Background: The ESCRT-0 complex proteins, Hrs and STAM, cooperatively recognize ubiquitylated endosomal cargo.
Results: Hrs is targeted to endosomes independently of STAM binding and dissociates from endosomes in a STAM binding-dependent manner.
Conclusion: Endosomal recruitment and dissociation cycle of the ESCRT-0 proteins is essential for efficient endosomal cargo sorting.
Significance: A crucial process in endosomal cargo sorting is identified.
Keywords: Endosome, Membrane Trafficking, Protein Sorting, Protein Targeting, Ubiquitylation (Ubiquitination), ESCRT-0, Hrs, STAM
Abstract
The ESCRT-0 complex, consisting of the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and the signal-transducing adaptor molecule (STAM) proteins, recognizes ubiquitylated cargo during the initial step of endosomal sorting. The endosomal accumulation of overexpressed Hrs has been reported previously to be associated with endosome enlargement. In this study, we have found that co-expressing exogenous STAM1 in Hrs-overexpressing cells leads to a diffuse localization for a large part of the Hrs accumulated on endosomes and a recovery of the impaired cargo protein degradation process, thus suggesting that exogenous STAM abrogates the abnormalities of the Hrs-positive endosomes. A fluorescently labeled Hrs, introduced into the cells by membrane permeabilization, exhibited endosomal localization in the absence of STAM1 and gradually dissociated from the endosomes upon the sequential addition of recombinant STAM1. Furthermore, when microinjected into cells, the fluorescently labeled Hrs also showed endosomal accumulation; however, ESCRT-0 complexes formed prior to the microinjection did not. Analysis of the state of the complex in HeLa cells using blue-native PAGE revealed that the membrane-associated Hrs exists partly as a monomer and not only in the STAM1-bound form. Thus, our data suggest that the membrane binding and dissociation cycle of the ESCRT-0 proteins on the endosomal membrane is a critical step during the cargo sorting process.
Introduction
Endocytosed membrane proteins are either recycled to the plasma membrane or transported to the lysosome for degradation. The ubiquitylation of membrane proteins acts as a sorting signal for their selective sequestration into an invaginated membrane and thus into multivesicular endosomes or bodies (MVBs).2 Ubiquitylation-dependent protein sorting on endosomes is mediated by the endosomal sorting complex required for transport (ESCRT) machinery, which was genetically identified as yeast vacuolar protein sorting class E mutants and classified as four different complexes, ESCRT-0, -I, -II, and -III. The ESCRT-0, -I, and -II complexes contain some ubiquitin-binding subunits (1). Early models of ESCRT-mediated cargo sorting suggested that ubiquitylated cargo is passed from one complex to another and hence is sorted into the intraluminal vesicles of MVBs (2). Further functional and structural studies proposed that the cooperative action of the ubiquitin-binding complexes overcomes the low affinity interactions of the individual ubiquitin-binding domains (UBDs) and the ubiquitin moieties, thus enabling selective recognition of the ubiquitylated cargo (1, 3). Reconstitution experiments using liposomes revealed the functional roles of ESCRT-III in membrane deformation and abscission of the bud neck, as driven by the spiral assembly of the ESCRT-III subunits (4, 5). However, other liposome reconstitution experiments using purified ESCRT proteins and model ubiquitylated cargo showed that, under physiological concentrations of ESCRT proteins, an ESCRT-I/-II super-complex, not ESCRT-III, induces bud formation (6).
The ESCRT-0 complex, which consists of the hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs) and the signal-transducing adaptor molecule (STAM) 1/2 proteins in metazoa, recognizes ubiquitylated cargo and concentrates it into endosomal microdomains via its interaction with clathrin (7). The Hrs and STAM proteins are structurally related, and both contain an N-terminal VHS (Vps27, Hrs, and STAM) domain, a ubiquitin-interacting motif (UIM), and a clathrin-binding motif. Both Hrs and STAM can interact with ubiquitin not only via their UIMs but also via their VHS domains (8). The crystal structures of Hrs and STAM, and the core complex of the yeast ortholog Vps27/Hse1, revealed that the ESCRT-0 proteins form two domain-swapped GAT (GGA1 and TOM) domains connected by an anti-parallel coiled coil (9, 10). The monomeric GAT domains of GGA1, GGA3, and TOM1 show ubiquitin binding properties (11), although the ubiquitin-binding ability of the Vps27/Hse1 coiled-coil core was undetectable (7). Thus, multiple UBDs in the Hrs·STAM complex seem to strengthen the overall affinity for ubiquitylated cargo and enable effective cargo sorting. Interestingly, in the absence of STAM, purified Hrs forms a cylindrical hexameric complex composed of three sets of antiparallel pairs (12). The multiple UBDs in the Hrs hexamer may also facilitate the binding to ubiquitylated cargo. The biological relationship between the Hrs·STAM heteromer and the homotypic Hrs hexamer in the cargo sorting process remains unresolved.
A reconstitution study also revealed that ESCRT-0 forms a cluster with ubiquitylated cargo in endosomal microdomains (6). The recruitment of this cluster to the bud necks via its interaction with ESCRT-I causes the confinement of cargo proteins within the buds (6). Meanwhile, endosomal accumulation of overexpressed Hrs causes a severe impairment in cargo degradation (13, 14). These experimental data suggest the functionally significant recruitment and dissociation cycle of ESCRT-0 on endosomes during cargo sorting. Here, we demonstrate that STAM mediates the dissociation of Hrs from the endosomal membrane. Prior to the dissociation process, the formation of the ESCRT-0 protein complex is not a prerequisite for endosomal targeting. The behavior of ESCRT-0 described in this study is required for the maintenance of normal endosomal morphology and the effective degradation of membrane proteins.
EXPERIMENTAL PROCEDURES
Plasmids
The expression vector pCXN2 has been described previously (15). The retroviral vector pMXs-puro and the lentiviral vector CSII-EF-MCS were kindly provided by Dr. T. Kitamura (Tokyo University, Tokyo, Japan) and Dr. H. Miyoshi (RIKEN BRC, Tsukuba, Japan), respectively. The cDNAs of HA-tagged Hrs, FLAG-tagged STAM1, V5-tagged STAM1, and FLAG-tagged associated molecule of the SH3 domain of STAM (AMSH) were introduced into these expression vectors. The mutant constructs were generated using PCR-based site-directed mutagenesis. All constructs were sequenced with a PRISM 3100 genetic analyzer (Applied Biosystems/Invitrogen). Bacterial expression vector pCold-I was purchased from TAKARA BIO (Shiga, Japan). pCO12-EGF receptor (EGFR) was obtained from the RIKEN BRC. pcDNA3-c-Cbl was a generous gift from Dr. Y. Yarden (Weizmann Institute of Science, Rehovot, Israel). pcDNA3-myc-ubiquitin was constructed by replacement of the HA tag in pcDNA3-HA-ubiquitin (a gift from Dr. K. Miyazono, Tokyo University, Tokyo, Japan) with the c-Myc epitope sequence using PCR-based mutagenesis. pEGFP-SARA-FYVE and pcDNA3-mRuby2-SARA-FYVE were constructed by the introduction of a cDNA fragment corresponding to the FYVE domain of human SARA into pEGFP-C1 (Clontech) and pcDNA3-mRuby2 (Addgene, Cambridge, MA).
Cell Culture
All cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. 2HP67 cells were generated by the retroviral transduction of HA-tagged Hrs into human embryonic kidney-derived 293T cells. The SL and DL cell lines were generated by lentiviral transduction of the wild-type STAM1 and the DSH3 mutant constructs, respectively, into 2HP67 cells. The mouse embryonic fibroblast cell lines HRSd and SSd were derived and cultured from Hrs−/− and STAM1−/−, STAM2−/− mice, respectively, as described previously (16). The SSdRe-S1, SSd-DSH3, and SSd-dCC sub-lines were generated by retrovirally transducing SSd cells with expression vectors for wild-type STAM1 or the STAM1 mutants DSH3 or dCC, respectively.
Antibodies
The rabbit polyclonal anti-Hrs antibody (M120) (17), the rat monoclonal anti-Hrs antibody (5G12) (16), the mouse monoclonal anti-STAM1 antibody (TUS1) (18), and the mouse monoclonal anti-AMSH antibody (9E521) (19) were generated as described previously. The mouse monoclonal anti-LAMP1 antibody was a generous gift from Dr. M. Fukuda (Burnham Institute for Medical Research, La Jolla, CA). The following antibodies were also used: anti-HA (Y-11; Santa Cruz Biotechnology, Dallas, TX), anti-FLAG (FLAGM2; Sigma), anti-EGFR (1005; Santa Cruz Biotechnology), anti-actin (AC-74; Sigma), anti-EEA1 (14/EEA1; BD Biosciences), and anti-Tsg101 (4A10; GeneTex, San Antonio, TX).
Immunofluorescence Microscopy
Cells grown on coverslips were fixed with ice-cold methanol for 3 min or 4% (w/v) paraformaldehyde for 10 min at room temperature. Following paraformaldehyde treatment, cells were permeabilized using 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 3 min at room temperature. The samples were incubated with the indicated primary antibodies (1 μg/ml) at 4 °C overnight, washed three times with PBS containing 0.1% bovine serum albumin (BSA), and incubated with the appropriate Alexa Fluor-conjugated secondary antibodies (1 μg/ml; Invitrogen) at 37 °C for 1 h. All antibodies were diluted into PBS containing 1% BSA. Fluorescence images were captured using a TCS SP2 AOBS confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany).
