Background: Ebola virus VP40 matrix protein is able to produce virus-like particles from cells.
Results: VP40 induces negative membrane curvature and scission from in vitro membranes.
Conclusion: Phosphatidylserine plays an important role in VP40-induced vesiculation.
Significance: Elucidation of VP40 interactions with lipids could lead to a better understanding of the Ebola virus replication cycle.
Keywords: Ebola Virus, Lipid Binding Protein, Phosphatidylserine, Viral Protein, Virus Assembly, VP40, Giant Unilammelar Vesicles, Matrix Protein, Membrane Budding, Viral Budding
Abstract
Ebola virus is from the Filoviridae family of viruses and is one of the most virulent pathogens known with ∼60% clinical fatality. The Ebola virus negative sense RNA genome encodes seven proteins including viral matrix protein 40 (VP40), which is the most abundant protein found in the virions. Within infected cells VP40 localizes at the inner leaflet of the plasma membrane (PM), binds lipids, and regulates formation of new virus particles. Expression of VP40 in mammalian cells is sufficient to form virus-like particles that are nearly indistinguishable from the authentic virions. However, how VP40 interacts with the PM and forms virus-like particles is for the most part unknown. To investigate VP40 lipid specificity in a model of viral egress we employed giant unilamellar vesicles with different lipid compositions. The results demonstrate VP40 selectively induces vesiculation from membranes containing phosphatidylserine (PS) at concentrations of PS that are representative of the PM inner leaflet content. The formation of intraluminal vesicles was not significantly detected in the presence of other important PM lipids including cholesterol and polyvalent phosphoinositides, further demonstrating PS selectivity. Taken together, these studies suggest that PM phosphatidylserine may be an important component of Ebola virus budding and that VP40 may be able to mediate PM scission.
Introduction
Ebola virus is a filamentous, lipid-enveloped virus from the Filoviridae family and is one of most virulent pathogens that can infect humans. Ebola virus causes viral hemorrhagic fever, with a fatality rate of ∼60%. With no current FDA approved vaccines or drugs, Ebola virus may pose a serious threat (1–3). Currently, an outbreak in Western Africa, the first large scale Ebola virus outbreak for this region; has caused more than 5800 infections and 2800 deaths as of September 2014 (4). Thus, there is urgency toward developing a treatment. Recently, significant strides have been made in development of vaccines (5), repositioning of previously approved FDA drugs (6), or monoclonal antibodies (7) that may be viable means of preventing or treating infections. The Ebola virus negative sense RNA genome encodes seven proteins including the transmembrane glycoprotein (8), which mediates Ebola virus entry into the host cell using the Niemann-Pick Type C1 cholesterol transporter (9–11). Although a number of studies have addressed the mechanism of cellular entry of the virus, less is known regarding how the virus replicates inside the host cell and forms an assembly site to egress from the inner leaflet of the host cell plasma membrane (PM).2
The Ebola virus matrix protein VP40 is a peripheral membrane protein that regulates budding and egress from the host cell PM (12–14). VP40, along with the minor matrix protein VP24, compose the layer that underlies the viral lipid envelope bridging the membrane embedded glycoprotein and the nucleocapsid (15, 16). VP40 can form VLPs when transfected into mammalian cells, which resemble authentic Ebola virions (12, 18). However, how VP40 binds the PM and promotes scission with or without host protein hijacking is still not clear. VP40 is a dimer (14) that has an ∼43 amino acid N-terminal region of unknown structure, a N-terminal domain that mediates dimerization (14) and a C-terminal domain that binds membranes (12, 14, 19–22) and mediates oligomerization (14). The N terminus of VP40 has two late domains (23, 24), a PTAP motif and a PPXY motif that are thought to mediate interactions with the cellular endosomal sorting complexes required for transport (ESCRT) machinery (25). Presumably VP40 late domains regulate membrane scission through interactions with the ESCRT machinery, but studies overall on the VP40 late domains suggest there may be ESCRT-dependent and/or independent forms of Ebola virus egress in cell culture (26).
Ebola virus VP40 has been shown to induce budding and scission from in vitro membranes, which is dependent upon insertion of a hydrophobic loop in its C-terminal domain (19). VP40 penetrates into the hydrocarbon core of membranes that recapitulate the PM lipid composition (19, 20). Single point mutations in a C-terminal domain hydrophobic patch significantly reduced both membrane penetration and viral egress (19). Inhibition of membrane penetration in cell culture resulted in a reduction in PM association of VP40, reduced VP40 oligomerization, and VLP production (19, 20). Penetration of VP40 more than halfway into one monolayer of the membrane bilayer along with VP40 oligomerization appears to be a driving force for regulating formation of VLPs (19, 20).
To better understand the process of VP40-mediated Ebola virus egress, it is crucial to understand how VP40 is able to interact with and bud from the PM to regulate formation of VLPs. Here we employ giant unilamellar vesicles (GUVs) as a model of viral budding to investigate the lipid selectivity of VP40-mediated budding. The GUVs have previously been utilized to understand membrane scission and to better understand the role of viral matrix proteins as well as peripheral proteins in membrane bending (19, 27–30). Here we varied the lipid compositions of GUVs to investigate the role of anionic lipids and cholesterol, which are enriched in the inner leaflet of the PM. VP40 selectively and rapidly induced budding and scission when PS was present at levels similar to the inner leaflet composition of the PM. Elucidating the effect of VP40 on GUVs composed of lipids that mimic biological membranes should help to reveal the mechanisms VP40 uses to remodel the host cell membrane.
