Background: Trp-Asp (WD) repeat protein 68 (WDR68) is a binding partner of dual specificity tyrosine phosphorylation-regulated protein kinase (DYRK1A), a kinase partly responsible for aspects of Down syndrome.
Results: The molecular chaperone T-complex protein 1 (TCP1) ring complex/chaperonin-containing TCP1 (TRiC/CCT) binds to WDR68 and modulates its structure, DYRK1A-binding, and cellular localization.
Conclusion: TRiC/CCT is essential for correct folding and function of WDR68.
Significance: TRiC/CCT may have a general role in forming the functional WD40 repeat seven-bladed β-propeller structure.
Keywords: Heat Shock Protein (HSP), Molecular Chaperone, Nuclear Translocation, Phosphoproteomics, Protein Phosphorylation, DYRK1A, TCP1, TRiC/CCT, WD40 Repeat, WDR68
Abstract
Trp-Asp (WD) repeat protein 68 (WDR68) is an evolutionarily conserved WD40 repeat protein that binds to several proteins, including dual specificity tyrosine phosphorylation-regulated protein kinase (DYRK1A), MAPK/ERK kinase kinase 1 (MEKK1), and Cullin4-damage-specific DNA-binding protein 1 (CUL4-DDB1). WDR68 affects multiple and diverse physiological functions, such as controlling anthocyanin synthesis in plants, tissue growth in insects, and craniofacial development in vertebrates. However, the biochemical basis and the regulatory mechanism of WDR68 activity remain largely unknown. To better understand the cellular function of WDR68, here we have isolated and identified cellular WDR68 binding partners using a phosphoproteomic approach. More than 200 cellular proteins with wide varieties of biochemical functions were identified as WDR68-binding protein candidates. Eight T-complex protein 1 (TCP1) subunits comprising the molecular chaperone TCP1 ring complex/chaperonin-containing TCP1 (TRiC/CCT) were identified as major WDR68-binding proteins, and phosphorylation sites in both WDR68 and TRiC/CCT were identified. Co-immunoprecipitation experiments confirmed the binding between TRiC/CCT and WDR68. Computer-aided structural analysis suggested that WDR68 forms a seven-bladed β-propeller ring. Experiments with a series of deletion mutants in combination with the structural modeling showed that three of the seven β-propeller blades of WDR68 are essential and sufficient for TRiC/CCT binding. Knockdown of cellular TRiC/CCT by siRNA caused an abnormal WDR68 structure and led to reduction of its DYRK1A-binding activity. Concomitantly, nuclear accumulation of WDR68 was suppressed by the knockdown of TRiC/CCT, and WDR68 formed cellular aggregates when overexpressed in the TRiC/CCT-deficient cells. Altogether, our results demonstrate that the molecular chaperone TRiC/CCT is essential for correct protein folding, DYRK1A binding, and nuclear accumulation of WDR68.
Introduction
The WD40 domain is a short stretch of ∼40 amino acids that is characterized by its tryptophan-aspartic acid (WD) dipeptide terminals (1, 2). More than 250 proteins containing tandem repeats of WD40 domains have been identified in the human genome (3). Structural analysis of the most extensively examined WD40 repeat protein, heterotrimeric G-protein β subunit, indicated that it forms a seven-bladed β-propeller circular structure (4, 5). The WD40 repeat facilitates protein-protein interactions coordinating multiprotein complex assemblies, where the repeating units serve as a rigid scaffold (1–3). Although the function of many WD40 proteins remains obscure, they have been suggested to play a role in a wide variety of cellular processes, including signal transduction, transcriptional regulation, cell cycle control, protein quality control, and cell apoptosis (3).
WDR682 with five WD40 repeats, also known as DCAF7 (Ddb1- and Cul4-associated factor 7 (6)), was originally identified in the petunia as a gene (AN11) encoding a protein that controls the pigmentation of flowers by stimulating the transcription of anthocyanin biosynthetic genes (7). Evolutionarily conserved homologs have been identified in many other species, including humans, that do not produce anthocyanin (7). All of the mammals studied to date from humans to opossums share exactly the same 342-amino acid sequence of WDR68, and the amino acid sequence of human WDR68 is 100 and 98% identical to avian and fish WDR68, respectively. Human WDR68 has 52% amino acid identity with petunia AN11 and partially rescues the an11 petunia mutant, showing the functional conservation (7). We and others have shown previously that WDR68 is indispensable for the proliferation and survival of mammalian cells (8, 9). Thus, WDR68 may play a fundamental role in cellular physiology, probably by facilitating protein-protein interactions, although the biochemical and physiological functions of WDR68 remain to be elucidated.
DYRK1A and DYRK1B are related mammalian protein kinases belonging to the dual specificity tyrosine phosphorylation-regulated protein kinase (DYRK) family (10, 11). We and others previously identified DYRK1A and DYRK1B as major cellular WDR68 binding partners (9, 12–14). The human gene for DYRK1A has received considerable attention because DYRK1A is encoded on chromosome 21 in the Down syndrome critical region (15, 16). Indeed, overexpression of DYRK1A is suggested to be responsible for part of the Down syndrome phenotype (17–21), possibly by antagonizing calcineurin-dependent dephosphorylation of NFAT (nuclear factor of activated T-cells) (22, 23). Overexpressed DYRK1A binds to WDR68 and disturbs its subcellular localization (9), and dysregulation of WDR68 subcellular localization contributes to the molecular mechanism for developmental craniofacial malformation (24), which is often observed in Down syndrome patients. WDR68 binds to other protein kinases MEKK1 and HIPK2 and thus facilitates protein complex formation and signaling from MEKK1 to HIPK2 (8). Taken together, WDR68 seems to function as a scaffold protein to control the cellular signal transduction systems mediated by protein kinases. Identification of the cellular WDR68-binding phosphoproteins will probably be essential to understand the physiological functions of WDR68.
The folding and activity of many proteins in cells are assisted by a set of specialized proteins called molecular chaperones. The mammalian molecular chaperone TCP1 (T-complex protein 1) ring complex (TRiC; also called CCT (chaperonin containing TCP1) or Hsp60 chaperone) is an ATP-dependent type II (group II) chaperonin, which encloses newly synthesized polypeptides for folding and thus prevents misfolding and aggregation during protein synthesis (25–27). TRiC/CCT is essential for cytosolic folding of many important functional cellular proteins involved in cell cycle regulation, cytoskeletal architecture formation, protein degradation, RNA and DNA processing, intracellular trafficking, metabolism, and signal transduction (25–27). TRiC/CCT consists of double-stacked rings, each one containing eight different radially arranged subunits of ∼60 kDa each (TCP1α to TCP1θ), forming a central cavity where protein folding takes place (28). TRiC/CCT substrate recognition depends mainly on exposed hydrophobic residues (27), but polar and charged residues also play a role (29). Although the molecular mechanism of substrate recognition and binding of TRiC/CCT is under investigation, each of the subunits may recognize different hydrophobic motifs within substrate proteins (29, 30). Potentially 5–10% of newly synthesized cytoplasmic polypeptides can associate with TRiC/CCT (31); however, only some of them have so far been shown to depend on this chaperonin for folding and function. TRiC/CCT prevents the aggregation of proteins with polyglutamine regions, suggesting its important role in neurodegenerative diseases (32–34).
In this study, we analyzed cellular WDR68 binding partners by a mass spectrometry-based proteomic approach. The major cellular binding proteins for WDR68 were identified as TRiC/CCT subunits. Our results show the functional importance of the chaperone activity of TRiC/CCT in the proper structure and function of WDR68 in cells.
MATERIALS AND METHODS
Antibodies and Plasmids
Anti-FLAG antibody (clone M2), HRP-conjugated anti-FLAG antibody, and anti-FLAG affinity resin were obtained from Sigma. Anti-HA (clone 12CA5), anti-TCP1α (clone 91a), and anti-TCP1β (clone PK/8/4/4i/2f) antibodies were obtained from Roche Applied Science and from StressGen-Enzo Life Sciences, respectively. The anti-Hsp90 antibody described previously (35, 36) was conjugated with HRP using the EZ-link Plus activated peroxidase kit (Pierce-Thermo Scientific). An affinity-purified antibody specific for WDR68 and mammalian expression plasmids encoding EGFP-WDR68, HA-WDR68, and 3×FLAG-DYRK1A were described previously (9).
Mammalian Cell Culture, Transfection, Cell Extraction, and Immunoprecipitation
COS7 and KB cells were cultured in DMEM supplemented with 10% FCS in humidified air containing 5% CO2. COS7 cells were transfected with plasmids by electroporation, and cell extracts were prepared as described previously (9, 37, 38). Extracts with equal amounts of protein were incubated with either anti-FLAG agarose, the antibody against WDR68, or anti-HA tag for 12 h at 4 °C. EZView Red protein G affinity gel (Sigma) was used for WDR68 immunoprecipitation. The immunocomplexes were extensively washed and analyzed as described previously (39, 40).
