Background: The HOPS tethering complex has five subunits with similar domain arrangements, but structural insights into these is scarce.
Results: The N-terminal domain of Vps18 reveals the structure of a β-propeller.
Conclusion: The β-propeller is dispensable for HOPS assembly but critical for stability and function.
Significance: Our data provide evidence of an evolutionarily conserved domain within HOPS.
Keywords: Crystal Structure, Endosome, Membrane Fusion, Rab, SNARE Proteins, HOPS, Tethering Complex, Vacuole
Abstract
Membrane fusion at the vacuole, the lysosome equivalent in yeast, requires the HOPS tethering complex, which is recruited by the Rab7 GTPase Ypt7. HOPS provides a template for the assembly of SNAREs and thus likely confers fusion at a distinct position on vacuoles. Five of the six subunits in HOPS have a similar domain prediction with strong similarity to COPII subunits and nuclear porins. Here, we show that Vps18 indeed has a seven-bladed β-propeller as its N-terminal domain by revealing its structure at 2.14 Å. The Vps18 N-terminal domain can interact with the N-terminal part of Vps11 and also binds to lipids. Although deletion of the Vps18 N-terminal domain does not preclude HOPS assembly, as revealed by negative stain electron microscopy, the complex is instable and cannot support membrane fusion in vitro. We thus conclude that the β-propeller of Vps18 is required for HOPS stability and function and that it can serve as a starting point for further structural analyses of the HOPS tethering complex.
Introduction
Protein and lipid transport between organelles of the endomembrane system relies on a conserved machinery. Cargoes are packaged into vesicles at a donor organelle and transported via the cytoskeleton to the target membrane. Here, vesicles are recognized by an interplay of membrane-resident Rab GTPases and tethering factors and ultimately fuse with the help of membrane-embedded SNARE proteins (1). Whereas the mechanism of SNARE-mediated fusion has been determined to some extent in molecular detail (2), the knowledge of the interplay between Rabs and tethering complexes is lagging behind. This is in part due to the lack of structural information on tethering complexes and their interaction with Rab GTPases (3).
Rabs are switch-like proteins with a C-terminal prenyl anchor (4, 5). In the GDP-bound form, Rabs are extracted and kept soluble with the help of Rab GDP dissociation inhibitors, whereas recruitment to the membrane and GTP loading of the Rab requires a specific guanine nucleotide exchange factor. The Rab-GTP complex then interacts with tethering factors to promote vesicle positioning and fusion (1, 6). Tethering factors come in two flavors. The CATCHR complexes, such as the exocyst involved in exocytosis or the COG complex in intra-Golgi transport, consist of central α-helical segments (3, 7). In contrast, the two large hexameric tethering complexes in the endolysosomal pathway, CORVET and HOPS, each contain five subunits that are structurally similar to COPII subunits, clathrin, and nuclear porins with predicted N-terminal β-propellers and C-terminal α-helical segments (6, 8). Only the Sec1/Munc18 (SM)3 protein Vps33 resembles other SM proteins as verified recently (9, 10). Although mainly characterized in yeast, both complexes seem to be conserved in metazoan cells (6, 11). Both complexes require Rab binding for their localization. CORVET binds the Rab5-like Vps21, whereas HOPS interacts with the Rab7-like Ypt7.
Recently, we could resolve the overall structure of the HOPS complex by negative stain electron microscopy (12) showing that the two Ypt7 binding subunits Vps41 and Vps39 reside at opposite ends of an elongated seahorse-shaped structure. In addition, the structure of the SM-protein Vps33 in complex with an α-helical segment of Vps16 was resolved (9, 10). The two remaining subunits, Vps11 and Vps18, have a similar domain arrangement to Vps16, Vps39, and Vps41 (6, 8).
Here, we demonstrate that the N-terminal domain of Vps18 indeed forms a bona fide seven-bladed β-propeller. We resolved its structure at 2.14 Å, determined its interactions within HOPS, and show that, although dispensable for HOPS assembly, the domain is required for stability and thus function.
EXPERIMENTAL PROCEDURES
Yeast Strains
Deletions of the Vps18 N-terminal domain were generated by insertion of the GAL1 promoter by homologous recombination. Likewise, C-terminal truncations were obtained (13). For overproduction of HOPS without the N-terminal domain, one copy of Vps18 was deleted, whereas the GAL1 promoter replaced the coding region of the N-terminal domain in the other allele. Either Vps41 or Vps18 was tagged C-terminally with the tandem affinity purification (TAP) tag. All other HOPS subunits were equally overproduced in the same diploid strain (12, 14). HOPS or HOPS subcomplexes (14) were purified via IgG beads and tobacco etch virus (TEV) protease cleavage as described (12).
Heterologous Protein Expression and Purification
Vps18NTD (residues 2–349) was subcloned from genomic Saccharomyces cerevisiae DNA into the pQlinkH vector (15) encoding an N-terminal hexahistidine tag and a TEV protease cleavage sequence using the forward primer 5′-GAAGA TCTAT AAAAA CACGT ATAGA GGAAG TTCAG TTAC-3′ and the reverse primer 3′-AGCCA TACCT TAAAT AATCA AGCCC TAACT CGCCG GCGTC AGTCA G-5′. Proteins were overexpressed in Escherichia coli RosettaTM 2(DE3) (Novagen) as follows; cells were grown in 2 × LB medium at 37 °C until A600 = 1.2 and induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside at 17 °C overnight. For preparation of a selenomethionine derivative, methionine biosynthesis was inhibited as described in Van Duyne et al. (16).
Bacterial lysate containing His-tagged protein was applied onto nickel iminodiacetic acid resin (Macherey-Nagel), washed with a step gradient (5 mm imidazole, 20 mm imidazole), and eluted with 250 mm imidazole in buffer A (10 mm Tris-HCl, pH 8.0, 300 mm NaCl). Fractions containing Vps18NTD were pooled and digested with His-tagged TEV protease while dialyzing against dialysis buffer (20 mm Tris, pH 8.0, 300 mm NaCl, 5% (v/v) glycerol). Non-cleaved protein and TEV protease were removed by passage over nickel iminodiacetic acid resin, and the concentrated sample was subjected to size-exclusion chromatography (Superdex 200, 26/60, GE Healthcare) in buffer containing 20 mm Tris acetate, pH 8.0, 150 mm NaCl. Purified protein was concentrated and flash-frozen in liquid nitrogen.