EXPERIMENTAL PROCEDURES
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (LRPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (POPI), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate, 18:1 PI(4,5)P2) (PI(4,5)P2), and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate, 18:1 PI(3,4,5)P3) (PI(3,4,5)P3) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) and used without further purification. The lipids were dissolved in solvents according the manufacturer's recommendation and stored as recommended unless specified. Cholesterol from ovine wool was also purchased from Avanti Polar Lipids, Inc. and used without further purification.
Protein Expression and Purification
VP40-WT was expressed and purified from Escherichia coli BL21(DE3) cells as previously described (20) with the following modifications. The purified GST-tagged protein was cleaved using a thrombin cleavage kit and the manufacturer's protocol (EMD Millipore). Protein concentrations were determined by the bicinchoninic acid (BCA) method and aliquots of 2 mg/ml were made using storage buffer (PBS buffer: pH 7.4, 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4). The lactadherin C2 domain construct was a kind gift of Dr. Sergio Grinstein (The Hospital for Sick Children, Toronto, Ontario, Canada) and was expressed and purified as previously described (31). The I-BAR domains of IRSp53 and MIM were gifts from Dr. Pekka Lappaiainen (University of Helsinki, Helsinki, Finland) and were expressed and purified as described previously (32).
GUV Formation and Quantification
An aliquot of varied lipid composition, prepared in a specific molar ratio, was suspended in chloroform and then used for GUV formation as previously described (33). Briefly, the desired ratio of varied lipids were mixed in an amber glass vial to give a total lipid concentration of 4 mm. The lipids were dried under a constant stream of nitrogen gas and then resuspended in 50 μl of chloroform resulting in a homogenously mixed lipid solution. Using a Hamilton glass syringe, the lipid solution was spotted on an ITO (indium tin oxide)-coated glass slide with an immobilized Teflon spacer. The organic solvent layer was allowed to dry completely. Carefully avoiding the formation of any air bubbles, the spacer chamber was then filled with freshly made 350 mm sucrose solution and covered with another ITO slide. Current was then passed through the set-up with the peak-to-peak voltage set to 3 V at a frequency of 20 Hz for 8–16 h. The GUVs were then carefully collected using a pipette, into glass vials, and used within 1 day. Control experiments were performed with slight changes in osmolarity in the absence of proteins. To demonstrate membrane bending specificity, varying concentrations of proteins were incubated with GUVs in Nunc 8-well plate Lab-Tek® chambered number 1.0 borosilicate coverglass system purchased from Thermo Fisher Scientific (Rochester, NY) and imaged using a confocal microscope. Quantification of GUVs was performed by imaging and counting at least 100 GUVs in each experiment. Experiments were repeated in triplicate to calculate a standard deviation and imaged as described previously (33).
Confocal Imaging
GUVs were imaged using a Zeiss LSM 710 confocal microscope with a Plan Apochromat ×63 1.4 NA oil objective. The 561 nm line of the argon ion laser was used for excitation of LRPE. The laser power was maintained at 1% throughout the experiment, and the emission was collected through a 565–610-nm filter.
Lipid Vesicles Preparation
POPC and POPS lipids dissolved in CHCl3 were mixed in a ratio of 80:20. The mixture was dried under nitrogen; the dried lipid was hydrated in PBS buffer, bringing the lipid concentration to 4 mm. The lipids were then incubated at 37 °C for 1 h, followed by extrusion 17 times through a 1-μm polycarbonate filter using the Mini Extruder (Avanti® Mini-Extruder). The vesicles were used for up to 7 days. During that time they were stored at 4 °C and routinely analyzed with dynamic light scattering to ensure their diameter was consistent.
Lipid Binding Assays
The liposome sedimentation assays were performed as previously described (34). Briefly, solutions of POPC, POPS, and various other lipids were mixed and dried down under nitrogen. The lipids were resuspended in 20 mm Tris, pH 7.4, containing 0.16 m KCl and incubated at 65 °C for 1 h. The lipids underwent three cycles of freezing in liquid nitrogen and thawing at 37 °C. Liposomes were then mixed with proteins in 100-μl reactions to yield solutions containing 1 mm total lipid and 5 μm protein. Reactions were incubated for 20 min at room temperature. Liposomes were collected by centrifugation for 20 min at 25,000 × g at room temperature. Supernatants were removed; pellets were resuspended in 100 μl of Laemmli buffer, resolved by SDS-PAGE, and developed with Coomassie Brilliant Blue dye.