EGFR Degradation Assay
Cells were transiently transfected with ubiquitin, c-Cbl, and EGFR expression vectors and then, 24 h later, treated with 100 ng/ml EGF for the indicated time periods at 37 °C. The cells were washed with PBS and lysed in lysis buffer (1% Nonidet P-40, 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm Na3VO4, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm phenylmethylsulfonyl fluoride, and 1 mm aprotinin). Cell extracts were solubilized in 4× Laemmli sample buffer (200 mm Tris-HCl, pH 6.8, 5% 2-mercaptoethanol, 8% SDS, 40% glycerol, and 1 μg/ml bromphenol blue) and then boiled for 5 min. Proteins were separated by SDS-PAGE and transferred onto Immobilon-P polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA). For immunoblotting analysis, membranes were blocked with 5% nonfat milk in TBS, 0.1% Tween 20 for 30 min at room temperature, and the indicated primary antibodies (1 μg/ml) at 4 °C overnight, and the relevant horseradish peroxidase-conjugated secondary antibodies (1 μg/ml; Cell Signaling Technology, Beverly, MA) for 1 h at room temperature. The immunoblots were developed using Immobilon Western HRP substrate (Millipore).
Cell Fractionation Assay and Complex State Analysis
Cells were washed twice with ice-cold PBS and homogenized in homogenization buffer (HB: 250 mm sucrose, Sigma mammalian protease inhibitor mixture, and either 20 mm HEPES, pH 7.2, or 20 mm MES, pH 5.5) by passing repeatedly through a 26-gauge needle. Membrane and cytosolic fractions from post-nuclear supernatants were prepared by ultracentrifugation at 100,000 × g for 15 min at 4 °C in an RP80AT rotor using a Himac CS100 ultracentrifuge (Hitachi Koki, Tokyo, Japan). The membrane pellets were dissolved in HB containing 0.5% Triton X-100 to a volume equal to that of the cytosolic supernatants. Membrane pellets were suspended in 1× Laemmli sample buffer and boiled for SDS-PAGE followed by immunoblotting.
To analyze the state of the ESCRT-0 complex, blue native (BN)-PAGE was used. HeLa cells were fractionated into membrane and cytosolic fractions at pH 5.5 or 7.2 using the ultracentrifuge technique, as described above. The cytosolic fraction at pH 5.5 was neutralized with 0.1 volumes of 500 mm HEPES, pH 7.2. Then, n-dodecyl-β-maltoside was added to all cell fractions at a final concentration of 0.2%. BN-PAGE was performed according to the method described by Fiala et al. (20). Samples were resolved on a 5–12% gradient BN-polyacrylamide gel. The protein-blotted PVDF membrane was treated with 25% methanol containing 10% acetic acid to remove the Coomassie Brilliant Blue G-250 dye and then subjected to immunoblotting analysis.
In Vitro Transport Assays Using a Semi-intact Cell System and Microinjection
His6-tagged ESCRT-0 proteins were expressed in Escherichia coli BL21(DE3) derivative, Nico21(DE3) cells (New England Biolabs, Ipswich, MA), by induction with 0.5 mm isopropyl thiogalactoside for 24 h at 15 °C. The proteins were affinity-purified using a His-Trap HP column (GE Healthcare) in accordance with the manufacturer's instructions. The purified Hrs was fluorescently labeled by incubating with equal mole amounts of Alexa Fluor 488 C5-maleimide (Molecular Probes/Invitrogen) for 1 h at 30 °C. The unreacted dye was then removed by passage through a Sephadex G-25 column. The labeling efficiencies were greater than 80%.
Hrs-deficient HRSd cells grown on a 35-mm glass bottom dish were permeabilized with 0.006% digitonin in Transport buffer (TB: 20 mm HEPES, pH 7.2, 110 mm potassium acetate, 5 mm sodium acetate, 2 mm magnesium acetate, 2 mm DTT) for 5 min at 37 °C. The permeabilized HRSd cells were washed twice with TB and then incubated in TB containing 10 μg/ml of the indicated proteins at 37 °C. Time-lapse images were collected at 5-min intervals with a Zeiss LSM7 LIVE laser-scanning microscope and analyzed using a ZEN software system (Carl Zeiss, Jena, Germany).
The microinjection was carried out using a Zeiss Axio Observer Z1 fluorescence microscope (Carl Zeiss) equipped with an Eppendorf Transfer-Man NK2 micromanipulator (Eppendorf AG, Hamburg, Germany). HRSd cells grown on a 35-mm glass bottom dish were injected with 1 mg/ml of the indicated proteins and loaded onto an Eppendorf FemtoTip microcapillary (Eppendorf AG). After 1 h of incubation at 37 °C, a fluorescence image of the HRSd cells injected with the Alexa Fluor 488-labeled Hrs was captured.
Detection of Ubiquitylated Hrs
The boiling SDS-lysis method described by Urbé et al. (21) was employed to detect ubiquitylated Hrs. Cells were transfected with expression plasmids for Myc-ubiquitin, c-Cbl, and EGFR. After a 24-h transfection, cells were stimulated with 100 ng/ml EGF for 10 min and then lysed in 400 μl of boiling lysis buffer (2% SDS, 1 mm EDTA, 50 mm NaF, 1 mm Na3VO4, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, and mammalian protease inhibitor mixture), transferred to 2-ml screw-cap tubes, and then incubated for 30 min at 110 °C. The lysates were diluted with 4 volumes of dilution buffer (2.5% Triton X-100, 12.5 mm Tris, pH 7.4, 187.5 mm NaCl, and mammalian protease inhibitor mixture) and centrifuged at 12,000 × g for 5 min at 4 °C. The supernatants were subjected to immunoprecipitation at 4 °C overnight with the anti-HA probe antibody and protein A-Sepharose (GE Healthcare). The immunoprecipitates were extensively washed with lysis buffer and boiled with 1× Laemmli sample buffer before SDS-PAGE followed by immunoblotting.
RESULTS
STAM1 Expression Leads to the Diffuse Localization of Overexpressed Hrs and Recovery from Hrs-induced Endosomal Swelling
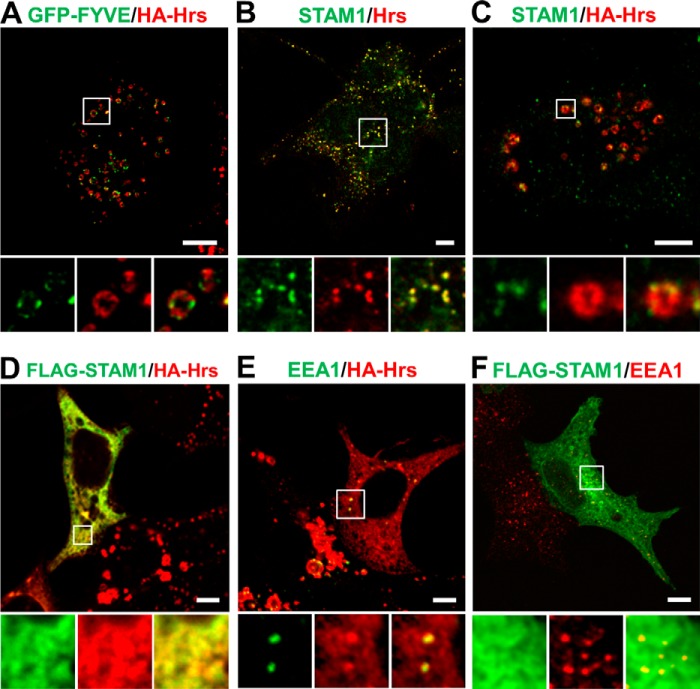
Overexpression of Hrs alone causes its accumulation on early endosomes, leading to the enlargement of the endosomes and the impairment of cargo sorting (13, 14). In the cell line 2HP67, which we established by stably transfecting HA-tagged Hrs into HEK293T cells, the overexpression of Hrs led to enlarged endosomes with a vesicular structure, with Hrs localized to microdomains distinct from the early endosomal marker SARA-FYVE-GFP (Fig. 1A). Endogenous STAM1 in normal HEK293T cells showed nearly the same punctate localization as Hrs (Fig. 1B), whereas in 2HP67 cells STAM1 was somewhat clustered in these Hrs-positive microdomains (Fig. 1C). Previous studies have reported that cooperative ubiquitin binding by the Hrs·STAM complex is required for endosomal sorting of the ubiquitylated cargo (8, 14). Moreover, there is a report that in Caenorhabditis elegans Hrs and STAM assemble into a heterotetrameric complex on the membrane both in vitro and in vivo (22). To investigate the effect of STAM co-expression on Hrs accumulation and endosome morphology, FLAG-tagged STAM1 was overexpressed in 2HP67 cells. We herein found that both Hrs and STAM1 exhibited a diffuse staining pattern (Fig. 1D). The same result was obtained in 2HP67 cells transfected with FLAG-tagged STAM2 (data not shown). Moreover, the effect of STAM1 expression on Hrs subcellular localization was also observed in HeLa cells (data not shown). Intriguingly, the Hrs-positive endosomes with the enlarged vesicular structures observed in the 2HP67 cells almost disappeared in the presence of exogenous STAM1, although punctate structures where Hrs and EEA1 co-localized were present (Fig. 1E). Overexpression of FLAG-tagged STAM1 in HEK293T cells did not affect the morphology of EEA1-positive endosomes (Fig. 1F). These results suggest that STAM1 can cause cells to recover from overexpressed Hrs-induced endosomal swelling.
FIGURE 1.
STAM1 expression leads to a diffuse localization of Hrs and a recovery from the endosome enlargement induced by Hrs overexpression. A, HA-Hrs-transduced HEK293T cell line, 2HP67, containing enlarged early endosomes. 2HP67 cells were transfected with GFP-SARA-FYVE (GFP-FYVE) as an early endosome marker and then immunostained for HA-Hrs. B, endogenous ESCRT-0 proteins in HEK293T cells. C, endogenous STAM1 localizes to Hrs-positive microdomains within enlarged endosomes. 2HP67 cells were immunostained for HA-Hrs and endogenous STAM1. D, STAM1 expression in Hrs-overexpressing cells results in a diffuse localization of Hrs and the disappearance of enlarged endosomes. 2HP67 cells were transfected with FLAG-STAM1 and then immunostained for HA-Hrs and FLAG-STAM1. E, morphology of early endosomes under the same experimental conditions as that of D. 2HP67 cells were transfected with FLAG-STAM1 and then immunostained for EEA1, as an early endosome marker, and HA-Hrs. F, overexpression of STAM1 has no effect on the morphology of EEA1-positive endosomes. HEK293T cells were transfected with FLAG-STAM1 and then immunostained for EEA1 and FLAG-STAM1. Scale bars, 5 μm.