Crystallization and Structure Determination
Native and selenomethionine derivate crystals of Vps18NTD were obtained using the hanging-drop vapor-diffusion method by mixing 1 μl of protein (11 mg/ml) in gel filtration buffer with 1 μl of a reservoir solution containing 25% (w/v) PEG 3350, 0.2 m (NH4)2SO4, 0.1 m Bis-Tris, pH 6.5, at room temperature. Diffraction data were collected at 100 K at beamlines 14.1 and 14.2 of BESSY II (Helmholtz-Zentrum Berlin für Materialien und Energie (17)). Selenomethionine derivative crystals were used for phasing by multiple-wavelength anomalous diffraction (MAD). Data were processed with XDS (18). Selenium atoms were located, and a model was obtained with SHELXC/D/E (19) and ARP/wARP (20) to generate an initial trace of the structure. This initial model was used for phasing by molecular replacement of the dataset obtained from the native crystal. The model was completed manually in COOT (21) and refined by alternating rounds of manual intervention and optimization in Refmac5 (22). TLS groups were assigned using the TLSMD web server (23). Model quality was evaluated by Molprobity (24). The final model was refined to Rwork/Rfree values of 20.1%/24.4%, respectively, at a resolution of 2.14 Å. No electron density was interpretable for amino acid residues 223–235, 252–260, and 295–308. PyMOL was used to prepare all structure figures (The PyMOL Molecular Graphics System, Version 1.7, Schrödinger LLC). The electrostatic surface potential was calculated using ABPS (25) as implemented in PyMOL. The hydrophobic surface potential was calculated according to Eisenberg et al. (26).
EM and Image Processing
Purified samples were negatively stained according to a modified protocol of (27). Briefly, a 4-μl drop of sample solution was applied to a glow-discharged carbon-coated copper grid washed with two 20-μl drops of deionized water and stained with two 10-μl drops of freshly prepared 0.035 mg/ml uranyl formate solution. Images were collected using a Jeol JEM-1400 transmission electron microscope equipped with a LaB6 cathode operating at 120 kV. Micrographs were taken with a CMOS camera (TVIPS TemCam F-416, 4kx4k). After manual selection of single particles, reference-free and reference-based alignments as well as K-means and iterative stable alignment and clustering (ISAC) classifications were performed within EMAN2 (28) and SPARX (29) as described (12).
GST Pulldown Assay
Vps18NTD (residues 2–349) was subcloned from genomic S. cerevisiae DNA into the pQlinkG vector (15) encoding an N-terminal GST tag and a TEV protease cleavage sequence using the same primers as for the His fusion protein (see above). Proteins were overexpressed in E. coli RosettaTM 2(DE3) (Novagen) as follows; cells were grown in 2 × LB medium at 37 °C until A600 = 1.2 and induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside at 17 °C overnight. Cell extract containing recombinant GST fusion protein was added without further purification to 25 μl of a slurry of pre-washed GSH beads (Qiagen) for 2 h at 4 °C to allow protein binding to the beads. Beads were washed 3 times with wash buffer (50 mm HEPES/KOH pH 7.5, 150 mm NaCl, 0.15% Nonidet P-40 (v/v; Igepal CA-630, Sigma), 1 mm DTT). Cell extracts containing recombinant His fusion protein were added without further purification and incubated for 3 h at 4 °C. Beads were again washed 8–15 times using wash buffer. 4 × Laemmli buffer (50 mm Tris-HCl, 100 mm DTT, 2% (w/v) SDS, 0.25% (w/v) bromphenol blue, 10% glycerol (w/v)) was added to the GSH beads, and bound protein was eluted by incubation at 95 °C for 5 min. Samples were analyzed by SDS-PAGE, Coomassie staining, and anti-His Western blotting.
Liposome Co-sedimentation Assay
Protein binding to liposomes was studied by a co-sedimentation assay using vesicles prepared from Folch extract (30) (Sigma). For preparation of liposomes, Folch extract dissolved in CHCl3 was dried under a stream of dry nitrogen gas and then exposed to high vacuum for at least 1 h to complete drying and to remove residual solvent. Lipids were rehydrated in 20 mm HEPES/KOH, pH 7.5, 150 mm KCl for 15 min at room temperature to give a final concentration of 4 mg/ml. After vigorous mixing, the rehydrated lipids were subjected to 10 freeze/thaw cycles, alternating between −78 °C and 37 °C. Unilamellar vesicles were prepared by extrusion (Mini-Extruder, Avanti Polar Lipids) using polycarbonate membranes (Avanti Polar Lipids) with decreasing pore sizes of 0.8, 0.2, 0.1, and 0.05 μm (21 passes each).
Centrifugation assay samples (40 μl) were prepared containing 5 μm purified protein with liposomes at varying concentrations (0.5–2.0 mg/ml) and different average diameters (50–800 nm). After incubation at room temperature for 10 min, samples were sedimented at 70,000 rpm for 15 min at 20 °C. Both supernatant and pellet were analyzed by SDS-PAGE using Coomassie staining as described (31).
RESULTS
Structure of the Vps18 N-terminal Domain
The HOPS complex consists of five subunits, each with predicted N-terminal β-propeller domains. We previously showed that CORVET assembly occurs in the absence of the N-terminal domains of CORVET Vps3 and Vps8, whereas the C-terminal domains are critical for assembly (32). In agreement with this, the in vitro assembly of the C-terminal predicted α-helical segments resulted in a HOPS core complex (33), suggesting that the β-propeller domains of all five similar subunits in HOPS and CORVET may be directed toward the cytosol. In an attempt to obtain insight into the domain architecture, we expressed distinct domains of all six subunits. Among those, the N-terminal domain of Vps18 was the best-behaving and thus chosen for further analysis.
Crystallization trials of different Vps18 truncation constructs resulted in crystals of a protein comprising the N-terminal domain (NTD) predicted to fold into a seven-bladed β-propeller and parts of the neighboring region predicted to form an α-solenoid structure (residues 2–349; Vps18NTD). Native Vps18NTD crystals diffracted to a resolution of 2.14 Å. The structure was determined by multiple-wavelength anomalous diffraction with a 2.4-Å dataset collected from a selenomethionine-substituted crystal (Table 1) and was refined to Rwork/Rfree values of 20.1%/24.4%, respectively. Residues 2–222, 234–252, 259–294, and 307–348 could be modeled into the electron density. The remaining loops and the C-terminal residue could not be modeled and are presumably disordered.