Deletion Mutants of WDR68
Deletion mutants encoding WDR68(1–217), WDR68(1–258), WDR68(1–304), WDR68(55–304), WDR68(150–342), WDR68(107–342), and WDR68(55–342) were described previously (9). Other WDR68 deletion mutants were produced by PCR amplification with the following primers. The antisense primers were as follows: WDR68(1–64), 5′-GCGGCCGCTTAGGTGTTTCTGCAAATAAACTC-3′; WDR68(1–108), 5′-GCGGCCGCTCACTCCAGCCTGGTCTCTGTTTC-3′; WDR68(1–164), 5′-GCGGCCGCCTAGCCAGACACGAGATTCACTCG-3′. A common sense primer (5′-CGATGCGGCCGCCATGTCCCTGCACGGCAAACGGA-3′) was used for the C-terminal deletion mutants. The sense primers were as follows: WDR68(299–342), 5′-GCGGCCGCCATGCCCCGAGCCATTGAGGAC-3′; WDR68(245–342), 5′-GCGGCCGCCATGGAGGTGGTGATTCTAGAT-3′; WDR68(203–342), 5′-GCGGCCGCCATGTTTGACCTCCGCCATCTA-3′. The same set of primers as above was used to generate fragments for WDR68(55–127), WDR68(55–164), WDR68(55–258), WDR68(107–217), WDR68(107–258), WDR68(107–304), WDR68(150–258), WDR68(150–304), and WDR68(203–304). A common antisense primer (5′-GCATGCGGCCGCCTACACTCTGAGTATCTCCAGGC-3′) was used for the N-terminal deletion mutants. All of the coding regions of WDR68 mutants were confirmed by direct sequencing.
Construction of Mammalian Expression Vectors
The NotI fragments of WDR68 and its deletion mutants were ligated into the NotI site of both p3×FLAG-CMV7.1 (Sigma) and pcDNA3HA (38) to obtain mammalian expression plasmids encoding WDR68 fusion proteins with a 3×FLAG and HA tag, respectively.
Large Scale Isolation and Mass Spectrometric Analysis of WDR68-binding Proteins
COS7 cells were transfected by electroporation with either a control empty vector or the plasmid encoding 3×FLAG-WDR68. Lysates were prepared from eight dishes (10-cm diameter) of control- or 3×FLAG-WDR68-transfected COS7 cells and mixed with 200 μl of anti-FLAG-agarose for 12 h at 4 °C. After extensive washing, bound proteins were eluted with 0.6 mg/ml of 3×FLAG peptide in washing buffer as described (9, 39, 40). Isolated proteins were concentrated by a Microcon centrifugal filter (10-kDa cut-off) and analyzed by SDS-PAGE followed by Coomassie Brilliant Blue protein staining. About 2 μg of either the WDR68 complex or the control sample was digested with trypsin. Peptides derived from the control immunoprecipitate and the 3×FLAG-WDR68 immunoprecipitate were labeled with iTRAQ reagent 114 and iTRAQ reagent 117 (AB Sciex), respectively, according to the manufacturer's instructions. Both samples were then combined. Phosphopeptides were enriched by immobilized metal affinity chromatography with PHOS-Select iron affinity gel (Sigma-Aldrich) according to the manufacturer's instructions. The iTRAQ-labeled phosphopeptide mixture was then dried, solubilized in 0.1% formic acid, and analyzed by a NanoLC-QSTAR Elite mass spectrometer (AB Sciex) with a NanoSpray III source. The ion source conditions were as follows: “ionspray voltage” = 1800 V; “curtain gas” = 20 p.s.i.; “declustering potential 1” = 60 V; “focusing potential” = 250 V; “declustering potential 2” = 15 V. The separation by nano-LC (KYA Technologies) was performed at a constant flow rate of 200 nl/min with a 170-min gradient. A QSTAR Elite mass spectrometer was used in standard MS/MS data-dependent acquisition mode. The 0.5-s survey MS spectra were collected (m/z 400–1800) followed by three MS/MS measurements on the most intense parent ions (20 counts/s threshold, 2–5 charge state, m/z 65–2000 mass range for MS/MS), using the manufacturer's “smart exit” setting 2. Previously targeted parent ions were excluded from repetitive MS/MS acquisition for 60 s (50 mDa mass tolerance). The reporter region was enhanced, and collision energy was adjusted by the manufacturer's setting, “Using iTRAQ Reagent.” The data files were processed by ProteinPilot version 2.0 software (AB Sciex) using the Paragon algorithm (41). All searches were performed against the Swissprot080221 database.
Immunofluorescent Staining
Immunofluorescent staining of both COS7 and KB cells was performed essentially as described (42). Alexa488-conjugated secondary antibody (Molecular Probes) was used. Fluorescent and phase-contrast images of cells were obtained with immersion oil using a fluorescent microscope (Zeiss Axiophoto).
RNA Interference Experiments
“Stealth Select” siRNAs (HSS110578 and HSS186251) specific for human TCP1α were obtained from Invitrogen. Stealth RNAi negative control duplexes (Invitrogen) were used as controls. siRNA transfection was performed using Lipofectamine RNAiMAX (Invitrogen) as described in the manufacturer's standard protocol with a final concentration of 12 nm siRNA. After 2 days, cells were transfected again with the same siRNAs. For siRNA/DNA co-transfection, Lipofectamine2000 (Invitrogen) was used according to the manufacturer's standard protocol with a final concentration of 32 nm siRNA and 1.6 μg/ml DNA. After an additional 2 days, cell extracts were prepared as described above, and the levels of proteins in the equal amount of the total proteins were examined by Western blotting analysis. The cellular distribution of WDR68 was examined by immunofluorescent microscopy as described above. The binding of WDR68 to DYRK1A was examined by co-immunoprecipitation experiments essentially as described previously (9, 39, 40). A modified buffer was used for cell extraction and immunoprecipitate washing (50 mm Tris-Cl, 10% glycerol, 150 mm NaCl, 2 mm EDTA, 1 mm DTT, 1% Nonidet P-40, supplemented with a 1:100 HALT phosphatase inhibitor mixture (Pierce) and 1:100 protease inhibitor mixture (Nacalai Tesque)).
Other Procedures
SDS-PAGE was performed with 10% acrylamide gels. BoltTM BisTris Plus neutral pH precast gradient (4–12%) gel electrophoresis with MES-SDS running buffer (Invitrogen) was performed according to the protocol of the manufacturer. Western blotting was performed using either horseradish peroxidase-conjugated secondary antibodies (GE Healthcare for both mouse and rabbit antibodies; Santa Cruz Biotechnology, Inc., for rat antibodies) or peroxidase-conjugated primary antibodies, and detection was performed using a chemiluminescent system as described (40).
RESULTS
Isolation and Mass Spectrometric Identification of Cellular WDR68-binding Proteins
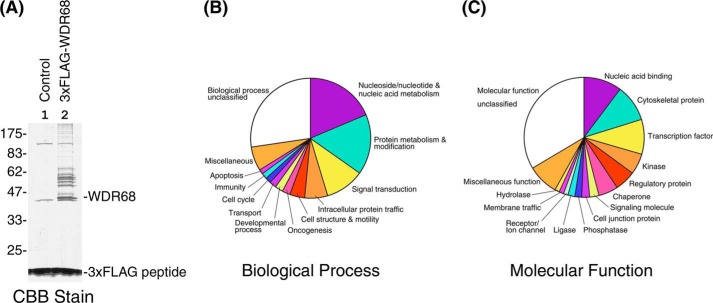
We searched for the cellular binding partners of WDR68 to understand the physiological role of WDR68. We took a phosphoproteomic approach, aiming to reveal WDR68-mediated kinase-substrate signaling pathways. A large scale isolation of WDR68-associated proteins was conducted by affinity purification of cellular proteins that bound to 3×FLAG-tagged WDR68 expressed in COS7 cells. A protein profile of the WDR68 complexes was visualized by SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 1A, lane 2), along with a control isolation from cells transfected with an empty vector (Fig. 1A, lane 1). 3×FLAG-WDR68 that migrated as a 42 kDa major band and a cluster of 50–60-kDa WDR68-binding proteins were detected (Fig. 1A, lane 2), which were not observed in the control sample (Fig. 1A, lane 1). In addition, many higher molecular weight proteins were observed specifically in the complex from the 3×FLAG-WDR68-expressing cell lysate but not from the mock-transfected control cell lysate.
FIGURE 1.
Isolation and identification of cellular WDR68-binding proteins. A, 3×FLAG-WDR68 and its binding proteins were isolated as a complex, and the protein profile of the complex in SDS-PAGE is shown by Coomassie Brilliant Blue (CBB) staining. Lane 1, proteins isolated from the mock-transfected lysate; lane 2, the complex isolated from the 3×FLAG-WDR68-expressing lysate. The positions of WDR68, 3×FLAG peptide, and molecular weight markers are shown. B, the proteomic analysis of the protein sample in A identified 250 cellular WDR68-binding protein candidates, and the PANTHER biological process classification of these proteins is shown. C, the PANTHER molecular function classification of 250 identified cellular WDR68-binding protein candidates is shown.
The mixture of WDR68-associated proteins was then trypsinized, and the obtained peptides were labeled with iTRAQ117 and mixed with iTRAQ114-labeled peptides derived from the control immunoprecipitate. After enrichment of phosphopeptides, the peptide mixture was examined by nano-LC/MS/MS analysis precisely as described above. The mass spectrum analyses were able to de novo sequence 2243 (70.5% of total) peptides, and 1926 distinct peptides were matched in the database and thus used for protein identification with 95% confidence. Candidates with a iTRAQ117/114 ratio (the WDR68 binding versus the control binding) less than 2 were eliminated as nonspecific contaminants. The analysis identified 250 cellular WDR68-binding protein candidates. As expected, WDR68 itself was identified, and 14 peptides were assigned to WDR68 with a sequence coverage of 50% (Table 1). Analysis of the isolated WDR68 peptide fragments identified three in vivo phosphorylation sites on WDR68: Ser-38, Ser-200, and Thr-257 (Table 1). DYRK1A (sequence coverage = 10.1%) and MEKK1 (sequence coverage = 19.2%), both previously shown to bind to WDR68 (8, 9, 12, 13), were also identified as WDR68 binding partners with the analysis, clearly indicating the validity of this screening method.
