Background: 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) activates transcription of CYP1A1.
Results: Ectopic expression of SIN3A reverses the inhibitory effect of SIN3A siRNAs on CYP1A1 induction. TCDD stimulates SIN3A binding to the CYP1A1 enhancer and promoter.
Conclusion: SIN3A coactivates TCDD induction of CYP1A1 transcription.
Significance: The anomalous stimulatory effect of SIN3A on transcription is extended to a gene that is rapidly and dramatically induced.
Keywords: Aryl Hydrocarbon Receptor (AHR), Chromatin Immunoprecipitation (ChIP), Gene Transcription, Transcriptional Coactivator, Xenobiotic
Abstract
CYP1A1 bioactivates several procarcinogens and detoxifies several xenobiotic compounds. Transcription of CYP1A1 is highly induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) via the aryl hydrocarbon receptor. We recently described an RNAi high throughput screening performed in the Hepa-1 mouse hepatoma cell line, which revealed that SIN3A is necessary for the induction of CYP1A1-dependent ethoxyresorufin-o-deethylase (EROD) enzymatic activity by TCDD. In the current studies, we sought to provide insight into the role of SIN3A in this process, particularly because studies on SIN3A have usually focused on its repressive activity on transcription. We report that ectopic expression of human SIN3A in Hepa-1 cells enhanced EROD induction by TCDD and efficiently rescued TCDD induction of EROD activity in cells treated with an siRNA to mouse SIN3A, thus validating a role for SIN3A in CYP1A1 induction. We demonstrate that SIN3A is required for TCDD induction of the CYP1A1 protein in Hepa-1 cells but not for expression of the aryl hydrocarbon receptor protein. In addition, siRNAs for SIN3A decreased TCDD-mediated induction of CYP1A1 mRNA and EROD activity in human hepatoma cell line Hep3B. We establish that TCDD treatment of Hepa-1 cells rapidly increases the degree of SIN3A binding to both the proximal promoter and enhancer of the Cyp1a1 gene and demonstrate that increased binding to the promoter also occurs in human Hep3B, HepG2, and MCF-7 cells. These studies establish that SIN3A physically interacts with the CYP1A1 gene and extends the transcriptional role of SIN3A to a gene that is very rapidly and dramatically induced.
Introduction
CYP1A1 is a phase I metabolizing enzyme involved in either the detoxification or bioactivation of xenobiotics and environmental pollutants, including certain polycyclic aromatic hydrocarbons and halogenated aromatic hydrocarbons (1). The expression of CYP1A1 is induced upon exposure to these polycyclic/halogenated aromatic hydrocarbons, such as benzo[a]pyrene, which can come from a number of environmental sources, including tobacco smoke, automobile exhaust, industrial by-products, and charbroiled foods (2). The most robust induction of CYP1A1, however, is elicited by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD),4 which is a persistent environmental pollutant and one of the most potent carcinogens known (3). The mechanism of induction, and thereby the proposed mechanism of toxicity, is through the aryl hydrocarbon receptor (AHR) pathway, where TCDD serves as a ligand for AHR. Ligand binding enables the translocation of AHR into the nucleus, where it can then form a heterodimer with the aryl hydrocarbon receptor nuclear translocator (ARNT) protein. The dimer subsequently binds xenobiotic response element core consensus sequences within genomic DNA to activate transcription (4, 5). There have been considerable advances in the field through the years, but further investigation is needed to completely understand the events governing CYP1A1 transcription (6–8).
In a recent study, we aimed to further elucidate the mechanism leading to induction of CYP1A1 by performing an RNAi high throughput screening (19). For these studies, we used the mouse hepatoma cell line Hepa1c1c7 (henceforth called Hepa-1), in which CYP1A1 mRNA is induced many hundred-fold within 24 h of treatment with TCDD (1). This screen revealed that the SIN3A protein is required for the TCDD induction of CYP1A1-dependent EROD activity in Hepa-1 cells. SIN3A is a transcriptional regulator and, as the core component of multiprotein complexes, serves as a platform for several protein interactions (9). The formation of diverse complexes from these various protein interactions permits SIN3A to be involved in an assortment of signaling pathways and associated biological processes, including chromatin modification, embryonic development, DNA repair, mitochondrial metabolism, and gene transcription (10). SIN3A itself has no known intrinsic enzymatic activity and is devoid of any known DNA-binding motifs, which makes its function dependent on its ability to bind multiple proteins and form specialized complexes (11). Recruitment of SIN3A to targeted genes is thereby mediated through its interaction with other DNA-binding factors, whereas the histone deacetylase enzymatic activity associated with SIN3A is mediated via its interaction with HDAC1/HDAC2 (9). The deacetylation activity of the SIN3A-HDAC complex serves to restore the basic charge of lysine necessary for histone compaction and is therefore thought to restrict access to the DNA and to ultimately inhibit transcription (12). Although SIN3A has thus been primarily identified as a transcriptional repressor, accumulating evidence indicates that SIN3A can also play a role in transcriptional activation in yeast, Drosophila, and mammalian cells (13).
In this study, we analyzed the role of SIN3A in AHR-mediated induction of CYP1A1. We firmly establish that SIN3A is required for TCDD induction of CYP1A1 in Hepa-1 cells; demonstrate that SIN3A is required for the induction of the CYP1A1 protein but not the AHR protein in these cells; reveal that SIN3A is also required for the induction of EROD activity and CYP1A1 mRNA in human cells; and, most interestingly, find that TCDD induces the recruitment of SIN3A to both the CYP1A1 proximal promoter and the enhancer region (containing a cluster of xenobiotic response elements) in Hepa-1 cells. We also confirm that binding to the promoter occurs in human cells. Transcriptional activation by SIN3A has not been as widely explored as transcriptional repression, and the results obtained from these studies shed light on this novel mechanism of action of SIN3A during the induction of AHR-mediated gene expression.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Cells were grown as a monolayer in α-minimal essential medium (Hepa1c1c7, Hep3B, and MCF-7) or in DMEM (HepG2), supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% fungizone (Invitrogen) at 37 °C in 5% CO2. Tissue culture dishes used to grow HepG2 cells were pretreated with 50 μg/ml poly-l-lysine. Treatments with TCDD (Wellington Laboratories, Guelph, Canada) dissolved in DMSO were at either 10 nm (Hepa-1) or 100 nm (Hep3B, HepG2, and MCF-7) concentrations, and the final concentration of DMSO in the medium was 0.1%.