TABLE 1.
X-ray data collection and structure refinement statistics
rms, root mean square.
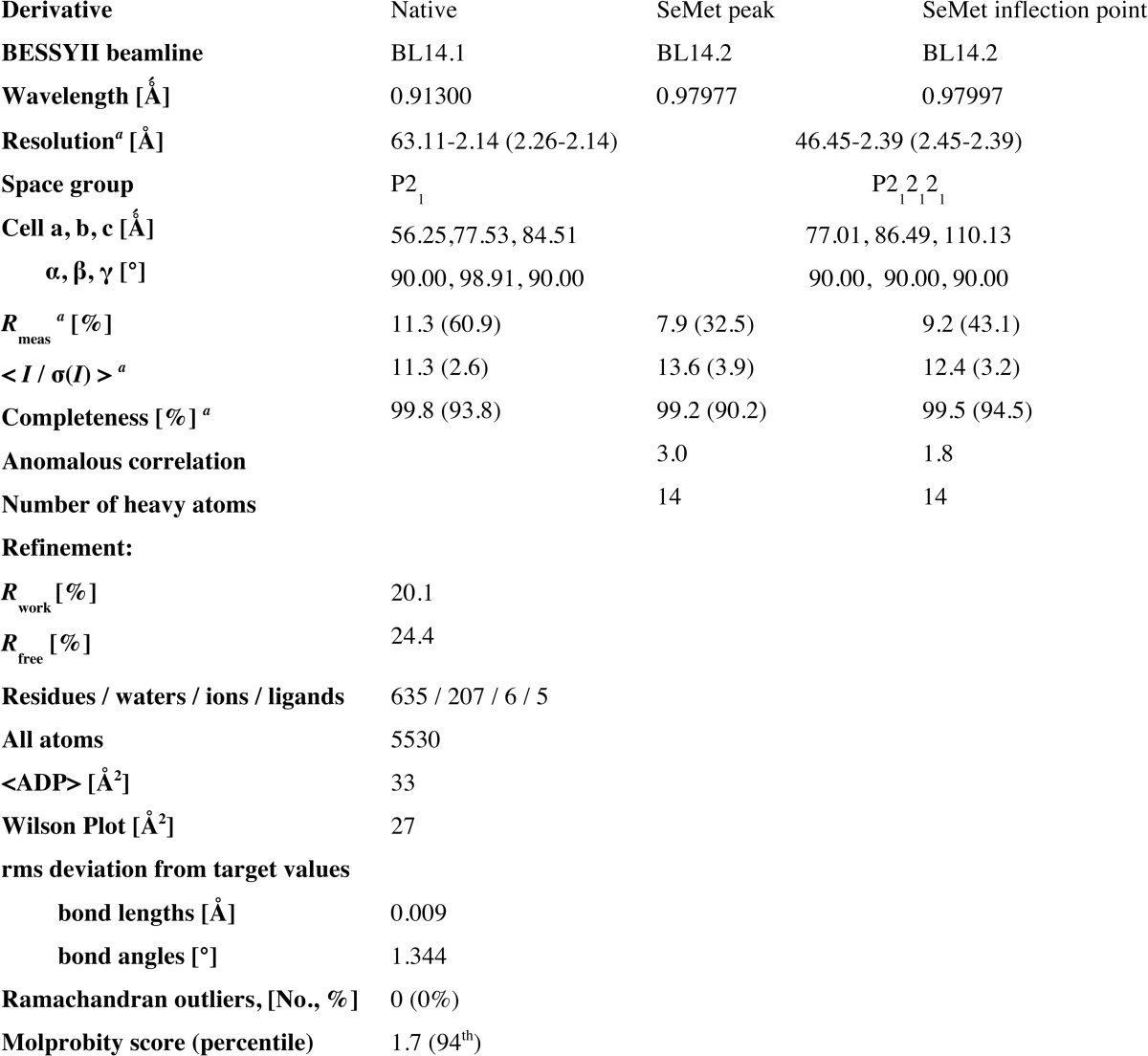
a The outer shell is in parentheses.
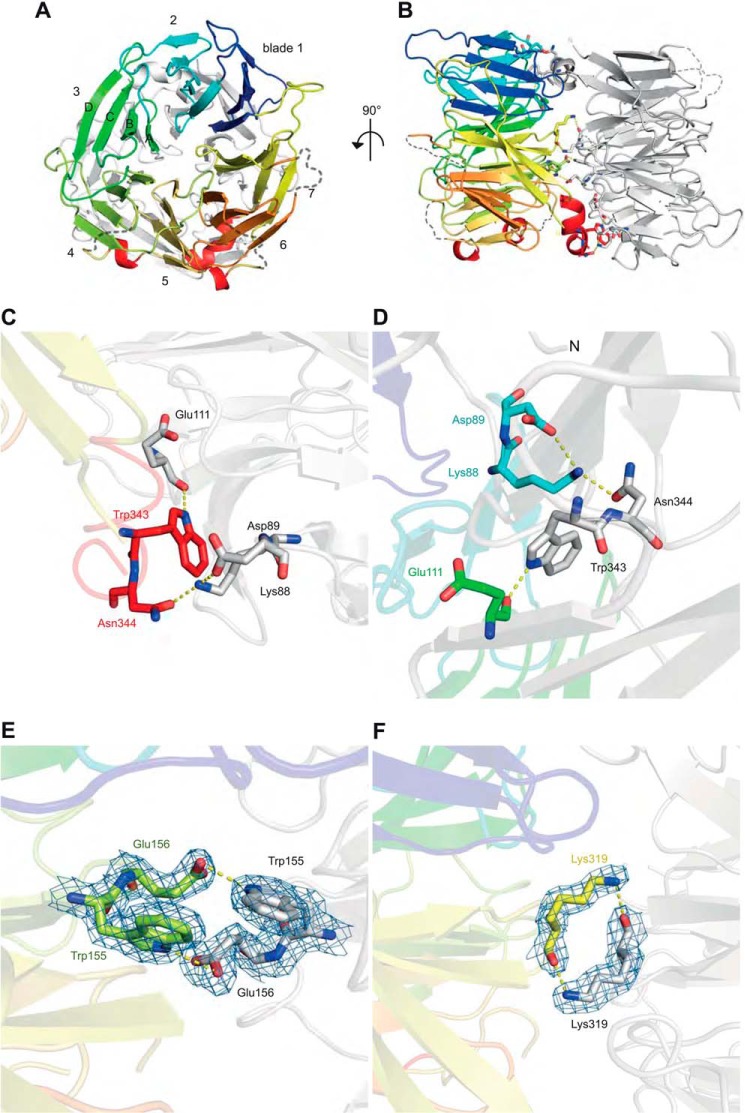
The asymmetric unit contains two highly similar copies of Vps18NTD (root mean square deviation 0.105 Å for matching α-carbons) related by a non-crystallographic dyad axis. Per protein chain, 1230 Å2 (8%) of the solvent-accessible surface are buried in the dimer interface, which is predicted by PISA to be unstable in solution (34). As predicted, the NTD of Vps18 forms a seven-bladed β-propeller (Fig. 1, A and B). Each blade consists of four antiparallel β-strands with the seventh blade formed by the first N-terminal strand (residues 3–9) and the last three C-terminal strands (residues 310–336). Except for the loop connecting strands C and D of blade 5 (residues 220–236), all strands are connected by short linkers. Longer loops are found between blades 5 and 6 (residues 249–259) and blades 6 and 7 (residues 294–309), both of which are disordered in the crystal structure. Vps18NTD is terminated by a C-terminal α-helix (residues 338–348), which belongs to the adjacent α-solenoid region.