RESULTS
VP40 Selectively Buds into PS-rich GUVs
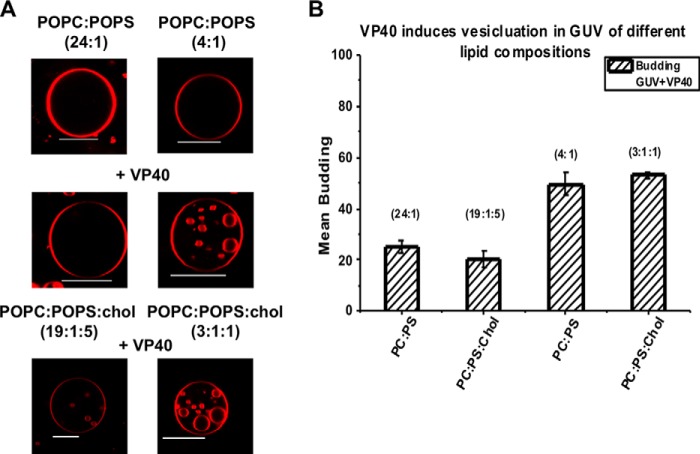
Ebola virus VP40 has been shown to insert into the inner leaflet of the PM using a hydrophobic loop in the C-terminal domain (19, 20, 22, 35) (see Fig. 1). These hydrophobic interactions with the PM have been shown to be a key step in Ebola virus egress (12, 19, 21) as truncation or mutation of hydrophobic residues that penetrate the PM abrogates viral budding. However, how VP40 senses the PM lipid environment and achieves budding from the host cell membrane is mostly unknown. To investigate the lipid selectivity of VP40 budding we examined the ability of VP40 to bud into GUVs harboring different lipid compositions, most notably using PM-enriched lipids. To confirm VP40 binding and budding into GUVs containing PS at a ratio similar to that in the PM, 5 μm VP40 was incubated with GUVs composed of either POPC:POPS (24:1) or POPC:POPS (4:1) containing 0.1% LRPE. In the presence of VP40, PC-rich GUVs (Fig. 2B) exhibited little detectable internal budding, whereas those containing 20% POPS (Fig. 2A) displayed a rapid and dramatic increase in the formation and scission of buds inside the GUVs. Due to their compromised stability at 5 μm VP40, the GUVs consequentially burst, releasing the vesicles (Fig. 2B). To capture this practically instantaneous effect of VP40 on the GUVs, the PS-rich GUVs were incubated with 10-fold less VP40 protein (Fig. 2C). The formation and scission of the buds off the GUV was still significant and allowed for capturing of images and quantitative analysis without compromising GUV stability.
FIGURE 1.
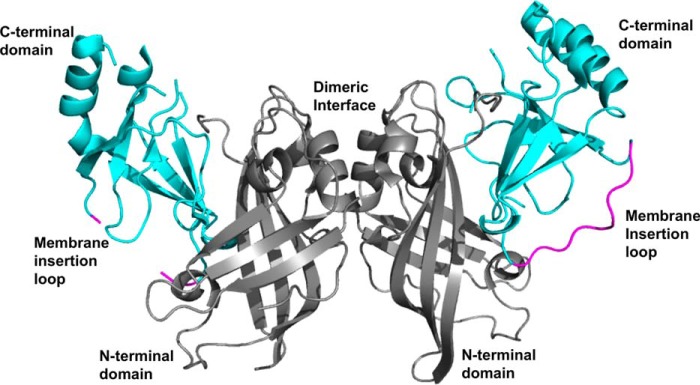
VP40 structure. VP40 (PDB code 4LDB) is a dimer with a N-terminal domain (gray) that mediates dimerization and a C-terminal domain (cyan) that binds membranes and regulates VP40 hexamerization. A hydrophobic loop in the VP40 C-terminal domain (magenta) has been shown to penetrate the hydrocarbon core of the plasma membrane to enhance VP40 membrane binding and plasma membrane localization. Note that some segments, for instance, the hydrophobic loop in one monomer, are missing from the dimeric crystal structure. The insertion of the C-terminal domain hydrophobic loop is also necessary for sufficient oligomerization of VP40 in cells and VLP formation.
FIGURE 2.
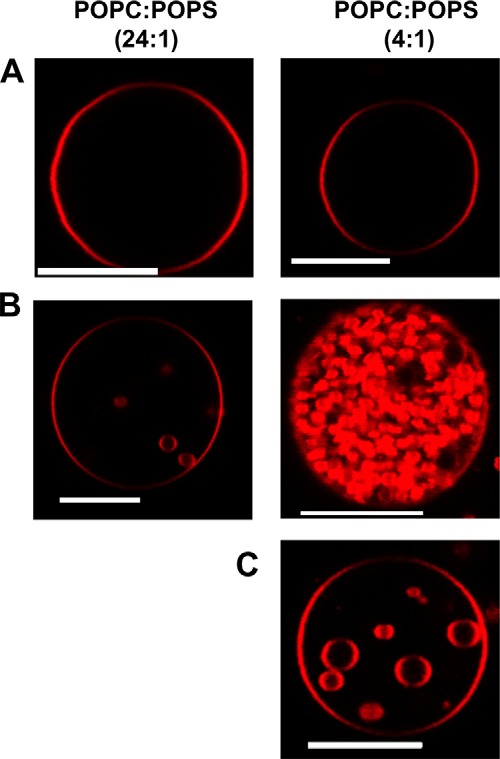
VP40 forms intraluminal vesicles and buds effectively into PS containing GUVs. A, POPC:POPS (24:1) and POPC:POPS (4:1) in GUV containing 0.1% LRPE were imaged without VP40 in PBS buffer, which lead to no detectable vesicle formation. B, incubation of the GUV with 5 μm VP40 for 20 min lead to intraluminal budding of vesicles into the GUV in PS-rich GUVs (20% PS, POPC:POPS 4:1), which is not evident in the low PS containing GUVs (4% PS). C, to diminish the effect of VP40 in the PS-rich GUVs so as to increase the ability to visualize and quantify intraluminal budding, the GUVs were incubated with 500 nm VP40. Scale bars = 10 μm.
