Background: The protein AID generates nucleotide insertions and deletions (indels) critical for antibody affinity maturation.
Results: The location, diversity, and evolution of indels were examined.
Conclusion: AID generates diverse sequence-related indels that localize to antigen binding regions in vitro and in vivo.
Significance: AID is sufficient to form indels that combine with point mutations to form a robust system for antibody evolution.
Keywords: Antibody, Antigen, Cellular Immune Response, Protein Evolution, X-ray Crystallography
Abstract
During somatic hypermutation (SHM), deamination of cytidine by activation-induced cytidine deaminase and subsequent DNA repair generates mutations within immunoglobulin V-regions. Nucleotide insertions and deletions (indels) have recently been shown to be critical for the evolution of antibody binding. Affinity maturation of 53 antibodies using in vitro SHM in a non-B cell context was compared with mutation patterns observed for SHM in vivo. The origin and frequency of indels seen during in vitro maturation were similar to that in vivo. Indels are localized to CDRs, and secondary mutations within insertions further optimize antigen binding. Structural determination of an antibody matured in vitro and comparison with human-derived antibodies containing insertions reveal conserved patterns of antibody maturation. These findings indicate that activation-induced cytidine deaminase acting on V-region sequences is sufficient to initiate authentic formation of indels in vitro and in vivo and that point mutations, indel formation, and clonal selection form a robust tripartite system for antibody evolution.
Introduction
The adaptive immune system combines multiple processes to generate a repertoire of diverse antibodies. Recombination of VDJ germ line segments results in a naive antibody repertoire with both sequence and length diversity in antigen contacting regions. B cell clones expressing naive antibodies with weak affinity for antigen are stimulated to express AID2 and initiate SHM. Maturation is accelerated by clonal expansion of cells expressing antibodies containing AID-mediated mutations that improve affinity (1).
Antibodies are modified by AID-mediated mutations, including point mutations and indels (2–5). Studies of in vivo derived immunoglobulin genes have highlighted the importance of indels in affinity maturation (5–8), and indels contribute to the diversification of the antibody repertoire (9–11). Indels generated in antibodies in vivo have been associated with SHM hotspots and are observed to localize predominantly in complementarity-determining regions (CDRs) (5). Antibodies containing SHM-derived indels have been demonstrated to play critical roles in antigen recognition during chronic infection (12–17).
The mechanisms underlying generation of indels during antibody affinity maturation are poorly understood, and examination has been hampered by their low in vivo frequency and the difficulties attending in situ monitoring of in vivo affinity maturation (9, 11). The diversity in CDR3 lengths introduced by V(D)J recombination makes the analysis of indels introduced in this region during in vivo SHM extremely challenging. Although technical advances have recently enabled an investigation of indels in vivo (5), questions remain regarding which components of the SHM machinery are critical for indel formation, the diversity of indels generated during maturation to an antigen, and the interplay between indels and single amino acid substitutions during subsequent maturation to improve affinity and specificity. Selection and expansion of cells producing antibodies containing indels are subject to a number of constraints, as expressed antibodies need to retain their overall structure and stability as well as improve antigen binding characteristics. In addition, antibodies that incorporate indels that result in increased nonspecific binding or in cross-reactivity to host tissues would likely be eliminated.
In this study, in vitro SHM was used to scrutinize the in situ creation, selection, and maturation of indels. Fifty three distinct antibodies were affinity matured against 21 different antigens, and our findings were compared with in vivo antibody repertoires. Indels observed during in vitro SHM were analyzed and found to significantly improve antibody affinity and function. Indels observed during in vitro affinity maturation were localized to regions likely to improve binding, in particular to CDR1 of the heavy chain (HC) and light chain (LC), similar to that observed in vivo. The crystal structures of a human antibody with and without an insertion derived from in vitro SHM were determined and compared with published antibody structures containing insertions. Multiple indels of related composition and origin were often observed for the same antibody during in vitro SHM, and secondary AID-mediated point mutations in and around the indel were found to further optimize antigen recognition. These findings suggest that AID expression in a heterologous context is sufficient to generate both indels and point mutations, and when combined with selection for improved antigen binding, it enables rapid evolution of naive antibody sequences to innumerable antigens.
EXPERIMENTAL PROCEDURES
In Vitro SHM Antibody Affinity Maturation
Antibody affinity maturation was conducted using in vitro somatic hypermutation as described previously (18–20). In short, the respective antibody was simultaneously displayed on the surface of, and secreted from, HEK293-c18 cells using an episomal vector system. After establishment of stable episomal cell lines, a vector for expression of AID was transfected into the cells to initiate somatic hypermutation. Cell populations co-expressing the antibody and AID were expanded to 2–4 × 107 cells, and fluorescence-activated cell sorting (FACS) was performed in the presence of fluorescently labeled antigen under increasingly stringent conditions. Iterative rounds of AID transfection and FACS selection, each isolating the brightest cells incubated in diminishing concentrations of fluorescent antigen, were used to enrich and identify cells expressing antibody variants with improved binding affinity for antigen. Cell pellets were collected in each round and submitted for antibody V-region sequencing by standard Sanger and/or next generation sequencing technology.
Sequencing and Preparation of PBMC cDNA
RNA from peripheral blood lymphocytes (PBMCs) from a total of 68 healthy donors was purchased from two sources as follows: seven donors from AllCells, Inc. (Alameda, CA), and 61 donors from HemaCare Corp. (San Fernando Valley, CA). Donors were from diverse ethnic backgrounds and of varying countries of origin. RNAs from an average of six donors were pooled for each polymerase chain reaction (PCR), and HC, κLC and λLC RT-PCRs were each run separately. Sequences for HC forward primers (5′ to 3′) are as follows: CCTATCCCCTGTGTGCCTTGGCAGTCTCAGggaggatcctcttyttggtggcagc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGgacctggaggatcctcttcttgstgg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGgggctgagctgggttttcctygttg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGggagtttgggctgagctggvttttyc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcggagtttgggctgagctgggttttc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctyctsctggtggargctcccagatg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcctcctggctgttctccaaggag; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctgtctccttcctcatcttcctgac; and CCTATCCCCTGTGTGCCTTGGCAGTCTCAGgacctggaggatcctcttcttggtg. Sequences for κLC forward primers (5′ to 3′) are as follows: CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcagctcctggggctcctgcwrctc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcagctyctggggctgctaatgctc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcttcctcctgctactctggctcccag; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctctgttgctctggatctctggtgcc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGccaggttcacctcctcagcttcctcc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctgctgctctgggttccagcctccag. Sequences for λLC forward primers (5′ to 3′) are as follows: CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcctcctcaccctcctcrytcactg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctcaccctcctcactcaggrcacag; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctctcctgctccccctcctcaytytc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcggcctcctctctcactgcacagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcccactcctcaacctctacacagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcgcagcctccttgctcactttacagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcggcgtccttgcttactgcacagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctcctcctctgcacagggtctctc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctgggtctccttctacctactgc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctctctcactgcacaggttccctc; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcaccctcctcgctcactgcacagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctgttcctcctcacttgctgcccagg; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctcggactccttgcttatggatcag; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGctcctcaccctcctcagtctcctcac; CCTATCCCCTGTGTGCCTTGGCAGTCTCAGcctgaccctcctcactcactctgcag. Sequences for HC reverse primers (5′ to 3′) are as follows: CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAGgaagacggatgggcccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAGgaagaccgatgggcccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAGGgaaaagggttggggcgga; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTGgaagacggatgggcccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTGgaagaccgatgggcccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTGGgaaaagggttggggcgga. Sequences for κLC reverse primers (5′ to 3′) are as follows: CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACActcatcagatggcgggaag; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTctcatcagatggcgggaag. λLC reverse primers (5′ to 3′) are as follows: CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAaccgtggggttggccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAaccgagggggcagccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGCTCGACAaccgatggggcagccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTaccgtggggttggccttg; CCATCTCATCCCTGCGTGTCTCCGACTCAGACGAGTGCGTaccgagggggcagccttg.