FIGURE 1.
Vps18NTD folds into a seven-bladed β-propeller. A schematic representation front view (A) and side view (B) of the crystal dimer is shown. The propeller blades of chain B are colored individually, whereas chain A is depicted in gray. Details are of the crystal dimer interface. Shown are stick representations of regions highlighted in Fig. 4. C and D corresponds to box D1 and D1′ of Fig. 4, E to D2, and F to D3. The blue mesh depicts the 2m|Fo| − D|Fc| electron density contoured at 1 σ. Yellow dashed lines indicate hydrogen bonds.
This terminal α-helix participates in two of the four intersubunit contacts of the crystal dimer, and it intercalates between the second and third blade of the opposing chain, resulting in a 180° rotation between the two protein copies in the asymmetric unit. In both copies the indole nitrogen of Trp-343 interacts with the backbone of Glu-111 via the carbonyl oxygen, and the amide side chain of Asn-344 forms hydrogen bonds to Lys-88 and Asp-89 (Fig. 1, C and D). The remaining two crystal interfaces contain a hydrogen bond between the indole nitrogen of Trp-155 and the carboxylate group of Glu-156 and another hydrogen bond between the amino group of Lys-319 in one copy and the backbone carbonyl group of Lys-319 in the other (Fig. 1, E and F).
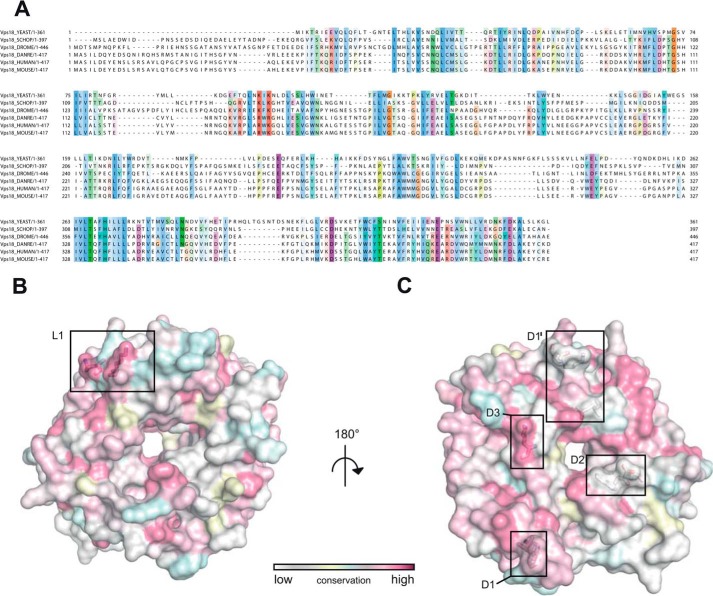
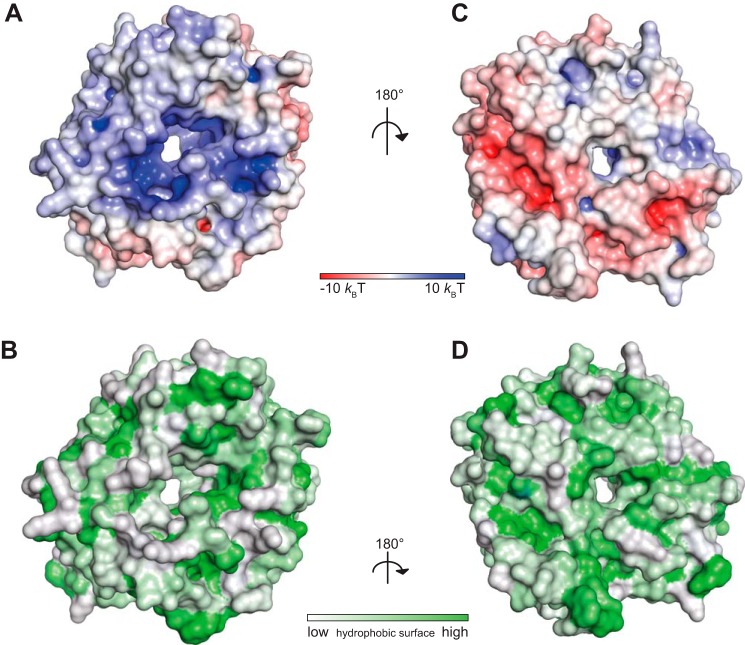
Interestingly, for five of the six HOPS subunits an N-terminal β-propeller domain followed by an α-solenoid region has been predicted (8), suggesting that the dimer observed in the crystal structure may be of physiological importance. However, only two residues participating in the dimer interface (Lys-319 and Asn-340) are conserved across species (see Fig. 4C). The interface itself is dominated by polar interactions (Fig. 2A) with only small patches of hydrophobic contact sites (Fig. 2B). The solvent-exposed side is predominantly positively charged, again with only minor patches of potential hydrophobic binding sites (Fig. 2, C and D).
