Background: Lys-63-linked ubiquitination in mitochondria occurs in PINK1/Parkin-mediated mitophagy, and its important roles have been proposed.
Results: The suppression of Lys-63-linked ubiquitination did not modulate PINK1/Parkin-mediated mitophagy and Drosophila mitochondrial phenotypes.
Conclusion: Lys-63-linked ubiquitination is dispensable for PINK1-Parkin pathway.
Significance: This is the first study to report the biological significance of Lys-63-linked ubiquitination in PINK1-Parkin pathway in vitro and in vivo.
Keywords: Mitochondria, Mitophagy, Parkin, PTEN-induced Putative Kinase 1 (PINK1), Ubiquitin, Ubc13
Abstract
PINK1/Parkin-mediated mitophagy is thought to ensure mitochondrial quality control in neurons as well as other cells. Upon the loss of mitochondrial membrane potential (ΔΨm), Lys-63-linked polyubiquitin chains accumulate on the mitochondrial outer membrane in a Parkin-dependent manner. However, the physiological significance of Lys-63-linked polyubiquitination during mitophagy is not fully understood. Here, we report that the suppression of Lys-63-linked polyubiquitination through the removal of Ubc13 activity essentially affects neither PINK1 activation nor the degradation of depolarized mitochondria. Moreover, the inactivation of Ubc13 did not modulate the mitochondrial phenotypes of PINK1 knockdown Drosophila. Our data indicate that the formation of Lys-63-linked polyubiquitin chains on depolarized mitochondria is not a key factor for the PINK1-Parkin pathway as was once thought.
Introduction
Mutations of the Parkin and PINK1 genes cause selective degeneration of the midbrain dopaminergic neurons in autosomal recessive juvenile Parkinson disease (1, 2). The Parkin and PINK1 genes encode a ubiquitin-ligase (E3)3 and a serine/threonine protein kinase, respectively (3–7). Loss of the Parkin and PINK1 genes in Drosophila leads to the degeneration of the mitochondria in tissues with high energy demands, such as the muscles and sperm, and genetic analysis has demonstrated that PINK1 is an upstream regulator of Parkin, suggesting an important role of Parkin and PINK1 in mitochondrial maintenance in the midbrain dopaminergic neurons that are affected in Parkinson disease (8–10).
A series of cell biological studies has provided strong evidence that Parkin cooperates with PINK1 to induce mitochondrial autophagy or mitophagy when the mitochondria are damaged (11–16). The reduction of ΔΨm leads to the accumulation and activation of PINK1 in the mitochondria (12, 17), which leads to the phosphorylation of a latent form of Parkin, priming its E3 activation (17, 18). PINK1 also phosphorylates ubiquitin (19–21), which in turn fully activates Parkin E3 activity, leading to Parkin translocation from the cytosol to the mitochondria and the subsequent ubiquitination of mitochondrial proteins (14, 15). Ubiquitin modification on the mitochondria induces the LC3-mediated autophagic elimination of the damaged mitochondria, a process known as mitophagy (11). The ubiquitination of mitochondrial proteins