TABLE 1.
Summary of mass spectrometric identification of WDR68 and its binding partner TRiC/CCT subunits and their phosphorylation sites
| Protein | Gene identifier | No. of identified peptides | Sequence coverage | Highest ratio over control | Average ratio | Phosphorylation sites |
|---|---|---|---|---|---|---|
| % | ||||||
| WDR68 | 48428729 | 14 | 50.0 | 261.0 | 84.5 | Ser-38, Ser-200, Thr-257 |
| TCP1α | 135538 | 11 | 25.5 | 70.0 | 45.4 | Ser-544 |
| TCP1β | 6094436 | 21 | 48.8 | 290.0 | 69.7 | Ser-3, Ser-54, Ser-260 |
| TCP1γ | 75077288 | 20 | 45.0 | 275.0 | 80.6 | Thr-86, Ser-170, Ser-297, Thr-512 |
| TCP1δ | 38455427 | 10 | 21.3 | 122.0 | 50.7 | Ser-9, Ser-381 |
| TCP1ϵ | 1351211 | 17 | 34.8 | 191.0 | 96.6 | Ser-51, Thr-473 |
| TCP1ζ | 730922 | 15 | 38.0 | 200.0 | 99.5 | Thr-6 |
| TCP1η | 3041738 | 13 | 29.7 | 187.0 | 72.3 | Thr-57, Ser-59, Thr-332, Thr-371, Thr-526, Thr-531 |
| TCP1θ | 75076052 | 14 | 30.3 | 203.0 | 62.7 | Thr-293, Thr-327, Thr-517 |
General Profiling of the WDR68-binding Protein Candidates
The PANTHER protein family classification (Biological Process) of the identified WDR68-binding protein candidates is shown in Fig. 1B. About a half of them could be classified into four categories that are involved in nucleotide and nucleic acid metabolism, protein metabolism and modification, signal transduction, and intracellular protein traffic, respectively. The molecular functions of these WDR68 binding partner candidates were analyzed according to the PANTHER protein profile classification and are shown in Fig. 1C. The biochemical functions of WDR68-binding proteins were diversified, but they were mainly classified as nucleic acid-binding proteins, cytoskeletal proteins, transcription factors, kinases, regulatory proteins, and chaperones. These analyses thus suggested that WDR68 may play roles in a wide variety of cellular processes, including especially DNA and protein metabolism, gene regulation, and signal transduction.
Identification of TRiC/CCT as a Cellular WDR68 Binding Partner
We noticed that all eight subunits of TRiC/CCT (TCP1α to TCP1θ) were included in the WDR68-binding protein list (Table 1). Because TRiC/CCT is composed of eight different subunits forming a double-stacked ring structure, this result suggests that the whole octagonal TRiC/CCT chaperone binds to WDR68. In addition, we detected in vivo phosphorylation sites on TRiC/CCT subunits, as shown in Table 1.
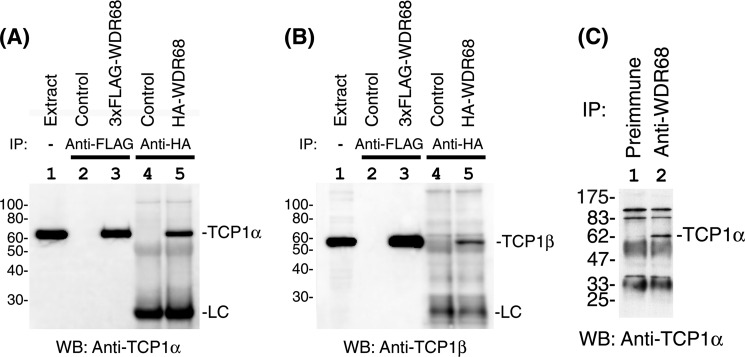
We then examined the binding between WDR68 and cellular TRiC/CCT by co-immunoprecipitation analysis. 3×FLAG-WDR68 expressed in COS7 cells was immunoprecipitated with anti-FLAG antibody, and the immunoprecipitates were examined for the presence of endogenous TCP1α, a subunit of TRiC/CCT. As shown in Fig. 2A, the WDR68 immunoprecipitate (lane 3) contained cellular TCP1α, but the control immunoprecipitate from the non-transfected cell lysate did not contain this protein (lane 2). Co-immunoprecipitation of TCP1α was also detected with HA-tagged WDR68 (Fig. 2A, lane 5) but not with the control immunoprecipitate (Fig. 2A, lane 4), indicating that the observed binding can be ascribed to WDR68 itself and not to the FLAG or HA tag. Moreover, we found the specific co-immunoprecipitation of endogenous TCP1β both with 3×FLAG-WDR68 (Fig. 2B, lanes 2 and 3) and HA-WDR68 (Fig. 2B, lanes 4 and 5). We have not examined the co-immunoprecipitation of other TRiC/CCT subunits with WDR68, but the proteomic identification of all eight subunits indicates that the double-octamer TRiC/CCT chaperone complex binds to WDR68. However, we cannot conclude that all eight TRiC/CCT subunits directly bind to WDR68.
FIGURE 2.
Association of TRiC/CCT with WDR68. A, tagged WDR68 was expressed in mammalian cultured cells, and the binding of TRiC/CCT was examined by co-immunoprecipitation analysis (IP). Lane 1, input (total cell extract); lane 2, control without 3×FLAG WDR68 expression; lane 3, 3×FLAG-WDR68 was expressed and immunoprecipitated; lane 4, control without HA-WDR68 expression; lane 5, HA-WDR68 was expressed and immunoprecipitated. The binding of endogenous TCP1α was examined by Western blotting (WB). B, the same set of samples as in A was examined for the binding to endogenous TCP1β by Western blotting. Lanes 1–5, same as in A. C, endogenous WDR68 was immunoprecipitated and the binding of cellular TCP1α was examined by Western blotting. Lane 1, control immunoprecipitation with preimmune antibody; lane 2, immunoprecipitation with anti-WDR68.
We next examined the endogenous binding between TRiC/CCT and WDR68. Endogenous WDR68 was immunoprecipitated with an anti-WDR68 antibody, and then the binding of endogenous TCP1α was examined by Western blotting. As shown in Fig. 2C, TCP1α was detected in the WDR68 immunoprecipitate (lane 2) but not in the control immunoprecipitate with a non-immune antibody (lane 1). Taken together, these results clearly indicate that TRiC/CCT binds to WDR68 in cells.
Structural Analysis of WDR68
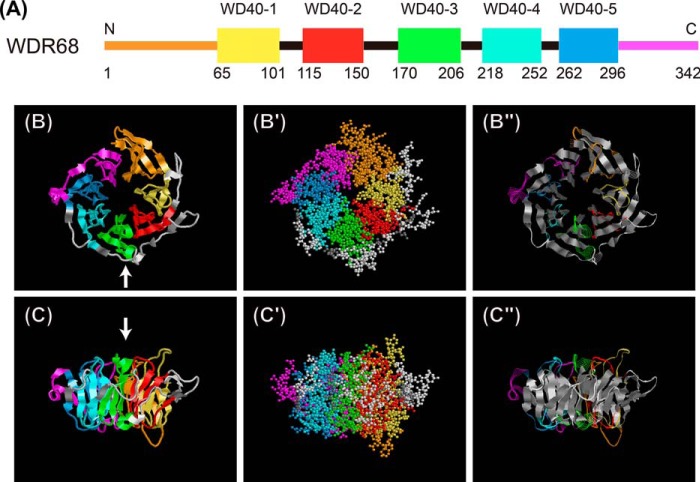
WDR68 contains five WD40 repeats (denoted here as WD40-1 to WD40-5) (Fig. 3A). On the other hand, the N- and C-terminal segments have no apparent sequence similarity with the WD40 repeats. Most of the structurally characterized WD40 proteins contain seven WD40 repeats and form seven-bladed β-propeller structures (3), suggesting that WDR68 probably contains at least five β-propeller blades. Because the three-dimensional structure of WDR68 had not been determined, we constructed a structural model of mammalian WDR68 by analyzing its amino acid sequence using Phyre2 computer software (43). The results indicated that the WDR68 structure can be modeled from the known three-dimensional structure of a histone-binding chaperone, rbbp7/RbAp46 (44) (fold library ID c3cfvA; Protein Data Bank code 3CFV) with a 100.0% confidence index and a 94% coverage. The predicted WDR68 structure shows a β-propeller with seven blades, five of which correspond to the five WD40 repeats (Fig. 3, A–C, yellow, red, green, cyan, and blue). Two additional β-propeller blades derive from both the N-terminal (Fig. 3, A–C, orange) and the C-terminal (Fig. 3, A–C, magenta) extensions, and they bind together to complete the seven-bladed ring (Fig. 3, B and C). Most of the model WDR68 structure is made from β-strands (Fig. 3, B″ and C″, shown in gray ribbons). We also analyzed the amino acid sequence of WDR68 using the I-TASSER computational program (45), and a predicted structural model was obtained based on the structure of a different template protein Pex7p (46) with a C-score = 0.50 and a TM-score = 0.78 ± 0.10. The overall seven-bladed β-propeller structure of the WDR68 model predicted by I-TASSER (data not shown) is highly similar to that obtained above with the Phyre2 program.
