Abstract
Downregulation and mutations of the nuclear-architecture proteins Lamin A and C cause misshapen nuclei and altered chromatin organization associated with cancer and laminopathies, including the premature-aging disease Hutchinson-Gilford progeria syndrome (HGPS). Here, we identified the small molecule “Remodelin” that improved nuclear architecture, chromatin organization and fitness of both human Lamin A/C depleted cells and HGPS-derived patient cells, and decreased markers of DNA damage in these cells. Using a combination of chemical, cellular and genetic approaches, we identified the acetyl-transferase protein NAT10 as the target of Remodelin that mediated nuclear shape rescue in laminopathic cells via microtubule reorganization. These findings provide insights into how NAT10 affects nuclear architecture, and suggest alternative strategies for treating laminopathies and aging.
Mutations in LMNA, which encodes Lamin A and C, cause pathologies known as laminopathies (1), including the accelerated-aging disease Hutchinson-Gilford progeria syndrome (HGPS) (2, 3). Deregulation of A-type lamins expression or localization also occurs in various cancers (4-6). Lamin A/C depletion or LMNA mutations cause enlarged, misshapen nuclei and altered chromatin organization (7). While some of these pathologies might reflect cell fragility, many likely result from downstream effects on chromatin structure, gene expression, DNA replication or DNA repair. Improving nuclear architecture of laminopathic cells can ameliorate defects in chromatin structure and other processes, thus improving some disease phenotypes (8, 9).
Lamin A/C depletion by small interfering RNA (siRNA) transfection (siLMNA) caused nuclear-shape defects (Fig. 1A, B, S1A, B), global chromatin relaxation and increased nuclear area (Fig. S1C, D). We therefore reasoned that modulating chromatin organization by lysine acetyltransferase (KAT) or lysine deacetylase (KDAC) inhibition might improve nuclear architecture defects of siLMNA cells. Compound screening identified the KAT inhibitor 4-(4-chlorophenyl)-2-(2-cyclopentylidenehydrazinyl)thiazole (1) (Fig. 1C) that restored nuclear circularity and global chromatin compaction in siLMNA cells (Fig. 1D, E, S1E-G). Although molecule 1 was identified in Saccharomyces cerevisiae as a GCN5 (General control of amino acid synthesis protein 5-like 2) network inhibitor (10), nuclear shape rescue was GCN5 independent, because the benchmark GCN5 inhibitor MB-3 had no effect on nuclear circularity. Complete nuclear-shape rescue occurred within 12 hours of treatment (Fig. 1F), independently of mitosis (Fig. S2A and Movie S1) and without markedly affecting the cell cycle (Fig. S2B, C). Moreover, compound 1 improved the nuclear morphology of several cancer cell lines displaying reduced Lamin A/C expression (Fig. S2D, E), indicating that its effects were not specific to siRNA-mediated Lamin A/C depletion.