FIGURE 4.
The dimer interface is less conserved than the residues involved in lipid binding. A, sequence alignment of the N-terminal domain of Vps18 across different eukaryotic species. Residues are colored according to the ClustalX scheme, which color codes conserved residues by their chemical nature: hydrophobic in blue, basic in red, polar uncharged in green, acidic in magenta, hydrophilic aromatic in cyan, cysteine in pink, glycine in orange, and proline in yellow. B and C, surface representation of chain B colored by evolutionary conservation. Key residues involved in lipid binding (B) and the crystal dimer interface (C) are highlighted and shown in detail in Fig. 1. Boxes D1 and D1′ correspond to panels C and D of Fig. 1, D2 to E, and D3 to F.
FIGURE 2.
The interface of the crystal dimer is dominated by polar interactions. A–D, surface representations of chain B colored by (A and C) electrostatic potential or (B and D) hydrophobic potential.
Interaction of Vps18NTD with Other HOPS Subunits and Lipids
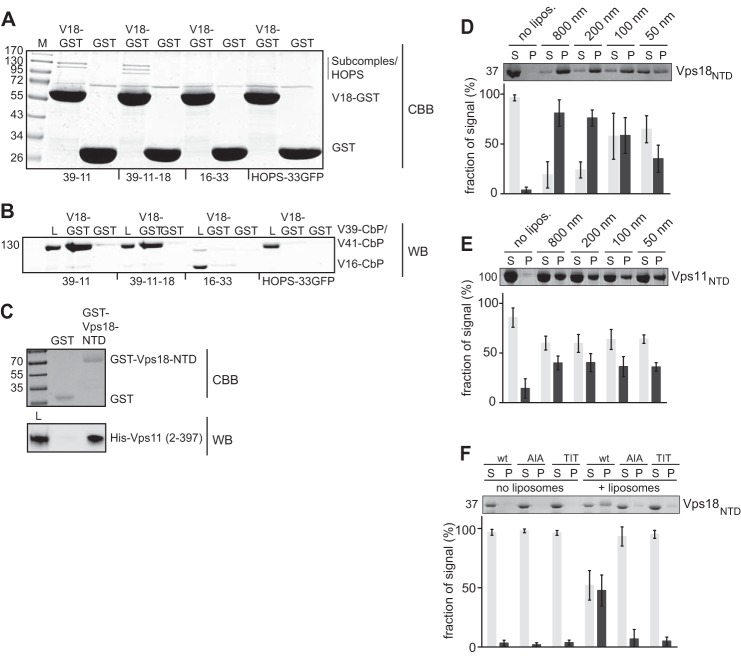
Our previous analysis of the HOPS overall structure showed that Vps18 is located in the large head of the seahorse-shaped complex, likely to be proximal to the Ypt7-interacting Vps41 subunit and the dimer of the SM protein Vps33 and Vps16 (12). The low resolution of the electron microscopy structure analysis precluded a more detailed localization. To characterize the binding mode of Vps18NTD further, we incubated stable HOPS subcomplexes purified from yeast with GST-Vps18NTD. We observed stable interaction both with the Vps39-Vps11 dimer and the Vps39-Vps11-Vps18 trimer (Fig. 3, A and B). No interactions were observed with the Vps33-Vps16 dimer or with the entire HOPS complex (Fig. 3, A and B). As Vps39 is found at the tail of HOPS, these data suggest that Vps18NTD interacts most likely directly with Vps11.
FIGURE 3.
The Vps18 β-propeller interacts with the predicted Vps11 propeller. A and B, interaction of Vps18NTD-GST with subcomplexes of HOPS. Vps18NTD-GST was incubated with the indicated Vps11-Vps39 dimer, the Vps11-Vps39-Vps18 trimer, with the Vps16-Vps33 dimer, and with the entire HOPS complex carrying a GFP tag on Vps33. The GST pulldown was conducted as before (14). Eluted proteins were analyzed on Coomassie-stained gels (CBB) or by Western blot (WB) against the calmodulin-binding protein (CbP) tag, which is part of the TAP tag. C, purified His-tagged Vps11 β-propeller was incubated with either GST or Vps18NTD-GST. Proteins were eluted by boiling and analyzed on Coomassie-stained gels or Western blot against the His-tag. D and E, Vps11NTD and Vps18NTD bound to membranes. Both domains were incubated with liposomes of different size and then pelleted by centrifugation (30 min at 100,000 g, 4 °C). S, supernatant (gray bars); P, pellet (dark gray bars). F, analysis of charge mutants in Vps18NTD was conducted as in F. AIA corresponds to K97A/K99A, and TIT corresponds to T97A/T99A.
Based on the observation of a dimer in the crystal structure, we considered that Vps11 might interact with Vps18 via its N-terminal β-propeller. Indeed, we observed the specific interaction of His-tagged Vps11NTD and GST-Vps18NTD (Fig. 3C).
Some β-propeller domains such as the autophagy protein Atg18, interact with lipids and thus support targeting to membranes (35). Indeed, when we incubated either the Vps11 or the Vps18 N-terminal domain with liposomes, we observed lipid binding for both constructs (Fig. 3, D and E). Furthermore, we detected that Vps18NTD shows a curvature sensitiveness, interacting stronger with larger than with smaller liposomes (Fig. 3D). Differently, for the Vps11 domain we did not observe any curvature preference (Fig. 3E). We noticed that Vps18NTD contains conserved lysines in positions 97 and 99. These residues are located in a loop between blades 2 and 3 (Fig. 4B, inset L1), they are conserved across species, and they represent the only positively charged residues on the surface of Vps18NTD (Fig. 4, A and B), which are conserved. Therefore, we mutated these residues and compared lipid binding to wild-type Vps18NTD. As shown in Fig. 3F, liposome binding was impaired in both mutants lacking the positive charges, suggesting that Vps18 may interact with lipids indeed also on endosomes and vacuoles as well.