Two μg of RNA from each pool were reverse-transcribed by priming with a mixture of oligo(dT) and random hexamers using a SuperScript® III first-strand synthesis system (Invitrogen) as per the manufacturer's protocol. One-tenth of each reaction was then amplified with HC-, κ-, or λ-specific internal oligonucleotides in a 50-μl reaction as follows: 95 °C for 7 min; 95 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 min for 20 cycles; 68 °C for 7 min and then 4 °C. Approximately 1/100th of each reaction was amplified for 25 cycles using “external” primers at 95 °C for 7 min; 95 °C for 30 s, 55 °C for 30 s, and 68 °C for 1 min; then 68 °C for 7 min and then 4 °C. 5 μl of each reaction were run on a 1% agarose gel, and bands of correct size were excised from the gel. DNA was recovered using a ZymocleanTM gel DNA recovery kit (Zymo Research, Irvine, CA). Recovered DNA was sent to 454 Life Sciences (Roche Applied Science) for sequencing.
Sequencing of in Vitro AID-mediated Affinity Maturation
For Sanger sequencing, oligonucleotide primers were used that encompass ∼140 nucleotides 5′ to the ATG start codon through 30 nucleotides 3′ to the junction of the variable/constant regions, to yield amplicons of ∼550 nucleotides in length for both HC and LC.
Next Generation Sequencing Analysis of in Vitro Antibody Repertoires
For next generation sequencing, ∼5 × 105 HEK293-c18 cells were washed once with PBS, spun down, and resuspended in 50 μl of Lyse-N-Go buffer (Fisher). After incubation at 95 °C for 10 min, cell lysates were centrifuged at 14,000 rpm for 10 min to remove cell debris. Antibody VH and VL open reading frames were amplified by PCR from their episomal expression vectors (18) using specific primers complementary to the CMV promoter region and to the heavy and light chain constant regions using Pfx high fidelity polymerase (Invitrogen): CMV forward primer is as follows: 5′-TACGGTGGGAGGTCTATATAAGCA-3′; HC reverse primer is as follows: 5′-CTGAGTTCCACGACACCGTCACAG-3′; and LC reverse primer is as follows: 5′-GTTACCCGATTGGAGGGCGTTATC-3′. PCR fragments were purified using the Qiagen PCR cleanup kit (Qiagen) and quantified using the Quant-iT PicoGreen dsDNA kit (Invitrogen) according to the manufacturers' protocols. Sequencing output was processed by the 454 Amplicon Default pipeline (Roche Applied Science). Typically ∼50,000–200,000 HC and/or LC reads were obtained per sample for a given experiment.
Analysis of in Vitro and in Vitro Sequencing Data
For the in vitro derived samples, reads were mapped to the respective known parental heavy chain and light chain sequences incorporated in the vectors using 454 GSMapper (Roche Applied Science). For in vivo derived samples, read mapping was carried out using IgBlast (21) with searching against germ line gene sequences and their allelic variants assembled from multiple database sources. Indels were called from pairwise alignments generated by GSMapper, and those transformed from IgBlast (for in vivo samples) using VarScan (22) with customized modification. The signal peptide sequence was excluded in this analysis. Indels shorter than three bases, those occurring at the boundaries of sequence reads (within 5 nucleotides), and those in or proximal to homopolymer sequences (A, T, C, or G; n ≥ 6) were excluded from consideration. High quality reads were then mapped to their originating HC or LC variable region immunoglobulin sequence (in vivo) or starting sequence (in vitro) by Smith-Waterman-based realignment (23). Indel-related motifs were detected by examining the base context of the parental sequences at the regions where indel occurred. For each insertion, 10 bp (five leading and five trailing) around the locus were extracted, and for each deletion, in addition to the five leading and five trailing pairs, the exact deleted sequences were also extracted.
SPR Affinity Measurements
Binding kinetics and affinities of IgG1 antibodies were determined by SPR on a BIAcore T200 instrument (GE Healthcare). Approximately 200–400 response units of the antibodies were captured on a CM5 sensor chip previously immobilized with 10,000 response units of an anti-human Fc-specific antibody (GE Healthcare). For the anti-human β-nerve growth factor (βNGF) antibodies, soluble hβNGF (R&D Systems) was diluted 3-fold from 500 to 6 nm with HBS-EP+ buffer. Each hβNGF concentration was then injected for 2 min at a flow rate of 30 μl/min and allowed to dissociate for 2 min. A similar protocol was followed for the anti-human cell line-derived neurotrophic factor receptor α1 (GFRα1) antibodies, where the soluble extracellular domain of human GFRα1 was diluted and used as the analyte. Each hGFRα1 concentration was then injected for 3 min at a flow rate of 30 μl/min and allowed to dissociate for 3 min. The surface was regenerated with 60 μl of 3 m MgCl2 after each cycle. Double reference subtracted sensorgrams were fit globally using a 1:1 binding model with mass transport with the BIAcore T200 evaluation software to determine dissociation constants (KD).
Protein Production
The Fab portions of the HC and LC of an anti-hβNGF antibody with and without a 9-amino acid insertion were cloned into pcDNA3.3 vectors and transiently expressed in Expi293F cells (Invitrogen) using ExpiFectamine reagent following the manufacturer's instructions. After 5–7 days of incubation at 37 °C and 5% CO2, cell culture supernatants were harvested. Fabs were captured using KappaSelect resin (GE Healthcare). The column was washed with 10 column volumes of 1× PBS, pH 7.4, and proteins were eluted with 100 mm glycine, pH 2.5. Eluted fractions containing purified Fab were buffer-exchanged into 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, and protein concentrations were determined by absorption at 280 nm.
Crystallization and Data Processing
Fab APE1531 (5.8 mg/ml) was crystallized from 12% PEG 20,000, 0.1 m NaCl, 0.1 m MES, pH 6.5, at 22 °C by sitting drop vapor diffusion by mixing 1.0 μl of protein solution with 0.5 μl of reservoir solution. Crystals were cryo-protected with 30% glycerol in reservoir solution and vitrified in liquid nitrogen. A complete data set to 1.75 Å was collected at the Stanford Synchrotron Radiation Lightsource beamline 11-1 (Palo Alto, CA), integrated with XDS (24), and scaled with SCALA (25).
Fab APE1551 (6.5 mg/ml) was crystallized from 16% PEG 6000, 0.1 m citric acid, pH 6–7, at 22 °C by sitting drop vapor diffusion by mixing 0.5 μl of protein solution with 0.5 μl of reservoir solution. Crystals were cryo-protected with 30% glycerol in reservoir solution and vitrified in liquid nitrogen. A complete data set to 1.60 Å was collected at the Stanford Synchrotron Radiation Lightsource beamline 12-2 and was integrated and scaled with HKL2000 (26).
