transferred to an FEI Titan Krios electron microscope operated at 300 kV. Images were collected using FEI automated single particle acquisition software (EPU) and recorded on a back-thinned FEI Falcon II detector at a calibrated magnification of 104,478 (yielding a pixel size of 1.34 Å). An in-house system (37) was used to intercept the videos from the detector at a rate of 17 frames for the 1 s exposures. Defocus values in the final data set ranged from 1.5-3.5 μm.
Image processing
323,292 particles were picked from 1,521 micrographs using semi-automated particle picking with EMAN2 (38). Contrast transfer function parameters were estimated using CTFFIND3 (39). All 2D and 3D classifications and refinements were performed using RELION (12). We used reference-free 2D class averaging to discard bad particles, and initiated 3D refinements with a 60 Å low-pass filtered cryo-EM reconstruction of the yeast mitoribosome. Intact human mitoribosomes were sorted from poorly aligned particles using 3D classification (Fig. S1). 107,679 particles were used for the final reconstruction. In order to correct for beam-induced movements, particle ‘polishing’ was performed using statistical movie processing in RELION (37, 40). For these calculations, we used running averages of 5 movie frames, and a standard deviation of 1 pixel for the translational alignment. To further increase the accuracy of the per-particle movement correction, we fit linear tracks through the optimal translations for all running averages, and included neighboring particles on the micrograph in these fits. In addition, we employed a resolution and dose-dependent model for the radiation damage, where each frame is weighted with a different Bfactor as estimated from single-frame reconstructions (40). This yielded a map with an overall resolution of 3.8 Å. A soft mask was applied to the mt-LSU to improve map quality and overall resolution to 3.4 Å (8). This in silico step is preferable to microscopy on the isolated subunit, which shows preferential orientation on the EM grids. It also reflects the conformation of the LSU in the intact ribosome. Local resolution was calculated using ResMap (41) (Fig. S2). Reported resolutions are based on the gold-standard FSC=0.143 criterion (42). FSC curves were corrected for the effects of a soft mask using high-resolution noise substitution (43). Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector, and then sharpened by applying a B-factor (of −90 Å2) that was estimated using automated procedures (44).
Model building and refinement
For the interpretation of the structure with a model, the revised Cambridge Reference Sequence (rCRS) (45) is used to number the mtDNA-encoded nucleic acid components of the mitoribosome. Ribosomal proteins are named following the new nomenclature (8, 46) and rRNA helices are numbered corresponding to those in Escherichia coli (47).
Initially, the model of Sus scorfa mt-LSU (PDB ID: 4CE4) was placed into the density map of human mt-LSU using Chimera (48). Unassigned helices present in the model were excluded. Each chain of the porcine model was subjected to Jiggle Fit in Coot v0.8 (8) to overcome local differences in protein and rRNA positions. The sequences of each of the porcine proteins were aligned with the sequences of the human homologs obtained from UniProt (49) using Clustal Omega (50). The porcine models were then mutated and renumbered to that of the human sequences. The same procedure was applied to conserved mitoribosomal proteins identified in the yeast mt-LSU (PDB ID: 3J6B), but not in the porcine mt-LSU (mL40, mL41, mL43, mL46 and mL50). The models were then manually rebuilt using sidechain information present in the density map and extensions built de novo in Coot (51). Initial models for the L7/L12 stalk proteins uL10 and uL11 were obtained from the structure of Thermus thermophilus 70S ribosome bound to EF-G (PDB ID: 4JUX) in which these proteins are ordered (52).
The sequence of mt-LSU (16S) rRNA was obtained from the revised Cambridge Reference Sequence (rCRS) for human mitochondrial DNA (NCBI ID: NC_012920.1) (45) and aligned with the rRNA sequence obtained from the model of porcine mt-LSU. Conserved regions without insertions or deletions were extracted from the porcine model, mutated and renumbered. These conserved sections were then connected by de novo building of RNA. The rRNA was then manually rebuilt in Coot and differences between the human and porcine rRNA made when necessary.
To locate supernumerary proteins not previously identified (mL37, mL42, mL51, mL48, mL53, mL63, CRIF1, mS30, and S18a), the remaining density was populated with idealized secondary structure elements (SSEs) that were subsequently connected by poly-alanine traces. The length and connectivity of SSEs were compared with predicted secondary structure patterns to unambiguously assign sequences. The positions of bulky sidechains were used both for determination and validation of the correct assignment.
