Abstract
We introduce the antibody landscape, a method for the quantitative analysis of antibody-mediated immunity to antigenically variable pathogens, achieved by accounting for antigenic variation among pathogen strains. We generated antibody landscapes to study immune profiles covering 43 years of influenza A/H3N2 virus evolution for 69 individuals monitored for infection over six years and for 225 individuals pre- and post-vaccination. On infection and vaccination titers increased broadly, including previously encountered viruses far beyond the extent of cross-reactivity observed after a primary infection. We explored implications for vaccination, and found that use of an antigenically advanced virus had the dual benefit of inducing antibodies against both advanced and previous antigenic clusters. These results indicate that pre-emptive vaccine updates may improve influenza vaccine efficacy in previously-exposed individuals.
Much of the global burden of infectious disease today is caused by antigenically variable pathogens, which escape immunity induced by prior infection or vaccination by changing the molecular structure recognized by antibodies. Human influenza viruses are notorious for their capacity to evolve and evade the adaptive immune response. This evolution has been progressive and step-wise (fig. S1)(1), with antigenically similar viruses circulating for a few years before strains with related but novel antigenic characteristics replace them (2). As a result, vaccine strain updates, based on analyses of circulating viruses, are necessary to maintain vaccine effectiveness.
The current vaccine strain selection strategy is to choose a virus that is antigenically representative of circulating viruses, mostly determined by testing a global selection of virus isolates against a panel of ferret antisera using the hemagglutination inhibition (HI) assay (3). The ferrets used in such studies are influenza-naïve prior to inoculation, and each antiserum has been raised by infection with only a single virus. Such post-inoculation ferret antisera provide well-understood data for the characterization of antigenic differences between influenza viruses (2, 4). However, this strategy does not account for the influence of prior immunity on the response induced by the vaccine when administered to humans.
The direct analysis of human serological data presents an opportunity to assess and understand immune responses in the context of differing background immunity and to use this information as the basis for improved vaccine strain selection and evaluation. Indeed, such data are used in the vaccine strain selection process. Unfortunately, immunological patterns in human serological data are difficult to interpret because of complex, and usually unknown, exposure histories and the confounding factor of cross-reactivity due to antigenic relationships among strains. As a result, in-depth analyses of serological data have been difficult and, despite excellent cross-sectional seroepidemiology (5), our understanding of the typical characteristics of the human serological response to infection and vaccination has remained limited.
Results from the original, and seminal, studies on the antibody-mediated immune response to influenza virus infection and vaccination in humans (6-9) have often been interpreted as “original antigenic sin” — a hypothesis that proposes an anamnestic reinforcement of the level of antibody to the strain that first infected the individual that dampens the serologic response to the current virus (9-11). This definition is, however, far from concrete and the historical literature on the effect of immune memory on the generation of responses to variant antigens has been particularly equivocal.