Structure Determination, Refinement, and Analysis
The structure of the APE1531 Fab was determined by molecular replacement (MR) to 1.75 Å resolution in monoclinic space group C2 (VM = 2.3 Å3/Da for one molecule per asymmetric unit). The Protein Data Bank (PDB) was searched for sequences with the highest identity to the individual Ig domains of APE1531. 3QOT for the VL:VH and 3SOB for CL:CH1 were then used as MR templates, and MR solutions were found using PHASER (27). The MR model was subjected to rigid body refinement and restrained all atom refinement with REFMAC5 (28). Further refinement was achieved by alternating cycles of model building with COOT (29) and refinement with REFMAC5. The final model was refined to Rcryst = 15.0% and Rfree = 18.1% (Table 1) and consists of one APE1531 Fab (chain L residues 1–214 and chain H residues 1–231), 2 MES buffer molecules, 4 acetate molecules, 1 short-chain PEG molecule, and 373 waters per asymmetric unit. The structure of the APE1551 Fab was determined by molecular replacement to 1.60 Å resolution in monoclinic space group C2 (VM = 2.5 Å3/Da for one molecule per asymmetric unit). The individual Ig domains of APE1531 were used as MR templates, and the MR solution was rebuilt and refined similarly as described above. The final model was refined to Rcryst = 16.6% and Rfree = 19.8% and consists of one APE1551 Fab (chain L residues 1–214 and chain H residues 2–230), and 434 waters per asymmetric unit. The final statistics for APE1531 and APE1551 are shown in Table 1. The quality of the structures was evaluated with the quality control features implemented in COOT, and superimpositions were done with SSM or LSQ, as implemented in COOT. Coordinates and structure factors have been deposited in the PDB Protein Data Bank with accession numbers 4NWT (APE1531) and 4NWU (APE1551).
TABLE 1.
Crystallography data collection and refinement statistics
Crystallographic data collection and refinement statistics for the structural determination of the anti-hβNGF antibody with (PDB code 4NWU) and without (PDB code 4NWT) a nucleotide insertion in H2.
| Data collection | APE1531 (4NWT) | APE1551 (4NWU) |
|---|---|---|
| Space group | C2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 127.72, 64.36, 63.81 | 127.05, 66.25, 63.88 |
| α, β, γ (°) | 90.00, 108.67, 90.00 | 90.00, 108.54, 90.00 |
| Wavelength (Å) | 0.97830 | 0.97950 |
| Resolution (Å) | 40-1.75 (1.80-1.75)a | 50-1.6 (1.63-1.60) |
| Rmerge (%) | 4.8 (60.4)a | 6.7 (62.3)a |
| 〈I/σΙ〉 | 25.3 (2.9)a | 23.9 (2.4)a |
| Completeness (%) | 98.5 (97.7)a | 97.9 (96.3)a |
| Unique reflections | 49,566 | 64,694 |
| Redundancy | 6.9 (6.6)a | 6.6 (5.0)a |
| Refinement | ||
| Resolution (Å) | 38.6-1.75 | 45.6-1.6 |
| No. of reflections work/test | 46,262/2452 | 61,414/3280 |
| Rwork/Rfree (%) | 15.0/18.1 | 16.6/19.8 |
| r.m.s.d. | ||
| Bond lengths (Å) | 0.019 | 0.019 |
| Bond angles (°) | 1.99 | 1.98 |
| Ramachandran statistics | ||
| Favored/allowed/outliers (%) | 97.4/2.3/0.2 | 96.5/3.5/0.0 |
a Highest resolution shell is shown in parentheses.
RESULTS
To characterize the indel repertoire of antibodies in vivo and in vitro, antibody V-regions from several populations were sequenced (Table 2). Normal human PBMC samples, obtained from 68 normal human donors and composed of both immature and mature B cells, were sequenced, and high quality V-region sequences were mapped to the closest human germ line V-region sequence. To investigate the in situ creation, selection and maturation of indels emanating from individual antibodies, an in vitro SHM system coupled with a mammalian cell display of full-length IgGs was used (18). In total, 39 human germ line antibodies and 14 CDR-grafted antibodies, directed against 21 unique antigens, were matured to high affinity via SHM in vitro. In addition, two in vitro SHM populations were collected for comparison in which antibodies were co-expressed with AID, without selection for improved antigen binding. Antibodies were also expressed in vitro in the absence of AID, to assess the spectrum of indels specific to sample handling and sequencing.
TABLE 2.
NGS and Sanger sequencing results for in vivo and in vitro derived antibodies
The number of samples, reads, and indels observed for in vivo and in vitro derived antibody samples is reported. The number of in-frame (e.g., n = 3,6,9, … ) indels observed is shown in parentheses. NA means not available.
| In vitro AID, no binding selection | In vitro no AID | In vitro AID, binding selection | In vivo | |
|---|---|---|---|---|
| HC | ||||
| Samples | 18 | 3 | 132 | 68 |
| Reads | 1,672,921 | 363,535 | 143,329 | 285,331 |
| Indels | 274 (113) | 0 | (54) | 1,497 (1,173) |
| LC | ||||
| Samples | 16 | NA | 132 | 1 |
| Reads | 1,419,998 | NA | 118,762 | 197,374 |
| Indels | 215 (91) | NA | (51) | 899 (658) |
Indels were detected in both in vivo and in vitro antibody sequences, but not in in vitro samples lacking AID (Table 2). Indels were found in 0.52 and 0.46% of all in vivo HC and LC sequences, respectively, with 0.41 and 0.33% of HC and LC containing in-frame indels. Indels were also detected in sequences from in vitro AID samples with or without selection for improved antigen binding, but not in sequences from in vitro samples lacking AID, indicating that AID is essential and sufficient for indel generation in non-B cells and that the techniques utilized for analysis and in vitro sample manipulation did not generate indels. The frequency of in-frame indels in in vitro AID samples was ∼0.05–0.1%, a rate 10-fold lower than the in vivo rate. Indels recovered from in vitro AID samples, with or without antigen binding selection, were produced exclusively by local sequence duplication (Fig. 1), consistent with indels observed in vivo here and elsewhere (11, 30).
FIGURE 1.

Examples of the indels observed for a single antibody HC. Examples of the indels observed for a single antibody HC obtained when the antibody was expressed in vitro with AID in the absence of selection for improved antigen binding are shown. The starting amino acid and DNA HC sequence (top) is shown together with five in-frame indels and one indel producing a frameshift (bottom). CDR H1 regions are highlighted with a box; deletions are indicated by dashes, and Kabat numbering is shown across the top. For insertions, the upstream sequence is shown in underlined light gray type, and the resulting duplicated sequence is highlighted in black.