Transfection of siRNA
Hepa-1 or Hep3B cells were transfected for 72 h with 25 nm siRNA (Thermo Scientific Dharmacon, Lafayette, CO) using Lipofectamine RNAiMax transfection reagent according to the manufacturer's protocol (Invitrogen).
Ethoxyresorufin-O-deethylase (EROD) Assay
To assay for CYP1A1 enzymatic activity, cells were treated with 10 μm ethoxyresorufin and 500 μm dicumarol (Sigma-Aldrich) in phenol red-free minimal essential medium supplemented with 10% FBS, 1% l-glutamate, and 1% penicillin-streptomycin. Phenol red-free medium reduces light scattering and background fluorescence, whereas dicumarol prevents further metabolic degradation of the fluorescent product, resorufin (530 nm excitation/590 nm emission). Reactions were incubated for the optimized time of 45–60 min at 37 °C, and resorufin-related fluorescence was detected in a Modulus Microplate fluorescence reader (Turner Biosystems, Madison, WI).
Quantitative PCR
RNA was isolated using RNEasy Mini columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA synthesis from 2 μg of total RNA was performed with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's protocols using random hexamers as primers (Invitrogen). Synthesized cDNA was used as a template for qPCR at a 1:10 dilution, and SYBR Green (Qiagen, Valencia, CA) was used according to standard protocols on an Applied Biosystems 7500 real-time PCR machine. Primers for qPCR were designed using Primer Express 3.0 software (Applied Biosystems) and were synthesized by Fisher. Quantities were normalized to those for the GADPH glycolytic housekeeping gene.
Western Blotting
Cellular lysates were resolved on a 4–12% SDS gel in a minigel apparatus (Invitrogen) and transferred to nitrocellulose membrane with a semidry apparatus (Thermo Scientific Pierce). Blots were then blocked with 3% nonfat milk and incubated with an antibody against CYP1A1 (Santa Cruz Biotechnology, Inc.) at 1:200, AHR (14) at 1:200, or GAPDH (Santa Cruz Biotechnology) at 1:2,000. Donkey anti-rabbit IgG or anti-mouse IgG conjugated with horseradish peroxidase was used as secondary antibody at 1:10,000 or 1:5,000, respectively. Horseradish peroxidase was then detected with a chemiluminescent kit (GE Healthcare), and densitometry calculations were performed using ImageJ software (National Institutes of Health).
Transduction for EROD Rescue
Retrovirus was made by cotransfection of 293T cells with the pMSCV-ires/GFP retroviral expression vector (15), containing the human SIN3A cDNA, and a pCL-Eco packaging plasmid, using the BioT (Bioland Scientific LLC, Paramount, CA) transfection reagent. The following day, transfected 293T cells were treated with 10 mm sodium butyrate for 9 h to enhance viral production, and after an additional 24 h, the viral suspension was harvested. Hepa-1 cells were treated with 2 ml of this viral suspension in 6-well culture dishes in α-minimal essential medium supplemented with 8 μg/ml Polybrene and then centrifuged at 37 °C for 1.5 h at 2,500 rpm. An additional 2 ml of fresh α-minimal essential medium was subsequently added, and after 48 h, the cells were trypsinized and plated in a 10-cm plate. Once confluent, cells successfully transduced were isolated using fluorescence-activated cell sorting (FACS) for GFP expression.
Chromatin Immunoprecipitation Assay
Confluent cells were treated with TCDD for the indicated times and then treated with a 1% formaldehyde solution for 10 min at 37 °C to cross-link DNA to proteins and proteins to proteins. All subsequent steps were performed at 4 °C. Cross-linking was stopped by the addition of 0.125 m glycine for 5 min. The cells were then rinsed twice and collected in ice-cold PBS supplemented with 1× protease inhibitor solution (Affymetrix/U.S. Biochemical Corp.). Centrifugation was performed at 1,500 × g for 10 min in a Beckman tabletop centrifuge, and the pelleted cells were resuspended in 420 μl of lysis buffer (Affymetrix/U.S. Biochemical Corp.). After a 10-min incubation on ice, cell lysates were sonicated using an ultrasonicator (Diagenode) at high amplitude with 30-s pulses for a total of 13 min to shear DNA fragments to sizes between 200 and 900 base pairs. Cellular debris was precipitated by centrifugation at 13,000 rpm for 10 min, and supernatants were diluted 1:5 in lysis buffer supplemented with 1× protease inhibitor solution. For immunoclearing, a 50% slurry of protein A-Sepharose beads in Tris-EDTA, 2.5 μg of sonicated salmon sperm DNA, bovine serum albumin solution was added to the supernatant and incubated on a rotator for 1 h. Supernatants were isolated, treated with 2 μg of the SIN3A antibody (K-20, Santa Cruz Biotechnology, Inc.) overnight on a rotator, and then treated with a 50% slurry of protein A-Sepharose beads in Tris-EDTA, 2.5 μg of sonicated salmon sperm DNA, bovine serum solution for 2 h on a rotator. The beads were pelleted and sequentially washed in buffers, and chromatin complexes were then eluted from the beads according to the manufacturer's protocols (Affymetrix/U.S. Biochemical Corp.). The cross-linking was reversed by incubating samples in high salt conditions at 65 °C overnight. DNA was extracted using DNeasy minicolumns (Qiagen) according to the manufacturer's protocols. Real-time PCR analysis of the DNA was performed using the methods described previously by our laboratory (16). Background signals represented by the IgG negative controls were subtracted from all samples, and analysis of the ChIP real-time PCR results was reported relative to that of total inputs. Primers used for the murine CYP1A1 proximal promoter region were 5′-AATGGAGGCCCCAGTACTTAC-3′ and 5′-AGAACTACCACCTTCAGGGTTAGG-3′ (17), those for the murine CYP1A1 enhancer region were 5-AGGCTCTTCTCACGCAACTC-3 and 5-TAAGCCTGCTCCATCCTCTG-3 (16), and the primers for the human CYP1A1 proximal promoter were 5′-CCCGCCTATAAAGGTGGCA-3′ and 5′-AGCAACTCACCTGAGGTACTG-3′ (18). To test for the specificity of the SIN3A antibody used in the ChIP analysis, the antibody was incubated with the peptide against which it was raised (K-20, Santa Cruz Biotechnology) or an unrelated peptide of similar length (CTSKSQESWRTTLSTSQETRK) in the lysis buffer at a ratio of 1:5 at 4 °C for 1 h on a rotator.