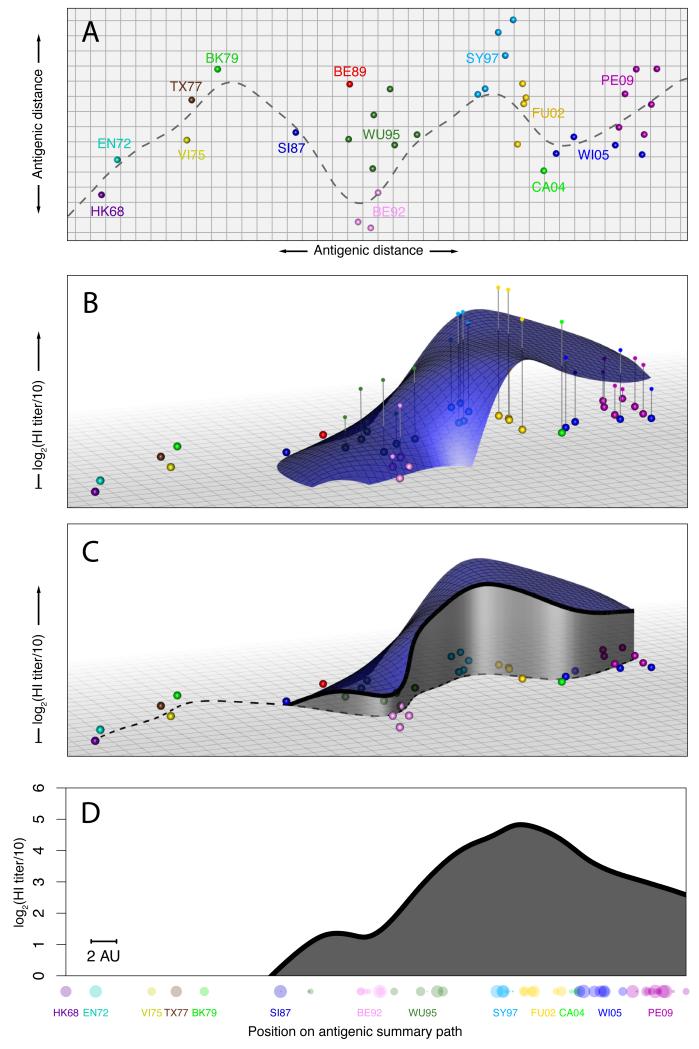
To increase our ability to quantitatively study human serological data of antigenically variable pathogens, we present a methodology that enables detailed analyses and visualization of complex serological data by plotting antibody-mediated immunity as a function of the antigenic relationships among viruses. To achieve this, we first used antigenic cartography (2) to determine the antigenic relationships among a selection of 81 viruses spanning 43 years of influenza A/H3N2 evolution, using HI titrations of first-infection ferret sera (Fig. 1A, fig. S2, Tables S1 and S8). Human serum samples were then titrated against the same viruses and their HI titers plotted in an extra dimension added to the antigenic map (Fig. 1B).
Fig. 1.
Creating an antibody landscape. (A) Antigenic map of A/H3N2 showing virus strains color-coded by antigenic cluster. Both axes represent antigenic distance, the spacing between grid lines is 1 antigenic unit, corresponding to a twofold dilution of antiserum in the HI assay. Two units correspond to fourfold dilution, three units to eightfold dilution, and so on (2). The gray line shows a path through the antigenic clusters in chronological order calculated by fitting a smoothing spline (1). (B) An additional dimension indicates the measured antibody titers as vertical impulses and a smooth surface is fitted using locally weighted multiple linear regression to create the antibody landscape within the convex hull bounded by the viruses titrated (RMSE of fit = 1.23 HI log2-units). (C) The height of the landscape along the path in (A) shows a slice through the landscape (1). (D) The height of the landscape along the antigenic summary path is plotted to create a rotation-independent 2D summary visualization of the landscape. Titrated virus strains are shown in their corresponding positions along the x-axis, symbol radius is inversely proportional to antigenic distance from the path, symbol color indicates antigenic cluster. The scale bar indicates 2 antigen units; each antigenic unit is a 2-fold dilution in the HI assay.
We found that HI titers of a given serum are related for antigenically similar viruses (fig. S3), and thus a representative smooth surface could be fitted through these HI titers. The resulting antibody landscape represents an immune profile for each serum with elevations corresponding to regions in the antigenic map with higher antibody levels (figs. S4-S5, S13). Since the landscape at any given point is a function of surrounding data points, antibody levels can be inferred for viruses not included in the titration set. For antibody landscapes of influenza A/H3N2 based on the HI assay, we found that the landscape predicted omitted HI titers with a root-mean-square error of 1.3 log2-units, compared to an estimated error arising from HI assay repeatability alone of 0.9 (Table S10, figs. S6-S11, S14).
To aid the visual comparison of multiple landscapes, we used a path on the antigenic map that passes through each antigenic cluster in chronological order (Fig. 1C). The corresponding values of the landscape along this summary path were used to represent the three-dimensional landscape in two dimensions (Fig. 1D and fig. S12).