Location of Indels in Vitro and in Vivo
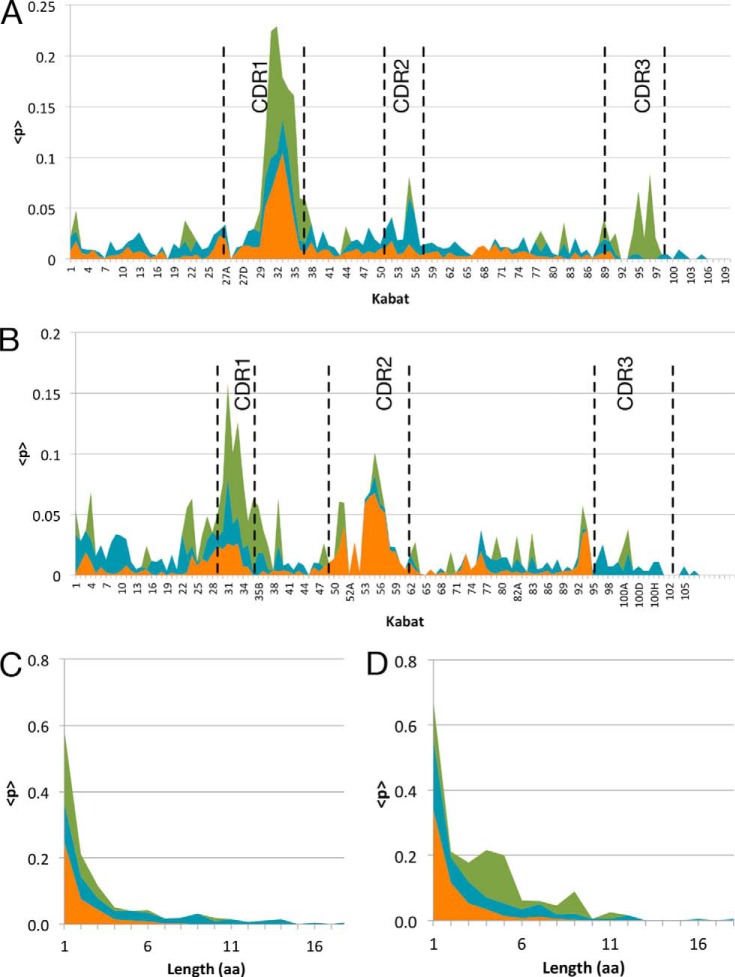
The location of indels in affinity-matured antibodies was comparable in vivo and in vitro. HC indels in vivo were strongly biased toward both HC CDR1 (i.e. CDRH1) and CDRH2, although LC indels preferentially occurred in CDRL1 (Fig. 2). This parallels previous findings, where 47% of insertions and 29% of deletions were found in CDRH1 and 13 and 7% in CDRH2 (5). Because the exact originating sequences of CDR3s in vivo are not known, indels could not be conclusively mapped for this region and were therefore omitted from in vivo analysis.
FIGURE 2.
Indel localization and length distribution in variable regions. Indel localization and length distribution in variable regions of antibodies subjected to in vitro AID without selection for improved antigen binding (blue), in vitro AID with selection for improved antigen binding (green), and in vivo antibodies obtained from immature and mature B cells (orange). Indel frequency for each Kabat position is shown in the LC (A) and in the HC (B). The frequency of observed amino acid lengths of deletions (C) and insertions (D) is shown, with same colors as above.
For antibody sequences co-expressed AID in vitro but in the absence of antigen binding selection, indels were distributed throughout the V-region and were only modestly enriched in CDR regions (Fig. 2). Affinity maturation in vitro led to significant enrichment of indels in CDR regions, most prominently for CDRH1 and CDRL1. Because the originating sequence is known for the in vitro samples, indels within HC and LC CDR3s were analyzed and observed to be enriched in CDRL3 and CDRH3.
The majority of in vivo in-frame indels were short (1 or 2 amino acids in length), with ∼90% of insertions and deletions being ≤3 amino acids. The longest observed insert was 9 amino acids, and the longest deletion was 9 amino acids. For in vitro derived and affinity-matured antibodies, the majority of in-frame indels was relatively short, with 50% comprising less than 4 amino acids (Fig. 2, C and D), although the indels observed in antibodies in the absence of antigen binding selection were significantly longer (up to 18 amino acids). This finding may indicate the increased likelihood of longer indels disrupting antibody structure and/or stability thereby interfering with functional selection. Insertions ranging from 3 to 11 amino acids in length were observed during in vitro maturation with functional selection. In contrast, deletions of 1 or 2 amino acids accounted for the majority of observed events in both the in vitro and in vivo samples (90% ≤ 3 amino acids).
In situ indel generation, selection, and evolution in individual antibodies was further examined using in vitro SHM (18). Fifty three antibodies were affinity-matured using iterative FACS selection (0.1–0.5%) of cells binding progressively lower concentrations of fluorescently labeled antigen, and sequencing was performed to identify point mutations and indels following each round of selection. Final affinity-matured antibodies contained a total of 358 point mutations, and nine of the antibodies contained one or more indels (n = 11).
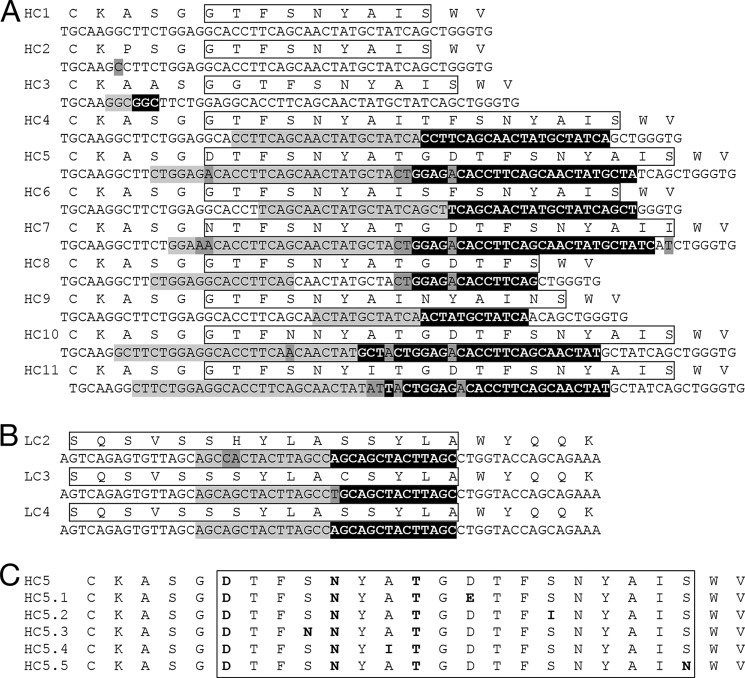
Numerous Sequence-related Indels Generated during Affinity Maturation
Related indels were frequently observed during in vitro maturation of an antibody to an antigen, and enriched insertions were further optimized by point mutations. A total of nine unique insertions were observed at the junction of framework region 1 (FW1)/CDRH1 (HC3–HC11, Fig. 3A) during initial maturation of a germ line antibody to hβNGF (19). All insertions were generated by local sequence duplication and resulted in a CDRH1 loop that is extended by 1–9 additional amino acids. The majority of the originating insertions were accompanied by point mutations, and the locations of these mutations appear to be semi-conserved among the nine unique sequences containing related indels (Fig. 3A). Only in one instance was the originating and duplicated sequence not immediately adjacent (HC8, Fig. 3B), consistent with reported mechanisms of indel formation in vivo (9–11).
FIGURE 3.