FIGURE 3.
A seven-blade β-propeller model of WDR68 based on computational structural analysis. A, schematic illustration of five WD40 repeats (depicted as WD40-1 to WD40-5 with their starting and ending amino acid numbers) in the WDR68 amino acid sequence. B and C, a three-dimensional model of WDR68 showing a seven-blade β-propeller structure was obtained by analyzing its amino acid sequence with the Phyre2 program. Colors used in the scheme (A) and the structure (B and C) are as follows: orange, aa 1–64 (N-terminal extension); yellow, aa 65–101 (WD40-1); red, aa 115–150 (WD40-2); green, aa 170–206 (WD40-3); cyan, aa 218–252 (WD40-4); blue, aa 262–296 (WD40-5); magenta, aa 297–342 (C-terminal extension). Ribbon models (B and C) and ball models (B′ and C′) are illustrated. Vertical views (B, B′, and B″) and side views (C, C′, and C″) are shown, and the arrows in B and in C indicate the direction from which the side and vertical view images are obtained, respectively. In the vertical view images (B), the amino acid sequence starts from the top of the image and runs clockwise to the C-terminal end. In B″ and C″, β-strands are shown in gray ribbons, and other regions are shown as colored wire strands.
Relationship between the WDR68 Structure and Its TRiC/CCT Binding
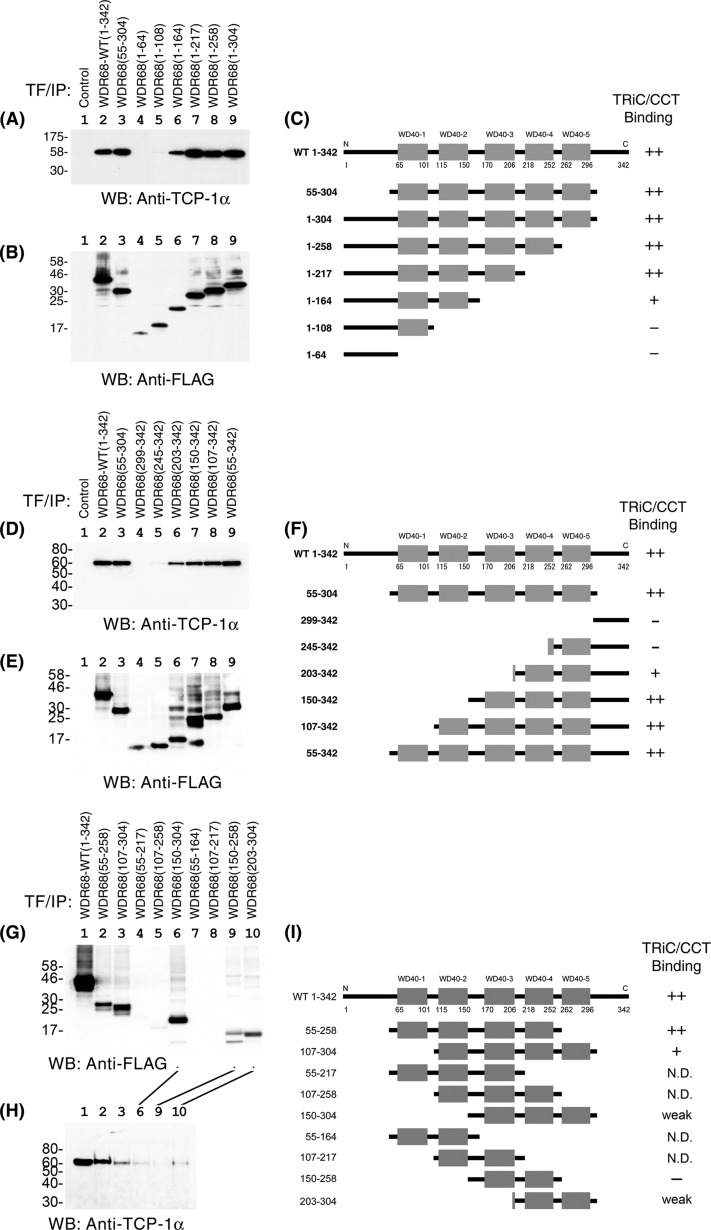
To identify which part of the molecule is responsible for the TRiC/CCT binding, we made various deletion mutants of WDR68 and examined their ability to bind to endogenous TRiC/CCT by co-immunoprecipitation experiments. First, we analyzed the binding of TRiC/CCT to the central core five WD40 repeats of WDR68. WDR68(55–304), containing the five WD40 repeats and lacking both the N-terminal and C-terminal extensions, bound to TCP1α equally well (Fig. 4A, lane 3) as wild type WDR68 (lane 2), indicating that the five WD40 repeats are sufficient for TRiC/CCT binding. We then examined the binding of TCP1α to various C-terminal deletion mutants of WDR68. WDR68(1–164), WDR68(1–217), WDR68(1–258), and WDR68(1–304) all efficiently bound to TCP1α, as shown in Fig. 4A (lanes 6–9; see their expressions in Fig. 4B and their structures in Fig. 4C). On the other hand, neither WDR68(1–108) with only one WD40 repeat nor WDR68(1–64) (the N-terminal extension) bound to TCP1α (Fig. 4A, lanes 4 and 5). We obtained the same conclusion by analyzing the association of HA-tagged WDR68 mutants with endogenous TCP1α (data not shown). The simplest interpretation of the results may be that the region with amino acids 109–164 containing WD40-2 is responsible for the binding to TRiC/CCT.
FIGURE 4.
Binding of deletion mutants of WDR68 to TRiC/CCT. A, binding of TCP1α to C-terminal deletion mutants of WDR68 was examined by co-immunoprecipitation experiments (IP). B, expression of WDR68 C-terminal deletion mutants was examined by Western blotting (WB). C, a schematic illustration of WDR68 C-terminal deletion mutants and their ability to bind to TRiC/CCT is presented. D, binding of TCP1α to N-terminal deletion mutants of WDR68 was examined by co-immunoprecipitation experiments. E, expression of WDR68 N-terminal deletion mutants was examined by Western blotting. F, a schematic illustration of WDR68 N-terminal deletion mutants and their ability to bind to TRiC/CCT. G, expression of WDR68 fragments without N- and C-terminal regions was examined by Western blotting. H, binding of TCP1α to the WDR68 deletion mutants without N- and C-terminal regions was examined by co-immunoprecipitation experiments. I, schematic illustration of additional WDR68 deletion mutants without N- and C-terminal regions and their ability to bind to TRiC/CCT. N.D., not determined; these mutants were not stably expressed, as shown in G. Representative co-immunoprecipitation results from three independent experiments are shown (A, D, and H).
We next examined the binding of TCP1α to various N-terminal deletion mutants of WDR68. Unexpectedly, WDR68(203–342), WDR68(150–342), WDR68(107–342), and WDR68(55–342), the first two of which lack WD40-2, bound efficiently to TCP1α (Fig. 4D, lanes 6–9; see their expressions in Fig. 4E and their structures in Fig. 4F). Therefore, the WD40-2 sequence containing amino acids 109–164 is not necessarily required for TRiC/CCT binding. On the other hand, neither WDR68(245–342) nor WDR68(299–342) could bind to TCP1α (Fig. 4D, lanes 4 and 5). All of these co-immunoprecipitation experiments were repeated three times, and the results were highly reproducible in independent experiments. Because the structural analysis indicated that both the N terminus and C terminus stretches form the β-blade (Fig. 3), these results with the N-terminal and C-terminal deletion mutants indicate that three of seven blades in WDR68, irrespective of the position, are required for the binding to TRiC/CCT.
We also made nine additional WDR68 deletion mutants that lack both N- and C-terminal extensions (Fig. 4I). Four of these mutants, WDR68(55–127), WDR68(107–258), WDR68(55–164), and WDR68(107–217), were not stable enough in transfected COS7 cells (Fig. 4G, lanes 4, 5, 7, and 8). We therefore analyzed the TCP1α-binding activity of the rest of these mutants, including WDR68(55–258), WDR68(107–304), WDR68(150–304), WDR68(150–258), and WDR68(203–304) (Fig. 4H). The result indicated that deletion mutants with three or four WD40 repeats, but not a mutant with two repeats, bind to TCP1α (Fig. 4H, lanes 2, 3, 6, and 9). The results were consistent with the above conclusion that three of seven β-blades in WDR68 are required for efficient TCP1α binding. The only exception was the weak TCP1α binding to WDR68(203–304) that contains only two full WD40 repeats (Fig. 4H, lane 10); however, this might be ascribed to a small fragment of WD40-3 in addition to WD40-4 and WD40-5 in this mutant (Fig. 4I).