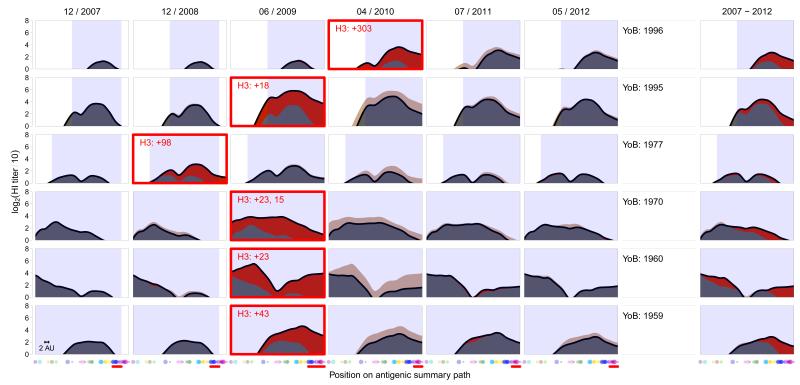
We used this methodology to study serological data we generated from samples taken annually between 2007 and 2012 from unvaccinated individuals in the Ha Nam household cohort study in Vietnam (12). More than 10,000 HI titrations were performed to construct a total of 324 landscapes for 69 individuals born between 1917 and 2005, allowing us to assess the serological changes over time (Fig. 2, Tables S3, S4, fig. S15). Titers were highest for influenza viruses that circulated when an individual was approximately 6 years old (figs. S42-S43), corresponding with the time-frame of first infection (13). Antibody levels against newly circulating viruses tended to be lower than against strains circulating earlier in an individual’s lifetime, as reported previously (5,7-9,11). In addition, previous results found some cross-reactivity to strains that circulated before an individual’s birth (5, 7-9,14) and based on the extent of detectable titers to viruses in circulation only before an individual’s birth, we quantified this antibody cross-reactivity to be 0-2 antigenic clusters (Table S11). There was substantial heterogeneity among the antibody landscapes of different individuals; however, each individual’s landscape shape was typically stable from one year to the next and had distinctive individual features (within-person r=0.86 (standard deviation±0.22), between-person r=0.28±0.21, figs. S16-S20).
Fig. 2.
Antibody landscapes from 2007-2012 for six individuals. The black line represents the landscape height for each position on the antigenic summary path through the antigenic clusters from Fig. 1A. The first sample taken after a confirmed A/H3N2 influenza virus infection is marked with a red box, and the red number gives the days from the start of influenza-like illness to serum collection. The red shading indicates increases, and beige decreases, compared to the previous year. The blue-shaded area indicates antigenic clusters that circulated during an individual’s lifespan until sample collection (Table S9). Dots along the x-axis indicate the subset of 30 viruses used to generate these landscapes - contemporary strains likely causing the infection are indicated with a red horizontal bar (Table S2). The rightmost column shows the difference between the landscape in 2012 compared to 2007. The scale bar indicates 2 antigenic units.
Infection with A/H3N2 resulted in a strikingly broad antibody response (Fig. 2 and figs. S21-S22) that was typically governed by the extent of the pre-exposure antibody landscape (fig. S45). This antibody response far exceeded the extent of cross-reactivity typically produced in the response following primary exposure with one of the circulating viruses (Fig S44, S47). For example, an individual born in 1970, infected in 2009 (Fig. 2, third row), had a substantial long-distance response back to the Hong Kong 1968 (HK68) antigenic cluster and all clusters in between, even though these older viruses had not circulated for decades. To illustrate the substantial breadth of this back-boost, there have been 13 antigenic cluster transitions from HK68 until Perth 2009 (PE09), each approximately 4.5 antigenic units (corresponding to a 24-fold dilution of antiserum in the HI assay). These antigenic changes have necessitated over 20 vaccine strain updates, and are the result of changes in 69 of the 346 amino acid positions in the HA1 domain of the hemagglutinin gene between HK68 and the PE09 vaccine strain, including substitutions in all of the seven key antigenic positions identified by Koel et al. (15).
Because of the range of this response, and its dependence on the pre-exposure antibody landscape, we call it a “back-boost”. The magnitude of back-boost response declined with antigenic distance from the likely infecting virus (fig. S46). Although the response to older viruses was substantial, titer increases were largest for viruses from the contemporary antigenic cluster, in contrast to a common interpretation of the original antigenic sin hypothesis (fig. S47). Polymerase chain reaction confirmed infections with influenza B, A/H1N1 and A/H1N1(pdm09) often caused negligible changes in the A/H3N2 antibody landscape (fig. S23), indicating that the back-boost is type and subtype-specific.