Requirement of Vps18NTD for HOPS Function in Vivo
To address the role of the Vps18NTD in CORVET and HOPS function, we deleted either the N- or C-terminal domain and compared viability of the resulting strains (Fig. 5A). Vps18 as a subunit of two tethering complexes is required for growth at higher temperatures, which is also observed if the C-terminal domain (ΔCTD) is deleted. Surprisingly, deletion of the entire N-terminal domain showed no effect on growth (Fig. 5A). We, therefore, analyzed vacuole morphology as an in vivo test of functionality. Vacuoles were stained with the lipophilic dye FM4-64. Round vacuoles as observed in wild-type cells were lost in both, vps18Δ and the ΔCTD mutant, but persisted in the ΔNTD mutant. This suggests that HOPS can indeed remain active without Vps18NTD. We have previously shown that impairment of the Rab interaction by tagging Ypt7 with N-terminal GFP can be used as a sensor of HOPS functionality (36). We thus tagged Ypt7 similarly in the Vps18ΔNTD mutant, and we obtained fragmented vacuoles, indicating that the N-terminal domain of Vps18 is indeed required for HOPS function. In agreement with this result, trafficking of the methionine transporter Mup1 from the cell surface to the vacuole resulted in frequent endosomal dots proximal to the vacuole (Fig. 5C), indicative of impaired fusion between late endosomes and the vacuole.
FIGURE 5.
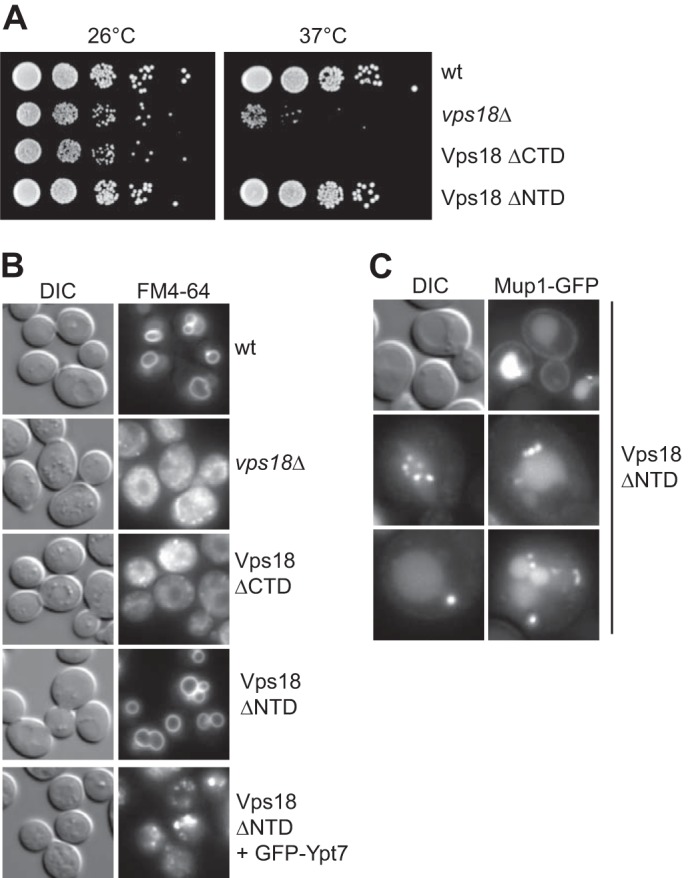
Role of Vps18NTD for endosomal function in yeast cells. A, growth assay of Vps18 mutants. The indicated strains carrying either a deletion of Vps18, wild-type Vps18, or C-terminal and N-terminal truncations were spotted onto yeast extract/peptone/dextrose plates and grown at 26 °C or 37 °C for up to 4 days. B, effect of mutations on vacuole morphology. Vacuoles in the respective strains were stained with 30 μm FM4-64 according to standard protocols (42), and cells were observed by fluorescence microscopy. DIC, differential interference contrast. C, cells lacking the Vps18NTD with endocytic defects. Mup1 was GFP-tagged and monitored upon methionine addition by fluorescence microscopy.
Localization of Vps18NTD and Function within HOPS
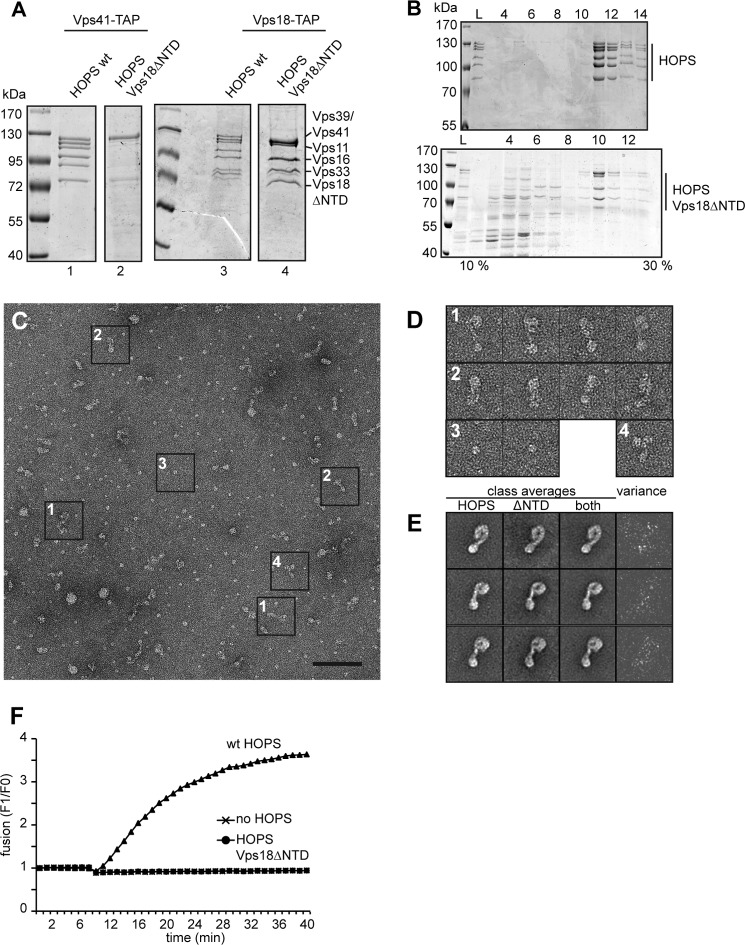
To localize Vps18NTD within HOPS and determine the functionality of the truncated complex, we co-overexpressed truncated Vps18 in the context of the remaining overproduced subunits. Whereas wild-type HOPS could be efficiently purified via TAP-tagged Vps41, HOPS with Vps18ΔNTD was poorly recovered (Fig. 6A). We suggest that tagging of Vps41 interferes with assembly as both subunits are proximal to each other (14). Indeed, after moving the TAP tag onto Vps18, we were able to purify the complex (Fig. 6A). We then used a glycerol gradient procedure to enrich HOPS for subsequent structural analyses, and we observed partial disassembly of HOPS with Vps18ΔNTD (Fig. 6B). As some intact HOPS remained, we investigated the ultrastructure of the complex by negative-stain electron microscopy (Fig. 6C). Besides the entire complex (Fig. 6, C and D, sample 1), we also observed multiple incomplete complexes (samples 2–4). To identify the relative position of the β-propeller domain, we compared the data of the β-propeller deletion mutant with data of wild-type HOPS (Fig. 6E). However, the difference map was too noisy to identify distinct alterations reliably and, thus, the exact position of the Vps18 β-propeller domain. For functional analysis of HOPS without the β-propeller of Vps18, we used a SNARE-dependent liposome fusion assay (37). Whereas wild-type HOPS was required for fusion and strongly stimulated the assay, no fusion signal was obtained with the mutant HOPS complex (Fig. 6F). This indicates that the purified complex is either too instable to support membrane fusion or non-functional. We conclude that the β-propeller of Vps18 has a critical role in the structural stability of the HOPS complex and may support membrane interactions by binding to the membrane directly.