Statistical Analysis
Student's two-tailed t test was performed to obtain statistical significance, and data are presented as mean ± S.D. Graphs and blots are representative of at least two separate experiments.
RESULTS
SIN3A Is Required for TCDD Induction of CYP1A1-dependent EROD Activity in the Mouse Hepatoma Cell Line, Hepa-1
We previously performed an RNAi high throughput screening to identify proteins necessary for induction of CYP1A1-dependent EROD enzymatic activity (19). In this screening, four individual siRNAs per target gene were tested in duplicate, and each candidate gene was then ranked according to both the quality of the assay and the quantity of hits. Sin3A ranked seventh out of the 5,600 different gene candidates tested, with all four of the siRNAs generating hits, giving a p value of 0.0002: a ranking higher than both AHR and ARNT. Studies with an endoribonuclease-prepared siRNA and with one of the same chemically synthesized siRNAs as used in the original screen confirmed the results from the original screen. Two independent SIN3A siRNAs also reduced the levels of the Sin3A mRNA by about 75% and reduced induced CYP1A1 mRNA levels by about 80 and 50% (19) (data not shown).
Ectopic Expression of Human SIN3A Demonstrates That the Effects of the siRNA to Mouse SIN3A Are Not Off-target Effects and Confirms That This Gene Product Is Required for CYP1A1 Induction by TCDD
To exclude the possibility that the observed results were due to off-target effects of the SIN3A siRNAs, we transduced Hepa-1 cells with the pMSCV-ires/GFP retroviral expression vector containing the human SIN3A cDNA or no insert (to serve as a negative control) and isolated successfully transduced cells by FACS of GFP-positive cells. These cells were then transfected with the murine siRNA counterpart. The murine siRNA sequence was BLASTed against the human genome to ensure that human SIN3A expression would be unaffected. Expression of human SIN3A efficiently rescued TCDD-induced EROD levels in cells treated with the mouse siRNA when compared with empty vector controls (Fig. 1). The SIN3A cDNA did not rescue cells treated with siRNAs to CYP1A1 or AHR, demonstrating the specificity of the effect of this cDNA. In addition, expressing SIN3A in cells not transfected with the corresponding siRNAs significantly increased TCDD-induced CYP1A1 EROD activity relative to the empty vector control, further validating the involvement of this protein in CYP1A1 expression.
FIGURE 1.
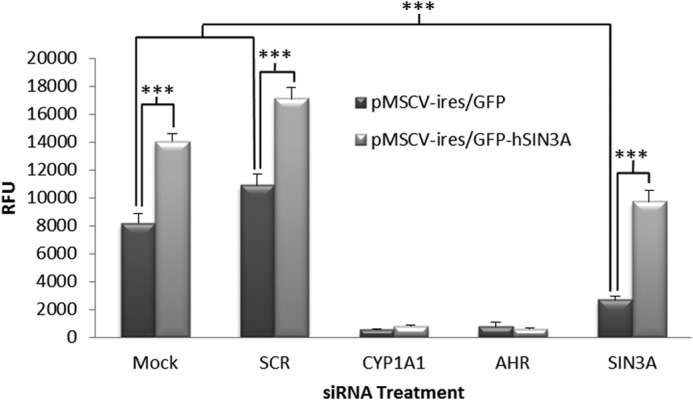
Ectopic expression of human SIN3A rescues the effects of siSIN3A on induction of CYP1A1-dependent EROD activity. Hepa-1 cells were retrovirally transduced with a pMSCV-ires/GFP expression vector containing a human SIN3A cDNA insert or transduced with the parental vector. Cells expressing GFP, and therefore successfully transduced, were isolated using FACS and subsequently transfected with siRNAs to the indicated mouse genes. The cells were treated with TCDD, and EROD activity was measured to determine whether overexpression of SIN3A was sufficient to overcome the effects of siSIN3A. *, p < 0.05; ***, p < 0.001. RFU, relative fluorescence units. Error bars, S.D.
SIN3A Is Necessary for TCDD Induction of the CYP1A1 Protein but Is Not Required for Expression of the AHR Protein
We next investigated whether CYP1A1 and AHR protein levels were affected by SIN3A siRNA treatment (Fig. 2). An siRNA to AHR was used as a positive control, and a scrambled siRNA was used as a negative control. A decrease in AHR protein level occurred in the TCDD-treated samples, consistent with the observations of other investigators that TCDD treatment leads to proteolysis of AHR (20–22). The siRNA to AHR inhibited both AHR expression and the induction of CYP1A1, whereas the scrambled siRNA did not have these effects. The siRNA to SIN3A decreased the TCDD-induced expression of the CYP1A1 protein by 85% but did not affect or only marginally affected the expression of the AHR protein, as compared with the scrambled siRNA control. Thus, SIN3A is required for TCDD induction of CYP1A1 mRNA, CYP1A1 protein, and CYP1A1 enzymatic activity in Hepa-1 cells, and its effects are not mediated by an effect on AHR expression.
FIGURE 2.
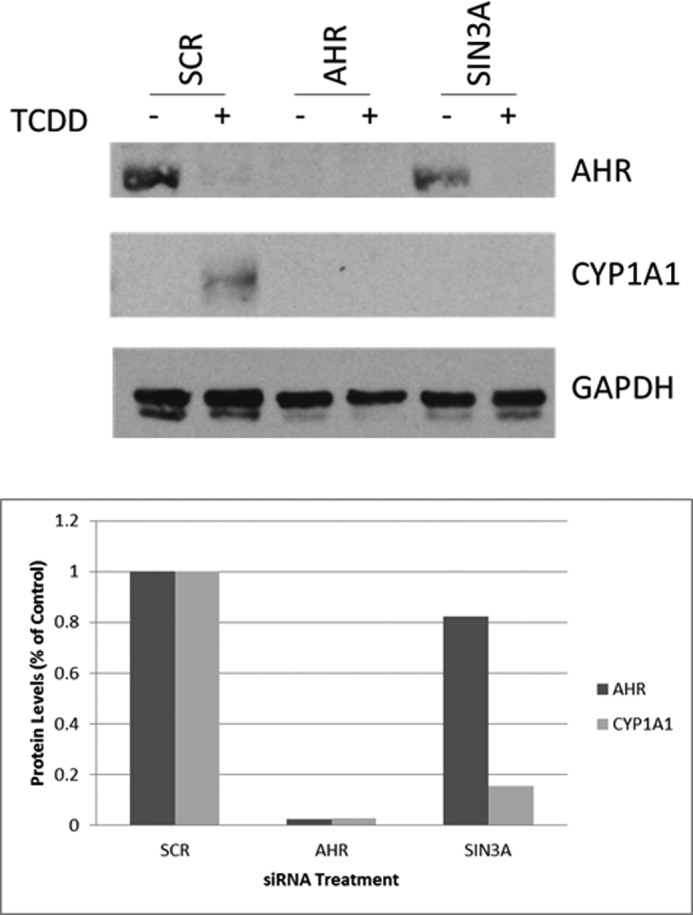
SIN3A is necessary for TCDD-induced CYP1A1 protein expression but not for AHR protein expression. a, Western blot analysis of the CYP1A1 and AHR proteins after transfection of Hepa-1 cells with siAHR, siSIN3A, or a scrambled siRNA in the absence (−) or presence (+) of TCDD. Densitometry quantification of the Western blot was normalized to GAPDH (b).