TRiC/CCT Is Required for WDR68 Binding to DYRK1A
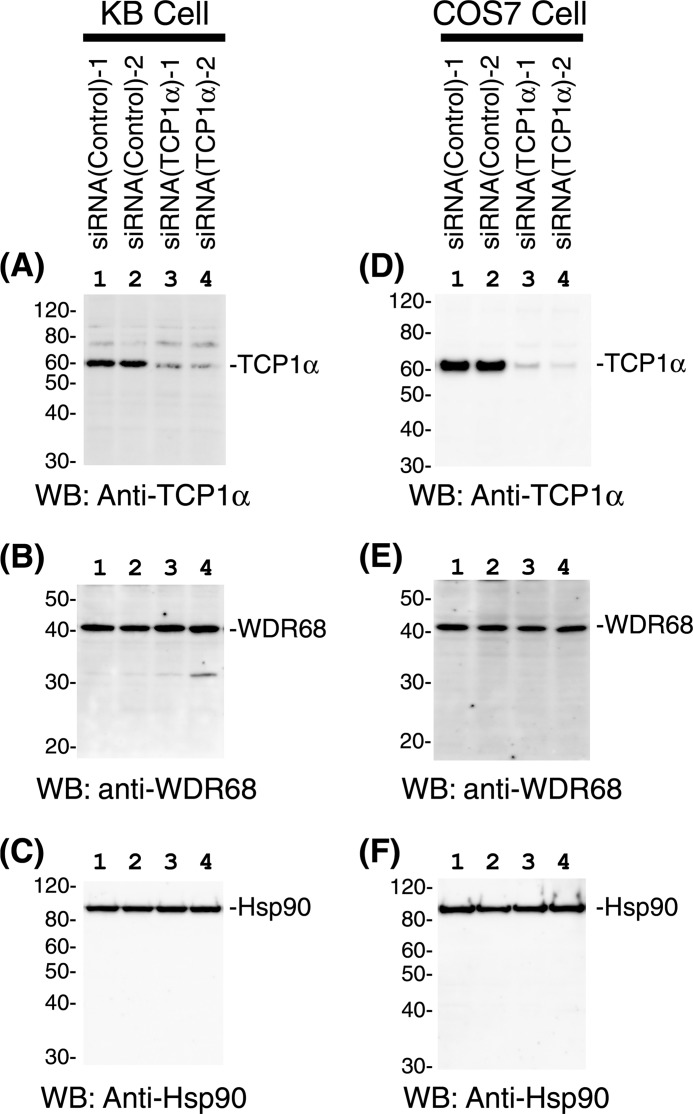
TRiC/CCT is often required for the cellular stability of its client proteins. To elucidate the functional importance of TRiC/CCT binding to WDR68, we conducted siRNA-induced suppression of TRiC/CCT. Two independent siRNAs specific for TCP1α, along with two control siRNAs, were introduced into both KB and COS7 cells. The amounts of endogenous TCP1α were examined by Western blotting using equal amounts of the total protein from the cell extracts. As shown in Fig. 5, A and D, a specific and significant reduction of TCP1α was observed with both siRNAs (lanes 3 and 4) as compared with controls (lanes 1 and 2) in both KB and COS7 cells. The reduction of TCP1α did not affect the cellular levels of either WDR68 (Fig. 5, B and E) or a control protein Hsp90 (Fig. 5, C and F). We concluded that TRiC/CCT is not required for either the cellular stability or the degradation of endogenous WDR68.
FIGURE 5.
Effect of siRNA-induced TCP1α suppression on the cellular levels of WDR68. KB (A–C) and COS7 (D–F) cells were transfected with either control siRNAs (lanes 1 and 2) or two independent TCP1α-specific siRNAs (lane 3, HSS110578; lane 4, HSS186251). After 48 h, cell extracts were prepared, and the amounts of TCP1α (A and D), WDR68 (B and E), and a control protein, Hsp90 (C and F), were examined by Western blotting (WB).
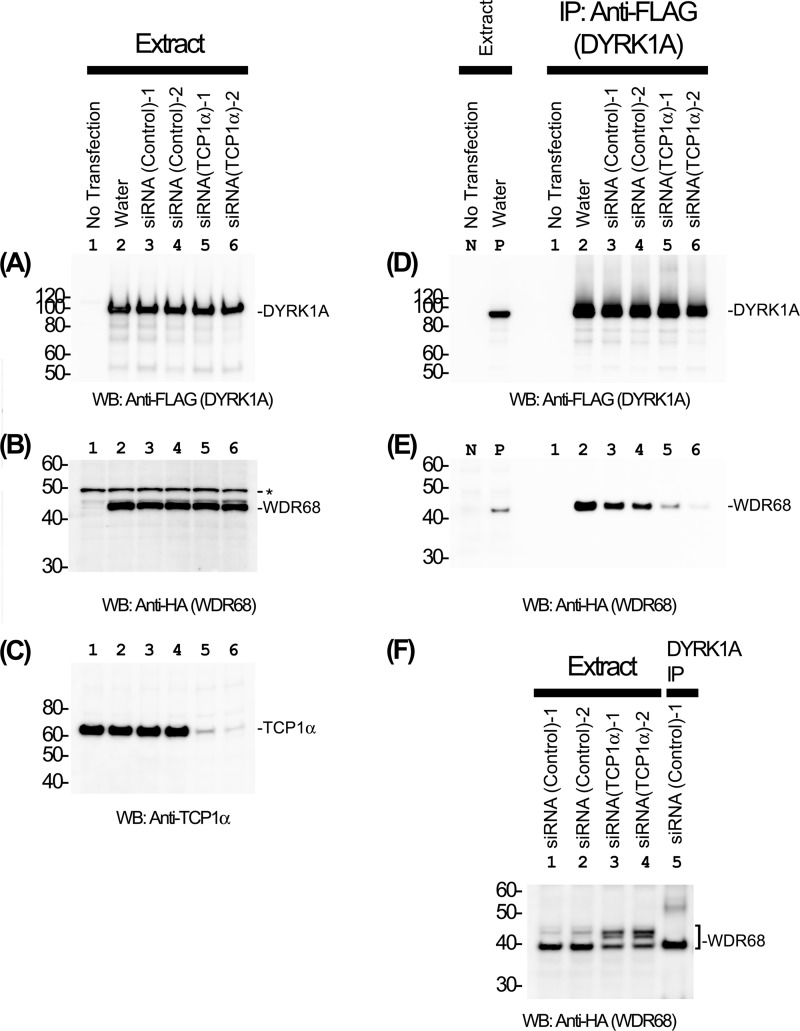
We then hypothesized that TRiC/CCT is essential for the function of WDR68. No enzymatic activity has been described for WDR68, although it is known to bind to several signaling protein kinases. We examined the DYRK1A-binding activity of WDR68 by co-expression/co-immunoprecipitation experiments. Either TCP1α-specific siRNAs or two control siRNAs were introduced into COS7 cells, and then 3×FLAG-DYRK1A and HA-WDR68 were expressed by transfection. TCP1α siRNAs (Fig. 6C, lanes 5 and 6), but not control siRNAs (lanes 3 and 4), specifically reduced endogenous TCP1α expression as expected. The expression levels of 3×FLAG-DYRK1A (Fig. 6A) and HA-WDR68 (Fig. 6B) were not affected by TCP1α siRNA transfection. Expressed DYRK1A was then immunoprecipitated (Fig. 6D), and the amounts of bound HA-WDR68 were examined by Western blotting (Fig. 6E). The result showed that the reduction of TCP1α resulted in a marked decrease in the binding of WDR68 to DYRK1A (Fig. 6E, compare lanes 3 and 4 with lanes 5 and 6), suggesting that TRiC/CCT is required for efficient binding of WDR68 to DYRK1A. The same samples were analyzed under a special gel electrophoresis condition where proteins were denatured under mild conditions and run in a gradient acrylamide gel in MES-based neutral pH. We noticed that a significant portion of WDR68 in TCP1α-deficient cells migrated more slowly as compared with WDR68 in normal cells (Fig. 6F, compare lanes 3 and 4 with lanes 1 and 2). The DYRK1A-binding form of WDR68 in the DYRK1A immunocomplex from the normal cell extract co-migrated with the faster migrating form of WDR68 (Fig. 6F, lane 5). These results suggest that WDR68 forms an aberrant structure (top band in Fig. 6F) when TCP1α levels are reduced, and only WDR68 with the normal structure in the presence of sufficient TCP1α (bottom band in Fig. 6F) has DYRK1A-binding activity. Taken together, our results suggest that TCP1α is essential for the structural integrity of WDR68 that has the ability to bind to its natural target protein DYRK1A. We reproducibly observed higher WDR68 in the water control than control siRNAs (Fig. 6E, lanes 2–4). The levels of expressed 3×FLAG-DYRK1A and HA-WDR68 were slightly but noticeably higher in the water control than siRNA controls (see Fig. 6, A, B, and D, lanes 2–4). In addition, cells looked slightly different under microscopic observation between the water control and the siRNA controls. We therefore could not exclude a possibility that the control siRNAs, despite their inability to suppress a specific gene, show certain nonspecific effects on cells.
FIGURE 6.