Unique insertions recovered during in vitro affinity maturation. Amino acid and nucleic acid V sequences containing unique insertions were recovered during in vitro affinity maturation of two representative human antibodies. A, HC of an anti-hβNGF antibody; B, LC of an anti-hGFRα1 antibody. The amino acid sequences are shown on the top, and the respective DNA sequence is below. CDRH1 and CDRL1 regions are highlighted with a box; originating sequence is shown in light gray; inserted sequence in black; and point mutations from the parental sequence shown in intermediate dark gray. C, mutations that enriched in the context of indel HC5 (A) during subsequent affinity maturation of the antibody are shown highlighted in bold.
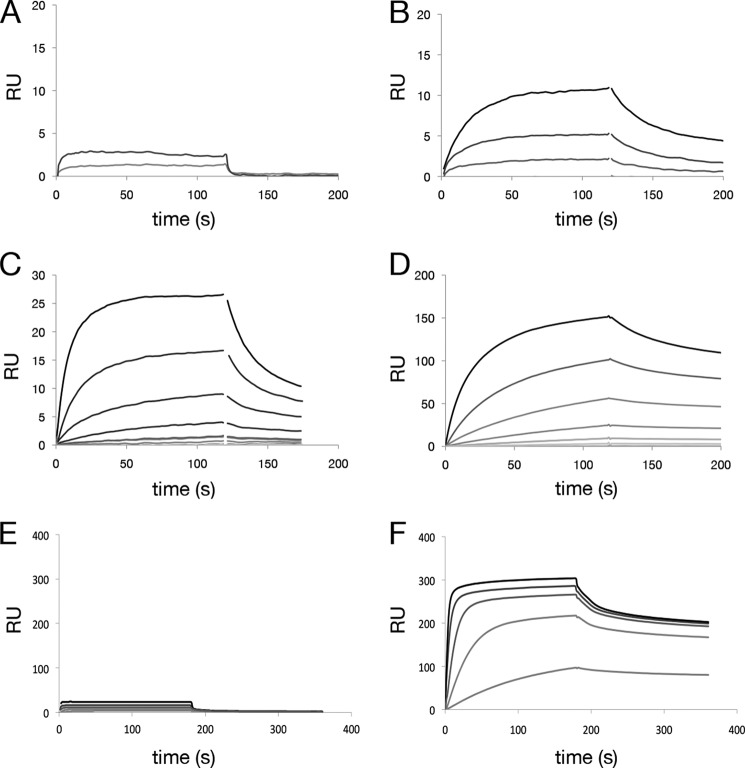
Several of the anti-hβNGF HC sequences containing an insertion were recovered, paired with the parental LC, and expressed as full-length IgGs, and binding kinetics were characterized using SPR. All of the HCs tested (HC3–6) improved affinity for hβNGF (KD = 900, 2.1, 20, and 6 nm, respectively) over that of the parental antibody (KD ≫1 μm) (Fig. 4) with corresponding improvements in bioactivity (19). During the course of affinity maturation, additional point mutations were observed within the FW1/CDRH1 region that improved binding affinity (Fig. 3), presumably by means of direct contacts between FW1/CDRH1 residues and the antigen.
FIGURE 4.
Improvement in binding affinity for antibodies containing insertions. Improvement of antigen binding affinity for anti-hβNGF and anti-hGFRα1 antibodies containing CDRH1 and CDRL1 insertions, respectively. SPR was carried out by antibody capture at low density on an anti-human IgG surface followed by flowing antigen over the surface. SPR sensorgrams are shown for an anti-hβNGF antibody containing two point mutations, S31N and L45F in CDRH1 and FW2 (A); the same antibody with additionally incorporated insertions derived from in vitro SHM with affinity improvements of >20-fold (corresponding to Fig. 3A HC4-HC6) is shown (B–D). SPR sensorgrams for an anti-hGFRα1 antibody containing no mutations are shown in E and for the same antibody with a 5-amino acid insertion in CDRL1 (LC3 in Fig. 3B paired with parental HC), which confers an >40-fold improvement in affinity, in F. RU, response unit.
Likewise, multiple point mutations as well as three unique insertions (LC2–LC4) in CDRL1 were observed during affinity maturation of an antibody to hGFRα1 (Fig. 3B). When tested by SPR, the most frequently observed insertion (LC3) improved antigen binding (KD = 26 nm) over that of the parental antibody (KD >1 μm) (Fig. 4, E and F).
Crystal Structures of an Antibody with and without an Indel
To examine the role of in vitro indels on antibody conformation and function, crystal structures of Fab fragments of the anti-hβNGF antibody before and after incorporation of the insertion in CDRH1 were determined. The structure of the originating antibody was determined to 1.75 Å (Table 1). AlaL51 is the only residue in the disallowed Ramachandran region but has very well defined electron density. L51 is in a conserved γ-turn in almost all other antibodies and often mis-assigned by analysis programs as an outlier (31).
The antibody resembles that of a typical Fab with the dominant features of the combining site being the long CDRH3 and CDRL1 loops that extend over the remainder of the CDR loops. Both the conformation and length of these CDRs are within the range of previously observed Fab crystal structures, as confirmed by three-dimensional structure searches for the individual VH and VL domains. All but two side chains in CDRH3, TrpH100c and ArgH100g, in the combining site display well defined electron density, including all residues of CDRH1 (Fig. 5, A and B). Elongated additional electron density in the center of the combining site was modeled as a short chain PEG located between the base of CDRH3, CDRH2, and the tip of the shorter CDRL3.
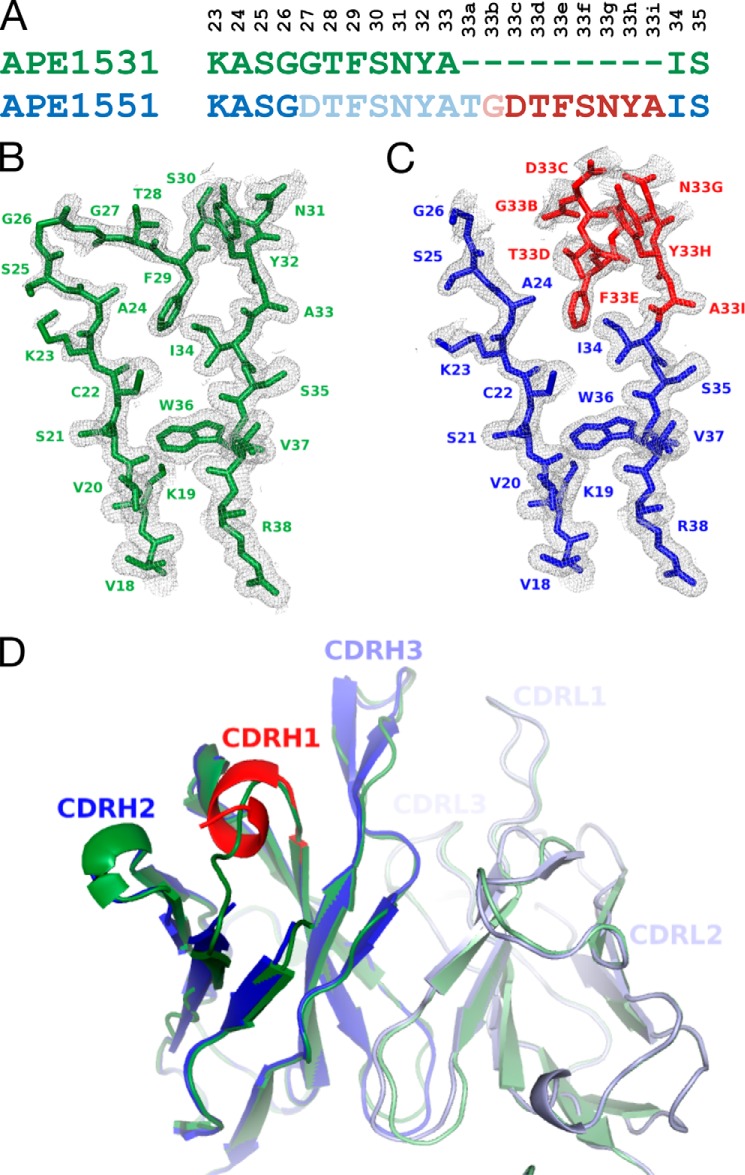
FIGURE 5.