SIN3A Is Necessary for TCDD Induction of CYP1A1-dependent EROD Activity and of CYP1A1 mRNA in Human Cells
An siRNA to human SIN3A significantly diminished TCDD-induced EROD activity in the human hepatoma cell line, Hep3B (Fig. 3). In addition, three different siRNAs diminished the levels of Sin3A by 70–80% in Hep3B cells and diminished the TCDD-induced CYP1A1 mRNA levels by an average of 40% (Fig. 4). These results demonstrate that SIN3A is required for the induction of CYP1A1 in human cells as well as in mouse cells.
FIGURE 3.
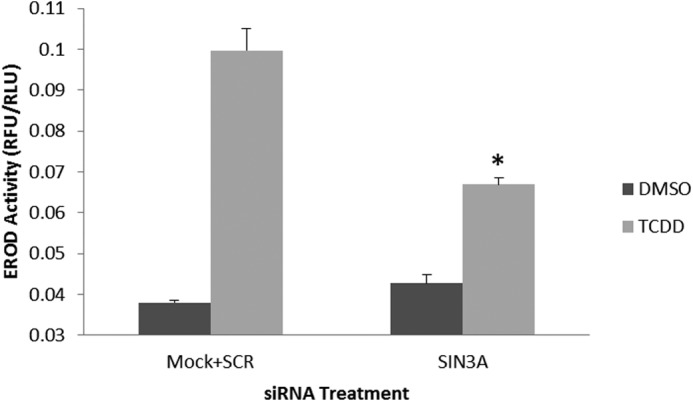
SIN3A is necessary for induction of Cyp1a1 EROD activity by TCDD in the human hepatoma cell line, Hep3B. Hep3B cells were transfected with an siRNA to SIN3A, treated with TCDD or vehicle (DMSO), and subsequently analyzed for TCDD-induced CYP1A1-dependent EROD activity. *, p < 0.05 compared with the average of cells treated with a scrambled siRNA (SCR) or untreated with any siRNA (Mock) (negative controls). Error bars, S.D.
FIGURE 4.
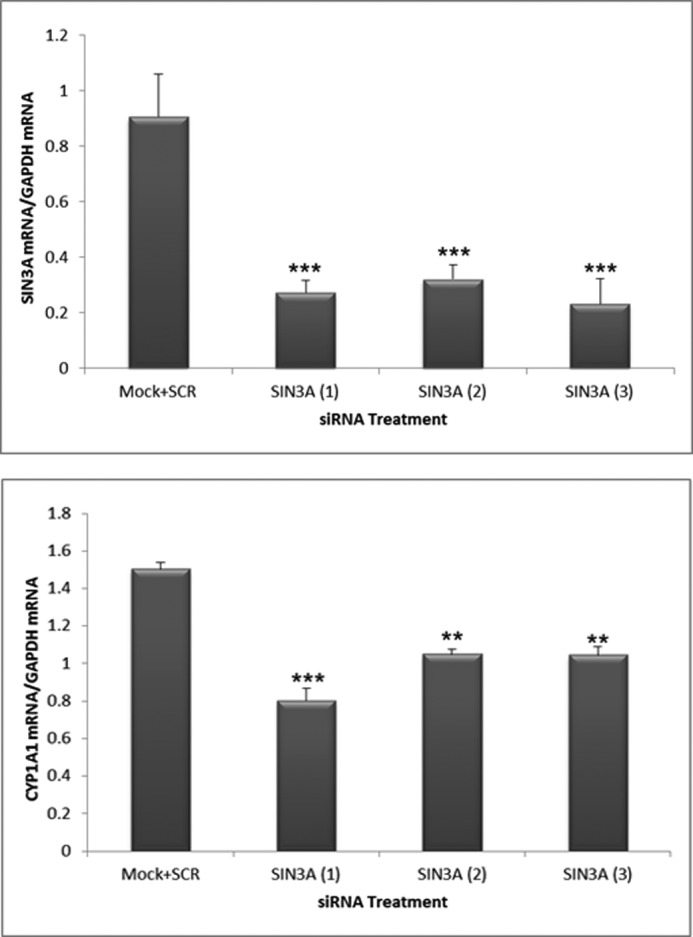
SIN3A is necessary for induction of CYP1A1 mRNA by TCDD in the human hepatoma cell line Hep3B. Hep3B cells were transfected with three different siRNAs to human SIN3A and subsequently analyzed for the expression of SIN3A and CYP1A1 mRNAs after TCDD treatment. The negative control is the average of cells treated with a scrambled siRNA (SCR) or untreated with any siRNA (Mock). Data were normalized to the levels of GAPDH mRNA. **, p < 0.01; ***, p < 0.001 compared with Mock + SCR controls. Error bars, S.D.