TRiC/CCT is required for the efficient binding of WDR68 to DYRK1A. COS7 cells were transfected with either control or TCP1α-specific siRNAs along with expression plasmids for 3×FLAG-DYRK1A and HA-WDR68. After 48 h, cell extracts were prepared, and the amounts of DYRK1A (A), WDR68 (B), and TCP1α (C) were examined by Western blotting (WB). Lane 1, no DNA transfection; lane 2, transfected with expression plasmids without siRNA; lanes 3 and 4, two independent control siRNAs; lanes 5 and 6, two independent TCP1α-specific siRNAs (lane 5, HSS110578; lane 6, HSS186251). D and E, the binding of WDR68 to DYRK1A was examined by co-immunoprecipitation experiments (IP), and the amounts of DYRK1A (D) and WDR68 (E) in the DYRK1A immunoprecipitates were examined by Western blotting. Lanes 1–6, same as in A–C; lane N, extract of cells without DNA transfection as a negative control; lane P, extract of cells transfected with DYRK1A and WDR68 as a positive control. F, WDR68 in cell extracts and in the immunoprecipitate was analyzed by neutral pH gradient gel electrophoresis followed by Western blotting. Lanes 1 and 2, control siRNA-transfected cell extracts; lanes 3 and 4, TCP1α-specific siRNA-transfected cell extracts; lane 5, DYRK1A immunoprecipitate from control siRNA-transfected cells. The positions of DYRK1A, WDR68, TCP1α, and molecular weight markers are shown in A–F. An asterisk in B indicates nonspecific bands.
TRiC/CCT Is Required for WDR68 Nuclear Translocation
We previously reported that the binding of DYRK1A to WDR68 induces nuclear accumulation of WDR68 (9). Therefore, the reduced DYRK1A-binding activity of WDR68 in the TCP1α down-regulated cells may affect subcellular localization of WDR68. We examined the intracellular localization of WDR68 by immunofluorescent microscopy after siRNA-mediated suppression of TRiC/CCT. In agreement with previous reports (9, 24), WDR68 was distributed throughout the cell after control siRNA treatment, and cytoplasmic staining was stronger than nuclear WDR68 in some cells, whereas in other cells, the staining predominated in the nucleus (Fig. 7A). This result suggests a dynamic shuttling of WDR68 between the cytoplasm and the nucleus. In contrast, when cellular TCP1α was reduced by two independent siRNAs, WDR68 was localized almost exclusively in the cytoplasm in all of the cells, and no nuclear accumulation of WDR68 was observed (Fig. 7B). The results indicate that TRiC/CCT is required for the nuclear accumulation of WDR68. The total staining intensities were not significantly changed by TCP1α suppression, in agreement with the results shown by Western blotting (Fig. 5B), suggesting that there was no change in WDR68 synthesis or degradation.
FIGURE 7.
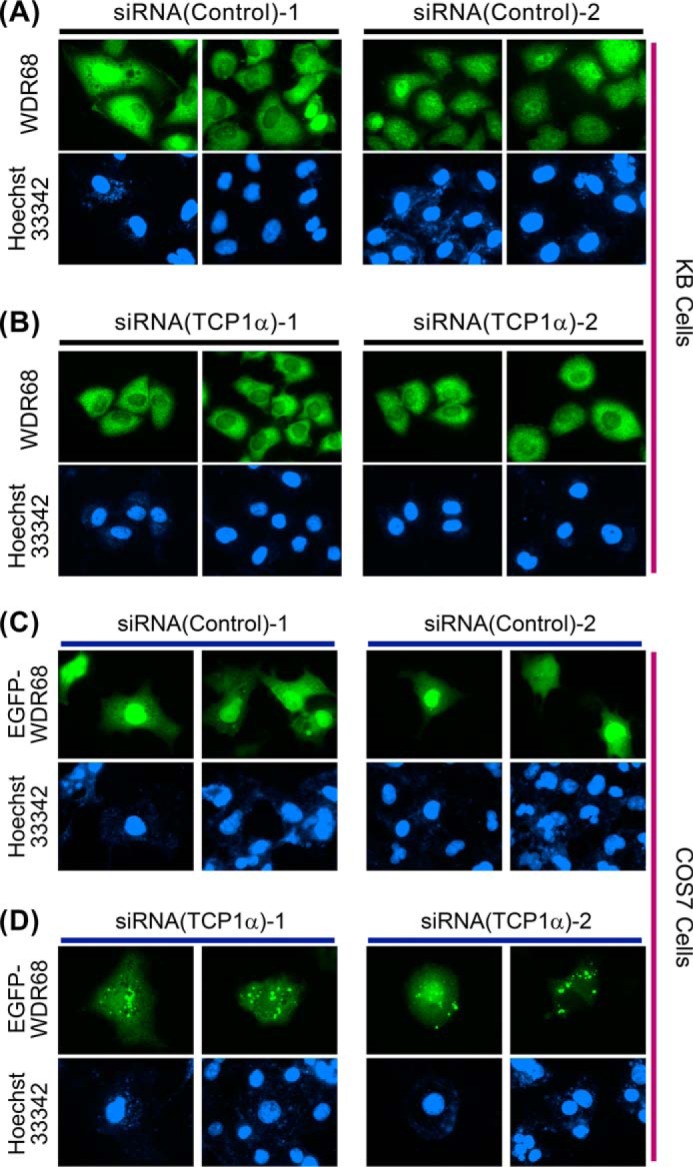
Effect of siRNA-induced suppression of TCP1α on the cellular distribution of endogenous and overexpressed WDR68. KB cells were transfected with either two control siRNAs (A, left and right) or two independent TCP1α-specific siRNAs (B, left (HSS110578) and right (HSS186251)). Cellular localization of endogenous WDR68 was examined by immunofluorescent microscopy. COS7 cells were transfected with either two control siRNAs (C, left and right) or two independent TCP1α-specific siRNAs (D, left (HSS110578) and right (HSS186251)) along with an expression plasmid for EGFP-WDR68. The cellular localization of EGFP-WDR68 was examined by fluorescent microscopy. Two representative green fluorescent images (top panels) are shown for each condition with blue nuclear images by Hoechst 33342 dye staining (bottom panels) (A–D).
The inability of WDR68 to accumulate in the nucleus in the TCP1α knockdown cells suggests that the structural maturation of WDR68 might require the sufficient chaperone function of TRiC/CCT. Finally, we examined the cytoplasmic form of overexpressed WDR68 in either the presence or absence of TRiC/CCT. GFP-tagged WDR68 was expressed in COS7 cells, and the intracellular distribution of GFP-WDR68 was examined by fluorescent microscopy. As shown in Fig. 7C, in the control siRNA-transfected cells, GFP-WDR68 was distributed throughout the cells uniformly in both the cytosol and the nucleus. In contrast, when TCP1α was suppressed by two independent siRNAs, GFP-WDR68 formed punctate dot structures in the cytoplasm (Fig. 7D). This result suggests that the WDR68 structure was disrupted under the condition of insufficient TRiC/CCT function, leading to the decrease in the cellular solubility of WDR68 and thus causing WDR68 aggregate formation. The total staining intensities were not significantly changed by TCP1α suppression. The WDR68 aggregates should be solubilized by detergent (1% Nonidet P-40) included in the buffer used for cell extract preparation, giving equal levels of WDR68 after TCP1α suppression (Fig. 5E). Therefore, our cell biological and biochemical data consistently indicate that TRiC/CCT is not involved in WDR68 translation or degradation. Altogether, our results show that TRiC/CCT binds to and chaperones WDR68 to form an appropriate mature structure that is essential for WDR68 solubility and nuclear translocation.
DISCUSSION
In this study, we identified TRiC/CCT as a major WDR68 binding partner by proteomic analysis and examined in detail the association between WDR68 and TRiC/CCT in mammalian cells by co-immunoprecipitation assays. Our results indicate the physiological importance of TRiC/CCT for correct folding, cytoplasmic solubility, DYRK1A-binding activity, and nuclear translocation of WDR68. The variety of identified WDR68 binding partners suggested that WDR68 has many physiological functions and is involved in multiple regulatory mechanisms. The relationship between WDR68 structure and its binding to TRiC/CCT was also evaluated and defined. We propose that TRiC/CCT chaperone is an important factor that facilitates the correct folding and function of WDR68 in cells.
WDR68 Structure and TRiC/CCT Binding
We analyzed the amino acid sequence to computationally predict its three-dimensional structure using protein folds of experimentally determined three-dimensional structures as templates. Two independent algorithms showed that WDR68 forms a seven-bladed β-propeller ring (Fig. 3), which is often observed in proteins with seven WD40 repeats. Coronin-1 forms a seven-bladed β-propeller, five blades of which are composed of its five WD40 repeats, and two additional blades are encoded in the sequences that lack any homology to the canonical WD40 motif (47). Similarly, WDR68 encodes five WD40 repeats forming five blades, and two additional blades of the structural model are made from the N- and C-terminal extensions that have no sequence similarity with the WD40 repeats. By assessing the binding of many WDR68 deletion mutants to TRiC/CCT, we concluded that three blades, irrespective of their position, are required and sufficient for TRiC/CCT binding (Fig. 4). This is in sharp contrast to the case of the binding of WDR68 to DYRK1A; our previous finding showed that even the loss of a single blade of WDR68 abolishes the binding to DYRK1A (9). A tethering function of WDR68 has been proposed (8); therefore, one WDR68 molecule should bind simultaneously to more than two target proteins. The predicted ring structure of WDR68 implies that two target proteins bind to both the top and bottom surfaces of the ring simultaneously.
The Role of TRiC/CCT in WD40 Folding
Several WD40 repeat-containing proteins, including trimeric G protein β (48, 49), COP1 (50), Cdc20, and Cdh1 (51), have been previously reported to bind to TRiC/CCT. Large scale protein-protein interaction maps were obtained by a proteome-wide mass spectrometric analysis in Saccharomyces cerevisiae (52), and the data analysis revealed that at least 21 proteins interact with TRiC/CCT, 16 of which have in common seven WD40 repeats (53). Analysis of the interaction with the G protein β subunit in vitro revealed that TRiC/CCT preferentially associates with the hydrophobic β-sheet region in proteins (49). Moreover, the TRiC/CCT interactome obtained by cDNA expression screening in a cell-free mammalian expression system indicated that the substrates of TRiC/CCT are enriched in multidomain proteins with a propensity for β-strands (31). These previous findings are consistent with the β-strand-rich structure of WDR68 obtained by our computational modeling (Fig. 3, B″ and C″). We propose that WD40 repeat proteins with the seven-bladed β-propeller structure generally depend on TRiC/CCT for their folding into the native conformation necessary for protein complex assembly in cells.