Typically, the broad initial response was followed by a period of titer decay during which antibody titers stabilized to form an altered antibody landscape over the course of approximately one year (fig. S24). Comparison of the antibody landscapes of 2007 and 2012 (Fig. 2) shows that the antigenic region for which increased titers were maintained long-term was substantially narrower than that of the initial response to infection. This long-term persistence of increased antibody titers was more specific to the antigenic region of the likely infecting strain, but still spanned multiple antigenic clusters (fig. S46).
Next, we investigated whether the back-boost observed following infection could be used to improve vaccine effectiveness. In the vaccine strain selection process, it is sometimes unclear whether currently circulating strains or antigenically novel strains are most likely to predominate in the next influenza season. The resulting dilemma is whether it is more beneficial to leave the vaccine strain unchanged, or to pre-emptively update the vaccine to match a novel strain, without certainty over which antigenic cluster of viruses will indeed circulate.
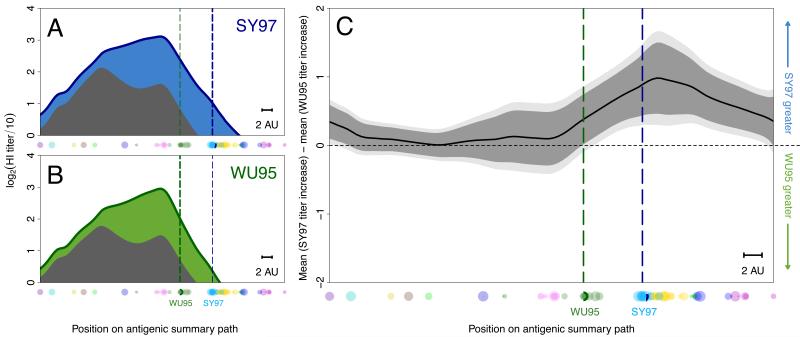
It would take a large, prospective, multi-year clinical trial comparing the two vaccination approaches to answer these questions definitively. However, we were able to retrospectively test the approach with the sera of 225 human vaccinees from two annual influenza vaccine re-registration studies, by identifying an antigenic cluster transition for which there was little circulation of the new cluster before a novel vaccine strain was first tested. Both groups had therefore received antigenically different vaccines, and yet there was no significant difference in the average pre-vaccination antibody landscapes of the two studies (figs. S30-S31). Individuals in the first study (n=102, Table S6), performed in 1997, received the A/Nanchang/933/95 vaccine from the Wuhan 95 (WU95) antigenic cluster to which there had been some prior exposure, whereas individuals in the 1998 study (n=123, Table S7) received the A/Sydney/5/97 vaccine from the antigenically advanced Sydney 97 (SY97) cluster to which there was substantially less pre-vaccination immunity – thus mimicking a pre-emptive update.
Individual antibody landscapes were constructed from serum samples taken pre-vaccination and four weeks post-vaccination (figs. S25-26, Table S5) and combined to give overall pre-vaccination and post-vaccination antibody landscapes (Fig. 3A,B). As expected following a vaccine update, average vaccination responses were significantly greater against later antigenic clusters following vaccination with the antigenically advanced SY97 strain (figs. S32-S35). The back-boost following infection was also observed for the vaccination studies, and interestingly, the magnitude and breadth of the response to infection and vaccination were comparable (figs. S27-S29). Indeed, the back-boost in the SY97-vaccine study resulted in a slightly larger response to WU95 viruses than the response in the WU95-vaccine study (Fig. 3C). These findings also held when studying only elderly individuals (fig. S36), and individuals with a low pre-vaccination titer against WU95, typically considered the most susceptible (fig S37-S38) (16). We further tested a subset of vaccination sera with a neutralization assay, and these data support the results from the HI assay (figs. S40-S41). Despite differences in pre-vaccination landscapes, a second study of the WI05-PE09 cluster transition also demonstrated a similar back-boost upon vaccination (fig. S39).