SH3 Domain of STAM Is Required for the Diffuse Localization of Hrs and the Rescue of Hrs-induced Endosomal Swelling
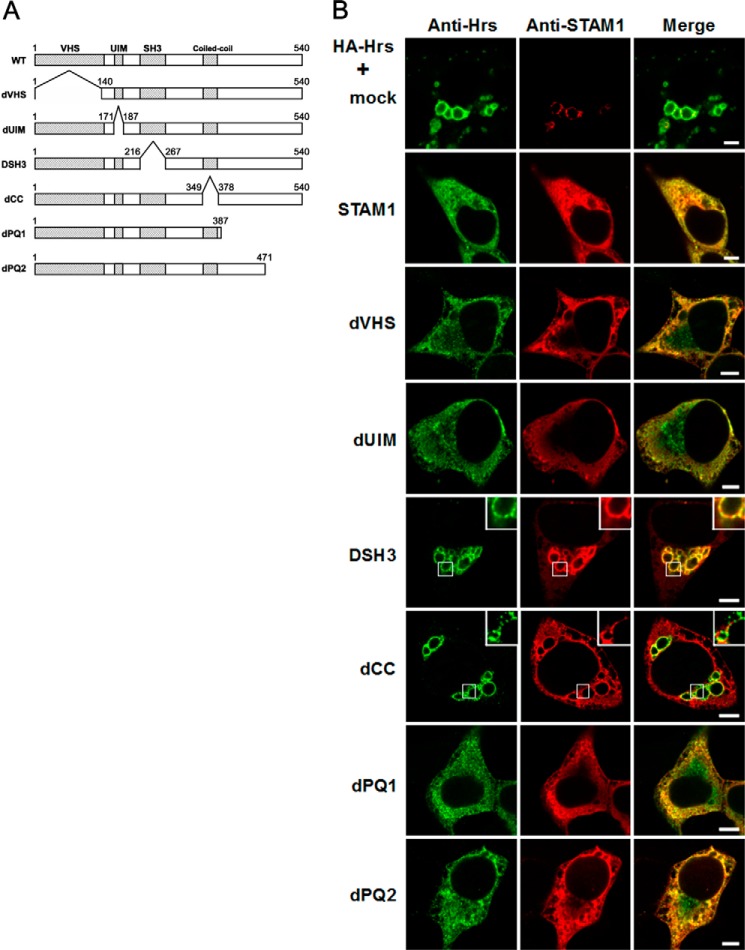
We next sought to determine the functional domain within STAM required for the diffuse localization by an immunostaining analysis on HEK293T cells transiently co-transfected with HA-tagged Hrs and wild-type STAM1 or a series of STAM1 deletion mutants (Fig. 2A). We often observed much larger endosomes in HEK293T cells transiently expressing HA-Hrs than the endosomes in 2HP67 cells (Fig. 2B). However, cells transiently co-transfected with wild-type STAM1 and HA-Hrs exhibited a diffuse staining for these proteins and an absence of enlarged endosomes (Fig. 2B) as well as STAM1-stably transfected 2HP67 cells. Co-expression of the dVHS(Δ1–139), dUIM(Δ170–186), dPQ1(Δ388–540), or dPQ2(Δ472–540) mutants with HA-Hrs produced almost the same results as co-expression of wild-type ESCRT-0 proteins (Fig. 2B). In contrast, co-expression of HA-Hrs and either the dCC(Δ350–377) or the DSH3(Δ217–266) STAM1 mutants that lack the coiled-coil region or the SH3 domain, respectively, led to Hrs-accumulated enlarged endosomes (Fig. 2B). This was also observed for cells where only HA-Hrs was introduced (Fig. 2B). The STAM1 dCC mutant staining pattern was diffuse in the cytoplasm and did not co-localize with Hrs on endosomes. Because the endosomal localization of STAM proteins is defined by their interaction with Hrs through their coiled-coil region (16, 23), it was expected that the Hrs binding-deficient dCC mutant would show a localization pattern distinct from that of Hrs. In contrast, the STAM1 DSH3 mutant predominantly localized to the Hrs-positive domains of the enlarged vesicular structures and concomitantly exhibited a diffuse weak cytoplasmic stain (Fig. 2B). The same effect of the STAM1 DSH3 mutant on Hrs-induced enlarged endosomes was observed in HeLa cells (data not shown). Thus, despite retaining its Hrs binding ability (17), expression of the STAM1 DSH3 mutant failed to reduce the endosomal accumulation of Hrs.
FIGURE 2.
Effect of STAM mutants on Hrs subcellular localization and endosomal morphology in Hrs-overexpressing cells. A, schematic representation of the structure of the STAM1 deletion mutants. The Vps27-Hrs-STAM (VHS), UIM, Src-homology 3 (SH3), and coiled-coil domains are indicated at the top of the diagram. B, expression of STAM1 mutants lacking the SH3 domain or the coiled-coil region fail to induce a diffuse localization of Hrs. HEK293T cells were transiently transfected with HA-Hrs and either an empty vector (mock), wild-type STAM1, or STAM1 deletion mutants and then immunostained with anti-Hrs and anti-STAM1 antibodies. Scale bars, 5 μm.
Next, when expressed at a physiological concentration, we examined whether the STAM1 DSH3 mutant could affect the localization of endogenous Hrs. SSd cells, which are embryonic fibroblasts derived from STAM1/2 double knock-out mice (Fig. 3) (23), were stably transduced with either wild-type STAM1 or the STAM1 dCC or DSH3 mutant constructs. Immunostaining for Hrs in wild-type STAM1-transduced (SSdRe-S1) or dCC mutant-transduced (SSd-dCC) cells showed essentially the same staining patterns as the parental SSd cells (Fig. 3). In contrast, in DSH3 mutant-transduced (SSd-DSH) cells, Hrs-positive endosomes were found to decrease in number and presented a swollen ring-shaped structure (Fig. 3), indicating that physiological levels of the STAM1 DSH3 mutant cause the accumulation of endogenous Hrs and the associated abnormalities in endosomal morphology. Although SSd cells exhibit no enlargement in endosome size, despite lacking STAM1/2, these cells appear to be a sensitive detector of the effects of mutant expression. Together, these findings indicate that the SH3 domain of STAM1 is a key functional domain for endosomal dissociation of Hrs and the maintenance of normal endosomal morphology. However, it is unclear whether the diffuse localization of ESCRT-0 results from a STAM-mediated endosomal dissociation of Hrs or a STAM-dependent restriction on the endosomal targeting of Hrs.
FIGURE 3.
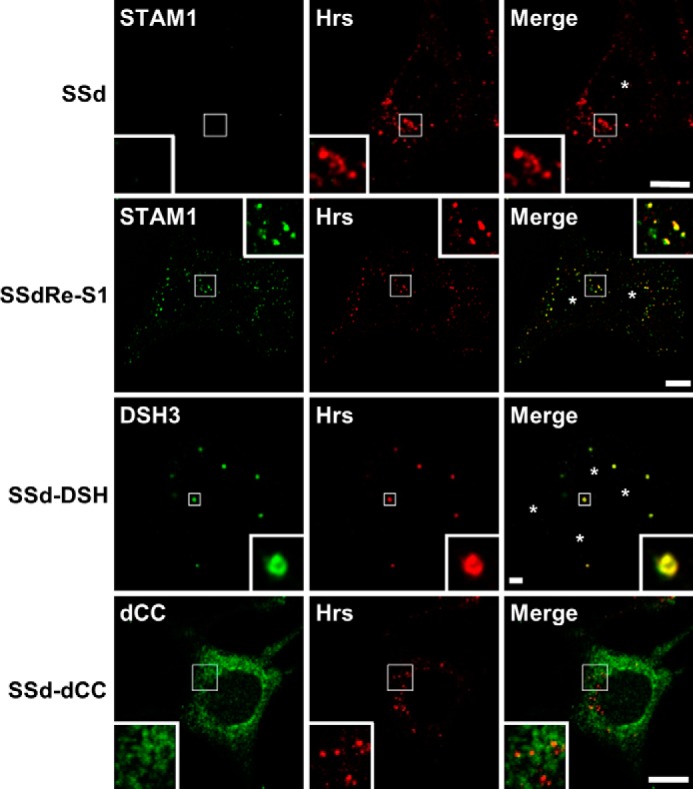
STAM1/2 double knock-out fibroblast cells transduced with the STAM1 DSH3 mutant exhibit enlarged endosomes. SSdRe-S1, SSd-DSH3, and SSd-dCC cells were created by stably transducing STAM1/2 double knock-out embryonic fibroblast-derived SSd cells with wild-type STAM1 or the STAM1 DSH3 or dCC mutant constructs, respectively. The cells were immunostained with anti-Hrs and anti-STAM1 antibodies. Asterisks represent nuclear localization. Scale bars, 5 μm.