SIN3A Binds to the Enhancer and the Proximal Promoter of the CYP1A1 Gene and Binding Is Enhanced by Treatment of the Cells with TCDD
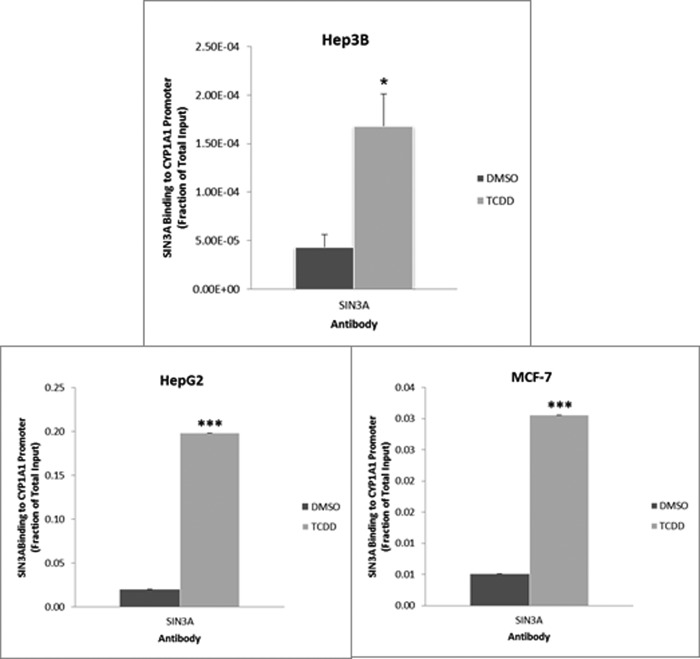
To define the role of SIN3A in the TCDD-induced expression of CYP1A1, we performed chromatin immunoprecipitation (ChIP) assays on the enhancer and proximal promoter regions of CYP1A1 in mouse Hepa-1 cells. In this analysis, we used primers flanking the TATA box in the proximal promoter of the gene. The enhancer is located ∼1,000 bp upstream of the TATA box and contains several xenobiotic responsive elements that are known to bind the AHR. Our protocols clearly distinguish binding at the proximal promoter from binding at the enhancer region of the gene (17, 18). We studied several durations of TCDD treatment because we and others have found that the degree of AHR binding to the enhancer region of TCDD- responsive genes can oscillate rapidly after TCDD treatment (16, 23–26). We found that SIN3A is recruited to both the proximal promoter and the enhancer of the CYP1A1 gene in Hepa-1 cells and that recruitment was significantly increased in cells treated with TCDD. Although there were some differences in the degree of recruitment at the different time points, we did not obtain convincing evidence for cycling (Fig. 5). To provide evidence for the specificity of the SIN3A antibody used for the ChIP assay, we preabsorbed the antibody against either the peptide to which the antibody was raised or an unrelated peptide. The SIN3A peptide eliminated TCDD induction of SIN3A binding to the CYP1A1 promoter and enhancer compared with the nonspecific peptide (Fig. 6). We also demonstrated that TCDD enhanced recruitment of SIN3A to the CYP1A1 promoter in the human hepatoma cell lines Hep3B and HepG2 and the human breast cancer cell line MCF-7 (Fig. 7), all of which are known to be inducible by TCDD for CYP1A1 (18).
FIGURE 5.
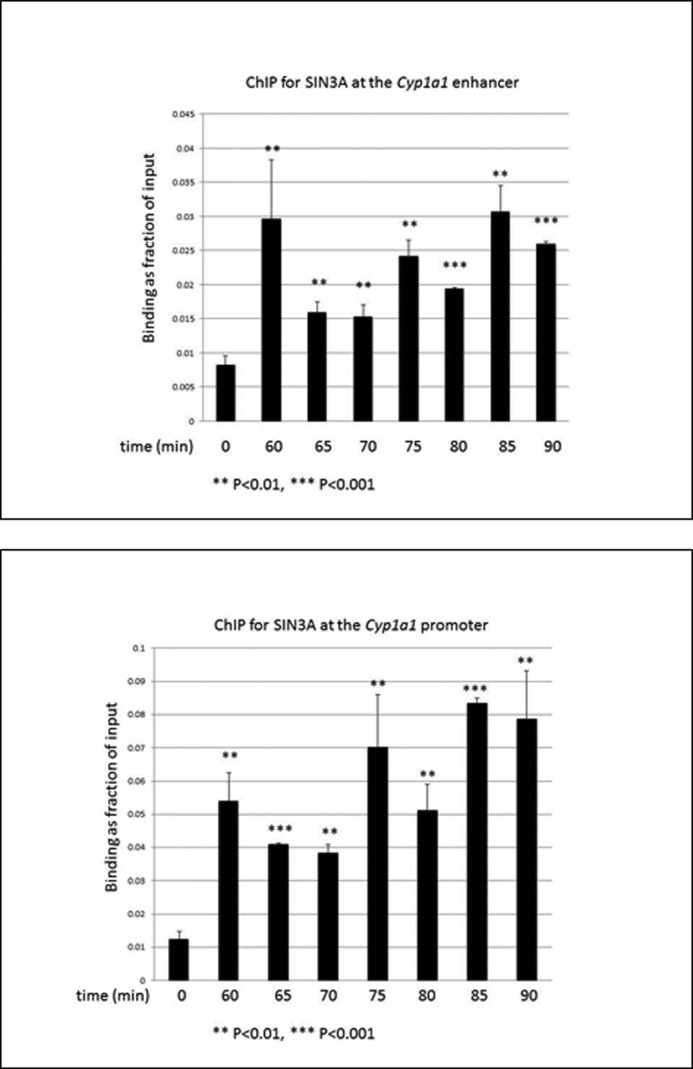
TCDD treatment enhances recruitment of SIN3A to the CYP1A1 enhancer and proximal promoter in Hepa-1 cells in a constant fashion. Cells were subjected to ChIP analysis after the indicated periods of time of treatment with 10 nm TCDD. The recruitment of SIN3A to the enhancer and proximal promoter of the Cyp1a1 gene was measured by real-time PCR of the ChIP samples (A and B). Data were normalized to the total input. **, p < 0.01; ***, p < 0.001 compared with (DMSO) control. Error bars, S.D.
FIGURE 6.