mainly produces Lys-63-linked polyubiquitin and only a small portion of Lys-48 linkages (22, 23). The Lys-63-linked polyubiquitin chain is proposed to activate PINK1 (24) and the mitochondrial translocation of Parkin (25). We examined the impact of Lys-63-linked polyubiquitination on PINK1/Parkin-mediated mitophagy in cells and mitochondrial maintenance in Drosophila and report that Lys-63-linked polyubiquitination is dispensable for PINK1 activation, mitochondrial clearance, and Drosophila mitochondrial homeostasis.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, Plasmids, and Cell Lines
The following antibodies were used in the Western blot analysis: anti-PINK1 (1:1,000 dilution; Novus Biologicals, BC100-494), anti-Mfn1 (1:1,000 dilution; Abnova, clone 3C9), anti-Ubc13 (1:1,000 dilution; Life Technologies, clone 4E11), anti-polyubiquitin (1:1,000 dilution; MBL International, clone FK2), anti-Lys-63-linked polyubiquitin (1:1,000 dilution; Cell Signaling Technology, clone D7A11), anti-Lys-48-linked polyubiquitin (1:1,000 dilution; Cell Signaling Technology, clone D9D5), anti-Tom20 (1:500 dilution; Santa Cruz Biotechnology, FL-145), anti-HA (1:1,000 dilution; Roche Applied Science, clone 3F10), anti-FLAG-HRP (1:2,000 dilution; Sigma-Aldrich, clone M2), anti-actin (1:10,000 dilution; Millipore, MAb1501), anti-Hsp60 (1:10,000 dilution; BD Biosciences, clone 24/Hsp60), anti-NDUFS3 (1:10,000 dilution; Abcam, 17D95), anti-Drosophila Hsp60 (1:1,000 dilution; Cell Signaling Technology, D307), and anti-Drosophila Mitofusin (dMfn) (1:2,000 dilution; made in-house). The following antibodies were used for immunocytochemistry analysis: anti-polyubiquitin (1:250 dilution; MBL International, clone FK2), anti-Lys-63-linked polyubiquitin (1:50 dilution; Millipore, clone Apu3), and anti-Tom20 (1:1,000 dilution; Santa Cruz Biotechnology, FL-145). Mouse embryonic fibroblasts (MEFs) harboring wild-type or homozygous loxP-flanked Ubc13 alleles (26) were stably transfected with Cre recombinase controlled by Tet-On systems. Ubc13 genes were floxed out following Cre-mediated excision by treatment with 1 μg/ml doxycycline (Dox) for 72 h to generate Ubc13−/− MEFs. Wild-type Ubc13 MEFs were also treated with Dox as a control. The plasmids encoding GFP-Parkin, HA-Parkin, and PINK1-FLAG have been described previously (15, 27). MEFs and HeLa cells were retrovirally transfected with pMXs-puro harboring PINK1-FLAG, HA-Parkin, and GFP-Parkin, and the infected cells were selected with 1 μg/ml puromycin. The mitochondrial uncoupler carbonyl cyanide m-chlorophenyl hydrazine (CCCP) and the ubiquitin-activating enzyme (E1)-specific inhibitor UBEI-41 were purchased from Sigma-Aldrich. The mitochondrial uncoupler valinomycin and TUBE1-agarose were obtained from Wako and LifeSensors, respectively.
Immunocytochemical and Biochemical Analyses