Restrained refinement was performed in REFMAC v5.8 modified for optimal fit to EM density maps (8, 53). For human mitoribosomal proteins with bacterial homologs, main-chain interatomic distance restraints were derived from a reference structure (PDB ID: 3V2D) using ProSMART (54). For all proteins, ProSMART was also used to generate idealized helical restraints and hydrogen-bond restraints for β-sheets. These reference and secondary structure restraints were maintained throughout refinement. Base pair and stacking restraints, generated using the program LIBG (8), were also applied. FSCaverage was monitored during refinement to follow the fit-to-density, and the final model was validated using MolProbity (55). Cross-validation against over-fitting was calculated as previously described (8, 56). Model building and refinement statistics are given in table S1.
Structure analysis
The intra-ribosomal protein-protein interaction network was mapped as an approximate two-dimensional representation of the three-dimension organization of the proteins in the human mt-LSU using CytoScape (57). Node sizes represent protein molecular weights calculated excluding predicted mitochondrial-targeting peptides. The edge thickness represents the buried surface area between proteins calculated using PDBePISA (58).
To visualize the exit tunnel, HOLLOW (59) was used to fill the channel volume with dummy atoms defined on a 0.5 Å grid. An interior probe of 2.5 Å was used to explore the cavity and a probe of 15 Å used to roll over the surface to define depressions.
Figures
All figures were generated using PyMOL (60) or Chimera (48). The secondary structure diagram of the human mitoribosomal RNA was prepared by extracting base pairs from the model using DSSR (61). The secondary structure diagram was drawn in VARNA (62) and finalized in Inkscape.
Supplementary Material
Acknowledgments
We thank Y. Gordiyenko for help with rRNA purification, S. Peak-Chew and M. Skehel for mass spectrometry, G. McMullan, X. Chen, and C. Savva for help with data collection, J. Grimmett and T. Darling for help with computing and I. S. Fernández and J. L. Llácer for stimulating discussions. This work was funded by grants from the UK Medical Research Council (MC_U105184332 to VR and MC_UP_A025_1013 to SHWS); a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute and the Jeantet Foundation (VR); fellowships from Human Frontiers Science Program (AA), and EU FP7 Marie Curie (XB). Cryo-EM density maps have been deposited with the EMDB (accession number EMD-2762) and coordinates have been deposited with the PDB (entry code 3J7Y).
References
- 1.O’Brien T. Properties of Human Mitochondrial Ribosomes. IUBMB Life. 2003;55:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 2.Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Bio Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 3.Skladal D. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- 4.Boczonadi V, Horvath R. Mitochondria: Impaired mitochondrial translation in human disease. Int J Biochem Cell Biol. 2014;48:77–84. doi: 10.1016/j.biocel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotgia F, et al. Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell cycle. 2012;11:4390–4401. doi: 10.4161/cc.22777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skrtić M, et al. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer cell. 2011;20:674–688. doi: 10.1016/j.ccr.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh R, Sripada L, Singh R. Side effects of antibiotics during bacterial infection: Mitochondria, the main target in host cell. Mitochondrion. 2014;16:50–54. doi: 10.1016/j.mito.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greber BJ, et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- 10.Kaushal PS, et al. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci USA. 2014;111:7284–7289. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SOM, See supplementary materials online.