The β-propeller structure composed of WD40 repeats is believed to serve as a scaffold for simultaneous interactions with different proteins (1, 2, 54). WDR68 is not an exception because it binds to multiple proteins, including DYRK1A (9, 12, 13), DYRK1B (14), MEKK1 (8), HIPK2 (8), CUL4-DDB1 (6), DIAP1 (13), Drosophila atypical cadherin Dachsous (55), and TRiC/CCT (this study). Our proteomic analysis also identified many more putative WDR68-binding proteins with a wide variety of biological functions. WDR68 may function as a scaffold for interaction or as a vehicle for the delivery of a wide array of cellular proteins.
Two related WD40 repeat proteins, Cdc20 and Cdh1, bind to anaphase-promoting complex (APC/C), an E3 ubiquitin ligase, and confer the substrate specificities for ubiquitin-dependent proteolysis (56, 57). WDR68 binds to an E3 ubiquitin ligase, CUL4 (6), and to a canonical mitogen-activated protein kinase kinase kinase, MEKK1 (8) (this study), which also acts as an E3 ubiquitin ligase (58). WDR68 thus may have a role in protein ubiquitination and degradation. TRiC/CCT was shown to promote activation of APC/C via the generation of functional forms of both Cdc20 and Cdh1 (51). By analogy, TRiC/CCT might facilitate activation of the CUL4-DDB1 complex by chaperoning the folding of functional WDR68.
Phosphoproteomic Analysis of the WDR68-TRiC/CCT Complex
In this study, we have identified eight TRiC/CCT subunits as WDR68 binding partners and their in vivo phosphorylation sites by phosphoproteomic analysis. Among the 25 TRiC/CCT phosphorylation sites we have identified here, 18 are novel modification sites that are not described in the PhosphoSitePlus database (http://www.phosphosite.org). The importance of post-translational modifications in chaperone function has been recently emphasized for Hsp90 and its co-chaperones (59–61); however, surprisingly little is known about the regulation of TRiC/CCT by phosphorylation. To our knowledge, only one paper has described the identification of a responsible kinase and physiological regulation of TRiC/CCT by phosphorylation (62). We have also identified three novel phosphorylation sites in WDR68 (Ser-38, Ser-200, and Thr-257). Functional regulation of WDR68 by phosphorylation needs to be investigated in the future. WDR68 has been suggested to play a role in tethering signaling molecules; therefore, we anticipated that WDR68 may facilitate phosphorylation of certain DYRK1A substrates by binding both DYRK1A and its substrates. WDR68-binding phosphoproteins we have identified in this study may thus be good candidates for such DYRK1A substrates and will be elucidated in future studies.
Nuclear Translocation of WDR68 and TRiC/CCT
WDR68 has been reported to be localized in either the nucleus (63), cytoplasm (7), or both (9, 13). WDR68 probably shuttles between the cytoplasm and the nucleus. Nuclear access of WDR68 was shown to be critical for its role in normal craniofacial development in zebrafish (24), suggesting a functional importance of the cellular localization of WDR68. Our results indicate that WDR68 cannot accumulate in the nucleus without the presence of sufficient amounts of TRiC/CCT (Fig. 7). We have previously shown that the binding of DYRK1A to WDR68 induces nuclear accumulation of WDR68 (9). We speculate that TRiC/CCT may facilitate the structural maturation of WDR68, which is necessary for DYRK1A recognition and association. Indeed, our results indicated that WDR68 forms an abnormal structure lacking DYRK1A-binding activity in TRiC/CCT-deficient cells (Fig. 6). TRiC/CCT is a general molecular chaperone whose function is required for many other cellular proteins; thus, knockdown of TRiC/CCT might disrupt the structure of a large number of cellular proteins. Therefore, the inability to bind DYRK1A might not be a single and direct cause of the exclusion of WDR68 from the nucleus.
Down Syndrome and the Physiological Function of WDR68 in Different Species
DYRK1A is encoded in the most critical region (21q-22) of human chromosome 21, and its aberrant overexpression contributes to mental retardation and to other developmental disorders observed in Down syndrome patients (15, 16). Previously, we found that overexpression of DYRK1A induced nuclear translocation of WDR68 in a kinase activity-independent manner (9). WDR68 regulates gene expression (7, 14, 63), so the binding of overexpressed DYRK1A to WDR68 in the nucleus might interfere with normal gene expression. This could partly explain how overexpressed DYRK1A affects gene expression and induces the pleiotropic phenotypes observed in Down syndrome patients. Specific facial features are among the characteristics of Down syndrome, and an important role in craniofacial development was suggested for DYRK1A in a mouse model (22) and also for WDR68 in zebrafish (24, 63), further supporting our findings that WDR68 works cooperatively with DYRK1A in cells.
WDR68 and its homologues are observed in a wide array of species with high sequence identity. WDR68 plays a role in anthocyanin synthesis in petunia flowers (7) and in phytomelanin pigmentation in the seed coats of the Japanese morning glory (64). WDR68 homologues in Arabidopsis (LWD1 and LWD2) regulate photoperiodic flowering by modulating the proper rhythmic expression of genes (65). Recently, WDR68 in the fruit fly (Riquiqui) was shown to regulate the Hippo pathway downstream of atypical cadherin Dachsous and to control tissue growth (55). In fish, WDR68 is involved in craniofacial development (24, 63). Due to these diversified functions and deficiency phenotypes of WDR68 observed in different species, it is challenging to attribute a simple physiological role to WDR68. Probably, WDR68 serves as a general scaffold for protein-protein interactions, and different cells in different species utilize the suitable surfaces of WDR68 for a particular protein-protein interaction in a context-dependent manner. Further identification of proteins interacting with WDR68 should be of particular importance to reveal the physiological function of WDR68 in each specific milieu.
Acknowledgments
We thank T. Sakabe, T. Aoki, and M. Nakagawa for excellent technical assistance. We thank Drs. N. Iwabe (Kyoto University) and K. Katoh (Osaka University) for help and discussion regarding the amino acid sequence analyses.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
- WDR68
- WD repeat protein 68
- DYRK
- dual specificity tyrosine phosphorylation-regulated protein kinase
- TCP1
- T-complex protein 1
- TRiC
- TCP1 ring complex
- CCT
- chaperonin containing TCP1
- Hsp
- heat shock protein
- HIPK
- homeodomain interacting protein kinase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- APC/C
- anaphase-promoting complex.