Crystal structures of an antibody with and without a 9-residue insertion. Crystal structures are shown of an anti-human βNGF antibody Fab with (PDB code 4NWU) and without (PDB code 4NWT) a 9-residue insertion in CDRH1. A, alignment of the CDRH1 antibody sequence with and without the insertion; Kabat numbering is listed vertically above. The insert is indicated in red, and residues that were not ordered in the crystal structure are shown with lighter shading. B, crystal structure and 2Fo − Fc electron density contoured at 1σ of CDRH1 and flanking residues of the parent; C, insertion containing antibody. Insert residues are shown in red, and original sequence is shown in blue. No electron density was observed for eight residues, AspH27–AlaH33 and the first residue of the insertion, ThrH33a. D, schematic representation of the superimposed structures of the variable domains with and without CDRH1 insertion, color-coded as above. There are no major structural differences except in CDRH1, where the C-terminal portion of the 9-amino acid insertion occupies the place of the original residues, although the N-terminal part undergoes local rearrangements and leads to disorder indicative of conformational flexibility.
The Fab structure containing the nine amino acid insertion in CDRH1 (HC5 in Fig. 3B) was crystallized in same space group with similar unit cell constants as the parent antibody, and its structure was determined to 1.6 Å resolution (Table 1). Most residues were well ordered with the exception of CDRH1. No electron density was observed for AspH27–AlaH33 and ThrH33a, which is the first residue of the insertion, although the remainder of the insertion (GlyH33b–AlaH33i) and residues C-terminal thereof had clearly defined electron density (Fig. 5C).
Aside from the major rearrangements in CDRH1, the overall structure and the conformation of the combining site of the Fab containing the insertion did not significantly differ from that of the originating antibody (Fig. 5D). All atoms of the two Fabs superimpose with an r.m.s.d. of 0.58 Å; for VL all atoms superimpose with an r.m.s.d. of 0.51 Å and for VH with an r.m.s.d. of 1.34 Å. The all-atom r.m.s.d. values for VH without CDRH1 residues LysH23–SerH35 is 0.46 Å, indicating that indeed the only major structural differences between the two Fabs can be attributed to the CDRH1 region (Fig. 5D). Well defined electron density was observed for the side chains of TrpH100c and ArgH100g, whereas the side chain of TyrL94 appears to exist in different rotamers, one of which occupies the position where the elongated ligand was observed in the parental antibody.
DISCUSSION
SHM in vitro mirrors key aspects of the adaptive immune response in B cells, including the selection of low affinity antibodies from libraries of human germ line V-gene segments with recombined (D)J regions, AID-induced SHM, and iterative selection of cells expressing antibodies with improved binding for antigen (18, 19). SHM in vitro has previously been shown to generate amino acid diversity closely resembling those observed in vivo with mutations preferentially localizing to known AID hot spots (18, 32–35). Nucleotide insertions and deletions have recently emerged as key mechanisms to enhance sequence diversity resulting in additional binding solutions to antigens. Large scale data analysis of in vivo indel diversity (5, 7) revealed an indel frequency of up to 6.5% in circulating B cells (5–9).
In this study, SHM in vitro was used to dissect the in situ creation, selection, and maturation of indels in antibodies, as compared with in vivo antibody repertoires. We found that heterologous expression of AID is sufficient to generate indels in vitro, which were generated throughout the V-region in the absence of antigen binding selection, and which subsequently were enriched in CDR regions during affinity maturation. Analysis of indel location after SHM and antigen selection in vitro is remarkably similar to that observed in in vivo derived antibodies. Multiple sequence-related indels were often generated for a maturing antibody, with indel enrichment and subsequent point mutations facilitating rapid optimization of the binding paratope. When tested, indels were found to improve both antibody affinity and function.
Indels originate from sequence duplication in vivo and in vitro and, due to the lack of a distinct editing mechanism and AID-mediated point mutations, the resulting sequence space that can be explored is vast. CDR loops have been shown to adopt a limited set of canonical conformations that are dependent on the side-chain packing, hydrogen bonding, or conformational preferences of key residues (36, 37). Indels preferentially map to CDRs (5) and thereby significantly expand the structural repertoire of individual CDRs through generation of a multitude of novel, unique structural solutions (38). Comparison of in vitro and in vivo derived antibodies containing SHM indels demonstrates a strong preference for the addition of indels at conserved sites proximal to the CDRH3/L3 regions (Fig. 6), where they are well positioned to extend and optimize the antigen recognition surface (14, 15).
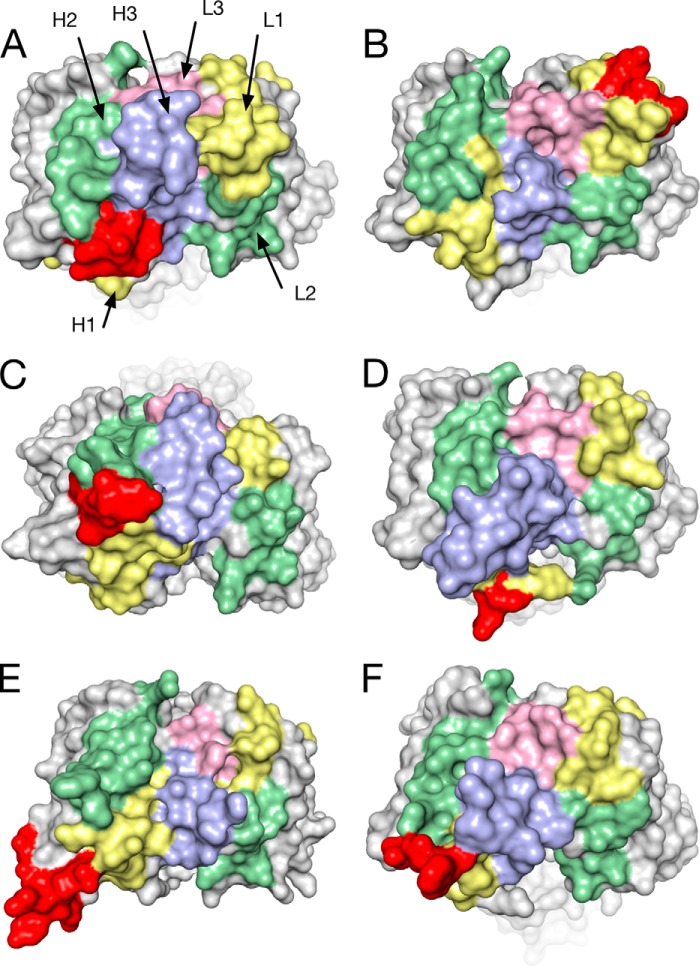
FIGURE 6.