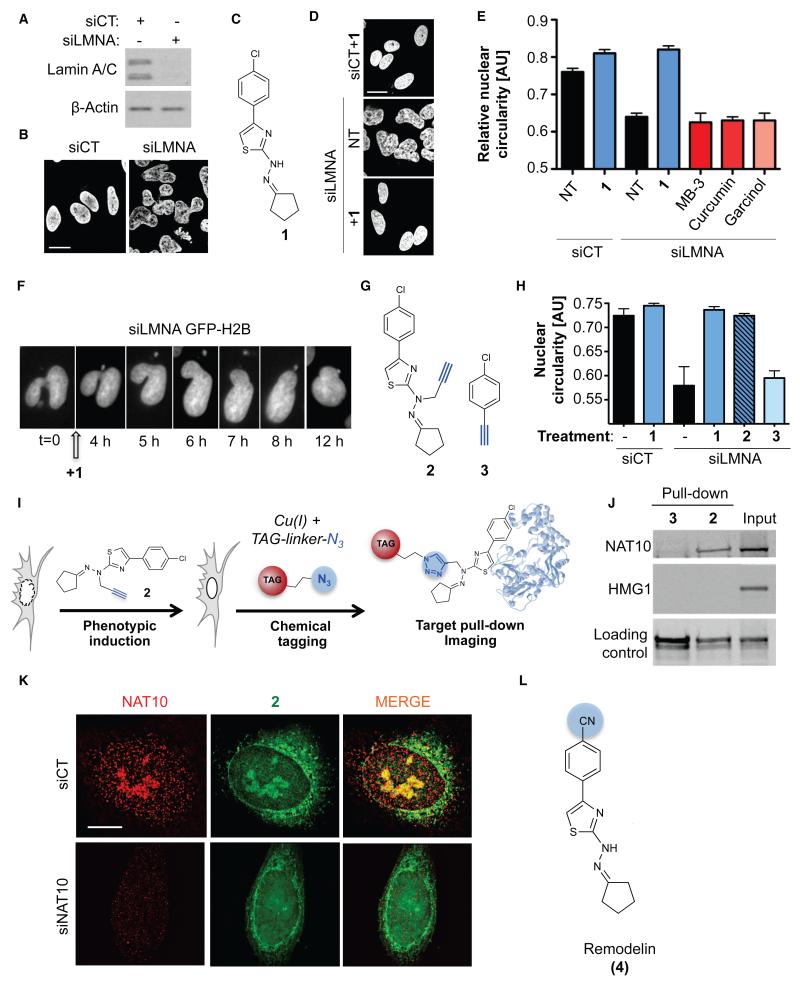
Figure 1. A small molecule restores nuclear shape in Lamin A/C depleted cells and targets the acetyltransferase NAT10.
A) Lamin A/C depletion (siLMNA) in U2OS cells compared to negative control (siCT). B) Nuclear shape observed by DAPI staining. C) Molecular structure of 4-(4-chlorophenyl)-2-(2-cyclopentylidenehydrazinyl)thiazole (1). D) Nuclear shape rescue observed by DAPI staining after treatment with 1. E) Quantification of nuclear circularity in non-treated (NT) cells or cells treated with the indicated compounds (means of three independent experiments with n>212 ± s.d.). F) Live imaging pictures of nuclear shape rescue in GFP-H2B expressing U2OS cells transfected with siLMNA and treated with 1. G) Molecular structure of clickable analogue 2 and clickable inactive control molecule 3. H) Quantification of U2OS nuclear circularity (means of three independent experiments with n>224 ± s.d.) I) Principle of click-chemistry strategy for small molecule tagging. J) Pull-down of clickable molecules 2 and 3 pre-incubated in U2OS cells and analysis of bound proteins. K) Representative high-resolution microscopy pictures of NAT10 (red) and fluorescently labeled 2 (green) in control or NAT10 depleted cells (siNAT10). Scale bars: 10 μm. L) Molecular structure of Remodelin (4), a stable and more potent analog of 1.
To identify putative targets of 1, we synthesized a “clickable” cell-active analog 2 and used inactive molecule 3 as a negative control (Fig. 1G, H). We then employed “click-chemistry” to retrieve and validate drug-associated proteins. The alkyne “click” moiety selectively reacts with an azide group upon copper exposure, allowing tagging of the “clickable” molecules in cells (Fig. 1I). First, we used a biotinylated derivative of 2 (Fig. S3A), retrieved associated proteins with streptavidin beads, and identified several protein species whose staining intensities were reduced by an excess of competitor 1 (Fig. S2B). These proteins were then identified by mass spectrometry LC-MS/MS (Fig. S2C). N-acetyltransferase 10 (NAT10) was the only KAT protein identified, thus being the only likely relevant target of 1. NAT10 was previously linked with the SUN1 nuclear envelope protein (11), whose depletion rescues nuclear shape in LMNA KO cells (8), and NAT10 KAT activity has been demonstrated towards microtubules and histones (12).