To examine the effect of varying VP40 concentration and understand the limits of the protein to induce vesicular structures and eventually undergo scission, GUVs were incubated with concentrations of VP40 ranging from 50 nm to 1 μm. Confocal images revealed that budding from POPC:POPS (4:1) GUVs occurred in a concentration-dependent manner (Fig. 3A). To quantify, 100 GUVs were imaged and counted to calculate the percent of GUVs exhibiting scission of intraluminal vesicles, referred to as budding henceforth. At low VP40 concentrations of 50 nm, ∼11% of the GUVs exhibited detectable budding, whereas for higher concentrations of 1 μm VP40, ∼85% of the GUVs revealed budding (Fig. 3A). With a higher concentration of VP40, the number of vesicles per GUV was also enriched, as seen in Fig. 2B. 5 μm VP40 protein with POPC:POPS (4:1) GUVs had nearly an instantaneous budding effect leading to bursting of GUVs. POPC:POPS GUVs, with 250 nm VP40, showed ∼50% budding response while maintaining the stability of GUVs throughout the duration of the experiment. Hence, 250 nm was chosen as the standard concentration for further experiments unless stated otherwise. This is also similar to the Kd value determined for VP40 binding to PS containing vesicles (19).
FIGURE 3.
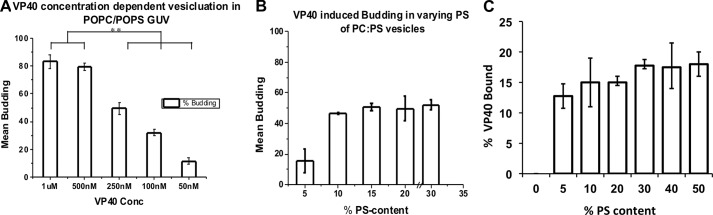
Effect of varying amount of PS or VP40 in GUVs. A, VP40 (50 nm-1 μm) was incubated with POPC:POPS (4:1) GUVs to monitor the amount of budding into the vesicles. A total of 100 GUVs were quantified for mean budding. B, GUVs with different concentrations of POPS were made and quantified for mean budding when incubated with 250 nm VP40. Standard error was determined for each experimental condition to yield the error bars as shown. C, the concentration dependence of VP40 binding to varying PS containing liposomes was performed using a vesicle sedimentation assay. Standard error was determined for each experimental condition to yield the error bars as shown. **, p < 0.005.
To further understand the role of PS on VP40 budding, GUVs were prepared with varying amounts of POPS, ranging from 5 to 30 mol % added at the expense of POPC. GUVs were incubated with 250 nm VP40 and imaged for further quantification. Fig. 3B displays the quantitative results, showing that the amount of budding, for concentrations of PS higher than 10 mol %, did not differ significantly and ∼50% of GUVs consistently displayed detectable scission of vesicles. We can conclude that the vesicles budding from the membrane reach saturation at PS concentrations greater than 10 mol % PS (Fig. 3B). Lipid sedimentation experiments with vesicles containing increasing concentrations of PS also showed a binding saturation at PS content greater than 10 mol % (Fig. 3C). Together these results suggest a minimum concentration of PS necessary for efficient VP40 binding and budding at the PM.
Effect of Cholesterol on VP40 Budding
To investigate the role of the abundant PM lipid cholesterol in VP40-induced viral budding we monitored VP40 budding into GUVs containing 20 mol % cholesterol. The GUVs composed of POPC:POPS:cholesterol (19:1:5) and POPC:POPS:cholesterol (3:1:1) were incubated with 250 nm VP40 and imaged after 20 min. Images were collected for PC-rich GUVs with and without cholesterol (Fig. 4A), which failed to exhibit any significant differences in budding (Fig. 4B). In the case of POPC:POPS:cholesterol (4:1:1) GUVs, the results were similar to that of POPC:POPS:cholesterol (19:1:5) GUVs. Quantification of the images revealed no significant difference in the amount of VP40 budding in the absence or presence of cholesterol for POPC:POPS (24:1) and POPC:POPS (4:1) GUVs (Fig. 4B).
FIGURE 4.
Effects of cholesterol on VP40 intraluminal budding from GUVs. A, GUVs incubated with 250 nm VP40 in both POPC and POPC:POPS vesicles without/with cholesterol presented with visually comparable results. Scale bars = 10 μm. B, quantification of a total of 100 GUVs for POPC:POPS:cholesterol (19:1:5) and POPC:POPS:cholesterol (3:1:1) was performed.