Hrs Is Targeted to Endosomes without Complex Formation with STAM1 and Dissociates from Endosomes in a STAM1 Association-dependent Manner
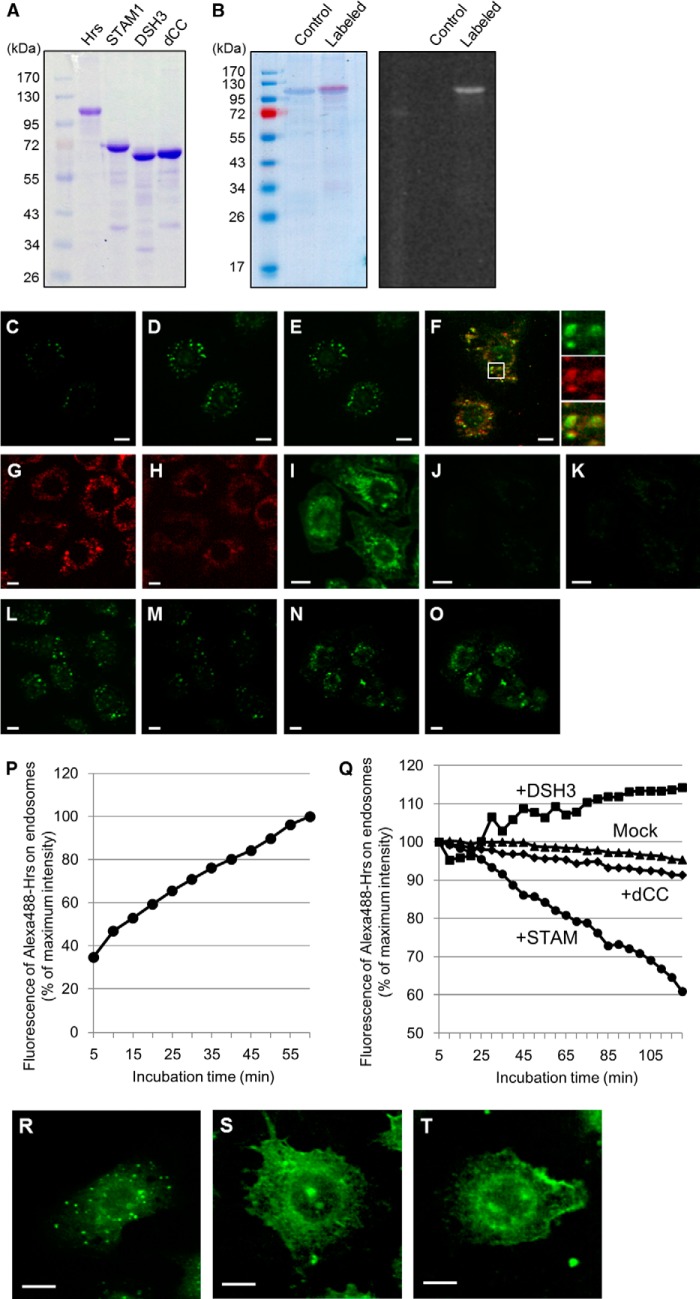
To examine the direct action(s) of STAM1 on endosome-localized Hrs, we used a combination of recombinant proteins and semi-intact cells. We prepared recombinant histidine-tagged Hrs, STAM1, STAM1 DSH3, and dCC mutants by expression in E. coli and purified the proteins on a metal-chelating column (Fig. 4A). The purified Hrs was fluorescently labeled with Alexa Fluor 488 dye (Fig. 4B). On loading of digitonin-permeabilized HRSd cells, embryonic fibroblasts derived from Hrs knock-out mice (16), with Alexa Fluor 488-labeled Hrs, a gradual increase of the fluorescence in the cytoplasmic punctate structures was observed (Fig. 4, C, D, and P). After replacing the culture medium, with the “mock” buffer solution not containing recombinant Hrs, 95% of the initial fluorescence signal was retained for up to 120 min (Fig. 4, D, E, and Q). To confirm that the fluorescently labeled Hrs localized structures are early endosomes, the permeabilized HRSd cells expressing the mRuby2-SARA-FYVE protein as an early endosome marker were loaded with the fluorescently labeled Hrs. Both the marker protein and the Hrs were significantly co-localized in intracellular puncta (Fig. 4F), representing an early endosomal targeting of the fluorescent-labeled Hrs. Lee et al. (24) reported that increasing the cytosolic pH weakened the binding affinity between the FYVE domain and phosphatidylinositol 3-phosphate (PI(3)P), a phospholipid enriched in the early endosome membranes, resulting in the dissociation of the FYVE domain-containing proteins from the endosomal membrane. Consistent with this, when HRSd cells expressing the FYVE domain-fused mRuby2 as an endosome-associated marker protein were treated with a buffer solution containing 100 mm HEPES-KOH, pH 7.2, the mRuby2 fluorescence on endosomal structures was clearly observed to decrease, whereas the diffuse cytoplasmic signal concomitantly increased (Fig. 4, G and H). Analysis using LysoSensor Green dye (Molecular Probes/Invitrogen), which becomes more fluorescent in acidic organelles, revealed that endosomes and lysosomes in semi-intact HRSd cells maintain their acidic environment (Fig. 4I). Most of the acidic organelle-probe fluorescence was lost following treatment with a buffer containing 100 mm HEPES-KOH, pH 7.2, representing a neutralization of the acidic organelles (Fig. 4J). When added subsequently to the buffer treatment, fluorescently labeled Hrs no longer showed its punctate pattern in the semi-intact HRSd cells (Fig. 4K). These data suggest that endosomal localization of a peripheral membrane protein Hrs depends on the acidic microenvironment on the endosomal surface. We next examined whether the subsequent addition of nonlabeled recombinant STAM1 affects the endosomal localization of the previously loaded fluorescent Hrs. The fluorescence of the labeled Hrs on endosomes decreased over time and had ≈60% of the initial intensity at 120 min after the input of the recombinant STAM1 (Fig. 4, L, M, and Q). As a control, the loading for 120 min of Hrs binding-deficient dCC mutant resulted in a similar change in the fluorescence of the endosomal Hrs after replacing the mock buffer solution (Fig. 4Q). In contrast, addition of the nonlabeled DSH mutants led to a small increase in the fluorescence of the labeled Hrs on endosomes (Fig. 4, N, O, and Q). We further investigated the requirement for the formation of the ESCRT-0 complex prior to its endosomal targeting. Loading of neither the Hrs-STAM1 nor the Hrs-DSH3 pre-formed complexes showed an endosomal localization in semi-intact HRSd cells (data not shown). However, because this may be due to the impermeability of the large ESCRT-0 complex to the digitonin-treated plasma membrane, we microinjected the recombinant proteins into HRSd cells. Microinjection of the Alexa Fluor 488-labeled Hrs alone resulted in the punctate fluorescent pattern, as observed for Hrs loading into the semi-intact cells (Fig. 4R). Conversely, neither the Hrs-STAM1 nor the Hrs-DSH3 pre-formed complexes showed endosomal accumulation (Fig. 4, S and T). Thus, complex formation of Hrs with STAM1 appears not to have a facilitatory but rather an inhibitory effect on their endosomal localization. When the Hrs-DSH3 pre-formed complex was microinjected into cells, it exhibited a diffuse cytoplasmic localization (Fig. 4T), whereas in the digitonin-permeabilized cells, a nonlabeled DSH3 mutant seemed to rather promote the accumulation of the Hrs pre-localized on endosomes (Fig. 4, N, O, and Q). These data suggest that the pre-existence of Hrs on endosomes is a prerequisite for the endosomal localization of STAM1. When taken together, our data suggest that Hrs may be targeted to endosomes without complex formation with STAM1 and associate with STAM1 on endosomes in vivo. However, the ESCRT-0 complex does eventually dissociate from endosomes.
FIGURE 4.
STAM-independent endosomal targeting and STAM-mediated endosomal dissociation of Hrs. A, purification of the recombinant histidine-tagged Hrs, STAM1, and STAM1 mutants. The purified proteins were analyzed by SDS-PAGE with Coomassie Brilliant Blue staining. B, fluorescent labeling of the recombinant Hrs. The purified Hrs was labeled with the Alexa Fluor 488 dye. Coomassie Brilliant Blue-stained gel (left panel) and a UV-transilluminated (right panel) SDS-polyacrylamide gel after electrophoresis are shown. C and D, digitonin-permeabilized HRS knock-out mouse embryonic fibroblast HRSd cells were incubated in transport buffer (TB) containing 20 mm HEPES, pH 7.2, with Alexa Fluor 488-labeled Hrs (Alexa488-Hrs) at 37 °C. C and D represent the fluorescence images after 5- and 60-min incubation periods, respectively. The mean fluorescence intensity of the Alexa488-Hrs on endosomal punctate structures in semi-intact cells was quantified from the fluorescence images every 5 min for 1 h after Hrs loading (P). E, semi-intact cells in D were further incubated in TB buffer in the absence of Alexa488-Hrs and images captured every 5 min for the subsequent 2 h. The relative values of the mean intensity of Alexa488-Hrs fluorescence were plotted as the Mock dataset in Q. F, Alexa488-Hrs is targeted to the early endosome marker mRuby2-SARA-FYVE (mRuby2-FYVE)-positive endosomes in semi-intact HRSd cells. HRSd cells expressing mRuby2-FYVE (red) were permeabilized with digitonin and then treated with the TB-containing Alexa488-Hrs (green). The fluorescence image was captured after a 60-min incubation. G and H, neutralization of the intracellular pH elicits dissociation of the FYVE domain-containing proteins from the endosomal surface. Permeabilized HRSd cells expressing mRuby2-FYVE were treated with TB containing 100 mm HEPES, pH 7.2. G and H represent the fluorescence images before and after a 60-min incubation, respectively. I, analysis using the acidic organelles probe LysoSensor dye demonstrates the acidic environment of endosomes in semi-intact HRSd cells. J, intracellular environment of the cells in I is neutralized by treatment with TB containing 100 mm HEPES, pH 7.2. K, endosome-neutralized semi-intact cells in J were incubated in a TB containing 20 mm HEPES, pH 7.2, with Alexa488-Hrs at 37 °C for 1 h. L–O, effect of STAM1- and DSH3 mutant-loading on the endosomal localization of Hrs. Alexa488-Hrs loaded semi-intact cells, as shown in D, were further incubated in a TB containing 20 mm HEPES, pH 7.2, with recombinant STAM1 (L and M) and DSH3 mutants (N and O). The fluorescence images after a 5-min (L and N) and a 120-min incubation (M and O) are presented. The relative values of the mean intensity of Alexa488-Hrs fluorescence in STAM1-, DSH3-, and dCC-loaded semi-intact cells were plotted as the +STAM, +DSH3, and +dCC datasets, respectively, in Q. R–T, requirement for the complex formation of ESCRT-0 proteins on their endosomal targeting. Alexa488-Hrs was mixed with nonlabeled STAM1 or DSH3 mutants, and then microinjected into HRSd cells. After a 1-h incubation, the fluorescence images of the cells microinjected with Alexa488-Hrs alone as a control (R), the Alexa488-Hrs·STAM1 complex (S), or the Alexa488-Hrs/DSH3 mutant (T) were acquired. Scale bars, 10 μm.