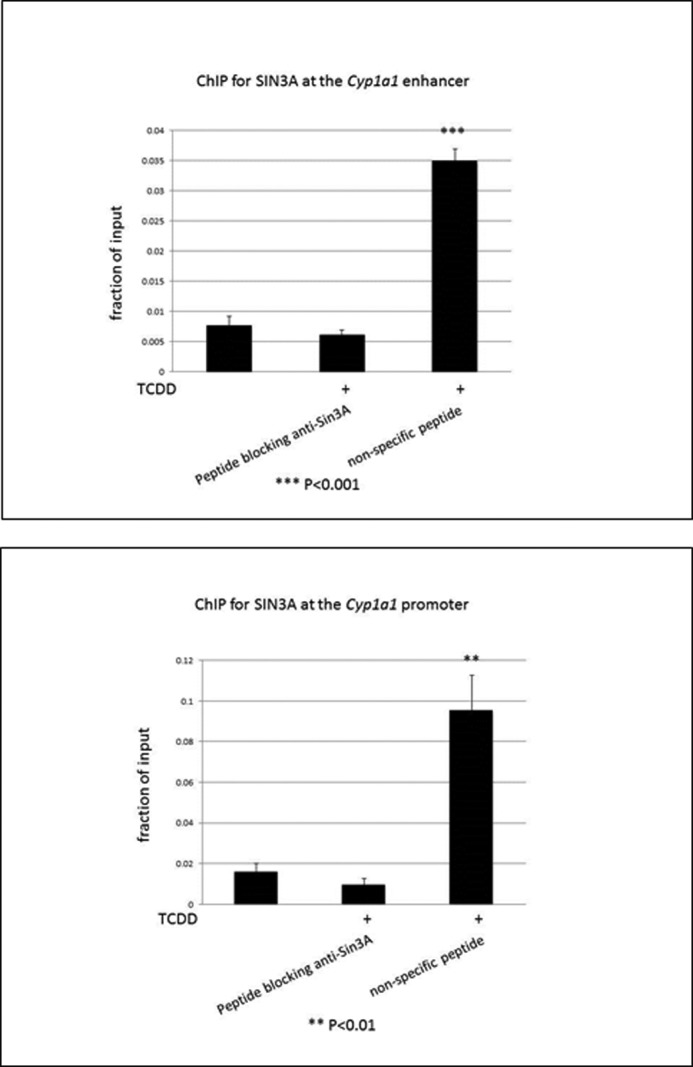
A blocking peptide for the SIN3A antibody eliminates TCDD treatment-enhanced recruitment of SIN3A to the CYP1A1 enhancer and proximal promoter in Hepa-1 cells. Cells were subjected to ChIP analysis after 1.5 h of 10 nm TCDD treatment with the use of a blocking peptide for the SIN3A antibody or a nonspecific peptide, as indicated. The recruitment of SIN3A to the enhancer and promoter of the CYP1A1 gene was measured by real-time PCR of the ChIP samples (A and B). Data were normalized to the total input. **, p < 0.01; ***, p < 0.001 compared with the (DMSO) control. Error bars, S.D.
FIGURE 7.
TCDD treatment enhances recruitment of SIN3A to the CYP1A1 proximal promoter in human Hep3B, HepG2, and MCF-7 cells. Cells were subjected to ChIP analysis after 1 h of 100 nm TCDD treatment. The recruitment of SIN3A to the promoter of the CYP1A1 gene was measured by real-time PCR of ChIP samples from Hep3B (a), HepG2 (b), and MCF7 (c) cells. Data were normalized to the total input. *, p < 0.05; ***, p < 0.001 compared with the corresponding (DMSO) control. Error bars, S.D.
DISCUSSION
In this study, we report on a novel role for SIN3A in induction of the CYP1A1 gene. CYP1A1 has been studied as the hallmark of AHR-mediated gene induction. CYP1A1 metabolically activates a variety of environmental carcinogens, and furthermore, its overexpression can lead to carcinogenesis (6). Understanding the regulation of CYP1A1 will therefore contribute not only to an understanding of the dysregulation of AHR target genes but also the resulting toxicity of this dysregulation. We previously reported results from an RNAi high throughput screening we performed that indicated that SIN3A is necessary for TCDD induction of CYP1A1 in murine Hepa-1 cells (19). In this paper, we also demonstrate that overexpression of human SIN3A in Hepa-1 cells transfected with siRNA to mouse SIN3A was sufficient to rescue CYP1A1-mediated EROD activity, which validates our previous results and provides convincing evidence that our observations are a result of effects of the siRNA on SIN3A and not due to off-target effects. We also demonstrate that the expression of SIN3A is necessary for the induction of both CYP1A1 enzymatic EROD activity and mRNA expression in human cells. Additionally, we show that SIN3A is necessary for the induction of the CYP1A1 protein but not the AHR protein in Hepa-1 cells, indicating that SIN3A does not affect upstream events but directly impacts the CYP1A1 gene itself. We next established a physical interaction between SIN3A at both the proximal promoter and the enhancer of the CYP1A1 gene in vivo. Most importantly, binding of SIN3A increased rapidly after TCDD treatment.
SIN3A null mice die early in gestation, pointing to the essential role of the SIN3A protein in early embryonic development (10). SIN3A is known to associate with several proteins to exert activities on transcription, primarily through nucleosomal remodeling (11). Although generally associated with repression, HDAC activity characteristic of the SIN3A-HDAC complex has been known to alter chromatin domains that can activate gene transcription (27), and several studies have demonstrated that SIN3A is required for the expression of particular genes (9, 13, 28–34). In some of these cases, SIN3A has been shown to bind the gene in question (29, 31–34). Our studies now demonstrate that rapid recruitment of SIN3A to the corresponding enhancer and proximal promoter regions occurs after transcriptional activation of a highly inducible gene (CYP1A1).
In our previous paper (19), we reported on experiments in which we cotransfected Hepa-1 cells with an siRNA to SIN3A and a pGL-CYP1A1 firefly luciferase reporter construct. After 24 h of treatment with TCDD, we looked at luciferase expression, which was under control of the CYP1A1 promoter/enhancer region, to determine whether SIN3A exerts its effects on the 5′ upstream regulatory region of CYP1A1. Luciferase expression was diminished significantly but only modestly in cells treated with the siRNA targeting SIN3A. Because the pGL-CYP1A1 construct is not in the native/endogenous chromatin configuration in transfected cells, the only modest siRNA-induced diminution in the luciferase signal that we observed may be a consequence of SIN3A or its downstream effectors being required for chromatin remodeling.