Investigation of VP40 Specificity in Vesicle Budding and Scission
VP40, in the presence of PS-rich membranes, is able to achieve both membrane deformation and subsequent budding off the membrane into the GUV. To elucidate the specificity of VP40 intraluminal budding with PS we used bovine lactadherin C2 (Lact C2) (31), which binds PS with extreme selectivity and nanomolar affinity (36), and is currently the protein of choice for Ca2+-independent detection of PS in vitro and in cells. Moreover, Lact C2 has a similar membrane affinity for PS vesicles (36) as that of VP40 (19). A high concentration of Lact C2, when incubated with POPC:POPS GUVs lead to vesicle formation (positive curvature) on the outer leaflet of the GUVs. The vesicles formed were attached to the GUVs and scission off the membrane was not evident (Fig. 4C). Although membrane bending has not been tested for Lact C2, these effects at high protein concentrations are not surprising as protein crowding on the membrane surface has been shown to be sufficient to bend membranes in vitro (37). Hence, to confirm that the membrane bending was due to protein crowding at the membrane interface, POPC:POPS GUVs were incubated with 250 nm Lact C2 (Fig. 5C) and no membrane deformation or vesicles (positive or negative curvature) were detected.
FIGURE 5.
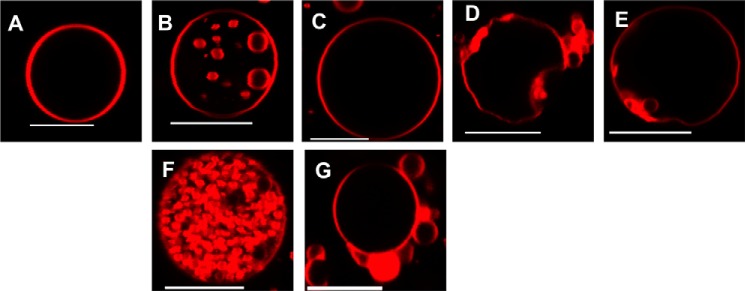
Specificity of VP40-mediated scission of buds from PS containing GUV membranes. POPC:POPS (4:1) GUVs were incubated with the respective protein for 20 min before imaging and the scission of membrane vesicles was then monitored for 100 GUVs. POPC:POPS (4:1) GUVs are shown 20 min post-addition of buffer or 250 nm protein. A, PBS; B, VP40; C, Lact C2; D, IRSp53 I-BAR; and E, MIM I-BAR. POPC:POPS (4:1) were also imaged 20 min post-addition of 5 μm (F) VP40 or (G) Lact C2. Only VP40 buds are completely cleaved into the GUV. Scale bars = 10 μm.
Insulin receptor tyrosine kinase substrate p53 (IRSp53) and missing-in-metastasis (MIM) contain inverse BAR (I-BAR) domains that have been shown to induce negative membrane curvature both in cells and in in vitro systems (30, 32) but not scission. We used purified I-BAR domains to further investigate the hypothesis that VP40 induces budding and scission in a PS selective manner. The effects of I-BAR domain proteins at low concentrations (250 nm) on POPC:POPS (4:1) GUVs were examined to demonstrate the PS specificity in scission of VP40. The GUV membranes were significantly deformed for both IRSp53 and MIM I-BAR domains, and there was formation of buds from the membrane into the GUVs (Fig. 5, D and E), however, for both IRSp53 and MIM, the vesicles remained attached to the membrane throughout the duration of the experiment. Thus, unlike for those observed for VP40, the buds were unable to undergo scission from the membrane. Hence, for quantification of the GUVs, even though vesicles formed at the membrane, if complete scission was not achieved they were not quantified as intraluminal vesicles (Fig. 6). From Fig. 6 we further speculate that, unlike the Lact C2 and I-BAR proteins, VP40 harbors scission-like activity to bud off the membrane in the absence of host cell proteins.
FIGURE 6.
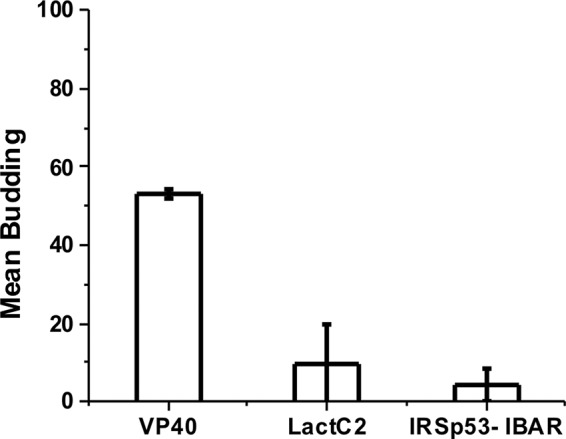
Protein-induced scission of vesicles is unique to VP40. Quantification of GUVs revealed VP40-induced vesicle scission in 50% of GUVs. The PS binding sensor Lact C2 does not induce intraluminal vesicles but does cause positive membrane curvature generation at high protein concentrations. Although the IRSp53 I-BAR domain induces negative curvature and bud formation, scission of vesicles was not detectable. Experiments were repeated in triplicate and standard error was calculated for each experiment to determine the error bars as shown.