Comparison of human antibody structures containing insertions. Molecular surface representation of the crystal structures of Fabs of human antibodies with different specificities featuring insertions ranging from 4 to 9 amino acids. CDRH1 (yellow), CDRH2 (green), CDRH3 (blue), CDRL1 (yellow), CDRL2 (green), CDRL3 (pink), and insertions (red) are shown in the same orientations in a top view onto the combining site. A, APE1551, an anti-human βNGF antibody with a 9-residue insert in CDRH1 (PDB code 4NWU). B, bH1, a dual specific antibody to HER2 and VEGF, 4-residue insert in CDRL1 (PDB code 3BDY) (47). C, PGT127, an anti-gp120 antibody that recognizes conserved glycan residues, 6-residue insert in CDRH1, 4-residue deletion in CDRL1 (PDB code 3TWC) (15). D, C05, an anti-influenza A hemagglutinin antibody, 5-residue insert in CDRH1 (PDB code 4FP8) (17). E, VRC06, an anti-gp120 antibody, 7-residue insert in heavy chain FR3, 1-residue deletion in CDRL1 (PDB code 4JB9) (48). F, PGT135, an anti-gp120 antibody, 5-residue insert in CDRH1 (PDB code 4JM2) (49).
Analysis of SHM in vitro allows facile analysis of the nature and composition of indels in the CDR3 region. Such analysis is difficult or impossible to carry out in antibodies derived from in vivo sources due to the nontemplated nature of the V(D)J rearrangements. The unique sequence composition of the CDR3s is thought to provide initial antigen recognition, with point mutations and indels modulating affinity and specificity by expanding the interaction interface or by remodeling the combining site (16). Notably, indels appear to play a prominent role in the optimization of antibodies to pathogens, which present rapidly evolving antigens (e.g. HIV), or to antigens whose topological, conformational, or surface chemistry properties (e.g. glycosylation) make generation of high affinity antibodies difficult. Significant mutation of framework regions and pronounced use of indels suggest that immune responses to these challenging antigens (1, 14–16, 39, 40) often require significant remodeling of regions peripheral to the combining site center. Previous analysis of non-IgG protein structures found that indel residues are disordered and preferentially occur in regions of increased disorder tolerant to accommodating new sequences (41–46).
This work extends previous reports demonstrating the importance of secondary mechanisms in antibody diversification beyond V(D)J recombination and AID-mediated single nucleotide mutations. Generation of multiple sequence-related indels and their subsequent optimization by SHM dramatically increases the sequence and structure space that can be explored by the adaptive immune system. Many parallels to protein evolution in general are apparent, including preferential location in regions of structural plasticity, intrinsic disorder, and conformational flexibility, maintenance of overall fold with local modification of secondary structures in the vicinity of the indel, and a “mutagenic” effect of indels on their flanking regions.
The atomic coordinates and structure factors (codes 4NWT and 4NWU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AID
- activation-induced cytidine deaminase
- CDR
- complementarity-determining region
- SHM
- somatic hypermutation
- HC
- heavy chain
- LC
- light chain
- SPR
- surface plasmon resonance
- hβNGF
- human β nerve growth factor
- CDRH1
- HC CDR1
- CDRH2
- HC CDR2
- CDRH3
- HC CDR3
- CDRL1
- LC CDR1
- CDRL2
- LC CDR2
- CDRL3
- LC CDR3
- hGFRα1
- human glial cell line-derived neurotrophic factor family receptor α
- PBMC
- peripheral blood leukocytes
- MR
- molecular replacement
- indel
- insertion and deletion
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Peled J. U., Kuang F. L., Iglesias-Ussel M. D., Roa S., Kalis S. L., Goodman M. F., Scharff M. D. (2008) The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26, 481–511 [DOI] [PubMed] [Google Scholar]
- 2. Klein F., Mouquet H., Dosenovic P., Scheid J. F., Scharf L., Nussenzweig M. C. (2013) Antibodies in HIV-1 vaccine development and therapy. Science 341, 1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Throsby M., van den Brink E., Jongeneelen M., Poon L. L., Alard P., Cornelissen L., Bakker A., Cox F., van Deventer E., Guan Y., Cinatl J., ter Meulen J., Lasters I., Carsetti R., Peiris M., de Kruif J., Goudsmit J. (2008) Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory cells. PLoS One. 3, e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ekiert D. C., Bhabha G., Elsliger M. A., Friesen R. H., Jongeneelen M., Throsby M., Goudsmit J., Wilson I. A. (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Briney B. S., Willis J. R., Crowe J. E., Jr. (2012) Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun. 13, 523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goossens T., Klein U., Küppers R. (1998) Frequent occurrence of deletion and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc. Natl. Acad. Sci. U.S.A. 95, 2463–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briney B. S., Crowe J. E., Jr. (2013) Secondary mechanisms of diversification in the human antibody repertoire. Front. Immunol. 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miura Y., Chu C. C., Dines D. M., Asnis S. E., Furie R. A., Chiorazzi N. (2003) Diversification of the Ig variable region gene repertoire of synovial B lymphocytes by nucleotide insertion and deletion. Mol. Med. 9, 166–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilson P. C., de Bouteiller O., Liu Y.-J., Potter K., Banchereau J., Capra J. D., Pascual V. (1998) Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J. Exp. Med. 187, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Wildt R. M., van Venrooij W. J., Winter G., Hoet R. M., Tomlinson I. M. (1999) Somatic insertions and deletions shape the human antibody repertoire. J. Mol. Biol. 294, 701–710 [DOI] [PubMed] [Google Scholar]
- 11. Reason D. C., Zhou J. (2006) Codon insertion and deletion functions as a somatic diversification in human antibody repertoires. Biol. Direct. 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou C., Jacobsen F. W., Cai L., Chen Q., Shen W. D. (2010) Development of a novel mammalian cell surface antibody display platform. mAbs 2, 508–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walker L. M., Phogat S. K., Chan-Hui P. Y., Wagner D., Phung P., Goss J. L., Wrin T., Simek M. D., Fling S., Mitcham J. L., Lehrman J. K., Priddy F. H., Olsen O. A., Frey S. M., Hammond P. W., Protocol G Principal Investigators, Kaminsky S., Zamb T., Moyle M., Koff W. C., Poignard P., Burton D. R. (2009) Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N. S., Louder M., McKee K., O'Dell S., Perfetto S., Schmidt S. D., Shi W., Wu L., Yang Y., Yang Z. Y., Yang Z., Zhang Z., Bonsignori M., Crump J. A., Kapiga S. H., Sam N. E., Haynes B. F., Simek M., Burton D. R., Koff W. C., Doria-Rose N. A., Connors M., NISC Comparative Sequencing Program, Mullikin J. C., Nabel G. J., Roederer M., Shapiro L., Kwong P. D., Mascola J. R. (2011) Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333, 1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pejchal