Pre-incubating clickable molecules with live cells followed by click pull-down identified NAT10 as a specific target of 2 in vivo (Fig. 1J), thereby establishing our protocol as a framework for identifying specific partners without photo-crosslinking agents (13). In parallel, we visualized sub-cellular localizations of clickable molecules by fluorescence microscopy (14) (Fig. 1K). This revealed that 2 specifically accumulated in nucleoli and also localized at the nuclear periphery and in the cytoplasm (Fig. 1K and Fig. S4A). Corroborating the binding studies, this distribution overlapped with that of NAT10, and moreover, NAT10 depletion (Fig. S4B) led to a marked reduction of molecule 2 in the nucleolus (Fig. 1K) without changes in nucleolar architecture (Fig. S4C reveals that NAT10 localization was not affected by treatment with 1 or siLMNA). Furthermore, we established direct physical interaction between 1 and NAT10 using circular dichroism spectroscopy (Fig. S4D). Because molecule 1 was rapidly degraded upon light or air exposure, we explored its structural derivatives for their ability to rescue nuclear shape of siLMNA cells (Fig. S5A). The most potent and stable analog identified (4; Fig. S5B) was named “Remodelin” based on its ability to remodel nuclear architecture of siLMNA cells (Fig. 1L).
Consistent with a model in which Remodelin impacts on siLMNA cells through NAT10 targeting, we found that NAT10 depletion (Fig. 2A) corrected the aberrant nuclear morphology of siLMNA cells (Fig. 2B). Because 1, the analog of Remodelin, is a KAT inhibitor, we assessed whether Remodelin affected NAT10 KAT activity. Aligning human NAT10 with the 3D structure of its bacterial homolog in complex with acetyl-CoA (15) led us to mutate conserved glycine-641 to glutamate (G641E), predicted to impair acetyl-CoA binding (Fig. 2C, D, S6A). Indeed, this mutation blocked NAT10 activity (Fig. 2E, S6B). Moreover, wild-type NAT10 activity was inhibited by Remodelin or clickable molecule 2, establishing Remodelin as a bona fide NAT10 inhibitor. Complementation assays with siRNA-resistant wild-type (NAT10 WT) or G641E mutant NAT10 (NAT10 MUT) (Fig. S6C, D) showed that rescue of nuclear circularity in siLMNA cells required NAT10 catalytic function (Fig. 2F). Thus, inactivating NAT10 KAT activity by mutation or by Remodelin restores normal nuclear morphology in siLMNA cells.
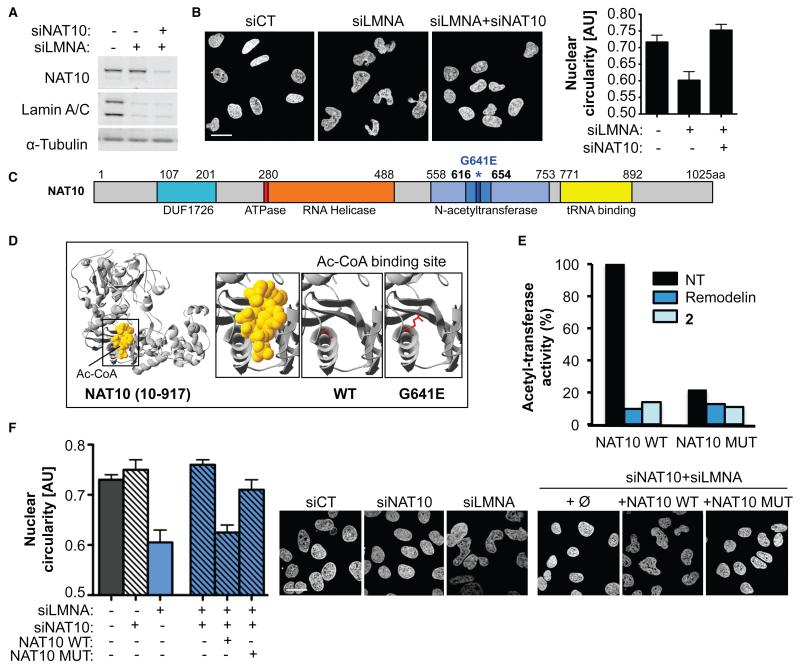
Figure 2. Inhibiting NAT10 activity by Remodelin mediates nuclear shape rescue of LMNA depleted cells.