REFERENCES
- 1. Smith T. F., Gaitatzes C., Saxena K., Neer E. J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 2. Li D., Roberts R. (2001) WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol. Life Sci. 58, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stirnimann C. U., Petsalaki E., Russell R. B., Müller C. W. (2010) WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574 [DOI] [PubMed] [Google Scholar]
- 4. Wall M. A., Coleman D. E., Lee E., Iñiguez-Lluhi J. A., Posner B. A., Gilman A. G., Sprang S. R. (1995) The structure of the G protein heterotrimer Giα1β1γ2. Cell 83, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 5. Sondek J., Bohm A., Lambright D. G., Hamm H. E., Sigler P. B. (1996) Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature 379, 369–374 [DOI] [PubMed] [Google Scholar]
- 6. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 7. de Vetten N., Quattrocchio F., Mol J., Koes R. (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11, 1422–1434 [DOI] [PubMed] [Google Scholar]
- 8. Ritterhoff S., Farah C. M., Grabitzki J., Lochnit G., Skurat A. V., Schmitz M. L. (2010) The WD40-repeat protein Han11 functions as a scaffold protein to control HIPK2 and MEKK1 kinase functions. EMBO J. 29, 3750–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miyata Y., Nishida E. (2011) DYRK1A binds to an evolutionarily conserved WD40-repeat protein WDR68 and induces its nuclear translocation. Biochim. Biophys. Acta 1813, 1728–1739 [DOI] [PubMed] [Google Scholar]
- 10. Becker W., Joost H.-G. (1999) Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid Res. Mol. Biol. 62, 1–17 [DOI] [PubMed] [Google Scholar]
- 11. Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F. J., Joost H.-G. (1998) Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 273, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 12. Skurat A. V., Dietrich A. D. (2004) Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 279, 2490–2498 [DOI] [PubMed] [Google Scholar]
- 13. Morita K., Lo Celso C., Spencer-Dene B., Zouboulis C. C., Watt F. M. (2006) HAN11 binds mDia1 and controls GLI1 transcriptional activity. J. Dermatol. Sci. 44, 11–20 [DOI] [PubMed] [Google Scholar]
- 14. Mazmanian G., Kovshilovsky M., Yen D., Mohanty A., Mohanty S., Nee A., Nissen R. M. (2010) The zebrafish dyrk1b gene is important for endoderm formation. Genesis 48, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galceran J., de Graaf K., Tejedor F. J., Becker W. (2003) The Mnb/Dyrk1A protein kinase: genetic and biochemical properties. J. Neural Transm. Suppl. 67, 139–148 [DOI] [PubMed] [Google Scholar]
- 16. Hämmerle B., Elizalde C., Galceran J., Becker W., Tejedor F. J. (2003) The Mnb/Dyrk1A protein kinase: neurobiological functions and Down syndrome implications. J. Neural Transm. Suppl. 67, 129–137 [DOI] [PubMed] [Google Scholar]
- 17. Altafaj X., Dierssen M., Baamonde C., Martí E., Visa J., Guimerà J., Oset M., González J. R., Flórez J., Fillat C., Estivill X. (2001) Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 10, 1915–1923 [DOI] [PubMed] [Google Scholar]
- 18. Martínez de Lagrán M., Altafaj X., Gallego X., Martí E., Estivill X., Sahún I., Fillat C., Dierssen M. (2004) Motor phenotypic alterations in TgDyrk1a transgenic mice implicate DYRK1A in Down syndrome motor dysfunction. Neurobiol. Dis. 15, 132–142 [DOI] [PubMed] [Google Scholar]
- 19. Park J., Yang E. J., Yoon J. H., Chung K. C. (2007) Dyrk1A overexpression in immortalized hippocampal cells produces the neuropathological features of Down syndrome. Mol. Cell Neurosci. 36, 270–279 [DOI] [PubMed] [Google Scholar]
- 20. Liu F., Liang Z., Wegiel J., Hwang Y. W., Iqbal K., Grundke-Iqbal I., Ramakrishna N., Gong C. X. (2008) Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J. 22, 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yabut O., Domogauer J., D'Arcangelo G. (2010) Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J. Neurosci. 30, 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arron J. R., Winslow M. M., Polleri A., Chang C. P., Wu H., Gao X., Neilson J. R., Chen L., Heit J. J., Kim S. K., Yamasaki N., Miyakawa T., Francke U., Graef I. A., Crabtree G. R. (2006) NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- 23. Gwack Y., Sharma S., Nardone J., Tanasa B., Iuga A., Srikanth S., Okamura H., Bolton D., Feske S., Hogan P. G., Rao A. (2006) A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature 441, 646–650 [DOI] [PubMed] [Google Scholar]
- 24. Wang B., Doan D., Roman Petersen Y., Alvarado E., Alvarado G., Bhandari A., Mohanty A., Mohanty S., Nissen R. M. (2013) WDR68 requires nuclear access for craniofacial development. PLoS One 8, e54363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 26. Spiess C., Meyer A. S., Reissmann S., Frydman J. (2004) Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 14, 598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 28. Dunn A. Y., Melville M. W., Frydman J. (2001) Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J. Struct. Biol. 135, 176–184 [DOI] [PubMed] [Google Scholar]
- 29. Spiess C., Miller E. J., McClellan A. J., Frydman J. (2006) Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell 24, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim S., Willison K. R., Horwich A. L. (1994) Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 19, 543–548 [DOI] [PubMed] [Google Scholar]
- 31. Yam A. Y., Xia Y., Lin H. T., Burlingame A., Gerstein M., Frydman J. (2008) Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kitamura A., Kubota H., Pack C. G., Matsumoto G., Hirayama S., Takahashi Y., Kimura H., Kinjo M., Morimoto R. I., Nagata K. (2006) Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 8, 1163–1170 [DOI] [PubMed] [Google Scholar]
- 33. Tam S., Geller R., Spiess C., Frydman J. (2006) The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 8, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Broadley S. A., Hartl F. U. (2009) The role of molecular chaperones in human misfolding diseases. FEBS Lett. 583, 2647–2653 [DOI] [PubMed] [Google Scholar]
- 35. Koyasu S., Nishida E., Kadowaki T., Matsuzaki F., Iida K., Harada F., Kasuga M., Sakai H., Yahara I. (1986) Two mammalian heat shock proteins, HSP90 and HSP100, are actin-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 83, 8054–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miyata Y., Yahara I. (2000) p53-independent association between SV40 large T antigen and the major cytosolic heat shock protein, HSP90. Oncogene 19, 1477–1484 [DOI] [PubMed] [Google Scholar]
- 37. Miyata Y., Chambraud B., Radanyi C., Leclerc J., Lebeau M.-C., Renoir J.-M., Shirai R., Catelli M.-G., Yahara I., Baulieu E.-E. (1997) Phosphorylation of the immunosuppressant FK506-binding protein FKBP52 by casein kinase II (CK2): regulation of HSP90-binding activity of FKBP52. Proc. Natl. Acad. Sci. U.S.A. 94, 14500–14505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miyata Y., Akashi M., Nishida E. (1999) Molecular cloning and characterization of a novel member of the MAP kinase superfamily. Genes Cells 4, 299–309 [DOI] [PubMed] [Google Scholar]
- 39. Miyata Y., Ikawa Y., Shibuya M., Nishida E. (2001) Specific association of a set of molecular chaperones including HSP90 and Cdc37 with MOK, a member of the MAP kinase superfamily. J. Biol. Chem. 276, 21841–21848 [DOI] [PubMed] [Google Scholar]
- 40. Miyata Y., Nishida E. (2004) CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone Cdc37. Mol. Cell Biol. 24, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 42. Miyata Y., Nishida E. (2005) CK2 binds, phosphorylates, and regulates its pivotal substrate Cdc37, an Hsp90-cochaperone. Mol. Cell. Biochem. 274, 171–179 [DOI] [PubMed] [Google Scholar]
- 43. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 44. Murzina N. V., Pei X. Y., Zhang W., Sparkes M., Vicente-Garcia J., Pratap J. V., McLaughlin S. H., Ben-Shahar T. R., Verreault A., Luisi B. F., Laue E. D. (2008) Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan D., Nakatsu T., Kato H. (2013) Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p. Nat. Struct. Mol. Biol. 20, 987–993 [DOI] [PubMed] [Google Scholar]
- 47. Appleton B. A., Wu P., Wiesmann C. (2006) The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure 14, 87–96 [DOI] [PubMed] [Google Scholar]
- 48. Wells C. A., Dingus J., Hildebrandt J. D. (2006) Role of the chaperonin CCT/TRiC complex in G protein βγ-dimer assembly. J. Biol. Chem. 281, 20221–20232 [DOI] [PubMed] [Google Scholar]
- 49. Kubota S., Kubota H., Nagata K. (2006) Cytosolic chaperonin protects folding intermediates of Gβ from aggregation by recognizing hydrophobic β-strands. Proc. Natl. Acad. Sci. U.S.A. 103, 8360–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yi C., Li S., Wang J., Wei N., Deng X. W. (2006) Affinity purification reveals the association of WD40 protein constitutive photomorphogenic 1 with the hetero-oligomeric TCP-1 chaperonin complex in mammalian cells. Int. J. Biochem. Cell Biol. 38, 1076–1083 [DOI] [PubMed] [Google Scholar]
- 51. Camasses A., Bogdanova A., Shevchenko A., Zachariae W. (2003) The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12, 87–100 [DOI] [PubMed] [Google Scholar]
- 52. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 53. Valpuesta J. M., Martín-Benito J., Gómez-Puertas P., Carrascosa J. L., Willison K. R. (2002) Structure and function of a protein folding machine: the eukaryotic cytosolic chaperonin CCT. FEBS Lett. 529, 11–16 [DOI] [PubMed] [Google Scholar]
- 54. van der Voorn L., Ploegh H. L. (1992) The WD-40 repeat. FEBS Lett. 307, 131–134 [DOI] [PubMed] [Google Scholar]
- 55. Degoutin J. L., Milton C. C., Yu E., Tipping M., Bosveld F., Yang L., Bellaiche Y., Veraksa A., Harvey K. F. (2013) Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat. Cell Biol. 15, 1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fang G., Yu H., Kirschner M. W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163–171 [DOI] [PubMed] [Google Scholar]
- 57. Yu H. (2007) Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell 27, 3–16 [DOI] [PubMed] [Google Scholar]
- 58. Lu Z., Xu S., Joazeiro C., Cobb M. H., Hunter T. (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9, 945–956 [DOI] [PubMed] [Google Scholar]
- 59. Miyata Y., Nakamoto H., Neckers L. (2013) The therapeutic target Hsp90 and cancer hallmarks. Curr. Pharm. Des. 19, 347–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mollapour M., Neckers L. (2012) Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta 1823, 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cloutier P., Coulombe B. (2013) Regulation of molecular chaperones through post-translational modifications: decrypting the chaperone code. Biochim. Biophys. Acta 1829, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Abe Y., Yoon S. O., Kubota K., Mendoza M. C., Gygi S. P., Blenis J. (2009) p90 ribosomal S6 kinase and p70 ribosomal S6 kinase link phosphorylation of the eukaryotic chaperonin containing TCP-1 to growth factor, insulin, and nutrient signaling. J. Biol. Chem. 284, 14939–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nissen R. M., Amsterdam A., Hopkins N. (2006) A zebrafish screen for craniofacial mutants identifies WDR68 as a highly conserved gene required for endothelin-1 expression. BMC Dev. Biol. 6, 28–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Park K. I., Hoshino A. (2012) A WD40-repeat protein controls proanthocyanidin and phytomelanin pigmentation in the seed coats of the Japanese morning glory. J. Plant Physiol. 169, 523–528 [DOI] [PubMed] [Google Scholar]
- 65. Wu J. F., Wang Y., Wu S. H. (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148, 948–959 [DOI] [PMC free article] [PubMed] [Google Scholar]