FIGURE 6.
Vps18NTD is required for HOPS stability. A, purification of HOPS without Vps18NTD. HOPS was purified on IgG beads via Vps41 (left) or Vps18 (right) from strains overexpressing all six subunits as described (12, 14). Eluates were applied to SDS-PAGE and Coomassie-stained. B, sizing of HOPS without Vps18NTD. HOPS-Vps18-TAP eluates of A were loaded onto 10–30% glycerol gradients and centrifuged for 18 h at 30,000 rpm in a SW40 rotor at 4 °C. 1-ml fractions were TCA-precipitated and analyzed by SDS-PAGE and Coomassie staining. C, representative electron micrograph of HOPS without Vps18NTD with examples for the different negatively stained particle types boxed. Scale bar, 100 nm. D, diverse particle sizes observed correspond to the dimensions of the complete HOPS without Vps18NTD (1), Vps18ΔNTD-Vps11-Vps39 (2), Vps41 (3), or Vps41- Vps18ΔNTD-Vps11-Vps39 (4). E, the datasets of HOPS-Vps39GFP and the complete HOPS without Vps18NTD were combined, aligned, and classified as described for HOPS-Vps39-GFP (12). Three representative class averages are shown in the third column (each of them containing about 110 particles). Averages calculated from the respective subclasses of HOPS-Vps39-GFP as well as HOPS without Vps18NTD are presented in the first and second columns, respectively. To analyze differences between both subclasses the variance (column 4) was calculated. F, liposome fusion requires the Vps18NTD within HOPS. Proteoliposomes bearing all four SNAREs (Vam3, Vam7, Vti1, and Nyv1) were prepared as described (37). One population carried both NBD (12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)))-phosphatidylethanolamine and rhodamine-phosphatidylethanolamine. Fusion was initiated by the addition of Sec17, Sec18, and HOPS (50 nm) and monitored by fluorescence dequenching (37).
DISCUSSION
The identification of the β-propeller domain in Vps18 provides strong evidence that five of the six HOPS subunits indeed contain a single β-propeller domain at their N termini. Like the predicted β-propellers of CORVET Vps8 and Vps3, the Vps18 β-propeller can be deleted, yet still allows for HOPS assembly (Figs. 5 and 6). Vps18NTD forms a dimer in the crystallographic asymmetric unit that might reflect its interaction within the HOPS complex. In line with this hypothesis, we observed interaction with the Vps11-Vps39 dimer, and we were able to narrow down the interface to the Vps11 β-propeller domain (Fig. 3). As noted above, four Class C proteins, Vps11, Vps18, Vps16, and Vps33, are shared between HOPS and CORVET. However, they do not form independent tetramers either during co-overexpression or as a consequence of deletion of any Rab binding subunit (14). This suggests that assembly of HOPS and CORVET depends on interactions among the Class C proteins that are complemented by the Rab binding subunits. In agreement with this, deletion of the HOPS subunit Vps41 results in disassembly of HOPS into a Vps11-Vps39 dimer, a Vps16-Vps33 dimer, and monomeric Vps18 (14).
Although deletion of Vps18NTD is well tolerated by yeast cells, additional mutations in Ypt7 and the accumulation of endocytic cargo indicate functional defects. Accordingly, HOPS without Vps18NTD was unstable and inactive in in vitro fusion assays. Apart from interacting with Vps11 and thus forming the linker between the two lobes of the HOPS complex, Vps18NTD binds lipids (Fig. 3) and may thus guarantee the proper orientation of HOPS in the interaction with Rab- and SNARE proteins. In addition, Vps18 binds to the Rab-binding protein Vps41 via its C-terminal domain, which positions it close to the Rab binding site of the large head within HOPS (9, 14). This adds Vps18 to proteins with distinct binding sites within HOPS such as the high curvature membrane-sensing amphipathic lipid packaging sensor motif in Vps41 (38), the mitochondrial binding site in the Vps39 β-propeller domain (39), the binding site for the N-terminal Habc domain of the SNARE Vam3, possibly Vps16,4 or the Vam7 binding sites (40), which have not been mapped yet. We consider it to be likely that these binding sites each have low affinity but support the recognition of the main HOPS interactors, the vacuolar SNAREs (via Vps33) and Ypt7 (via Vps41 and 39), by coincidence detection. In combination, these multiple additional binding sites along the entire complex demonstrate the complexity of HOPS function in fusion.
In addition to the structural characterization of the SM protein Vps33 together with Vps16 (9, 10), our study now provides solid evidence that Vps18, and thus five of six HOPS subunits, show an arrangement of an N-terminal β-propeller domain and a C-terminal α-solenoid. Indeed, the segment of Vps16 identified in co-crystals with Vps33 has a very similar fold like COPII subunits that were previously characterized (9, 10). Evolutionary analysis of HOPS and CORVET (41) as well as our psi-BLAST analysis indicate that the HOPS and CORVET subunits with this domain architecture are the product of gene duplications. One interesting aspect is that, unlike nuclear pore complexes or coats, HOPS forms a single particle (12). Further structural and genetic analyses will be necessary to unravel the precise arrangement of the subunits within HOPS and to dissect the role of the individual segments in HOPS function during tethering and subsequent fusion.