- 12.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voorhees RM, Fernández IS, Scheres SHW, Hegde RS. Structure of the Mammalian Ribosome-Sec61 Complex to 3.4 Å Resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lightowlers RN, Rozanska A, Chrzanowska-Lightowlers ZM. Mitochondrial protein synthesis: Figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014;588:2496–2503. doi: 10.1016/j.febslet.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koc EC, et al. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Letters. 2001;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 16.Koc EC. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. Journal of Biological Chemistry. 2001;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 17.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 18.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 19.Liao D. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. Journal of Molecular Evolution. 2000;51:305–317. doi: 10.1007/s002390010093. [DOI] [PubMed] [Google Scholar]
- 20.D’Onorio De Meo P, et al. MitoZoa 2.0: a database resource and search tools for comparative and evolutionary analyses of mitochondrial genomes in Metazoa. Nucleic acids research. 2012;40:D1168–D1172. doi: 10.1093/nar/gkr1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodin MH, Rakauskaite R, Dinman JD. The central core region of yeast ribosomal protein L11 is important for subunit joining and translational fidelity. Mol Genet Genomics. 2011;285:505–516. doi: 10.1007/s00438-011-0623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noeske J, Cate JHD. Structural basis for protein synthesis: snapshots of the ribosome in motion. Current Opinion in Structural Biology. 2012;22:743–749. doi: 10.1016/j.sbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helm M, et al. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17:1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 26.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann J, Jossinet F, Gautheret D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res. 2013;41:5494–5502. doi: 10.1093/nar/gkt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J Mol Biol. 2007;371:1047–1059. doi: 10.1016/j.jmb.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 30.Chung HK, Spremulli LL. Purification and characterization of elongation factor G from bovine liver mitochondria. J Biol Chem. 1990;265:21000–21004. [PubMed] [Google Scholar]
- 31.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci U S A. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. http://hannonlab.cshl.edu/fastx_toolkit/
- 34.Flicek P, et al. Ensembl 2012. Nucleic acids research. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jühling F, et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic acids research. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai X-C, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature genetics. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 46.Ban N, et al. A new system for naming ribosomal proteins. Current Opinion in Structural Biology. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maly P, Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Research. 1983;11:7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.U. Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Research. 2013;41:D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta crystallographica. Section D, Biological crystallography. 2012;68:404–417. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez IS, Bai X-C, Murshudov G, Scheres SHW, Ramakrishnan V. Initiation of Translation by Cricket Paralysis Virus IRES Requires Its Translocation in the Ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeLano WL. The PyMOL molecular graphics system. 2002. [Google Scholar]
- 61.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nature protocols. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica. Section D, Biological crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 64.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Research. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Surovtseva YV, et al. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Nat Acad Sci. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoo YA, et al. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Molecular and cellular biology. 2005;25:6603–6616. doi: 10.1128/MCB.25.15.6603-6616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conde JA, et al. Identification of a motif in BMRP required for interaction with Bcl-2 by site-directed mutagenesis studies. Journal of cellular biochemistry. 2012;113:3498–3508. doi: 10.1002/jcb.24226. [DOI] [PubMed] [Google Scholar]
- 68.Koc EC. The Large Subunit of the Mammalian Mitochondrial Ribosome. ANALYSIS OF THE COMPLEMENT OF RIBOSOMAL PROTEINS PRESENT. Journal of Biological Chemistry. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 69.Polianskyte Z, et al. LACTB is a filament-forming protein localized in mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18960–18965. doi: 10.1073/pnas.0906734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang HJ, Hong YB, Kim HJ, Bae I. CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J Biol Chem. 2010;285:21258–21268. doi: 10.1074/jbc.M109.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suh JH, Shong M, Choi HS, Lee K. CR6-interacting factor 1 represses the transactivation of androgen receptor by direct interaction. Mol Endocrinol. 2008;22:33–46. doi: 10.1210/me.2007-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kwon MC, et al. Crif1 is a novel transcriptional coactivator of STAT3. EMBO J. 2008;27:642–653. doi: 10.1038/sj.emboj.7601986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung HK, et al. CR6-interacting factor 1 interacts with Gadd45 family proteins and modulates the cell cycle. J Biol Chem. 2003;278:28079–28088. doi: 10.1074/jbc.M212835200. [DOI] [PubMed] [Google Scholar]
- 74.Kwon MC, et al. Essential role of CR6-interacting factor 1 (Crif1) in E74-like factor 3 (ELF3)-mediated intestinal development. J Biol Chem. 2009;284:33634–33641. doi: 10.1074/jbc.M109.059840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.2013 http://www.mitomap.org.