Cells plated on 3.5-mm glass-bottom dishes (MatTek) were fixed with 4% paraformaldehyde in PBS and permeabilized with 50 μg/ml digitonin in PBS. The cells were stained with anti-Tom20 or anti-ubiquitin antibodies. The cells were imaged using laser-scanning microscope systems (LSM510 META, Carl Zeiss). Phos-tag (Wako Pure Chemical Industries) Western blotting was performed as described previously (18).
Drosophila Genetics
Fly experiments were performed as described (28). The w1118 (w−) line was used as a wild-type genetic background. The Ubc13 RNAi line was obtained from the Vienna Drosophila RNAi Center and was characterized in Ref. 29. Other fly stocks used in this study have been described previously (8).
RESULTS AND DISCUSSION
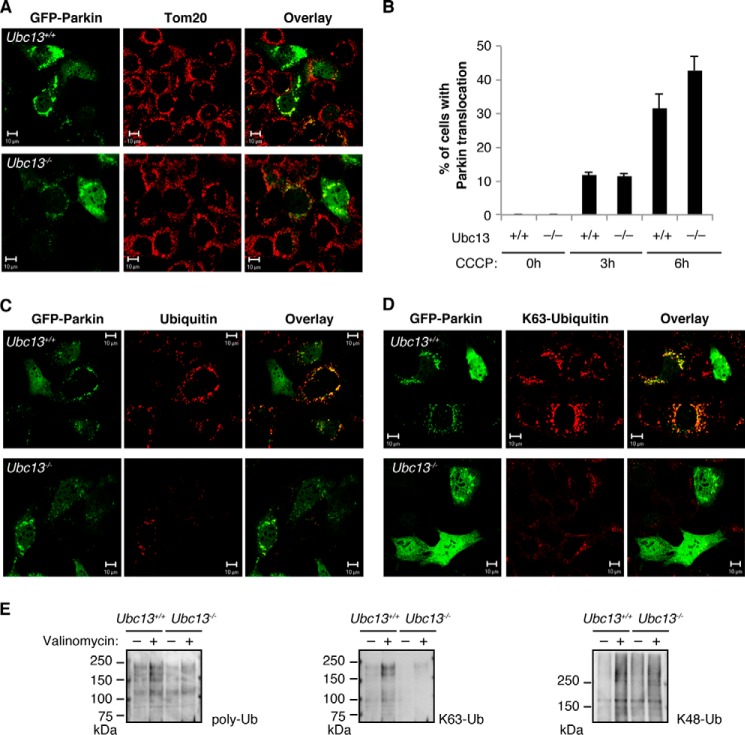
Because Ubc13 is an E2 enzyme crucial for generating Lys-63-linked chains (30), we tested PINK1/Parkin-mediated mitophagy in Ubc13 mutant cells to estimate the effects of Lys-63-linked polyubiquitin chain formation. In the Ubc13 mutant cells harboring the loxP-flanked Ubc13 gene, Ubc13 can be inactivated by Dox-induced flox-out. We inactivated Ubc13 by Dox treatment and induced the mitochondrial translocation of GFP-Parkin and the accumulation of ubiquitin chains using CCCP. The mitochondrial translocation of GFP-Parkin occurred with similar efficiency (Fig. 1, A and B). In contrast, the accumulation of total ubiquitin (Fig. 1, C and E) as well as Lys-63-linked polyubiquitin (Fig. 1, D and E) in the mitochondria was dramatically reduced in the absence of Ubc13 activity. Accumulation of Lys-48-linked polyubiquitin in the mitochondrial fractions was similar between Ubc13+/+ and Ubc13−/− MEFs expressing GFP-Parkin (Fig. 1E).
FIGURE 1.
The loss of Ubc13 activity impairs the accumulation of Lys-63-linked ubiquitin chains during Parkin-mediated mitophagy. A, MEFs retrovirally introduced with GFP-Parkin were treated with Dox to remove Ubc13 genes and then treated with 30 μm CCCP for 6 h. Parkin and mitochondria were visualized with GFP fluorescence (green) and anti-Tom20 (red), respectively. B, the mitochondrial translocation efficiency of Parkin treated as in A was graphed. The values represent the means ± S.E. of the percentages of cells exhibiting mitochondrial recruitment in three independent experiments. The translocation efficiency was similar in Ubc13+/+ and Ubc13−/− (3 h, p < 0.8024; 6 h, p < 0.1309 by Student's t test). C, ubiquitin accumulation was detected with anti-polyubiquitin (red) in cells treated as in A. D, accumulation of a Lys-63-linked ubiquitin (K63-Ubiquitin) chain was detected with anti-Lys-63 linkage-specific ubiquitin antibody (red) in cells treated as in A. Scale bars = 10 μm. E, accumulation of Lys-63-linked polyubiquitin (K63-Ub) but not of Lys-48-linked polyubiquitin (K48-Ub) was reduced in the absence of Ubc13 activity. Crude mitochondrial fractions from MEFs expressing GFP-Parkin (1 × 106) treated with (+) or without (−) 30 μm valinomycin for 6 h were prepared. Polyubiquitin purified with TUBE1-agarose in the mitochondrial fractions was detected by Western blot. poly-Ub, polyubiquitin. All experiments were repeated at least three times in A–D and two times in E, and representative results were shown.