Lipid Selectivity of VP40 Membrane Budding and Scission
To test whether VP40 membrane budding was PS specific and thus not due only to electrostatic interactions, anionic lipids including phosphatidylinositol (PI) and phosphoinositides (PIPs), which are enriched in the cytoplasmic PM leaflet (39, 40) were used in the GUV assays. GUVs composed of POPC:POPS:POPI:cholesterol (16.5:1:5:5) were incubated for 20 min with a low concentration (250 nm) of VP40. The images and quantification revealed a significant reduction of VP40 budding and scission from the GUV membranes (Fig. 7A). GUVs composed of phosphorylated derivatives of PI found in the PM were also used to further investigate electrostatic interactions as many peripheral proteins selectively interact with these lipids (40, 41). GUVs composed of POPC:POPS:PI(4,5)P2:cholesterol (18:1:0.75:5) and POPC:POPS:PIP3:cholesterol (18:1:0.75:5) were incubated with VP40 and imaged under the same conditions as that of the POPC:POPS:POPI:cholesterol GUV membrane. The confocal images taken for both PI(4,5)P2 and PIP3 resembled the images taken for GUVs containing PI; budding and scission were nearly undetectable (Fig. 7A). The quantification of GUVs showed that VP40 was poorly able to bud into the membrane of the GUVs containing PI, PI(4,5)P2, or PIP3 (Fig. 7B). In comparison, 250 nm VP40 displayed more budding into POPC:POPS (24:1) GUVs than into GUVs containing PI, PI(4,5)P2, or PIP3 (Fig. 7B). POPC:POPS (4:1) GUVs with low concentrations of VP40 (250 nm) yielded significantly higher budding when compared with any other GUV composition (Fig. 7B). This led us to conclude that budding for VP40 is PS selective rather than dependent on the anionic charge of the membrane.
FIGURE 7.
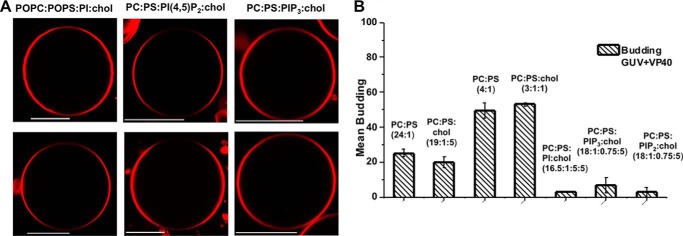
VP40 budding and scission is specific for PS in comparison to other PM anionic lipids. A, confocal images of GUVs composed of POPC:POPS:POPI:cholesterol (16.5:1:5:5), POPC:POPS:PI(4,5)P2:cholesterol (18:1:0.75:5), and POPC:POPS:PIP3:cholesterol (18:1:0.75:5) with 0.1% LRPE before (top panel) and after (bottom panel) 250 nm VP40. Scale bars = 10 μm. VP40 induced budding was significantly reduced for PI and PIP containing GUVs compared with those of POPC:POPS (24:1), POPC:POPS (4:1), POPC:POPS:cholesterol (19:1:5), or POPC:POPS:cholesterol (3:1:1). B, quantification of 100 GUVs for budding off the membrane in the presence of VP40 for each lipid composition. Experiments were repeated in triplicate and standard error was calculated for each experiment for the error bars as shown.
DISCUSSION
The PM is an asymmetric bilayer where the outer leaflet is composed mainly of PC, sphingomyelin, and glycosphingolipids and the inner leaflet is enriched with PE, PS, PI, and PIPs (42). Additionally, the PM is enriched in cholesterol although the distribution of cholesterol among the outer and inner leaflets is not as well understood. PS is the most abundant anionic lipid in the cytosolic leaflet at ∼15–20% (43, 44) and contributes to the recruitment of polycationic proteins as well as proteins containing PS binding domains (45). PS has also been shown to interact with several viral matrix proteins (46, 47) as well as VP40 (19–21, 48). However, the lipid specificity of VP40 has not been explored in much detail (22) and more rigorous analysis of VP40 lipid interactions is warranted.
Here we demonstrate that VP40 selectively induces membrane budding and scission from membranes enriched with PS. VP40 also requires ∼10 mol % or more PS in the membrane to effectively associate with vesicles and induce vesiculation into GUVs compared with that of a PC background. This binding selectivity may further help explain VP40 localization and assembly at the PM inner leaflet where PS is enriched compared with other cytoplasmic sites (49). To demonstrate the selectivity of VP40 budding, other anionic lipids that are found at the PM inner leaflet were used including: PI, PI(4,5)P2, and PIP3. Budding of VP40 into these GUVs was nearly undetectable under all conditions employed. It should also be noted that even when approximately equimolar anionic charge was compared (e.g. 20% PI versus 20% PS, or 3% PIP3 versus 10% PS), VP40 was still highly selective for PS-dependent budding. Furthermore, cholesterol did not play a major role in the in vitro budding system as it did not enhance nor inhibit PS-dependent budding and scission.
VP40 budding and scission effects were specific as the robust PS sensor Lact C2 did not induce detectable changes in GUVs at similar concentrations (e.g. 250 nm). At higher concentrations (5 μm Lact C2) positive membrane curvature changes were observed, which is consistent with membrane crowding effects (37). Additionally, the I-BAR domains of IRSp53 and MIM, which induced negative membrane curvature changes but not scission in our GUV assays were consistent with the previous report (32). Taken together, VP40 selectively induces membrane vesiculation and scission in a PS-dependent manner. It should also be noted that in our study bacterially expressed VP40 was used. In human cells, VP40 has been shown to be Tyr phosphorylated (50) and may also undergo other post-translational modifications that regulate the cellular membrane association and budding of viral particles.