A) NAT10 and Lamin A/C depletion in U2OS cells. B) Nuclear shape visualized by DAPI staining (left) and quantification of nuclear circularity (right; means of three independent experiments with n>267 ± s.d.). Scale bar: 20 μm. C) Representation of NAT10 with its known domains. The G641E mutation identified in D) is indicated in dark blue and asterisked. D) Modelled 3D structure of human NAT10 residues 10-917 showing the acetyl-CoA binding site (left) and disruption of Ac-CoA binding by NAT10 G641E mutation (right) visualized with Swiss-Prot PDB Viewer. E) In vitro acetylation assay showing the activity of NAT10 towards tubulin. F) Quantification of nuclear circularity (left) in cells stably expressing siRNA-resistant FLAG-NAT10 WT or FLAG-NAT10 G641E (NAT10 MUT; means of three independent experiments with n>198 ± s.d.) and nuclear shape visualized by DAPI staining (right). Scale bar: 20 μm.
We next investigated the effects of Remodelin on cells derived from LMNA-mutated HGPS patients (AG11498 and AG06297). HGPS cells accumulate Progerin, a permanently farnesylated, truncated form of Lamin A, leading to nuclear membrane folding and nuclear blebbing (Fig. 3A) that contribute to the premature-aging phenotypes of HGPS patients. Remodelin significantly reduced the prevalence of misshapen nuclei in HGPS cells (Fig. 3B) as well as in primary MRC5 fibroblasts aged in culture (Fig. S7A, B), which also accumulate Progerin upon extended passaging (16). By contrast, Remodelin had no effect on non-laminopathic Werner syndrome cells (Fig. S7C, D). Farnesyltransferase inhibitors (FTIs) prevent accumulation of farnesylated Progerin at the nuclear membrane, thus reducing HGPS nuclear blebbing (17). Unlike FTIs, however, Remodelin did not trigger pre-Lamin A accumulation (Fig. S8A), indicating that Remodelin is not an FTI. Remodelin and FTI did not act synergistically on nuclear shape improvement, however, suggesting that they impact on a common pathway (Fig. S8B, C). Unlike Remodelin, FTI did not improve nuclear morphology in siLMNA cells. Furthermore, although FTI improves HGPS nuclear shape, it had the opposite effect on Progerin-negative cells, (Fig S8B, C) probably by causing accumulation of unprocessed Lamin A and B and centrosome separation defects (18-20). By contrast, Remodelin prevented FTI-induced nuclear shape defects in normal fibroblasts and U2OS cells (Fig. S8B-D). The proportion of misshapen nuclei in HGPS cells was similarly decreased by Remodelin treatment or NAT10 depletion (Fig. 3C and Fig. S9A), implying that NAT10 inhibition by Remodelin mediates nuclear shape normalization in HGPS cells.