Fig. 3.
Comparison of two different vaccines. (A) The mean pre-vaccination landscape (gray) and landscape after vaccination with A/Sydney/5/97 (blue) in the 1998 study (123 individuals), or (B) with A/Nanchang/933/95 (green) in the 1997 study (102 individuals) for each position on the antigenic summary path. Dots along the x-axis indicate the subset of 70 viruses used to generate these landscapes. The vertical dotted lines indicate the position of the SY97 (blue) and WU95 (green) wild type vaccine viruses. (C) Comparison of titer increase after vaccination with A/Nanchang/933/95 or A/Sydney/5/97 for each position along the antigenic summary path. Above the horizontal midpoint indicates higher response to the A/Sydney/5/97 vaccine, below to the A/Nanchang/933/95 vaccine. Data were calculated from the average titer increase between each individual’s paired post-vaccination and pre-vaccination titers, with 95% (dark gray) and 99% (light gray) t-test based confidence intervals. The scale bar indicates 2 antigenic units.
The mechanism behind the broad back-boost is currently unknown, but we considered several hypotheses (1). In summary, rather than resulting from the production of novel antibodies with extensive cross-reactivity, the back-boost appears most consistent with memory-cell stimulation and antibody recall. This pattern of recall is consistent with raw data from the mid-20th century studies on the response to infection or vaccination where studies on antigenically different A/H1N1 strains also show a broad sub-type specific back-boost (6, 8-9). However, this phenomenon was never quantified and put in relation to the antigenic difference among the viruses.
Whether the original antigenic sin hypothesis refers to higher pre-exposure antibody titers, or also to a higher response to the first infecting virus is unclear, and both interpretations have been used over the past 60 years (17). We found no evidence for a predisposition in the antibody response towards the likely first infecting strain, and instead, we demonstrate that the increase in antibody titers is greatest to the most recently encountered strain. We do, however, corroborate the finding that pre-exposure antibody reactivity tends to be highest against strains encountered earlier in life (fig. S37) (5, 7-9, 11). The presence of higher pre-exposure static titers, but not higher dynamic responses, to the first infecting strain may explain seemingly contradictory reports whereby cross-sectional studies have tended to describe a serological bias supportive of the original antigenic sin dogma (5, 11) while investigations into actual responses upon exposure frequently oppose it (17,18).
These findings also shed light on the growth of the serological immunity over time. Although responses were often present against the oldest strains, these long-distance back-boosts were typically not maintained beyond a year (Fig 2. right panel, fig S24). This is evidence against the hypothesis of long-term and progressive “reinforcement” of antibody titers against earlier viruses upon exposure to each subsequent antigenic variant over time. Instead, the pattern of higher static titers against antigenic clusters encountered early in life may also be explained if the immune response to primary exposure is larger than the responses to subsequent exposures (Fig S48).
As others have speculated, it is plausible that the decreased antibody responses to subsequent exposures may be a result of “antigen trapping”, a hypothesis according to which binding of antigen by pre-existing cross-reactive antibodies and memory-cells decreases the antigenic load available for priming naïve B-cells and leads to a diminished novel response (5, 7, 10, 19-20). This would also explain why the closest antigenic match between the vaccine strain and the circulating strains does not necessarily generate the best antibody response against the corresponding cluster: the mismatch of an antigenically advanced strain is compensated for by a greater novel response, as a result of reduced antigen trapping (21). The extent of interference by antigen trapping on the novel antibody response depends on the degree of antigenic relatedness and prior immunity (22). Note, when individuals have no prior immunity to a subtype, such as young children, or in a pandemic, the best vaccine is likely the closest antigenic match as there will be no prior immunity to avoid and no back-boost to exploit.