R., Doores K. J., Walker L. M., Khayat R., Huang P. S., Wang S. K., Stanfield R. L., Julien J. P., Ramos A., Crispin M., Depetris R., Katpally U., Marozsan A., Cupo A., Maloveste S., Liu Y., McBride R., Ito Y., Sanders R. W., Ogohara C., Paulson J. C., Feizi T., Scanlan C. N., Wong C. H., Moore J. P., Olson W. C., Ward A. B., Poignard P., Schief W. R., Burton D. R., Wilson I. A. (2011) A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krause J. C., Ekiert D. C., Tumpey T. M., Smith P. B., Wilson I. A., Crowe J. E., Jr. (2011) An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody. mBio 2, e00345–e00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ekiert D. C., Kashyap A. K., Steel J., Rubrum A., Bhabha G., Khayat R., Lee J. H., Dillon M. A., O'Neil R. E., Faynboym A. M., Horowitz M., Horowitz L., Ward A. B., Palese P., Webby R., Lerner R. A., Bhatt R. R., Wilson I. A. (2012) Cross-neutralization of influenza A viruses mediated by a single antibody. Nature 489, 526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowers P. M., Horlick R. A., Kehry M. R., Neben T. Y., Tomlinson G. L., Altobell L., Zhang X., Macomber J. L., Krapf I. P., Wu B. F., McConnell A. D., Chau B., Berkebile A. D., Hare E., Verdino P., King D. J. (2014) Mammalian cell display for the discovery and optimization of antibody therapeutics. Methods 65, 44–56 [DOI] [PubMed] [Google Scholar]
- 19. Bowers P. M., Horlick R. A., Neben T. Y., Toobian R. M., Tomlinson G. L., Dalton J. L., Jones H. A., Chen A., Altobell L., 3rd., Zhang X., Macomber J. L., Krapf I. P., Wu B. F., McConnell A., Chau B., Holland T., Berkebile A. D., Neben S. S., Boyle W. J., King D. J. (2011) Coupling mammalian cell surface display with somatic hypermutation for the discovery and maturation of human antibodies. Proc. Natl. Acad. Sci. U.S.A. 108, 20455–20460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horlick R. A., Macomber J. L., Bowers P. M., Neben T. Y., Tomlinson G. L., Krapf I. P., Dalton J. L., Verdino P., King D. J. (2013) Simultaneous surface display and secretion of proteins from mammalian cells facilitates efficient in vitro selection and maturation of antibodies. J. Biol. Chem. 288, 19861–19869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye J., Ma N., Madden T. L., Ostell J. M. (2013) IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koboldt D. C., Chen K., Wylie T., Larson D. E., McLellan M. D., Mardis E. R., Weinstock G. M., Wilson R. K., Ding L. (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith T. F., Waterman M. S. (1981) Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 28. Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sale J. E., Neuberger M. S. (1998) TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutation B cell line. Immunity 9, 859–869 [DOI] [PubMed] [Google Scholar]
- 31. Stanfield R., Cabezas E., Satterthwait A., Stura E., Profy A., Wilson I. (1999) Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing fabs. Structure 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 32. Rogozin I. B., Diaz M. (2004) Cutting Edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172, 3382–3384 [DOI] [PubMed] [Google Scholar]
- 33. Dörner T., Foster S. J., Farner N. L., Lipsky P. E. (1998) Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur. J. Immunol. 28, 3384–3396 [DOI] [PubMed] [Google Scholar]
- 34. Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., Alt F. W. (2003) Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730 [DOI] [PubMed] [Google Scholar]
- 36. Chothia C., Lesk A. M., Tramontano A., Levitt M., Smith-Gill S. J., Air G., Sheriff S., Padlan E. A., Davies D., Tulip W. R. (1989) Conformations of immunoglobulin hypervariable regions. Nature 342, 877–883 [DOI] [PubMed] [Google Scholar]
- 37. Martin A. C., Thornton J. M. (1996) Structural families of loops in homologous proteins: automatic classification, modelling, and application to antibodies. J. Mol. Biol. 263, 800–815 [DOI] [PubMed] [Google Scholar]
- 38. Wang F., Ekiert D. C., Ahmad I., Yu W., Zhang Y., Bazirgan O., Torkamani A., Raudsepp T., Mwangi W., Criscitiello M. F., Wilson I. A., Schultz P. G., Smider V. V. (2013) Reshaping antibody diversity. Cell 153, 1379–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou T., Georgiev I., Wu X., Yang Z. Y., Dai K., Finzi A., Kwon Y. D., Scheid J. F., Shi W., Xu L., Yang Y., Zhu J., Nussenzweig M. C., Sodroski J., Shapiro L., Nabel G. J., Mascola J. R., Kwong P. D, (2010) Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Breden F., Lepik C., Longo N. S., Montero M., Lipsky P. E., Scott J. K. (2011) Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS ONE 3, e16857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsing M., Cherkasov A. (2008) Indel PDB: A database of structural insertions and deletions derived from sequence alignments of closely related proteins. BMC Bioinformatics 9, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim R., Guo J. (2010) Systematic analysis of short internal indels and their impact on protein folding. BMC Struct. Biol. 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Light S., Sagit R., Ekman D., Elofsson A. (2013a) Long indels are disordered: a study of disorder and indels in homologous eukaryotic proteins. Biochim. Biophys. Acta 1834, 890–897 [DOI] [PubMed] [Google Scholar]
- 44. Light S., Sagit R., Sachenkova O., Ekman D., Elofsson A. (2013) Protein expansion is primarily due to indels in intrinsically disordered regions. Mol. Biol. Evol. 30, 2645–2653 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z., Huang J., Wang Z., Wang L., Gao P. (2011) Impact of indels on the flanking regions in structural domains. Mol. Biol. Evol. 28, 291–301 [DOI] [PubMed] [Google Scholar]
- 46. Zhang Z., Xing C., Wang L., Gong B., Liu H. (2012) IndelFR: a database of indels in protein structures and their flanking regions. Nucleic Acids Res. 40, D512–D518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bostrom J., Yu S. F., Kan D., Appleton B. A., Lee C. V., Billeci K., Man W., Peale F., Ross S., Wiesmann C., Fuh G. (2009) Variants of the antibody Herceptin that interact with HER2 and VEGF at the antigen binding site. Science 323, 1610–1614 [DOI] [PubMed] [Google Scholar]
- 48. Georgiev I. S., Doria-Rose N. A., Zhou T., Kwon Y. D., Staupe R. P., Moquin S., Chuang G. Y., Louder M. K., Schmidt S. D., Altae-Tran H. R., Bailer R. T., McKee K., Nason M., O'Dell S., Ofek G., Pancera M., Srivatsan S., Shapiro L., Connors M., Migueles S. A., Morris L., Nishimura Y., Martin M. A., Mascola J. R., Kwong P. D. (2013) Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756 [DOI] [PubMed] [Google Scholar]
- 49. Kong L., Lee J. H., Doores K. J., Murin C. D., Julien J. P., McBride R., Liu Y., Marozsan A., Cupo A., Klasse P. J., Hoffenberg S., Caulfield M., King C. R., Hua Y., Le K. M., Khayat R., Deller M. C., Clayton T., Tien H., Feizi T., Sanders R. W., Paulson J. C., Moore J. P., Stanfield R. L., Burton D. R., Ward A. B., Wilson I. A. (2013) Supersite of immune vulnerability on the glycosylated face HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 20, 796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]