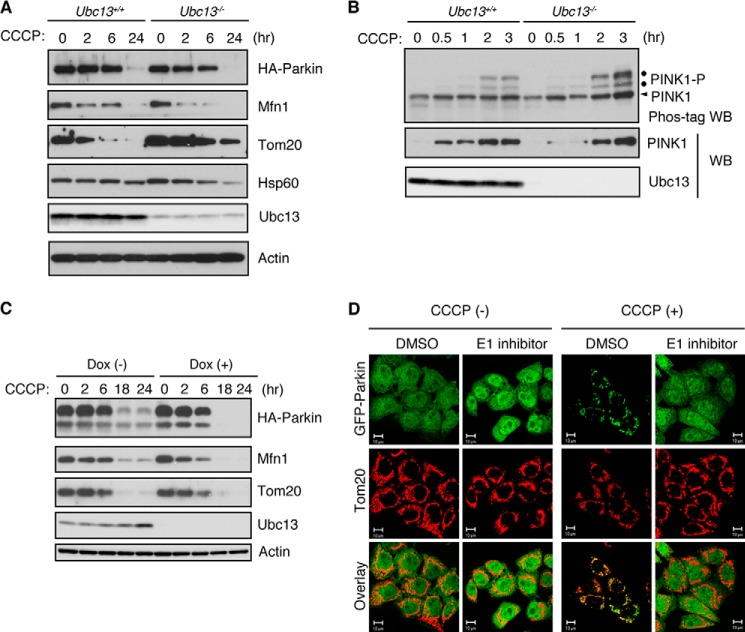
Polyubiquitination induces the degradation of mitochondria outer membrane proteins through the proteasome and recruits LC3-mediated autophagy machinery (22). To test whether autophagy is altered in Ubc13−/− MEFs, we examined the levels of Mfn1, a known substrate of Parkin E3; a mitochondrial outer membrane protein, Tom20; and a matrix protein, Hsp60. The time-dependent degradation of Mfn1, Tom20, and Hsp60 in Ubc13−/− MEFs was comparable with that in Ubc13+/+ MEFs (Fig. 2A). When Parkin is activated upon CCCP treatment, Parkin is subjected to autodegradation by the proteasome (18). The degradation efficiency of HA-tagged Parkin was similar between Ubc13+/+ and Ubc13−/− MEFs, suggesting that the formation of Lys-63-linked polyubiquitin affects neither the activation of Parkin nor the autophagic clearance of mitochondria.
FIGURE 2.
Suppression of Lys-63-linked ubiquitin chain formation does not affect PINK1 activation or mitochondrial clearance. A, MEFs expressing HA-Parkin were treated with 30 μm CCCP for up to 24 h and subjected to Western blot analysis. Mfn1 and Tom20 were used as markers of mitochondrial outer membrane proteins. Hsp60 was used as a marker of mitochondrial matrix proteins. Actin was used as a loading control. B, MEFs expressing PINK1-FLAG were treated with 30 μm CCCP as in A. The autophosphorylation of PINK1 and accumulation of PINK1 were estimated by Phos-tag Western blot with anti-PINK1 (Phos-tag WB) and conventional Western blot with anti-FLAG (WB). C, MEFs harboring loxP-flanked Ubc13 were treated with (+) or without (−) Dox for 72 h and were further treated with CCCP for the indicated time periods. The degradation of Parkin, Mfn1, and Tom20 was analyzed by Western blot analysis. D, HeLa cells stably expressing GFP-Parkin were pretreated with 60 μm UBEI-41 (E1 inhibitor) or dimethyl sulfoxide (DMSO) solvent for 1 h and were further treated with or without 20 μm CCCP for 3 h. GFP-Parkin and mitochondria were visualized with GFP signal (green) and anti-Tom20 (red), respectively. Scale bars = 10 μm. All experiments were repeated at least three times in A–C and two times in D.
It has been proposed that Lys-63-linked ubiquitination of PINK1 by TRAF6 is required for the mitochondrial accumulation of PINK1 and mitochondrial translocation of Parkin upon a reduction of ΔΨm (24). PINK1 stabilization on the mitochondrial outer membrane stimulates its dimerization and is closely correlated with its autophosphorylation at Ser-228 and Ser-402 in an intermolecular fashion (31), through which PINK1 kinase activity is thought to be activated (32). We estimated the extent of PINK1 accumulation and PINK1 autophosphorylation by conventional Western blot and Phos-tag Western blot analyses, respectively (Fig. 2B). However, there was no evidence that PINK1 accumulation and autophosphorylation were altered in the absence of Ubc13 activity, suggesting that the formation of the Lys-63-linked polyubiquitin chain is not a key factor in PINK1 regulation in mitophagy.
Because MEFs are derived from a heterogeneous population of cells, the response to PINK1/Parkin-mediated mitophagy might differ among different batches of cells. To exclude this possibility, we used the same batch of Ubc13 mutant cells, which were treated with or without Dox. PINK1/Parkin-mediated mitophagy was induced by CCCP treatment for up to 24 h. We again confirmed that the efficiency of the degradation of HA-Parkin, Mfn1, and Tom20 is comparable between Dox-treated and untreated cells (Fig. 2C).