These findings highlight potentially important differences between the two types of vaccine mismatch in populations with prior immunity. Following a mismatch due to a delayed vaccine update (in which the vaccine strain, selected 10-14 months before the season in which it is used, lags behind influenza virus evolution), neither pre-existing nor newly induced antibodies provide immunity against the novel strains. Consequently, such vaccines have poor effectiveness in this mismatch situation (23-26). However, if there were a vaccine mismatch due to an incorrectly timed, pre-emptive antigenic update of the vaccine, then the data from our retrospective surrogate study indicate that the extensive back-boost would still induce equivalent titers against previous antigenic strains. Such vaccines would have the dual advantage of being effective against the antigenically novel viruses to which they were targeted while remaining effective, or being even more effective, for contemporary viruses if they continued to circulate.
Our results underscore the importance of accounting for antigenic variation to better understand multi-exposure sera, and provide a methodology for the direct visualization of otherwise complex serological patterns, allowing basic insights into the breadth of the adaptive humoral immune response to influenza and other antigenically variable pathogens. Antibody landscapes will be useful for the evaluation of evolutionary selection pressures (fig. S49) and the evaluation of different vaccination techniques, including the effect of adjuvants, vaccine composition, dose sparing, and the durability, breadth and magnitude of responses to universal vaccines. Our results indicate that pre-emptive vaccine updates may substantially improve influenza vaccine effectiveness in previously-exposed individuals. Prospective clinical trials will further test the breadth and longevity of the immunological response and protection provided by antigenically advanced vaccine strains.
Supplementary Material
Acknowledgments
We thank R. Bodewes, J. Bryant, D. Burke, N. Lewis, E. Selkov, B. Mühlemann, G. de Mutsert and F. Pistoor. We also thank the staff of the Ha Nam Provincial Preventive Medicine Centre, the Hamlet health workers, the National Institute for Hygiene and Epidemiology, Viet Nam for their support in conducting the fieldwork. We are indebted to the cooperation of the Ha Nam cohort and vaccine study participants. JMF is supported an Medical Research Council Fellowship grant MR/K021885/1 and a Junior Research Fellowship from Homerton College, LCK by the Gates-Cambridge Scholarship and NIH Oxford-Cambridge Scholars program, CAR by a Royal Society URF (RG55423). We acknowledge the NIAID-NIH Centers of Excellence for Influenza Research and Surveillance contracts HHSN266200700010C and HHSN272201400008C, Nederlandse Organisatie voor Wetenschappelijk Onderzoek VICI grant 91896613, the European Union FP7 programs EMPERIE (223498) and ANTIGONE (278976), Human Frontier Science Program grant P0050/2008, Wellcome Trust (WT087982MA) and NIH Director’s Pioneer Award DP1-OD000490-01. The Melbourne WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health. ADMEO (as Chief Scientific Officer of Viroclinics Biosciences BV) has advisory affiliations with GlaxoSmithKline, Novartis, and Roche. Sequences of the influenza viruses used in this study will be made available on Genbank.
Footnotes
References and Notes
- 1.Materials and methods are available as supplementary materials on Science Online.
- 2.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 3.Hirst GK. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 1942;75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr IG, et al. Epidemiological, antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: Basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009-2010 Northern Hemisphere season. Vaccine. 2010;28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 5.Lessler J, et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in Southern China. PLoS Pathog. 2012;8:e1002802. doi: 10.1371/journal.ppat.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horsfall FL, Rickard ER. Neutralizing antibodies in human serum after influenza A. The lack of strain specificity in the immunological response. J. Exp. Med. 1941;74:433–439. doi: 10.1084/jem.74.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennessy AV, Davenport FM, Francis T., Jr. Studies of antibodies to strains of influenza virus in persons of different ages in sera collected in a postepidemic period. J. Immunol. 1955;75:401–409. [PubMed] [Google Scholar]