How does VP40 selectively induce budding and scission on PS-enriched membranes? Membrane bending and formation of a vesicle from a planar membrane requires a large degree of thermal energy (51). Tight protein packing on the membrane interface creates a highly dense surface where the energetic requirement to bend the membrane into a sphere is significantly lowered (29). Indeed, VP40 tightly packs on the membrane bilayer of VLPs (38), where VP40 dimers rearrange into hexamers that can concatenate into long filaments (14). VP40 has been shown to associate with PS containing liposomes (21), which can induce VP40 oligomerization (48) through structural rearrangement to a zig-zagging filament that may be able to interact both laterally and longitudinally with other VP40 filaments (14, 17, 20). This structural rearrangement likely requires the membrane penetration of the C-terminal hydrophobic loop of VP40 (19, 20). VP40 protein insertion in the PS containing membrane combined with the orientation and shape of the VP40 filament that forms might increase the curvature stress and provide sufficient thermal energy to bend membranes. Thus, as VP40 structures have not yet been solved in the presence of membrane, there may be distinct structures of membrane-bound VP40 that form increasing selectivity for PS-dependent scission.
Acknowledgment
We thank Erica Ollmann Saphire for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant AI081077, the Center for Rare and Neglected Diseases at the University of Notre Dame, and grants from the Indiana University School of Medicine-South Bend Imaging and Flow Cytometry Core Facility (to R. V. S.).
- PM
- plasma membrane
- BAR
- Bar/Amiphysin/Rvs
- ESCRT
- endosomal sorting complexes required for transport
- IRSp53
- insulin receptor tyrosine kinase substrate p53
- ITO
- indium tin oxide
- GUV
- giant unilamellar vesicle
- Lact C2
- lactadherin C2 domain
- LRPE
- 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)
- MIM
- missing-in-metastasis
- PI(4,5)P2
- 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate)
- PIP3
- 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′,4′,5′-trisphosphate)
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPI
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylserine
- VLPs
- virus-like particles
- VP40
- viral protein 40.
REFERENCES
- 1. Ksiazek T. G. (1999) Clinical virology of Ebola hemorrhagic fever (EHF): virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J. Infect. Dis. 179, S177–S187 [DOI] [PubMed] [Google Scholar]
- 2. Johnson K. M., Lange J. V., Webb P. A., Murphy F. A. (1977) Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet 1, 569–571 [DOI] [PubMed] [Google Scholar]
- 3. Feldmann H. (2014) Ebola: a growing threat? N. Engl. J. Med. 371, 1375–1378 [DOI] [PubMed] [Google Scholar]
- 4. Farrar J. J., Piot P. (2014) The Ebola emergency: immediate action, ongoing strategy. N. Engl. J. Med. 371, 1481–1495 [DOI] [PubMed] [Google Scholar]
- 5. Blaney J. E., Marzi A., Willet M., Papaneri A. B., Wirblich C., Feldmann F., Holbrook M., Jahrling P., Feldmann H., Schnell M. J. (2013) Antibody quality and protection from lethal Ebola virus challenge in nonhuman primates immunized with Rabies virus based bivalent vaccine. PLoS Pathog. 9, e1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johansen L. M., Brannan J. M., Delos S. E., Shoemaker C. J., Stossel A., Lear C., Hoffstrom B. G., DeWald L. E., Schornberg K. L., Scully C., Lehár J., Hensley L. E., White J. M., Olinger G. G. (2013) FDA-approved selective estrogen receptor modulators inhibit Ebola virus infection. Sci. Transl. Med. 5, 190ra179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qiu X., Wong G., Audet J., Bello A., Fernando L., Alimonti J. B., Fausther-Bovendo H., Wei H., Aviles J., Hiatt E., Johnson A., Morton J., Swope K., Bohorov O., Bohorova N., Goodman C., Kim D., Pauly M. H., Velasco J., Pettitt J., Olinger G. G., Whaley K., Xu B., Strong J. E., Zeitlin L., Kobinger G. P. (2014) Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. E., Fusco M. L., Hessell A. J., Oswald W. B., Burton D. R., Saphire E. O. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miller E. H., Obernosterer G., Raaben M., Herbert A. S., Deffieu M. S., Krishnan A., Ndungo E., Sandesara R. G., Carette J. E., Kuehne A. I., Ruthel G., Pfeffer S. R., Dye J. M., Whelan S. P., Brummelkamp T. R., Chandran K. (2012) Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 31, 1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Côté M., Misasi J., Ren T., Bruchez A., Lee K., Filone C. M., Hensley L., Li Q., Ory D., Chandran K., Cunningham J. (2011) Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., Dal Cin P., Dye J. M., Whelan S. P., Chandran K., Brummelkamp T. R. (2011) Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jasenosky L. D., Neumann G., Lukashevich I., Kawaoka Y. (2001) Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75, 5205–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCarthy S. E., Johnson R. F., Zhang Y. A., Sunyer J. O., Harty R. N. (2007) Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J. Virol. 