It has been reported that Parkin is also involved in xenophagy for Mycobacterium tuberculosis, in which the co-localization of a Lys-63-linked ubiquitin chain with phagosomes containing M. tuberculosis was observed (33). Because the formation of Lys-63-linked ubiquitination, the subsequent accumulation of the ubiquitin adaptors, and the autophagy machinery are Parkin-dependent, Lys-63-linked ubiquitination likely mediates the recruitment of autophagy-related proteins, as proposed in studies of mitophagy (14, 22). Lys-63-linked ubiquitination is also observed in Salmonella xenophagy (34). However, the recruitment of the autophagy machinery occurred with the same efficiency in Salmonella xenophagy (35). The results describing both mitophagy and xenophagy suggest that the autophagy machinery can recognize other polyubiquitin linkages in addition to Lys-63 or that Lys-63 linkage is not involved in this step. Although Lys-63-linked ubiquitination is not essentially required for mitochondrial translocation of Parkin, the inhibition of all of ubiquitination reactions by an E1-specific inhibitor completely suppresses Parkin translocation, suggesting that ubiquitination is part of the regulation in Parkin translocation (Fig. 2D).
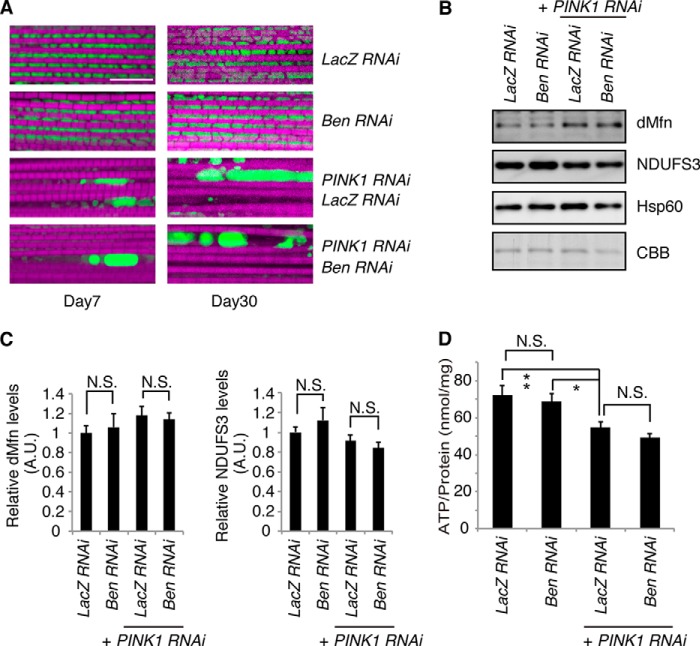
The formation of Lys-63-linked polyubiquitination by Ubc13 and Uev1a is involved in the TNF signaling in both mammals (36) and Drosophila (29). Knockdown of Bendless (Ben), an ortholog of Ubc13, suppresses TNF signaling in Drosophila, suggesting that the formation of Lys-63-linked polyubiquitination is inhibited (29). Muscular mitochondria in the thorax, in which Ben was inactivated, showed a normal gross morphology, implying that Lys-63-linked ubiquitination is dispensable for mitochondrial maintenance under steady-state conditions (Fig. 3A). In contrast, PINK1 activity is essential for maintaining mitochondrial homeostasis because inactivation of PINK1 largely leads to mitochondrial degeneration, as described previously (Fig. 3A) (8). The mitochondrial degeneration by PINK1 inactivation was no longer modulated by the suppression of Ben activity, even in old flies (Fig. 3A). Consistent with the histochemical analysis, levels of a mitochondrial outer membrane protein Mitofusin, which is a ubiquitination substrate of Parkin, as well as the mitochondrial complex I subunit NDUFS3, were not altered by Ben inactivation (Fig. 3, B and C). In addition, the absence of Ben did not affect mitochondrial ATP production (Fig. 3D).
FIGURE 3.