- 8.Davenport FM, Hennessy AV, Francis T., Jr. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 1953;98:641–656. doi: 10.1084/jem.98.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davenport FM, Hennessy AV. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J. Exp. Med. 1956;104:85–97. doi: 10.1084/jem.104.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazekas de St. Groth S, Webster RG. Disquisitions on original antigenic sin; I. Evidence in man. J. Exp. Med. 1966;124:331–345. doi: 10.1084/jem.124.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis T., Jr. On the doctrine of original antigenic sin. P. Am. Philos. Soc. 1960;104:572–578. [Google Scholar]
- 12.Horby P, et al. The epidemiology of interpandemic and pandemic influenza in Vietnam, 2007-2010: the Ha Nam household cohort study I. Am. J. Epidemiol. 2012;175:1062–1074. doi: 10.1093/aje/kws121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodewes R, et al. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 2011;18:469–476. doi: 10.1128/CVI.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davenport FM, Hennessy AV, Stuart-Harris CH, Francis T. Epidemiology of influenza. Comparative serological observations in England the United States. Lancet. 1955;266:469–747. doi: 10.1016/s0140-6736(55)93328-6. [DOI] [PubMed] [Google Scholar]
- 15.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 16.Coudeville L, et al. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010;10 doi: 10.1186/1471-2288-10-18. doi:10.1186/1471-2288-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donnell CD, et al. Humans and ferrets with prior H1N1 influenza virus infections do not exhibit evidence of original antigenic sin after infection or vaccination with the 2009 pandemic H1N1 influenza virus. Clin. Vaccine Immunol. 2014;21:737–746. doi: 10.1128/CVI.00790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JH, Skountzou I, Compans R, Jacob J. Original antigenic sin responses to influenza viruses. J. Immunol. 2009;183:3294–3301. doi: 10.4049/jimmunol.0900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MS, et al. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci. Transl. Med. 2013;5:198ra107. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14001–14006. doi: 10.1073/pnas.96.24.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan K. Understanding original antigenic sin in influenza with a dynamical system. PLoS ONE. 2011;6:e23910. doi: 10.1371/journal.pone.0023910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis T, Salk JE, Quilligan JJ. Experiences with vaccination against influenza in the spring of 1947: a preliminary report. Am. J. Public Health Nations Health. 1947;37:1013–1016. doi: 10.2105/ajph.37.8.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belongia EA, et al. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004-2005 season to the 2006-2007 season. J. Infect. Dis. 2009;199:159–167. doi: 10.1086/595861. [DOI] [PubMed] [Google Scholar]
- 25.Skowronski DM, et al. Component-specific effectiveness of trivalent influenza vaccine as monitored through a sentinel surveillance network in Canada. J. Infect. Dis. 2009;199:168–179. doi: 10.1086/595862. [DOI] [PubMed] [Google Scholar]
- 26.Bridges CB, et al. Effectiveness and cost-benefit of influenza vaccination of healthy working adults; a randomized controlled trial. J. Am. Med. Assoc. 2000;284:1655–1663. doi: 10.1001/jama.284.13.1655. [DOI] [PubMed] [Google Scholar]
- 27.Beyer WEP, Palache AM, Lüchters G, Nauta J, Osterhaus ADME. Seroprotection rate, mean fold increase, seroconversion rate: which parameter adequately expresses seroresponse to influenza vaccination? Virus Res. 2004;103:125–132. doi: 10.1016/j.virusres.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Kennard RW, Stone LA. Computer aided design of experiments. Technometrics. 1969;11:137–148. [Google Scholar]
- 29.Cauchemez S, et al. Influenza infection rates, measurement errors and the interpretation of paired serology. PLoS Pathog. 2012;8:e1003061. doi: 10.1371/journal.ppat.1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hannoun C, Megas F, Piercy J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res. 2004;103:133–138. doi: 10.1016/j.virusres.2004.02.025. [DOI] [PubMed] [Google Scholar]
- 31.WHO Collaborating Centres for Reference and Research on Influenza, Atlanta, London and Melbourne, Influenza: antigenic analysis of recent influenza virus isolates and influenza activity in the southern hemisphere. Weekly Epidemiological Record. 1997;72:293. [PubMed] [Google Scholar]
- 32.World Health Organization Recommended composition of influenza virus vaccines for use in the 2010 influenza season (southern hemisphere winter) Weekly Epidemiological Record. 2009;84:421–432. [PubMed] [Google Scholar]
- 33.Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webster RG, Laver WG, Air GM, Schild GC. Molecular mechanisms of variation in influenza viruses. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 35.Fitch WM, Leiter JME, Li X, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.