Membrane Binding of Hrs Is Controlled by Environmental pH and STAM Expression
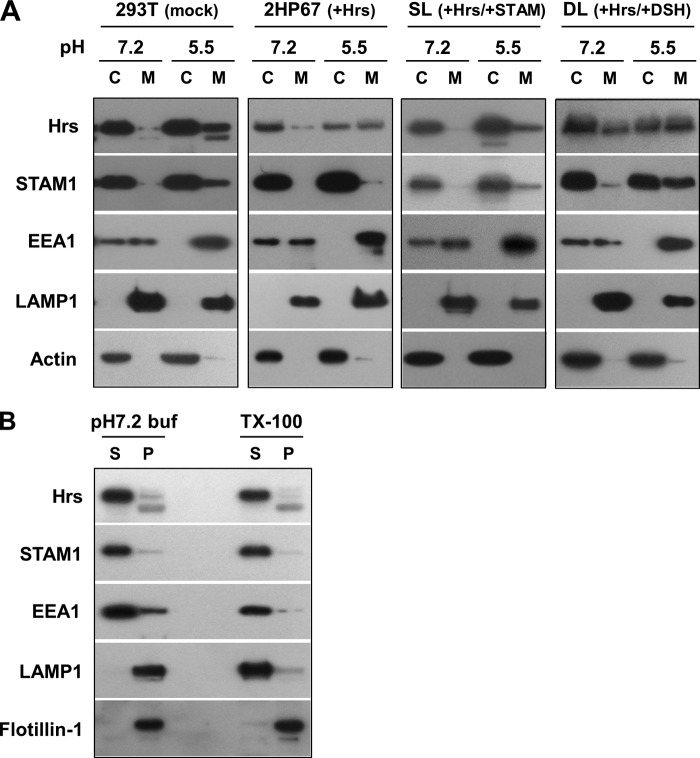
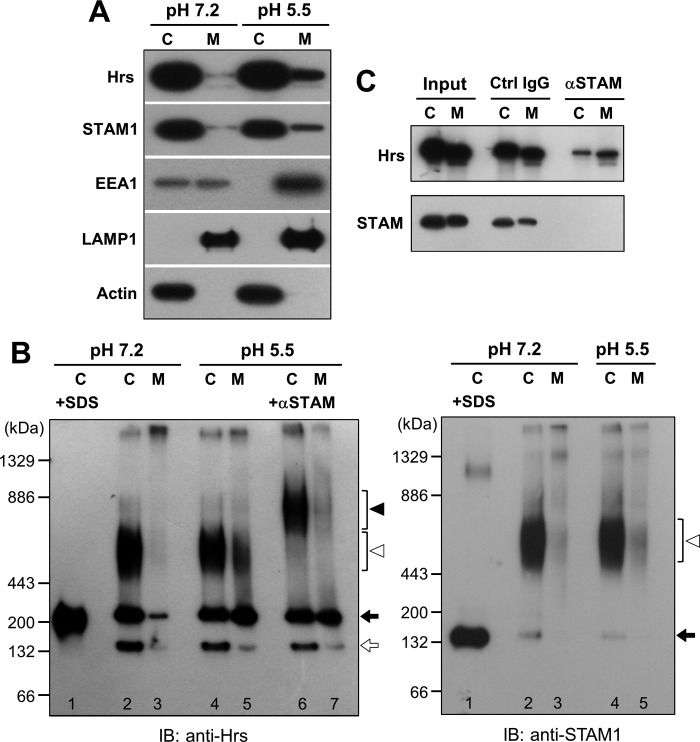
Most immunostaining studies in normal cells show a punctate staining pattern for Hrs and STAM, representing their endosomal localization. However, subcellular fractionation assays in previous reports have demonstrated that Hrs and STAM are predominantly localized to the cytoplasm (14, 21, 25). Considering an acidic pH-dependent interaction of the FYVE domain with the endosomal lipid PI(3)P (24), membrane-bound forms of Hrs and STAM are probably underestimated, because of the membrane dissociation that occurs during cell fractionation under neutral pH conditions. We therefore fractionated HEK293T cells, using a centrifugation technique, into cytosolic and membrane fractions at pH 5.5 and 7.2. The FYVE protein EEA1 was predominantly found in the membrane fraction at pH 5.5 but was distributed equally in both fractions at pH 7.2 (Fig. 5A), providing assurance that when cell fractionation is performed under acidic pH conditions it does indeed show a pH-dependent membrane binding for a FYVE protein. However, only 10% of endogenous Hrs, which is a member of the FYVE protein family, was recovered in the membrane fraction at pH 5.5, whereas almost all Hrs proteins were found in the cytoplasmic fraction at pH 7.2 (Fig. 5A). Thus, Hrs exhibits a pH-dependent membrane association, apparently with a lower affinity than EEA1. Endogenous STAM1 was distributed between both fractions in almost the same manner as Hrs (Fig. 5A). We then validated whether the ESCRT-0 proteins and EEA1 that were observed in the membrane fraction at pH 5.5 are not their aggregates due to the acidic solution. When the membrane fraction at pH 5.5 was subjected to 1% Triton X-100 protein extraction followed by ultracentrifugation, most of Hrs, STAM1, and EEA1 were recovered in the supernatants (Fig. 5B). Furthermore, the treatment with a 50 mm HEPES, pH 7.2, buffer led to detachment of a large part of these proteins from the membrane prepared under acidic conditions at pH 5.5 (Fig. 5B). As a control, we verified the distribution of lysosomal integral membrane protein LAMP1 and Triton X-100-insoluble raft-associated protein flotillin-1. LAMP1 was solubilized when treated with Triton X-100, whereas flotillin-1 was recovered in the precipitate following both treatments (Fig. 5B). These results indicate that distributions of the ESCRT-0 proteins and EEA1 to the membrane fraction at acidic pH are due to their pH-dependent membrane associations and not to aggregation of these proteins. Fractionation of 2HP67 cells overexpressing HA-tagged Hrs showed that half of the total Hrs was recovered in the membrane fraction at pH 5.5 (Fig. 5A), reflecting the endosomal accumulation of overexpressed Hrs. Unlike in HEK293T cells, endogenous STAM1 in 2HP67 cells was scarcely membrane-associated upon fractionation under acidic pH (Fig. 5A). Both Hrs and STAM1 in SL cells, established by the stable transfection of FLAG-tagged STAM1 into 2HP67 cells, have a fractionation pattern similar to that of the endogenous ESCRT-0 proteins in HEK293T cells (Fig. 5A). This indicates that the STAM1-mediated control of the subcellular localization of Hrs results in the diffuse staining pattern of the ESCRT-0 proteins, as shown in Fig. 1D. The Hrs fractionation pattern at pH 5.5 from the cell line DL, stably transfected with the FLAG-tagged DSH mutants, was very similar to that of 2HP67 cells (Fig. 5A). However, accumulation of the DSH mutant in the membrane fraction at pH 5.5 (40% of total mutant protein) is consistent with the data from the immunostaining of HEK293T cells overexpressing either Hrs or the DSH mutant (Figs. 2B and 5A), suggesting a requirement for the STAM1 SH3 domain in the control of Hrs localization. As mentioned above, fluorescently labeled Hrs also targets to endosomes in a pH-dependent manner (Fig. 4K). These data indicate that Hrs exhibits binding to endosomal membranes by a mechanism dependent on its acidic environment, whereas Hrs tends to be localized to the cytoplasm in the presence of STAM1.
FIGURE 5.
Environmental pH and STAM expression control the subcellular localization of Hrs. A, cell fractionation of HEK293T, 2HP67, SL, and DL cells. Cells were homogenized at the indicated pH. Cytosolic (C) and membrane (M) fractions from post-nuclear supernatants were prepared by ultracentrifugation at 100,000 × g for 15 min at 4 °C and subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. B, ESCRT-0 proteins fractionated in the membrane fraction at pH 5.5 are not aggregates. The cell membrane prepared at pH 5.5 as shown in A was suspended in 50 mm HEPES, pH 7.2 buffer (pH 7.2 buf), or solubilized with 1% Triton X-100 (TX-100), and then separated into supernatant (S) and precipitate (P) by ultracentrifugation. The samples were subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. The distribution of lysosomal integral membrane protein LAMP-1 and Triton X-100-insoluble raft-associated protein flotillin-1 were verified as the control.
STAM1-unbound Form of Hrs Exists on Endosomes in Vivo
The distributions of the ESCRT-0 subunit proteins between the cytosolic and membrane fractions from HeLa cells were similar to those from HEK293T cells (Figs. 5A and 6A). We next analyzed the state of the Hrs and STAM1 complexes in the cytosolic and membrane fractions by employing BN-PAGE. In the following immunoblot, an anti-Hrs antibody gave three major signals, at ≈140, ≈250, and ≈600 kDa, among all the fractions (Fig. 6B, left panel, lanes 2–5). BN-PAGE/immunoblotting of a cytoplasmic fraction isolated from cells at pH 7.2, which had been pre-denatured by SDS, showed an Hrs-positive band almost corresponding to the ≈250-kDa band of the native protein (Fig. 6B, left panel, lane 1), indicating that the ≈250-kDa band is likely to be a monomer of Hrs, whereas the ≈140-kDa band is probably a degraded product. The broad signal at ≈600 kDa was observed on the immunoblot against STAM1 as well (Fig. 6B, right panel, lane 2–5). When the fractions at pH 5.5 were pretreated with an anti-STAM1 antibody, the Hrs-positive signal at ≈600 kDa was shifted to a position equivalent to 800 kDa, whereas the signals at positions of ≈250 kDa did not change (Fig. 6B, left panel, lanes 6 and 7). This result indicates that the ≈600-kDa signal represents a complex formed between Hrs and STAM1, whereas the lower bands do not. The STAM1-positive band at the position of ≈150 kDa was observed in both the SDS-denatured and native samples (Fig. 6B, right panel, lanes 1, 2, and 4), representing a monomeric form of STAM1. Comparison between these two STAM1-positive signals revealed that a large proportion of STAM1 in HeLa cells forms a complex with Hrs, appearing as a ≈600-kDa signal on the BN-polyacrylamide gel (Fig. 6B, right panel). However, a remarkable proportion of Hrs exists in a STAM1-unbound, probably monomeric, form, as described above. We noticed that the relative abundance of STAM1-unbound Hrs in the membrane fraction at pH 5.5 appeared to be higher than that in the cytoplasmic fraction (Fig. 6B, left panel, lanes 4 and 5). To confirm this, STAM1-unbound Hrs was estimated by depleting the STAM1-bound Hrs pool using anti-STAM1 antibody-immobilized beads (Fig. 6C). After depletion of STAM1-bound Hrs, the residual Hrs remaining in the membrane fraction at pH 5.5 was 30%, with <5% in the cytoplasmic fraction (Fig. 6C). Thus, the ESCRT-0 complex consisting of Hrs and STAM1 is observed in both the cytoplasmic and membrane fractions. However, membrane-associated Hrs exists to some extent as a STAM1-unbound form, supporting the notion of the STAM-independent endosomal targeting activity of Hrs in vivo.