Inhibition of Ubc13 does not modulate the mitochondrial phenotypes caused by PINK1 inactivation. A, fluorescent images of the indirect flight muscle in 7- and 30-day-old adult flies expressing the indicated shRNAs are shown. To visualize the mitochondria, the mitoGFP (green) transgene was co-expressed, and the muscle tissue was counterstained with phalloidin (magenta). Representative images from three independent samples in each genotype are shown. Experiments were repeated two times. Scale bar = 10 μm in the fluorescent images. B, the protein levels of dMfn, complex I subunit NDUFS3, and Hsp60 from the thoraxes of 7-day-old adult flies were analyzed by Western blot. Coomassie Brilliant Blue (CBB) staining around the dMfn migration position confirms that approximately equivalent amounts of protein were loaded. C, the band intensities of dMfn and NDUFS3 were normalized to each Coomassie Brilliant Blue signal. The values (arbitrary units (A.U.)) represent the means ± S.E. from 4–5 independent samples as in B. Although dMfn and NDUFS3 levels showed increasing and decreasing tendencies, respectively, with PINK1 inactivation as reported (28), there were no statistical differences between any combinations. N.S., not significant. n = 4–5. D, ATP contents of thorax muscle tissues of 7-day-old adult flies were measured. ATP contents were normalized against the protein levels. The values represent the means ± S.E. from five independent samples. *, p < 0.05, **, p < 0.01 by Tukey-Kramer test. Fly genotypes used in A–D are as follows: UAS-mitoGFP/UAS-LacZ RNAi; MHC-GAL4/+ (LacZ RNAi), UAS-mitoGFP/UAS-Ben RNAi; MHC-GAL4/+ (Ben RNAi), UAS-mitoGFP/UAS-LacZ RNAi; MHC-GAL4, UAS-PINK1 RNAi/+ (PINK1 RNAi, LacZ RNAi), UAS-mitoGFP/UAS-Ben RNAi; MHC-GAL4, UAS-PINK1 RNAi/+ (PINK1 RNAi, Ben RNAi).
In conclusion, this study revealed that Lys-63-linked ubiquitination is dispensable for the PINK1-Parkin pathway. Although Lys-63-linked ubiquitination by Parkin has been suggested to be important for the suppression of protein toxicity by Parkin, further investigations will be required to determine whether specific roles of Lys-63-linked ubiquitination in the PINK1-Parkin pathway exist (37, 38).
Acknowledgments
We thank Drs. S. Akira and M. Yamamoto for Ubc13 mutant cells, and T. Arano and T. Imura for technical assistance.
Addendum
After submission of this study, two studies using siRNA against Ubc13 reported that Ubc13 has a role for the autophagy process of Parkin-mediated mitophagy (39) and Parkin translocation (40). As we also observed some delay in mitophagy in our initial study using siRNA, we feel that certain sequences of siRNA affect mitophagy.
This work was supported by grants from the Takeda Science Foundation, (to Y. I.), the Life Science Foundation of Japan (to Y. I.), the Daiichi-Sankyo Foundation for Life Science (to Y. I.), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to Y. I.), and Otsuka Pharmaceutical (to N. H. and Y. I.).
- E1
- ubiquitin-activating enzyme
- E2
- ubiquitin-conjugating enzyme
- E3
- ubiquitin-ligase
- MEF
- mouse embryonic fibroblast
- Mfn1
- Mitofusin1
- Dox
- doxycycline
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazine
- dMfn
- Drosophila Mitofusin.