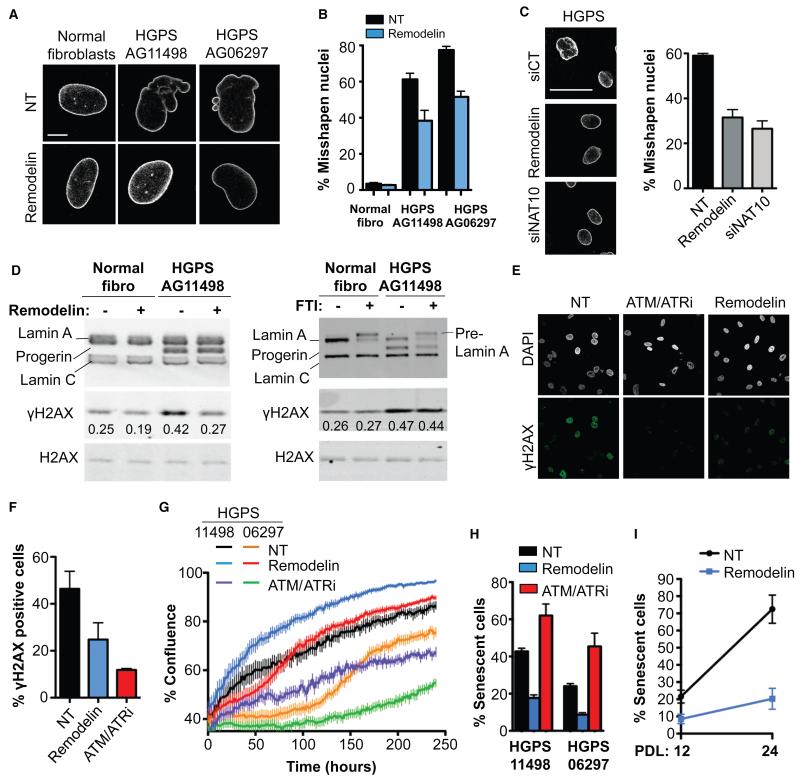
Figure 3. Remodelin targets NAT10 to improve nuclear shape and fitness of HGPS cells.
A) Representative immunofluorescence (IF) pictures of Lamin A/C in HGPS cell lines compared to matched normal fibroblasts at the same population doubling. Scale bar: 10 μm. B) Quantification of misshapen nuclei upon Remodelin treatment (means of three independent experiments with n>213 ± s.e.m.). C) Lamin A/C staining in HGPS AG11498 cells (left) and quantification of misshapen nuclei (right; means of three independent experiments with n>176 ± s.e.m.). Scale bar: 50 μm. D) Western blotting analysis of γH2AX after Remodelin or FTI treatment. E) Immunofluorescence analysis of γH2AX staining upon Remodelin or ATM/ATR inhibition (ATM/ATRi). F) Quantification of γH2AX positive cells observed by IF (means of three independent experiments with n>127 ± s.e.m.). G) HGPS proliferation upon Remodelin or ATM/ATR-inhibitor treatment (means of nine replicates ± s.e.m). H) Quantification of senescence-associated β-galactosidase positive cells (means of three independent experiments with n>257 ± s.e.m.). I) Quantification of senescence-associated β-galactosidase positive cells in HGPS AG11498 after 8 days of Remodelin treatment at population doubling 12 (PDL 12) and after several weeks of Remodelin treatment and 12 cell divisions (PDL 24) (means of two independent experiments with n>298 ± s.d.).
While nuclear shape improvement is not always associated with overall amelioration of HGPS cell phenotypes (21), Remodelin improved global HGPS-cell fitness as observed by decreased steady-state levels of the DNA double-strand break markers γH2AX and autophosphorylated ATM, decreased DNA damage signaling (Fig. 3D, S9B-D), improved chromatin and nucleolar organization (assessed by histone H3K9me3 and NAT10 staining), and decreased SUN1 accumulation at the nuclear envelope (Fig. S9E-G). Blocking DNA damage signaling by inhibiting the apical DNA-damage response kinases ATM and ATR decreased γH2AX (Fig. 3E, F). However, unlike Remodelin - which improved DNA replication (Fig. S9H), enhanced cell proliferation capacity (Fig. 3G) and decreased senescence (Fig. 3H; note other KAT inhibitors did not decrease senescence, Fig. S9I) - inhibiting ATM and ATR decreased proliferation and induced senescence (Fig. 3G, H). Similar effects were observed upon p53-pathway inhibition (Fig. S9I). Thus, Remodelin appears to reduce the amount of DNA damage in HGPS cells, while damage is still present but no longer signaled properly upon ATM/ATR inhibition, leading to cell growth arrest and senescence. Moreover, Remodelin conferred long-term benefits to HGPS cells, as seen by it decreasing senescence even after several weeks of treatment (Fig. 3I).