FIGURE 6.
Hrs exists partly as a monomer on endosomes in vivo. A, cell fractionation of HeLa cells. HeLa cells were homogenized at the indicated pH. Cytosolic (C) and membrane (M) fractions from post-nuclear supernatants were prepared by ultracentrifugation at 100,000 × g for 15 min at 4 °C and subjected to SDS-PAGE followed by immunoblotting with the indicated antibodies. B, BN-PAGE analysis of the endogenous Hrs·STAM1 complex. Cell fractions from HeLa cells, as shown in A, were separated by BN-PAGE followed by immunoblotting (IB) with anti-Hrs (left panel) and anti-STAM1 antibodies (right panel). SDS-pretreated cytosolic fractions at pH 7.2 (+SDS) were loaded into the 1st lane of each panel. From the cell fractions loaded from lanes 2–5 of the left panel, Hrs-positive signals were mainly detected at ≈140 (white arrow), ≈250 (black arrow), and ≈600 kDa (white arrowhead). Anti-STAM antibody pretreatment of cell fractions at pH 5.5 (+αSTAM) was loaded into lanes 6 and 7 of the left panel, in which the ≈600-kDa signal was shifted to a higher apparent molecular mass (≈800 kDa, black arrowhead) by complex formation with the antibody. In the right panel, STAM1-positive signals were mainly detected at ≈150 (black arrow) and ≈600 kDa (white arrowhead) positions. C, analysis of STAM-unbound Hrs by STAM protein depletion. Cell fractions at pH 5.5 from HeLa cells, as shown in A, were prepared. The neutralized cytosolic fractions and the 6-fold enriched membrane fractions were treated with control mouse IgG- or anti-STAM mouse IgG-conjugated Sepharose. Samples were separated by SDS-PAGE followed by immunoblotting with anti-Hrs (upper panel) and anti-STAM1 antibodies (lower panel).
Expression of STAM1, but Not a STAM1 DSH3 Mutant, Rescues Hrs Overexpression-induced Impairment of EGFR Degradation
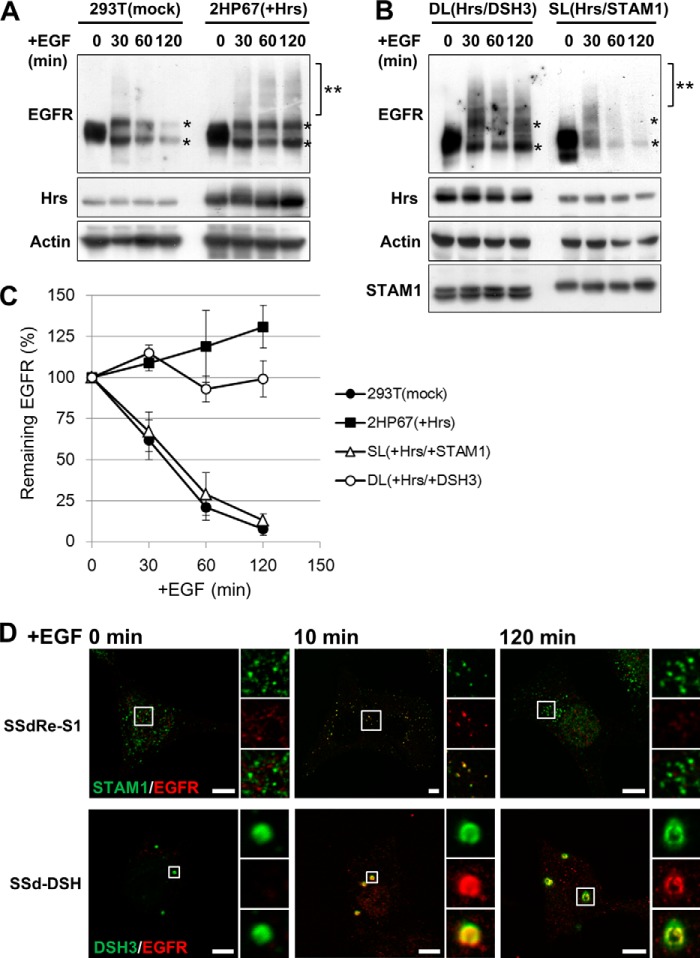
Concomitant with the morphological changes in endosomes, Hrs overexpression also inhibits the degradation of ubiquitylated cargo (13). Thus, we investigated whether expression of STAM1 can rescue the inhibitory effect of overexpressed Hrs on the EGFR degradation. In this assay, we used cells exogenously expressing ubiquitin and c-Cbl, thereby promoting EGFR degradation in HEK293T cells (26). In control cells, EGFR was almost completely degraded 120 min after stimulation (Fig. 7, A and C). In contrast, EGFR did not appear to be degraded in 2HP67 cells, indicating an inhibitory effect of Hrs overexpression on endosomal cargo sorting (Fig. 7, A and C). In SL cells, made by stably transducing wild-type STAM1 into Hrs-overexpressing 2HP67 cells, the EGFR degradation rate recovered, as compared with that seen in control cells, indicating that expression of wild-type STAM1 can abrogate the inhibitory effect of overexpressed Hrs on EGFR degradation. DL cells, made by stably transducing the DSH mutant into Hrs-overexpressing 2HP67 cells, retained the impairment in EGFR degradation, similar to 2HP67 cells (Fig. 7, B and C). Immunofluorescence analysis showed that endocytosed EGFRs in both SSdRe-S1 and SSd-DSH cells after 10 min of stimulation were delivered to STAM1- or DSH3 mutant-positive endosomes (Fig. 7D). At 120 min after EGF stimulation, EGFR remained localized in DSH3-positive endosomes in SSd-DSH cells, which contrasts with the disappearance of those in SSdRe-S1 cells, indicating the expression of the STAM1 DSH mutant at physiological levels causes impairment of EGFR degradation (Fig. 7D). These results suggested that the SH3 domain is required for the proper function of STAM1 in endosomal cargo sorting. Taken together, the effects of STAM1 and the DSH3 mutant co-expression on the impairment in EGFR degradation in Hrs-overexpressing cells correlate with those describing the abnormalities in endosomal morphology observed in the immunostaining analysis shown in Fig. 2B. Thus, our data suggest that the quantitative control of endosomal Hrs via STAM1-mediated endosomal dissociation is required for both proper cargo sorting and normal endosome morphology.
FIGURE 7.
Expression of wild-type STAM1, but not the DSH3 mutant, rescues the impairment of EGFR degradation in Hrs-overexpressing cells. A, impairment of EGFR degradation in Hrs-overexpressing 2HP67 cells. Cells were transfected with ubiquitin, c-Cbl, and EGFR for 24 h and then stimulated with EGF (100 ng/ml) for the indicated times at 37 °C. Lysates were analyzed by SDS-PAGE followed by immunoblotting with the indicated antibodies. After EGF stimulation, EGFR doublets (asterisk) and the ubiquitylated form (double asterisk) were observed. B, expression of wild-type STAM1, but not the DSH3 mutant, rescues EGFR degradation in Hrs-overexpressing cells. SL and DL cell lines were generated by stable transduction of 2HP67 cells with lentiviral vectors encoding either FLAG-STAM1 or the FLAG-DSH3 mutant. EGF stimulation and SDS-PAGE were followed by immunoblotting carried out as described in A. C, relative abundance of the remaining EGFR at the indicated times. Data represent the means ± S.D. from three independent experiments. D, impairment of EGFR degradation in the DSH3 mutant-transduced STAM1/2 knock-out cells. SSdRe-S1 and SSd-dCC cells were immunostained with anti-STAM1 and anti-EGFR antibodies at the indicated times after EGF stimulation (100 ng/ml). Scale bars, 5 μm.