- 76.Tiranti V, et al. A novel mutation in the mitochondrial tRNAVal gene associated with a complex neurological presentation. Annals of Neurology. 1998;43:98–101. doi: 10.1002/ana.410430116. [DOI] [PubMed] [Google Scholar]
- 77.Sacconi S, et al. Complex neurologic syndrome associated with the G1606A mutation of mitochondrial DNA. Archives of neurology. 2002;59:1013–1015. doi: 10.1001/archneur.59.6.1013. [DOI] [PubMed] [Google Scholar]
- 78.McFarland R, et al. Multiple neonatal deaths due to a homoplasmic mitochondrial DNA mutation. Nature genetics. 2002;30:145–146. doi: 10.1038/ng819. [DOI] [PubMed] [Google Scholar]
- 79.Schaefer AM, McFarland R, Blakely EL, He L. Prevalence of mitochondrial DNA disease in adults. Annals of Neurology. 2008;63:35–39. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 80.Arredondo JJ, et al. Mitochondrial tRNA valine as a recurrent target for mutations involved in mitochondrial cardiomyopathies. Mitochondrion. 2012;12:357–362. doi: 10.1016/j.mito.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 81.Horváth R, et al. Heteroplasmic mutation in the anticodon-stem of mitochondrial tRNA(Val) causing MNGIE-like gastrointestinal dysmotility and cachexia. Journal of neurology. 2009;256:810–815. doi: 10.1007/s00415-009-5023-8. [DOI] [PubMed] [Google Scholar]
- 82.Glatz C, D’Aco K, Smith S, Sondheimer N. Mutation in the mitochondrial tRNAVal causes mitochondrial encephalopathy, lactic acidosis and stroke-like episodes. Mitochondrion. 2011;11:615–619. doi: 10.1016/j.mito.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Coo IF, et al. A mitochondrial tRNA(Val) gene mutation (G1642A) in a patient with mitochondrial myopathy, lactic acidosis, and stroke-like episodes. Neurology. 1998;50:293–295. doi: 10.1212/wnl.50.1.293. [DOI] [PubMed] [Google Scholar]
- 84.Taylor RW, et al. MELAS associated with a mutation in the valine transfer RNA gene of mitochondrial DNA. Annals of Neurology. 1996;40:459–462. doi: 10.1002/ana.410400318. [DOI] [PubMed] [Google Scholar]
- 85.del Mar O’Callaghan M, et al. New mitochondrial DNA mutations in tRNA associated with three severe encephalopamyopathic phenotypes: neonatal, infantile, and childhood onset. Neurogenetics. 2012;13:245–250. doi: 10.1007/s10048-012-0322-0. [DOI] [PubMed] [Google Scholar]
- 86.Chalmers RM, et al. A mitochondrial DNA tRNAVal point mutation associated with adult-onset Leigh syndrome. Neurology. 1997;49:589–592. doi: 10.1212/wnl.49.2.589. [DOI] [PubMed] [Google Scholar]
- 87.Mostafaie N, et al. Mitochondrial genotype and risk for Alzheimer’s disease: cross-sectional data from the Vienna-Transdanube-Aging “VITA” study. Journal of neural transmission. 2004;111:1155–1165. doi: 10.1007/s00702-004-0161-8. [DOI] [PubMed] [Google Scholar]
- 88.Menotti F, et al. A novel mtDNA point mutation in tRNA(Val) is associated with hypertrophic cardiomyopathy and MELAS. Italian heart journal. 2004;5:460–465. [PubMed] [Google Scholar]
- 89.Tanji K, et al. A novel tRNAVal mitochondrial DNA mutation causing MELAS. Journal of the Neurological Sciences. 2008;270:23–27. doi: 10.1016/j.jns.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blakely EL, et al. Childhood neurological presentation of a novel mitochondrial tRNA(Val) gene mutation. Journal of the Neurological Sciences. 2004;225:99–103. doi: 10.1016/j.jns.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 91.Fliss MS, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 92.Jones JB, et al. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 93.Nishigaki Y, Marti R, Copeland WC, Hirano M. Site-specific somatic mitochondrial DNA point mutations in patients with thymidine phosphorylase deficiency. J Clin Invest. 2003;111:1913–1921. doi: 10.1172/JCI17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taylor RW, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Polyak K, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 96.Liu Z, et al. The novel mitochondrial 16S rRNA 2336T>C mutation is associated with hypertrophic cardiomyopathy. J Med Genet. 2014;51:176–184. doi: 10.1136/jmedgenet-2013-101818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang S, Batra A, Zhang Y, Ebenroth ES, Huang T. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion. 2010;10:350–357. doi: 10.1016/j.mito.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 98.Porcelli AM, et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet. 2010;19:1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- 99.Tang J, Qi Y, Bao XH, Wu XR. Mutational analysis of mitochondrial DNA of children with Rett syndrome. Pediatr Neurol. 1997;17:327–330. doi: 10.1016/s0887-8994(97)00151-3. [DOI] [PubMed] [Google Scholar]