NAT10 localizes mainly in the nucleolus, which has a known role in maintaining nuclear shape (22, 23). For example, depleting the nucleolar protein NPM1 changes microtubule stability (24) and affects nuclear shape via connections between the cytoskeleton and nuclear envelope. Because tubulin is a known NAT10 substrate, we examined the microtubule network and observed network reorganization upon Remodelin treatment, NAT10 depletion, or mutational inactivation of NAT10 (Fig. 4A). In accord with there being a functional link between microtubule reorganization and nuclear shape rescue by NAT10 inhibition, the microtubule destabilizing drugs nocodazole and colchicine also rescued the nuclear shape defects of siLMNA cells (Fig. 4B) and HGPS cells (Fig. S10A), while latrunculin A, an actin polymerization inhibitor, increased nuclear distortion. Remodelin effects were not linked to Golgi apparatus fragmentation or tubulin depolymerization because, in contrast to nocodazole or colchicine, Remodelin did not affect Golgi apparatus integrity or tubulin polymer assembly (Fig. 4C and D), and did not cause cells to accumulate in mitosis.
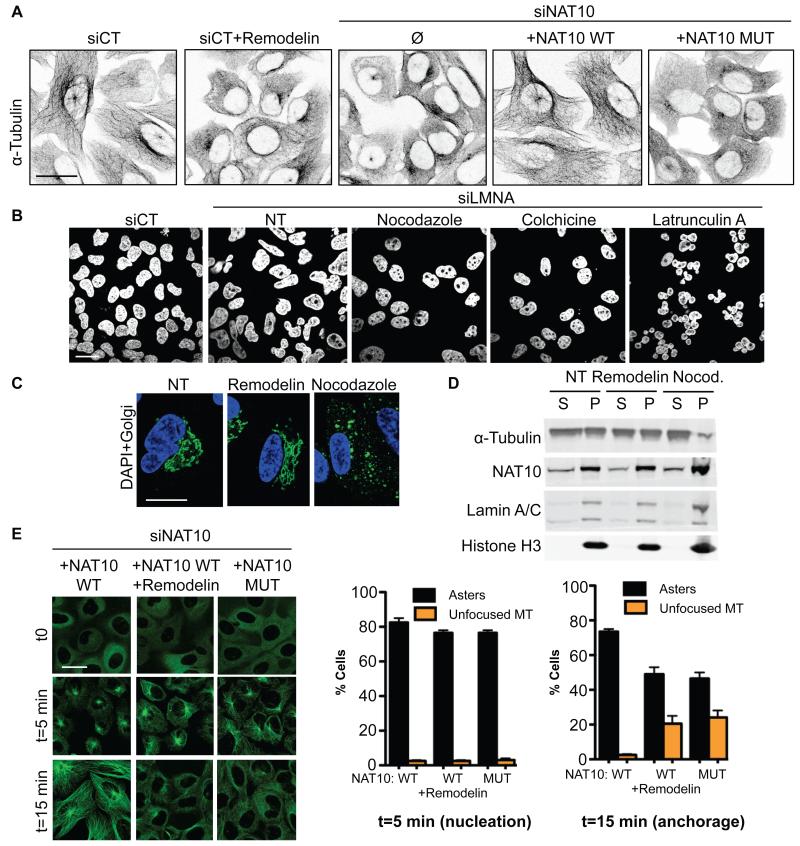
Figure 4. Inhibiting NAT10 acetyltransferase activity modifies microtubule organisation to rescue nuclear shape defects.