REFERENCES
- 1. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 2. Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., González-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- 3. Imai Y., Soda M., Takahashi R. (2000) Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 [DOI] [PubMed] [Google Scholar]
- 4. Shimura H., Hattori N., Kubo S. i., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 5. Beilina A., Van Der Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. (2005) Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl. Acad. Sci. U.S.A. 102, 5703–5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. (2005) Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 14, 3477–3492 [DOI] [PubMed] [Google Scholar]
- 7. Sim C. H., Lio D. S., Mok S. S., Masters C. L., Hill A. F., Culvenor J. G., Cheng H. C. (2006) C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 15, 3251–3262 [DOI] [PubMed] [Google Scholar]
- 8. Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. (2006) Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U.S.A. 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 10. Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- 11. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S., Magrané J., Moore D. J., Dawson V. L., Grailhe R., Dawson T. M., Li C., Tieu K., Przedborski S. (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 107, 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 15. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawajiri S., Saiki S., Sato S., Sato F., Hatano T., Eguchi H., Hattori N. (2010) PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 584, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 17. Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H. I., Campbell D. G., Gourlay R., Burchell L., Walden H., Macartney T. J., Deak M., Knebel A., Alessi D. R., Muqit M. M. (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating serine 65. Open Biol. 2, 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., Hattori N. (2012) PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2, 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D. G., Ritorto M. S., Hofmann K., Alessi D. R., Knebel A., Trost M., Muqit M. M. (2014) Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kane L. A., Lazarou M., Fogel A. I., Li Y., Yamano K., Sarraf S. A., Banerjee S., Youle R. J. (2014) PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., Endo T., Fon E. A., Trempe J.-F., Saeki Y., Tanaka K., Matsuda N. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 [DOI] [PubMed] [Google Scholar]
- 22. Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L., Hess S., Chan D. C. (2011) Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haddad D. M., Vilain S., Vos M., Esposito G., Matta S., Kalscheuer V. M., Craessaerts K., Leyssen M., Nascimento R. M., Vianna-Morgante A. M., De Strooper B., Van Esch H., Morais V. A., Verstreken P. (2013) Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol. Cell 50, 831–843 [DOI] [PubMed] [Google Scholar]
- 24. Murata H., Sakaguchi M., Kataoka K., Huh N. H. (2013) SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol. Biol. Cell 24, 2772–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng X., Hunter T. (2013) Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 23, 886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamamoto M., Okamoto T., Takeda K., Sato S., Sanjo H., Uematsu S., Saitoh T., Yamamoto N., Sakurai H., Ishii K. J., Yamaoka S., Kawai T., Matsuura Y., Takeuchi O., Akira S. (2006) Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7, 962–970 [DOI] [PubMed] [Google Scholar]
- 27. Shiba K., Arai T., Sato S., Kubo S., Ohba Y., Mizuno Y., Hattori N. (2009) Parkin stabilizes PINK1 through direct interaction. Biochem. Biophys. Res. Commun. 383, 331–335 [DOI] [PubMed] [Google Scholar]
- 28. Shiba-Fukushima K., Inoshita T., Hattori N., Imai Y. (2014) PINK1-mediated phosphorylation of Parkin boosts Parkin activity in Drosophila. PLoS Genet. 10, e1004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma X., Huang J., Yang L., Yang Y., Li W., Xue L. (2012) NOPO modulates Egr-induced JNK-independent cell death in Drosophila. Cell Res. 22, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hofmann R. M., Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 31. Okatsu K., Uno M., Koyano F., Go E., Kimura M., Oka T., Tanaka K., Matsuda N. (2013) A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J. Biol. Chem. 288, 36372–36384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M., Shiba-Fukushima K., Sato S., Shimizu H., Fukunaga Y., Taniguchi H., Komatsu M., Hattori N., Mihara K., Tanaka K., Matsuda N. (2012) PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 3, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manzanillo P. S., Ayres J. S., Watson R. O., Collins A. C., Souza G., Rae C. S., Schneider D. S., Nakamura K., Shiloh M. U., Cox J. S. (2013) The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501, 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Wijk S. J., Fiskin E., Putyrski M., Pampaloni F., Hou J., Wild P., Kensche T., Grecco H. E., Bastiaens P., Dikic I. (2012) Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell 47, 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fujita N., Morita E., Itoh T., Tanaka A., Nakaoka M., Osada Y., Umemoto T., Saitoh T., Nakatogawa H., Kobayashi S., Haraguchi T., Guan J. L., Iwai K., Tokunaga F., Saito K., Ishibashi K., Akira S., Fukuda M., Noda T., Yoshimori T. (2013) Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 203, 115–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 37. Olzmann J. A., Li L., Chudaev M. V., Chen J., Perez F. A., Palmiter R. D., Chin L. S. (2007) Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lim G. G., Chew K. C., Ng X. H., Henry-Basil A., Sim R. W., Tan J. M., Chai C., Lim K. L. (2013) Proteasome inhibition promotes Parkin-Ubc13 interaction and lysine 63-linked ubiquitination. PLoS One 8, e73235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Geisler S., Vollmer S., Golombek S., Kahle P. J. (2014) The ubiquitin-conjugating enzymes UBE2N, UBE2L3 and UBE2D2/3 are essential for Parkin-dependent mitophagy. J. Cell Sci. 127, 3280–3293 [DOI] [PubMed] [Google Scholar]
- 40. Fiesel F. C., Moussaud-Lamodière E. L., Ando M., Springer W. (2014) A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J. Cell Sci. 127, 3488–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]