Acknowledgments
We thank Angela Perz, Christina Iwert, and Johanna Kuchta for technical support and Anja Schütz for help during construct design.
This work was supported by the Deutsche Forschungsgemeinschaft (UN111/5-3 (to C. U.) and RA1781/2-3 (to S. R.)), by the Hans-Mühlenhoff foundation (to C. U.), and the Leibniz Graduate School of Molecular Biophysics, Berlin (to H. B. and U. H.).
The atomic coordinates and structure factors (code 4uuy) have been deposited in the Protein Data Bank (http://wwpdb.org/).
A. Lürick and C. Ungermann, unpublished observation.
- SM
- Sec1/Munc18
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- NTD
- N-terminal domain
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Kümmel D., Ungermann C. (2014) Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr. Opin. Cell Biol. 29, 61–66 [DOI] [PubMed] [Google Scholar]
- 2. Jahn R., Fasshauer D. (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu I.-M., Hughson F. M. (2010) Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156 [DOI] [PubMed] [Google Scholar]
- 4. Zhang X., He X., Fu X.-Y., Chang Z. (2006) Varp is a Rab21 guanine nucleotide exchange factor and regulates endosome dynamics. J. Cell Sci. 119, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 5. Lachmann J., Ungermann C., Engelbrecht-Vandré S. (2011) Rab GTPases and tethering in the yeast endocytic pathway. SMALL GTPases 2, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balderhaar H. J., Ungermann C. (2013) CORVET and HOPS tethering complexes: coordinators of endosome and lysosome fusion. J. Cell Sci. 126, 1307–1316 [DOI] [PubMed] [Google Scholar]
- 7. Richardson B. C., Smith R. D., Ungar D., Nakamura A., Jeffrey P. D., Lupashin V. V., Hughson F. M. (2009) Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc. Natl. Acad. Sci. U.S.A. 106, 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nickerson D. P., Brett C. L., Merz A. J. (2009) Vps-C complexes: gatekeepers of endolysosomal traffic. Curr. Opin. Cell Biol. 21, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graham S. C., Wartosch L., Gray S. R., Scourfield E. J., Deane J. E., Luzio J. P., Owen D. J. (2013) Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proc. Natl. Acad. Sci. U.S.A. 110, 13345–13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker R. W., Jeffrey P. D., Hughson F. M. (2013) Crystal structures of the Sec1/Munc18 (SM) protein Vps33, alone and bound to the homotypic fusion and vacuolar protein sorting (HOPS) subunit Vps16*. PLoS ONE 8, e67409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Solinger J. A., Spang A. (2013) Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 280, 2743–2757 [DOI] [PubMed] [Google Scholar]
- 12. Bröcker C., Kuhlee A., Gatsogiannis C., Balderhaar H. J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., Raunser S. (2012) Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. U.S.A. 109, 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 14. Ostrowicz C. W., Bröcker C., Ahnert F., Nordmann M., Lachmann J., Peplowska K., Perz A., Auffarth K., Engelbrecht-Vandré S., Ungermann C. (2010) Defined subunit arrangement and rab interactions are required for functionality of the HOPS tethering complex. Traffic 11, 1334–1346 [DOI] [PubMed] [Google Scholar]
- 15. Scheich C., Kümmel D., Soumailakakis D., Heinemann U., Büssow K. (2007) Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 35, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 17. Mueller U., Darowski N., Fuchs M. R., Förster R., Hellmig M., Paithankar K. S., Pühringer S., Steffien M., Zocher G., Weiss M. S. (2012) Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron. Radiat. 19, 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Painter J., Merritt E. A. (2005) A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr. D Biol. Crystallogr. 61, 465–471 [DOI] [PubMed] [Google Scholar]
- 24. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eisenberg D., Schwarz E., Komaromy M., Wall R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 [DOI] [PubMed] [Google Scholar]
- 27. Ohi M., Li Y., Cheng Y., Walz T. (2004) Negative staining and image classification: powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 29. Hohn M., Tang G., Goodyear G., Baldwin P. R., Huang Z., Penczek P. A., Yang C., Glaeser R. M., Adams P. D., Ludtke S. J. (2007) SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 [DOI] [PubMed] [Google Scholar]
- 30. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 31. Narayan K., Lemmon M. A. (2006) Determining selectivity of phosphoinositide-binding domains. Methods 39, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Epp N., Ungermann C. (2013) The N-Terminal Domains of Vps3 and Vps8 are critical for localization and function of the CORVET tethering complex on endosomes. PLoS ONE 8, e67307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo Z., Johnston W., Kovtun O., Mureev S., Bröcker C., Ungermann C., Alexandrov K. (2013) Subunit organisation of in vitro reconstituted HOPS and CORVET multisubunit membrane tethering complexes. PLoS ONE 8, e81534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Rieter E., Vinke F., Bakula D., Cebollero E., Ungermann C., Proikas-Cezanne T., Reggiori F. (2013) Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 126, 593–604 [DOI] [PubMed] [Google Scholar]
- 36. Cabrera M., Ostrowicz C. W., Mari M., LaGrassa T. J., Reggiori F., Ungermann C. (2009) Vps41 phosphorylation and the Rab Ypt7 control the targeting of the HOPS complex to endosome-vacuole fusion sites. Mol. Biol. Cell 20, 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mima J., Hickey C. M., Xu H., Jun Y., Wickner W. (2008) Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 27, 2031–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cabrera M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H.-J., Reggiori F., Engelbrecht-Vandré S., Ungermann C. (2010) Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 191, 845–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hönscher C., Mari M., Auffarth K., Bohnert M., Griffith J., Geerts W., van der Laan M., Cabrera M., Reggiori F., Ungermann C. (2014) Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86–94 [DOI] [PubMed] [Google Scholar]
- 40. Stroupe C., Collins K. M., Fratti R. A., Wickner W. (2006) Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 25, 1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klinger C. M., Klute M. J., Dacks J. B. (2013) Comparative genomic analysis of multi-subunit tethering complexes demonstrates an ancient pan-eukaryotic complement and sculpting in Apicomplexa. PLoS ONE 8, e76278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. LaGrassa T. J., Ungermann C. (2005) The vacuolar kinase Yck3 maintains organelle fragmentation by regulating the HOPS tethering complex. J. Cell Biol. 168, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]