A) Microtubule network visualisation by inverted IF pictures of α-tubulin. B) Nuclear shape visualisation after treating cells with microtubule or actin cytoskeleton disrupting agents. C) Visualisation of nuclear shape (DAPI, blue) and Golgi (anti-Giantin, green) integrity in siLMNA cells. D) Fractionation of polymerized (P) and soluble (S) tubulin upon Remodelin or nocodazole (Nocod.) treatment. E) Microtubule regrowth in cells transfected with siNAT10 and expressing the indicated siRNA-resistant constructs. α-tubulin IF staining (left) shows nucleation phase: t=5 min and microtubule anchorage (t=15 min). Right: quantification of cells with indicated patterns (means of three independent experiments with n>103 ± s.e.m.).
To gain insight into how NAT10 modifies microtubule organization, we analysed microtubule regrowth dynamics. While initial microtubule regrowth (nucleation phase) appeared normal after NAT10 inhibition in siLMNA cells (Fig. 4E t=5 min) and HGPS cells (Fig. S10C), microtubule anchorage to centrosomes was affected by Remodelin in both siLMNA cells (Fig. 4E) and HGPS cells (Fig. S10B), or by the NAT10 G641E mutation (Fig. 4E t=15 min), indicating that NAT10 KAT activity promotes microtubule anchorage. These results suggest that inhibiting NAT10 KAT activity in laminopathic cells reduces microtubule anchorage, thereby releasing an external force on the nuclear envelope, thus contributing to nuclear shape rescue and global enhancement of cellular fitness (Fig. S11). This is in line with studies showing that releasing microtubule forces on the nucleus by growing them on a “soft” substrate normalizes the nuclear shape of laminopathic cells (25), and helps explain why Remodelin, like other microtubule reorganizing agents (26), corrects FTI-induced nuclear shape defects in non-progeric cells (Fig. S8).
Here, we have shown that the small molecule Remodelin improves nuclear shape and fitness of both progeric and Lamin A/C depleted cells via inhibiting NAT10. While these effects appear to be connected to NAT10 organizing the microtubule network, we cannot rule out that inhibiting additional nuclear functions of NAT10, related to chromatin, might also contribute to global improvement of cellular fitness. Small molecule inhibitors of NAT10 may thus provide additional ways to study laminopathy-associated processes with spatial and temporal resolution, as well as opportunities for alleviating dystrophic and premature-ageing diseases.
Supplementary Material
Acknowledgements
We thank all members of the Jackson laboratory for help and support, J. Forment for discussions, K. Dry and J. Travers for discussions and critical reading of the manuscript. We thank Rimma Belotserkovskaya for the GFP-H2B U2OS cell line and Sonja Heidorn and Mathew Garnett (Wellcome Trust Sanger Institute) for providing us with the cancer cells used in the Lamin A/C expression screen. We thank Imagif and Institut de Chimie des Substances Naturelles, centre de recherche CNRS de Gif-sur-Yvette, France, for proteomic analysis. Research in the Jackson laboratory is funded by Cancer Research UK program grant C6/A11224, the European Research Council, and the European Community Seventh Framework Programme grant agreement no. HEALTH-F2-2010-259893 (DDR). Core funding is provided by CRUK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J. receives his salary from the University of Cambridge, UK, supplemented by CRUK. D.L is funded by an EMBO Long-term fellowship ALTF 834-2011 and by a Project Grant from the Medical Research Council, UK MR/L019116/1, S.B. was funded by an EMBO Long-Term fellowship ALTF 93-2010 and Cancer Research UK. R.R. is supported by the Centre National de la Recherche Scientifique. M.D. is supported by the European Research Council grant DDREAM. The data in this publication form part of the subject matter of a patent application number 1405991.9.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and notes
- 1.Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res. 2007 Jun 10;313:2121. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Sandre-Giovannoli A, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003 Jun 27;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003 May 15;423:293. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broers JL, et al. Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am J Pathol. 1993 Jul;143:211. [PMC free article] [PubMed] [Google Scholar]