81, 11452–11460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bornholdt Z. A., Noda T., Abelson D. M., Halfmann P., Wood M. R., Kawaoka Y., Saphire E. O. (2013) Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154, 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olejnik J., Ryabchikova E., Corley R. B., Mühlberger E. (2011) Intracellular events and cell fate in filovirus infection. Viruses 3, 1501–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harty R. N. (2009) No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res. 81, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adu-Gyamfi E., Digman M. A., Gratton E., Stahelin R. V. (2012) Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys. J. 102, 2517–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Licata J. M., Johnson R. F., Han Z., Harty R. N. (2004) Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78, 7344–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soni S. P., Adu-Gyamfi E., Yong S. S., Jee C. S., Stahelin R. V. (2013) The Ebola virus matrix protein deeply penetrates the plasma membrane: an important step in viral egress. Biophys. J. 104, 1940–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adu-Gyamfi E., Soni S. P., Xue Y., Digman M. A., Gratton E., Stahelin R. V. (2013) The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 288, 5779–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ruigrok R. W., Schoehn G., Dessen A., Forest E., Volchkov V., Dolnik O., Klenk H. D., Weissenhorn W. (2000) Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300, 103–112 [DOI] [PubMed] [Google Scholar]
- 22. Stahelin R. V. (2014) Membrane binding and bending in Ebola VP40 assembly and egress. Front. Microbiol. 5, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin-Serrano J., Zang T., Bieniasz P. D. (2001) HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 24. Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R. W., Weissenhorn W. (2003) Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326, 493–502 [DOI] [PubMed] [Google Scholar]
- 25. Henne W. M., Buchkovich N. J., Emr S. D. (2011) The ESCRT pathway. Dev. Cell 21, 77–91 [DOI] [PubMed] [Google Scholar]
- 26. Neumann G., Ebihara H., Takada A., Noda T., Kobasa D., Jasenosky L. D., Watanabe S., Kim J. H., Feldmann H., Kawaoka Y. (2005) Ebola virus VP40 late domains are not essential for viral replication in cell culture. J. Virol. 79, 10300–10307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Solon J., Gareil O., Bassereau P., Gaudin Y. (2005) Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J. Gen. Virol. 86, 3357–3363 [DOI] [PubMed] [Google Scholar]
- 29. Shnyrova A. V., Ayllon J., Mikhalyov I. I., Villar E., Zimmerberg J., Frolov V. A. (2007) Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J. Cell Biol. 179, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P. K., Kinnunen P. K., Butcher S. J., Lappalainen P. (2009) Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95–107 [DOI] [PubMed] [Google Scholar]
- 31. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 32. Mattila P. K., Pykäläinen A., Saarikangas J., Paavilainen V. O., Vihinen H., Jokitalo E., Lappalainen P. (2007) Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ward K. E., Ropa J. P., Adu-Gyamfi E., Stahelin R. V. (2012) C2 domain membrane penetration by group IVA cytosolic phospholipase A2 induces membrane curvature changes. J. Lipid Res. 53, 2656–2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He J., Vora M., Haney R. M., Filonov G. S., Musselman C. A., Burd C. G., Kutateladze A. G., Verkhusha V. V., Stahelin R. V., Kutateladze T. G. (2009) Membrane insertion of the FYVE domain is modulated by pH. Proteins 76, 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stahelin R. V. (2014) Could the Ebola virus matrix protein VP40 be a drug target? Expert Opin. Ther. Targets 18, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kay J. G., Koivusalo M., Ma X., Wohland T., Grinstein S. (2012) Phosphatidylserine dynamics in cellular membranes. Mol. Biol. Cell 23, 2198–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stachowiak J. C., Schmid E. M., Ryan C. J., Ann H. S., Sasaki D. Y., Sherman M. B., Geissler P. L., Fletcher D. A., Hayden C. C. (2012) Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 [DOI] [PubMed] [Google Scholar]
- 38. Bharat T. A., Noda T., Riches J. D., Kraehling V., Kolesnikova L., Becker S., Kawaoka Y., Briggs J. A. (2012) Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. U.S.A. 109, 4275–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stahelin R. V., Scott J. L., Frick C. T. (2014) Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem. Phys. Lipids 182, 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moravcevic K., Oxley C. L., Lemmon M. A. (2012) Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vance J. E., Steenbergen R. (2005) Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 44, 207–234 [DOI] [PubMed] [Google Scholar]
- 44. Vance J. E., Tasseva G. (2013) Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta 1831, 543–554 [DOI] [PubMed] [Google Scholar]
- 45. Cho W., Stahelin R. V. (2006) Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761, 838–849 [DOI] [PubMed] [Google Scholar]
- 46. Dick R. A., Goh S. L., Feigenson G. W., Vogt V. M. (2012) HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc. Natl. Acad. Sci. U.S.A. 109, 18761–18766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zakowski J. J., Petri W. A., Jr., Wagner R. R. (1981) Role of matrix protein in assembling the membrane of vesicular stomatitis virus: reconstitution of matrix protein with negatively charged phospholipid vesicles. Biochemistry 20, 3902–3907 [DOI] [PubMed] [Google Scholar]
- 48. Scianimanico S., Schoehn G., Timmins J., Ruigrok R. H., Klenk H. D., Weissenhorn W. (2000) Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19, 6732–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fairn G. D., Schieber N. L., Ariotti N., Murphy S., Kuerschner L., Webb R. I., Grinstein S., Parton R. G. (2011) High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J. Cell Biol. 194, 257–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. García M., Cooper A., Shi W., Bornmann W., Carrion R., Kalman D., Nabel G. J. (2012) Productive replication of Ebola virus Is regulated by the c-Abl1 tyrosine kinase. Sci. Transl. Med. 4, 123ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Helfrich W. (1973) Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C 28, 693–703 [DOI] [PubMed] [Google Scholar]