Structural analysis of SIN3A has revealed the presence of four paired α-helices (PAH), a histone interaction domain, and a highly conserved region. As a scaffold protein, different interaction regions on SIN3A have been established for various proteins. HDACs and SIN3A-associated proteins bind to the histone interaction domain motif of SIN3A, whereas the highly conserved region motif is considered important for binding to proteins necessary for repression, such as the nuclear receptor hormone corepressor Alien (35, 36). The PAH domains, however, are thought to represent the main interacting domains of SIN3A, and PAH2 has been identified as being able to bind transcription factors. SIN3A could associate indirectly with the AHR/ARNT dimer via one or more of the many transcriptional coactivator proteins known to interact with this dimer (8, 37). Alternatively, SIN3A could directly bind AHR and/or ARNT. Of interest, the structurally similar PAH motifs of SIN3A and the basic helix-loop-helix domains of certain transcription factors mediate their interaction. For example, HERP1 and HERP2 interact with SIN3A and HDAC1 through their basic helix-loop-helix domains (38). These data suggest the possibility that SIN3A may also bind AHR and/or ARNT, via the latter proteins' basic helix-loop-helix motifs. Supporting this notion, previous studies by Lee et al. (39) found that they could co-immunoprecipitate SDS3, a component of the SIN3-HDAC complex, using an anti-ARNT antibody and that this association occurred only in cells treated with TCDD. Additional experiments could confirm whether a similar interaction occurs between SIN3A and AHR.
Our siRNA screening library contained siRNAs to several subunits of the SIN3A complex, including Rbbp4, Rbbp7, Sap18, and Sap30. However, these siRNAs did not significantly reduce induction of CYP1A1 EROD activity (19). It is possible that these subunits are not essential for CYP1A1 induction. Alternatively, the corresponding siRNAs may not have been effective in the screen, as indicated, for example, by our data obtained for Rbbp4 and Rppb7, for which relatively low Z-prime values (representing the quality of the data in the original siRNA screen) were obtained. Our library also contained siRNAs targeting HDAC1 and HDAC2, and despite their high Z-prime values of 0.71 and 0.75, none of the four siRNAs to each of these proteins represented a hit in our screening, suggesting that these proteins may not be involved in CYP1A1 induction. Interestingly, Schnekenburger et al. (40) demonstrated that knocking down HDAC1 had no effect on induced or uninduced CYP1A1 expression. Despite these observations, they found that HDAC1 bound to the proximal promoter of the CYP1A1 gene in Hepa-1 cells and that this binding decreased on AHR agonist treatment (40). These last data, however, conflict with those of Suzuki and Nohara (41), who demonstrated that upon exposure to TCDD, the level of HDAC1 slightly increased at the promoter of CYP1A1 in Hepa-1 cells. Besides HDACs, SIN3A has been shown to form complexes with several other enzymatic proteins exhibiting histone methylation, DNA methylation, chromatin remodeling, or N-acetylglucoseamine transferase activities (42). Thus, it is possible that the role of SIN3A in TCDD induction of CYP1A1 is a result of enzymatic activity other than HDAC. Furthermore, two independent laboratories found that SIN3A associates with subunits of the BRG-based SWI/SNF chromatin remodeling complex (43, 44). We previously reported the functional involvement of BRG-1 in TCDD induction of CYP1A1 transcription (45). The association of BRG-1 with SIN3A could be the means by which SIN3A is required for CYP1A1 induction. In summary, this study establishes an essential role for SIN3A in AHR-mediated CYP1A1 induction and demonstrates that SIN3A can act as a transcriptional coactivator of a highly inducible gene.
Acknowledgments
We thank Drs. Ilona Bebenek, Peter Bui, and Sudheer Beedanagari for advice. FACS was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and the Center for AIDS Research Flow Cytometry Core Facility, which is supported by National Institutes of Health Grants CA-16042 and AI-28697, the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA28868 and R01ES015384 (to O. H.).
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- AHR
- aryl hydrocarbon receptor
- ARNT
- aryl hydrocarbon nuclear translocator
- EROD
- ethoxyresorufin-o-deethylase
- PAH
- paired α-helices
- HDAC
- histone deacetylase.
REFERENCES
- 1. Hankinson O. (1995) The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340 [DOI] [PubMed] [Google Scholar]
- 2. Ma Q., Lu A. Y. (2007) CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab. Dispos. 35, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 3. Mandal P. K. (2005) Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J. Comp. Physiol. B 175, 221–230 [DOI] [PubMed] [Google Scholar]
- 4. Monostory K., Pascussi J. M., Kóbori L., Dvorak Z. (2009) Hormonal regulation of CYP1A expression. Drug Metab. Rev. 41, 547–572 [DOI] [PubMed] [Google Scholar]
- 5. Nukaya M., Bradfield C. A. (2009) Conserved genomic structure of the Cyp1a1 and Cyp1a2 loci and their dioxin responsive elements cluster. Biochem. Pharmacol. 77, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Androutsopoulos V. P., Tsatsakis A. M., Spandidos D. A. (2009) Cytochrome P450 CYP1A1: wider roles in cancer progression and prevention. BMC Cancer 9, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujii-Kuriyama Y., Kawajiri K. (2010) Molecular mechanisms of the physiological functions of the aryl hydrocarbon (dioxin) receptor, a multifunctional regulator that senses and responds to environmental stimuli. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 40–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hankinson O. (2005) Role of coactivators in transcriptional activation by the aryl hydrocarbon receptor. Arch. Biochem. Biophys. 433, 379–386 [DOI] [PubMed] [Google Scholar]
- 9. Silverstein R. A., Ekwall K. (2005) Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17 [DOI] [PubMed] [Google Scholar]
- 10. Dannenberg J. H., David G., Zhong S., van der Torre J., Wong W. H., Depinho R. A. (2005) mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 19, 1581–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grzenda A., Lomberk G., Zhang J. S., Urrutia R. (2009) Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta 1789, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 13. Kadamb R., Mittal S., Bansal N., Batra H., Saluja D. (2013) Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 92, 237–246 [DOI] [PubMed] [Google Scholar]
- 14. Wang F., Shi S., Zhang R., Hankinson O. (2006) Identifying target genes of the aryl hydrocarbon receptor nuclear translocator (Arnt) using DNA microarray analysis. Biol. Chem. 387, 1215–1218 [DOI] [PubMed] [Google Scholar]