Defective STAM1-associated Deubiquitylating Enzyme Activity Does Not Lead to Endosomal Swelling
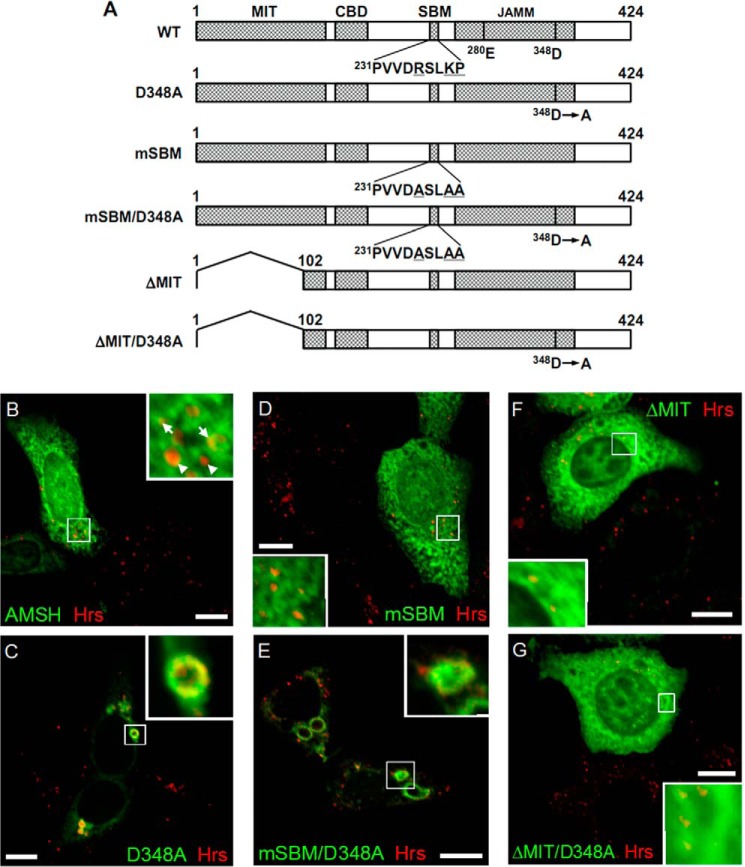
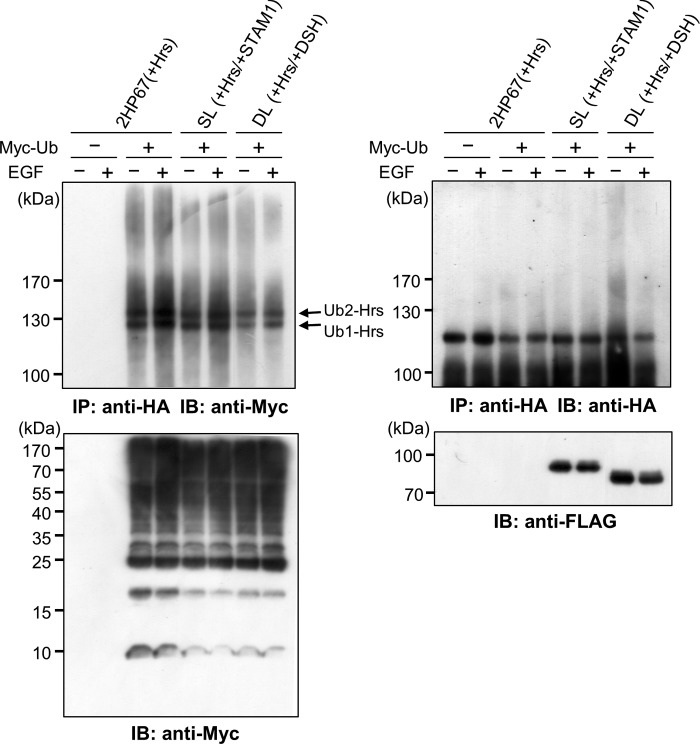
Previous studies have reported that the STAM-associated deubiquitylating enzymes AMSH and UBPY, which interact with the SH3 domain of STAM proteins, promote the down-regulation of ubiquitylated cell surface receptors (27). Expression of either AMSH or UBPY catalytically inactive mutants leads to endosomal swelling and the accumulation of endosomal ubiquitylated cargo (28–30). Therefore, a similar dominant negative effect of the STAM DSH3 mutant may be attributed to a deficiency in STAM-associated deubiquitylating activity. Consistent with a previous report (28), expression of AMSH-D348A (Fig. 8A) caused an accumulation of endogenous Hrs on enlarged AMSH-D348A-positive endosomes (Fig. 8C). Expression of wild-type AMSH had no effect on Hrs-positive endosomes (Fig. 8B). AMSH and UBPY are known to be associated with the STAM SH3 domain via a STAM-binding motif (SBM), PX(V/I)(D/N)RXXKP (31, 32). We found that a STAM binding-defective AMSH mutant, AMSH-mSBM (Fig. 8A), localized to the cytoplasm and not to Hrs-positive endosomes (Fig. 8D). In contrast, AMSH carrying mutations in both the SBM and the catalytic site, AMSH-mSBM/D348A (Fig. 8A), led to an enlargement of endosomes, on which Hrs accumulated (Fig. 8E), indicating that the dominant negative effect of inactive AMSH-D348A is independent of STAM binding ability. Considering that AMSH interacts with the ESCRT-III complex through its N-terminal microtubule interacting and transport domains (32–34), the endosome swelling caused by the expression of inactive AMSH-D348A may be due to a failure in the sorting process downstream of ESCRT-0 function. This is supported by the findings that AMSH-ΔMIT/D348A, lacking both the ESCRT-III interacting and deubiquitylating activities (Fig. 8A), as well as an enzymatically active AMSH-ΔMIT mutant (Fig. 8A), had no effect on the morphology of Hrs-positive endosomes, although they did co-localize with Hrs on endosomes (Fig. 8, F and G). Thus, the dominant negative effects on endosome morphology and endosomal cargo sorting by the expression of the DSH3 mutant are likely to be independent of the defect in STAM-associated deubiquitylating activity. We further analyzed the effect of STAM1 expression on the ubiquitylation state of Hrs. We prepared cell lysates from 2HP67, SL, and DL cells, which were transiently transfected with Myc-tagged ubiquitin, c-Cbl, and EGFR, and then immunoprecipitated Hrs, according to the previously described boiling SDS-lysis method (21). Immunoblotting with an anti-Myc antibody revealed mono- and di-ubiquitylated forms of Hrs, as well as poly-ubiquitylated Hrs appearing as a smear in all Hrs-overexpressing cell lines (Fig. 9). Hrs ubiquitylation after EGF stimulation exhibited only a slight increase (Fig. 9), a result that is consistent with those of Urbé et al. (21). It is noteworthy that the Hrs ubiquitylation was similar in all cell lines (Fig. 9), indicating that STAM1 expression has no effect on the level of Hrs ubiquitylation. Together with the immunostaining and cell fractionation analysis (Figs. 2B and 5), the subcellular localization of Hrs appears to be independent of the levels of Hrs ubiquitylation.
FIGURE 8.
STAM-mediated endosomal dissociation of Hrs is independent of STAM-associated deubiquitylating activity. A, schematic representation of the structure of the AMSH deletion mutants. The microtubule interacting and transport (MIT) domain, clathrin-binding domain (CBD), STAM-binding motif (SBM), and JAB1/MPN/Mov34 metalloprotease (JAMM) domains are indicated at the top of the diagram. The putative catalytic (Glu280) and metal-binding (Asp348) residues in the active site are indicated. The mutated residues in the SBM are underlined. B–G, HEK293T cells were transfected with wild-type AMSH, AMSH-D348A, AMSH-mSBM, AMSH-mSBM/D348A, AMSH-ΔMIT, or AMSH-ΔMIT/D348A and were then immunostained for endogenous Hrs and AMSH or the AMSH mutants. B, AMSH partially co-localizes with endogenous Hrs but does not induce morphological changes in Hrs-positive endosomes. Arrows in the inset indicate Hrs-positive endosomes co-localizing with AMSH, and arrowheads indicate Hrs-positive but AMSH-negative endosomes. C, deubiquitylating activity-deficient AMSH (AMSH-D348A) induces endosomal swelling. D, STAM1-binding-deficient AMSH (AMSH-mSBM) does not co-localize with Hrs. E, expression of the AMSH mSBM and the D348A double mutant (AMSH-mSBM/D348A) leads to the enlargement of Hrs-positive endosomes. F and G, both ESCRT-III binding-deficient AMSH (AMSH-ΔMIT) and ΔMIT and the D348A double mutant (AMSH-ΔMIT/D348A) co-localize with endogenous Hrs but have no effect on endosomal morphology. Scale bars, 10 μm.
FIGURE 9.
Ubiquitylation state of Hrs is independent of STAM1 expression. 2HP67, SL, and DL cells were transfected with Myc-ubiquitin, c-Cbl, and EGFR for 24 h and then stimulated with EGF (100 ng/ml) for 10 min at 37 °C. Cell lysates were prepared according to the boiling SDS-lysis method (21). Immunoprecipitates (IP) of HA-Hrs were separated by SDS-PAGE and then subjected to immunoblot (IB) analyses with anti-Myc (upper left panel) and anti-HA antibodies (upper right panel). The levels of Myc-ubiquitin (lower left panel), FLAG-STAM1, and FLAG-DSH3 (lower right panel) in the total lysate were examined by immunoblotting. Mono- (Ub1-Hrs) and di (Ub2-Hrs)-ubiquitylated forms of Hrs are indicated.
DISCUSSION
In this study, we have shown that the ESCRT-0 subunit Hrs is targeted to endosomes independent of STAM binding and dissociates from endosomes following its association with its counterpart protein STAM1. These processes probably maintain the consistent endosomal/cytoplasmic distribution of the ESCRT-0 complex, and their deficiency consequently leads to abnormal endosome morphology and an impairment of endosomal cargo sorting.
The interaction of FYVE domains with phosphatidylinositide PI(3)P-enriched endosomal membranes is regulated by intracellular acidic pH (24). The Hrs FYVE domain fused with GST, as well as the EEA1 FYVE domain, has been shown previously to exhibit intensive binding to PI(3)P-containing liposomes at pH 5.5 (24). We also showed that targeting of Hrs to the endosomal membrane is affected by environmental pH (Figs. 4 and 5). The kinetics study revealed that binding of the EEA1 FYVE domain to PI(3)P-containing vesicles at pH 6.0 is a 24-fold higher affinity (dissociation constant Kd at pH 6.0 = 49 nm) than that at pH 8.0 (35). The subsequent NMR studies revealed that the pH dependence of interaction between the FYVE domain and PI(3)P is attributed to a protonation of two histidine residues conserved in the FYVE domain sequences (24, 35). Combined with the crystal structure data (36), positively charged histidine residues that are protonated in an acidic environment interact with the negatively charged head groups of PI(3)P. These data thus suggest that endosomal targeting of a peripherally membrane-associated protein Hrs requires acidification of the cytoplasmic surface of endosomal membrane. The proton fluxes induce a transmembrane pH gradient across the lipid bilayer (37), thus providing a possible mechanism for this acidification. Concomitantly, proton leakage from the endosomal lumen by Na+/H+ exchangers may contribute to the formation of the acidic microenvironment. Mitsui et al. (38) demonstrated that the pH of the cytoplasmic surface of the endosomes in yeast cells is lower than that in the cytoplasm using a pH-sensitive fluorescent protein fused with Vps27, and the yeast Na+/H+ exchanger Nhx1/Vps44 activity is required for endosomal targeting of Vps27. When Hrs was associated with STAM1 prior to its introduction into cells, it no longer accumulated at endosomes (Fig. 4). In the absence of STAM proteins, purified mouse Hrs forms a hexameric homo-oligomer, which is able to bind to endosomal membranes in vitro (12). In contrast, we found for the first time that STAM-unbound Hrs exists predominantly as a monomer in vivo (Fig. 6). This finding indicates that complex formation between Hrs and STAM is not constitutive, and it potentially supports the hypothesis of endosomal targeting of Hrs independent of STAM binding.