- 100.Bayat A, et al. Identification of a novel mitochondrial mutation in Dupuytren’s disease using multiplex DHPLC. Plast Reconstr Surg. 2005;115:134–141. [PubMed] [Google Scholar]
- 101.Anderson ER, Burmester JK, Caldwell MD. Evaluation of a mitochondrial DNA mutation in maternally inherited and sporadic cases of Dupuytren disease. Clinical medicine and research. 2012;10:122–126. doi: 10.3121/cmr.2012.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeronimo C, et al. Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene. 2001;20:5195–5198. doi: 10.1038/sj.onc.1204646. [DOI] [PubMed] [Google Scholar]
- 103.Lorenc A, et al. Homoplasmic MELAS A3243G mtDNA mutation in a colon cancer sample. Mitochondrion. 2003;3:119–124. doi: 10.1016/S1567-7249(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 104.Zaki EA, et al. Two common mitochondrial DNA polymorphisms are highly associated with migraine headache and cyclic vomiting syndrome. Cephalalgia. 2009;29:719–728. doi: 10.1111/j.1468-2982.2008.01793.x. [DOI] [PubMed] [Google Scholar]
- 105.Coulbault L, et al. A novel mutation 3090 G>A of the mitochondrial 16S ribosomal RNA associated with myopathy. Biochem Biophys Res Commun. 2007;362:601–605. doi: 10.1016/j.bbrc.2007.08.040. [DOI] [PubMed] [Google Scholar]
- 106.Hsieh RH, Li JY, Pang CY, Wei YH. A novel mutation in the mitochondrial 16S rRNA gene in a patient with MELAS syndrome, diabetes mellitus, hyperthyroidism and cardiomyopathy. J Biomed Sci. 2001;8:328–335. doi: 10.1007/BF02258374. [DOI] [PubMed] [Google Scholar]
- 107.Shoffner JM, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 108.Galmiche L, et al. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Human mutation. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- 109.Serre V, et al. Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochimica et Biophysica Acta. 2013;1832:1304–1312. doi: 10.1016/j.bbadis.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carroll CJ, et al. Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. Journal of medical genetics. 2013;50:151–159. doi: 10.1136/jmedgenet-2012-101375. [DOI] [PubMed] [Google Scholar]
- 111.Nolden M, et al. The m-AAA Protease Defective in Hereditary Spastic Paraplegia Controls Ribosome Assembly in Mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 112.Valles I, et al. Identification of novel deregulated RNA metabolism-related genes in non-small cell lung cancer. PLoS One. 2012;7:e42086. doi: 10.1371/journal.pone.0042086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ribeiro R, et al. Obesity and prostate cancer: gene expression signature of human periprostatic adipose tissue. BMC medicine. 2012;10:108. doi: 10.1186/1741-7015-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim JC, et al. Gene expression profiling: canonical molecular changes and clinicopathological features in sporadic colorectal cancers. World journal of gastroenterology: WJG. 2008;14:6662. doi: 10.3748/wjg.14.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Best CJM, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clinical Cancer Research. 2005;11:6823–6834. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang X, et al. Cell cycle arrest of Jurkat cells by leukemic bone marrow stromal cells: possible mechanisms and involvement of CRIF1. Transplant Proc. 2011;43:2770–2773. doi: 10.1016/j.transproceed.2011.05.048. [DOI] [PubMed] [Google Scholar]
- 117.Huang CC, et al. Concurrent gene signatures for han chinese breast cancers. PLoS One. 2013;8:e76421. doi: 10.1371/journal.pone.0076421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Y, et al. Up-regulation of mitochondrial antioxidation signals in ovarian cancer cells with aggressive biologic behavior. Journal of Zhejiang University SCIENCE B. 2011;12:346–356. doi: 10.1631/jzus.B1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lin Y-W, Aplan PD. Gene expression profiling of precursor T-cell lymphoblastic leukemia/lymphoma identifies oncogenic pathways that are potential therapeutic targets. Leukemia. 2007;21:1276–1284. doi: 10.1038/sj.leu.2404685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Casari G, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.