- 5.Moss SF, et al. Decreased and aberrant nuclear lamin expression in gastrointestinal tract neoplasms. Gut. 1999 Nov;45:723. doi: 10.1136/gut.45.5.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venables RS, et al. Expression of individual lamins in basal cell carcinomas of the skin. Br J Cancer. 2001 Feb;84:512. doi: 10.1054/bjoc.2000.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galiova G, Bartova E, Raska I, Krejci J, Kozubek S. Chromatin changes induced by lamin A/C deficiency and the histone deacetylase inhibitor trichostatin A. Eur J Cell Biol. 2008 May;87:291. doi: 10.1016/j.ejcb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, et al. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012 Apr 27;149:565. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toth JI, et al. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc Natl Acad Sci U S A. 2005 Sep 6;102:12873. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chimenti F, et al. A novel histone acetyltransferase inhibitor modulating Gcn5 network: cyclopentylidene-[4-(4′-chlorophenyl)thiazol-2-yl)hydrazone. J Med Chem. 2009 Jan 22;52:530. doi: 10.1021/jm800885d. [DOI] [PubMed] [Google Scholar]
- 11.Chi YH, Haller K, Peloponese JM, Jr., Jeang KT. Histone acetyltransferase hALP and nuclear membrane protein hsSUN1 function in de-condensation of mitotic chromosomes. The Journal of biological chemistry. 2007 Sep 14;282:27447. doi: 10.1074/jbc.M703098200. [DOI] [PubMed] [Google Scholar]
- 12.Shen Q, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009 Jun 10;315:1653. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci U S A. 2009 Mar 24;106:4617. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012 Mar;8:301. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chimnaronk S, et al. RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. Embo J. 2009 May 6;28:1362. doi: 10.1038/emboj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006 May 19;312:1059. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capell BC, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2005 Sep 6;102:12879. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstraeten VL, et al. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci U S A. 2011 Mar 22;108:4997. doi: 10.1073/pnas.1019532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glynn MW, Glover TW. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum Mol Genet. 2005 Oct 15;14:2959. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Blocking farnesylation of the prelamin A variant in Hutchinson-Gilford progeria syndrome alters the distribution of A-type lamins. Nucleus. 2012 Sep-Oct;3:452. doi: 10.4161/nucl.21675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibrahim MX, et al. Targeting isoprenylcysteine methylation ameliorates disease in a mouse model of progeria. Science. 2013 Jun 14;340:1330. doi: 10.1126/science.1238880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin MA, et al. Fibrillarin, a nucleolar protein, is required for normal nuclear morphology and cellular growth in HeLa cells. Biochem Biophys Res Commun. 2007 Aug 24;360:320. doi: 10.1016/j.bbrc.2007.06.092. [DOI] [PubMed] [Google Scholar]
- 23.Amin MA, Matsunaga S, Uchiyama S, Fukui K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J. 2008 Nov 1;415:345. doi: 10.1042/BJ20081411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, et al. Nucleophosmin/B23 inhibits Eg5-mediated microtubule depolymerization by inactivating its ATPase activity. The Journal of biological chemistry. 2010 Jun 18;285:19060. doi: 10.1074/jbc.M110.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamiello C, et al. Soft substrates normalize nuclear morphology and prevent nuclear rupture in fibroblasts from a laminopathy patient with compound heterozygous LMNA mutations. Nucleus. 2013 Jan-Feb;4:61. doi: 10.4161/nucl.23388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki N, Del Villar K, Tamanoi F. Farnesyltransferase inhibitors induce dramatic morphological changes of KNRK cells that are blocked by microtubule interfering agents. Proc Natl Acad Sci U S A. 1998 Sep 1;95:10499. doi: 10.1073/pnas.95.18.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.