The overexpressed ESCRT-0 proteins, Hrs and STAM1, do not only co-localize on endosomes but also exhibit a remarkably diffuse localization (Fig. 1, D and E). Their intracellular distributions may be attributed to the STAM-dependent dissociation of endosome-localized Hrs (Fig. 4). In vitro reconstitution studies of MVB biogenesis in giant liposomes, using the purified yeast ESCRT proteins, clearly demonstrated that the ESCRT-0 complex clusters ubiquitylated cargo (6). The same group also demonstrated that the downstream ESCRT-I/-II supercomplex induces membrane bud formation, independent of cargo cluster formation by the ESCRT-0 complex. The subsequent entrapment of the cargo in the buds is required for the interaction between the yeast Hrs homolog, Vps27, in ESCRT-0 and the yeast Tsg101 homolog, Vps23, in ESCRT-I (6). However, ESCRT-0 is not entrapped in the buds formed by ESCRT-I/-II, suggesting that a process in which ESCRT-0 is released from endosomes following a handover of the ubiquitylated cargo to ESCRT-I. Overexpression of wild-type Hrs causes a severe impairment of endosomal cargo sorting (Fig. 7A) (13). Rescue of this Hrs-induced sorting defect by STAM1 co-expression, as shown in Fig. 7B, may indicate that the STAM1-mediated membrane dissociation of Hrs is an indispensable event for cargo migration from the ESCRT-0/ubiquitylated cargo clusters to the buds.
Acknowledgments
We thank the Instrumental Analysis Research Center for Human and Environmental Science at Shinshu University for technical assistance with DNA sequencing and immunostaining analyses.
This work was supported in part by Grant-in-aid for Scientific Research C_25461586 (to T. T.).
- MVB
- multivesicular body
- AMSH
- associated molecule with the SH3 domain of STAM
- EGFR
- epidermal growth factor receptor
- ESCRT
- endosomal sorting complex required for transport
- Hrs
- hepatocyte growth factor-regulated tyrosine kinase substrate
- BN-PAGE
- blue native-PAGE
- STAM
- signal-transducing adaptor molecule
- UIM
- ubiquitin-interacting motif
- PI(3)P
- phosphatidylinositol 3-phosphate
- UBD
- ubiquitin-binding domain
- SH3
- Src homology 3
- SBM
- STAM-binding motif.
REFERENCES
- 1. Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 2. Hurley J. H., Emr S. D. (2006) The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nickerson D. P., Russell M. R., Odorizzi G. (2007) A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 8, 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saksena S., Wahlman J., Teis D., Johnson A. E., Emr S. D. (2009) Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wollert T., Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Raiborg C., Bache K. G., Gillooly D. J., Madshus I. H., Stang E., Stenmark H. (2002) Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4, 394–398 [DOI] [PubMed] [Google Scholar]
- 8. Ren X., Hurley J. H. (2010) VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren X., Kloer D. P., Kim Y. C., Ghirlando R., Saidi L. F., Hummer G., Hurley J. H. (2009) Hybrid structural model of the complete human ESCRT-0 complex. Structure 17, 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prag G., Watson H., Kim Y. C., Beach B. M., Ghirlando R., Hummer G., Bonifacino J. S., Hurley J. H. (2007) The Vps27/Hse1 complex is a GAT domain-based scaffold for ubiquitin-dependent sorting. Dev. Cell 12, 973–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hurley J. H., Lee S., Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pullan L., Mullapudi S., Huang Z., Baldwin P. R., Chin C., Sun W., Tsujimoto S., Kolodziej S. J., Stoops J. K., Lee J. C., Waxham M. N., Bean A. J., Penczek P. A. (2006) The endosome-associated protein Hrs is hexameric and controls cargo sorting as a “master molecule.” Structure 14, 661–671 [DOI] [PubMed] [Google Scholar]
- 13. Bishop N., Horman A., Woodman P. (2002) Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bache K. G., Raiborg C., Mehlum A., Stenmark H. (2003b) STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513–12521 [DOI] [PubMed] [Google Scholar]
- 15. Takeshita T., Arita T., Higuchi M., Asao H., Endo K., Kuroda H., Tanaka N., Murata K., Ishii N., Sugamura K. (1997) STAM, signal transducing adaptor molecule, is associated with Janus kinases and involved in signaling for cell growth and c-myc induction. Immunity 6, 449–457 [DOI] [PubMed] [Google Scholar]
- 16. Kobayashi H., Tanaka N., Asao H., Miura S., Kyuuma M., Semura K., Ishii N., Sugamura K. (2005) Hrs, a mammalian master molecule in vesicular transport and protein sorting, suppresses the degradation of ESCRT proteins signal transducing adaptor molecule 1 and 2. J. Biol. Chem. 280, 10468–10477 [DOI] [PubMed] [Google Scholar]
- 17. Asao H., Sasaki Y., Arita T., Tanaka N., Endo K., Kasai H., Takeshita T., Endo Y., Fujita T., Sugamura K. (1997) Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J. Biol. Chem. 272, 32785–32791 [DOI] [PubMed] [Google Scholar]
- 18. Takeshita T., Arita T., Asao H., Tanaka N., Higuchi M., Kuroda H., Kaneko K., Munakata H., Endo Y., Fujita T., Sugamura K. (1996) Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem. Biophys. Res. Commun. 225, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 19. Tanaka N., Kaneko K., Asao H., Kasai H., Endo Y., Fujita T., Takeshita T., Sugamura K. (1999) Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J. Biol. Chem. 274, 19129–19135 [DOI] [PubMed] [Google Scholar]
- 20. Fiala G. J., Schamel W. W., Blumenthal B. (2011) Blue native polyacrylamide gel electrophoresis (BN-PAGE) for analysis of multiprotein complexes from cellular lysates. J. Vis. Exp. 48, e2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Urbé S., Sachse M., Row P. E., Preisinger C., Barr F. A., Strous G., Klumperman J., Clague M. J. (2003) The UIM domain of Hrs couples receptor sorting to vesicle formation. J. Cell Sci. 116, 4169–4179 [DOI] [PubMed] [Google Scholar]
- 22. Mayers J. R., Fyfe I., Schuh A. L., Chapman E. R., Edwardson J. M., Audhya A. (2011) ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J. Biol. Chem. 286, 9636–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanazawa C., Morita E., Yamada M., Ishii N., Miura S., Asao H., Yoshimori T., Sugamura K. (2003) Effects of deficiencies of STAMs and Hrs, mammalian class E Vps proteins, on receptor downregulation. Biochem. Biophys. Res. Commun. 309, 848–856 [DOI] [PubMed] [Google Scholar]
- 24. Lee S. A., Eyeson R., Cheever M. L., Geng J., Verkhusha V. V., Burd C., Overduin M., Kutateladze T. G. (2005) Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. U.S.A. 102, 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bache K. G., Brech A., Mehlum A., Stenmark H. (2003) Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J. Cell Biol. 162, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 27. Clague M. J., Urbé S. (2006) Endocytosis: the DUB version. Trends Cell Biol. 16, 551–559 [DOI] [PubMed] [Google Scholar]
- 28. McCullough J., Clague M. J., Urbé S. (2004) AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mizuno E., Kobayashi K., Yamamoto A., Kitamura N., Komada M. (2006) A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic 7, 1017–1031 [DOI] [PubMed] [Google Scholar]
- 30. Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 31. Kato M., Miyazawa K., Kitamura N. (2000) A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275, 37481–37487 [DOI] [PubMed] [Google Scholar]
- 32. McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., Urbé S. (2006) Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr. Biol. 16, 160–165 [DOI] [PubMed] [Google Scholar]
- 33. Agromayor M., Martin-Serrano J. (2006) Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281, 23083–23091 [DOI] [PubMed] [Google Scholar]
- 34. Kyuuma M., Kikuchi K., Kojima K., Sugawara Y., Sato M., Mano N., Goto J., Takeshita T., Yamamoto A., Sugamura K., Tanaka N. (2007) AMSH, an ESCRT-III associated enzyme, deubiquitinates cargo on MVB/late endosomes. Cell Struct. Funct. 31, 159–172 [DOI] [PubMed] [Google Scholar]
- 35. He J., Vora M., Haney R. M., Filonov G. S., Musselman C. A., Burd C. G., Kutateladze A. G., Verkhusha V. V., Stahelin R. V., Kutateladze T. G. (2009) Membrane insertion of the FYVE domain is modulated by pH. Proteins 76, 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dumas J. J., Merithew E., Sudharshan E., Rajamani D., Hayes S., Lawe D., Corvera S., Lambright D. G. (2001) Multivalent endosome targeting by homodimeric EEA1. Mol. Cell 8, 947–958 [DOI] [PubMed] [Google Scholar]
- 37. Miedema H., Staal M., Prins H. B. (1996) pH-induced proton permeability changes of plasma membrane vesicles. J. Membr. Biol. 152, 159–167 [DOI] [PubMed] [Google Scholar]
- 38. Mitsui K., Koshimura Y., Yoshikawa Y., Matsushita M., Kanazawa H. (2011) The endosomal Na+/H+ exchanger contributes to multivesicular body formation by regulating the recruitment of ESCRT-0 Vps27p to the endosomal membrane. J. Biol. Chem. 286, 37625–37638 [DOI] [PMC free article] [PubMed] [Google Scholar]