- 15. Wang F., Zhang R., Shi S., Hankinson O. (2007) The effect of aromatic hydrocarbon receptor on the phenotype of the Hepa 1c1c7 murine hepatoma cells in the absence of dioxin. Gene Regul. Syst. Bio. 1, 49–56 [PMC free article] [PubMed] [Google Scholar]
- 16. Beedanagari S. R., Taylor R. T., Hankinson O. (2010) Differential regulation of the dioxin-induced Cyp1a1 and Cyp1b1 genes in mouse hepatoma and fibroblast cell lines. Toxicol. Lett. 194, 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang S., Ge K., Roeder R. G., Hankinson O. (2004) Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J. Biol. Chem. 279, 13593–13600 [DOI] [PubMed] [Google Scholar]
- 18. Beedanagari S. R., Bebenek I., Bui P., Hankinson O. (2009) Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol. Sci. 110, 61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Solaimani P., Damoiseaux R., Hankinson O. (2013) Genome-wide RNAi high-throughput screen identifies proteins necessary for the AHR-dependent induction of CYP1A1 by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 136, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davarinos N. A., Pollenz R. S. (1999) Aryl hydrocarbon receptor imported into the nucleus following ligand binding is rapidly degraded via the cytosplasmic proteasome following nuclear export. J. Biol. Chem. 274, 28708–28715 [DOI] [PubMed] [Google Scholar]
- 21. Giannone J. V., Li W., Probst M., Okey A. B. (1998) Prolonged depletion of AH receptor without alteration of receptor mRNA levels after treatment of cells in culture with 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem. Pharmacol. 55, 489–497 [DOI] [PubMed] [Google Scholar]
- 22. Ma Q., Renzelli A. J., Baldwin K. T., Antonini J. M. (2000) Superinduction of CYP1A1 gene expression. Regulation of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced degradation of Ah receptor by cycloheximide. J. Biol. Chem. 275, 12676–12683 [DOI] [PubMed] [Google Scholar]
- 23. Hestermann E. V., Brown M. (2003) Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol. Cell Biol. 23, 7920–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matthews J., Wihlén B., Heldring N., MacPherson L., Helguero L., Treuter E., Haldosén L. A., Gustafsson J. A. (2007) Co-planar 3,3′,4,4′,5-pentachlorinated biphenyl and non-co-planar 2,2′,4,6,6-pentachlorinated biphenyl differentially induce recruitment of oestrogen receptor α to aryl hydrocarbon receptor target genes. Biochem. J. 406, 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taylor R. T., Wang F., Hsu E. L., Hankinson O. (2009) Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicol. Sci. 107, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wihlén B., Ahmed S., Inzunza J., Matthews J. (2009) Estrogen receptor subtype and promoter-specific modulation of aryl hydrocarbon receptor-dependent transcription. Mol. Cancer Res. 7, 977–986 [DOI] [PubMed] [Google Scholar]
- 27. Fischle W., Wang Y., Allis C. D. (2003) Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183 [DOI] [PubMed] [Google Scholar]
- 28. Yoshimoto H., Ohmae M., Yamashita I. (1992) The Saccharomyces cerevisiae GAM2/SIN3 protein plays a role in both activation and repression of transcription. Mol. Gen. Genet. 233, 327–330 [DOI] [PubMed] [Google Scholar]
- 29. De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., Posas F. (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374 [DOI] [PubMed] [Google Scholar]
- 30. Farhana L., Dawson M. I., Dannenberg J. H., Xu L., Fontana J. A. (2009) SHP and Sin3A expression are essential for adamantyl-substituted retinoid-related molecule-mediated nuclear factor-kappaB activation, c-Fos/c-Jun expression, and cellular apoptosis. Mol. Cancer Ther. 8, 1625–1635 [DOI] [PubMed] [Google Scholar]
- 31. Baltus G. A., Kowalski M. P., Tutter A. V., Kadam S. (2009) A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. J. Biol. Chem. 284, 6998–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Oevelen C., Bowman C., Pellegrino J., Asp P., Cheng J., Parisi F., Micsinai M., Kluger Y., Chu A., Blais A., David G., Dynlacht B. D. (2010) The mammalian Sin3 proteins are required for muscle development and sarcomere specification. Mol. Cell Biol. 30, 5686–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ythier D., Larrieu D., Binet R., Binda O., Brambilla C., Gazzeri S., Pedeux R. (2010) Sumoylation of ING2 regulates the transcription mediated by Sin3A. Oncogene 29, 5946–5956 [DOI] [PubMed] [Google Scholar]
- 34. Das T. K., Sangodkar J., Negre N., Narla G., Cagan R. L. (2013) Sin3a acts through a multi-gene module to regulate invasion in Drosophila and human tumors. Ocogene 32, 3184–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fleischer T. C., Yun U. J., Ayer D. E. (2003) Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell Biol. 23, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moehren U., Dressel U., Reeb C. A., Väisänen S., Dunlop T. W., Carlberg C., Baniahmad A. (2004) The highly conserved region of the co-repressor Sin3A functionally interacts with the co-repressor Alien. Nucleic Acids Res. 32, 2995–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hankinson O. (2012) in The AH Receptor in Biology and Toxicology, 1st Ed (Pohjanvirta R., ed) pp. 93–100, John Wiley & Sons, Inc., New York [Google Scholar]
- 38. Iso T., Sartorelli V., Poizat C., Iezzi S., Wu H. Y., Chung G., Kedes L., Hamamori Y. (2001) HERP, a novel heterodimer partner of HES/E(spl) in Notch signaling. Mol. Cell Biol. 21, 6080–6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee D. M., Lee S. H., Jeong K. T., Hwang S. J., Park J. H. (2012) SDS3 interacts with ARNT in an AhR ligand-specific manner regulating expression of cKrox and S100A4 in CD4+CD8+ DPK thymocytes differentiation. Environ. Toxicol. Pharmacol. 34, 858–868 [DOI] [PubMed] [Google Scholar]
- 40. Schnekenburger M., Peng L., Puga A. (2007) HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim. Biophys. Acta 1769, 569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki T., Nohara K. (2007) Regulatory factors involved in species-specific modulation of arylhydrocarbon receptor (AhR)-dependent gene expression in humans and mice. J. Biochem. 142, 443–452 [DOI] [PubMed] [Google Scholar]
- 42. Ellison-Zelski S. J., Alarid E. T. (2010) Maximum growth and survival of estrogen receptor-α positive breast cancer cells requires the Sin3A transcriptional repressor. Mol. Cancer 9, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuzmichev A., Zhang Y., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell Biol. 22, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sif S., Saurin A. J., Imbalzano A. N., Kingston R. E. (2001) Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15, 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang S., Hankinson O. (2002) Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J. Biol. Chem. 277, 11821–11827 [DOI] [PubMed] [Google Scholar]