Summary
Impaired carbon precursor supply through cpPDC is detrimental for TAG biosynthesis in Chlamydomonas reinhardtii under conditions of photoautotrophy and nitrogen starvation.
Key words: Acetyl-CoA, chlorophyll fluorescence transients, fatty acid synthesis, lipid, microalgae, photosystem II, pyruvate dehydrogenase.
Abstract
The chloroplast pyruvate dehydrogenase complex (cpPDC) catalyses the oxidative decarboxylation of pyruvate forming acetyl-CoA, an immediate primer for the initial reactions of de novo fatty acid (FA) synthesis. Little is known about the source of acetyl-CoA in the chloroplasts of photosynthetic microalgae, which are capable of producing high amounts of the storage lipid triacylglycerol (TAG) under conditions of nutrient stresses. We generated Chlamydomonas reinhardtii CC-1618 mutants with decreased expression of the PDC2_E1α gene, encoding the putative chloroplast pyruvate dehydrogenase subunit E1α, using artificial microRNA. A comparative study on the effects of PDC2_E1α silencing on FAs and TAG production in C. reinhardtii, grown photoautotrophically and mixotrophically, with and without a nitrogen source in the nutrient medium, was carried out. Reduced expression of PDC2 _E1α led to a severely hampered photoautotrophic growth phenotype with drastic impairment in TAG accumulation under nitrogen deprivation. In the presence of acetate, downregulation of PDC2_E1α exerted little to no effect on TAG production and photosynthetic activity. In contrast, under photoautotrophic conditions, especially in the absence of a nitrogen source, a dramatic decline in photosynthetic oxygen evolution and photosystem II quantum yield against a background of the apparent over-reduction of the photosynthetic electron chain was recorded. Our results suggest an essential role of cpPDC in the supply of carbon precursors for de novo FA synthesis in microalgae under conditions of photoautotrophy. A shortage of this supply is detrimental to the nitrogen-starvation-induced synthesis of storage TAG, an important carbon and energy sink in stressed Chlamydomonas cells, thereby impairing the acclimation ability of the microalga.
Introduction
Elucidation of the storage lipid metabolism in oleaginous photosynthetic microalgae is attracting major attention from the research community, given the potential capacity of these organisms to produce precursors for the biodiesel industry and the recently revealed peculiar traits of microalgal lipid metabolism (Liu and Benning, 2013; Goodenough et al., 2014; Li et al., 2014). This focus on microalgal lipid metabolism necessitates a deep understanding of the key regulatory factors and the major pathways implementing carbon allocation to fatty acid (FA) synthesis in the chloroplast, and subsequent processing towards the assembly of reserve lipids, mainly triacylglycerols (TAG). Enzymes involved in the direct conversion of pyruvate, originating from glycolysis, and some coupled reactions utilizing pyruvate and phosphoenolpyruvate (PEP)—the so-called pyruvate hub enzymes—have been proposed as promising targets for metabolic engineering in microalgae to shuffle the carbon flux towards FA and TAG biosynthesis (Smith et al., 2012; Jinkerson et al., 2013). In higher plants (Chapman et al., 2013), as well as in microalgae (Klein, 1986; Smith et al., 2012), both cytosolic and plastidic glycolytic pathways are incomplete, and hence they complement each other and depend on metabolite trafficking across the plastid membrane (Facchinelli and Weber, 2011). Notably, the green microalga Chlamydomonas reinhardtii lacks the chloroplast pyruvate kinase, generating pyruvate from PEP (Klein, 1986; Terashima et al. 2011). The chloroplast pyruvate dehydrogenase complex (cpPDC) is known to convert pyruvate into acetyl-CoA to be used in FA synthesis.
The oxidative decarboxylation of pyruvate to acetyl-CoA occurs along with the commensurate production of NADH. PDC consists of three main components: E1 or pyruvate dehydrogenase (PDH; EC 1.2.4.1), E2 (dihydrolipoamide acetyltransferase, EC 2.3.1.12), and E3 (dihydrolipoamide dehydrogenase, EC 1.6.4.3). E1 is composed of alpha (E1α) and beta (E1β) subunits. The pyruvate decarboxylation reaction, catalysed by PDH, is considered to be a rate-limiting step in the reaction sequence mediating PDC activity (Reid et al., 1977).
Acetyl-CoA is an important metabolite that participates in numerous anabolic reactions, as well as in protein acetylation, pinpointing the essential role played by PDC in overall cellular metabolism. Regarding the lipid biosynthesis pathways, much evidence suggests that plastid PDC plays a dominant role in providing acetyl-CoA for FA synthesis in the cells of oleaginous plant tissues and organs (Ke et al., 2000; Lin et al., 2003; Bourgis et al., 2011). Direct evidence of the critical role of plastid PDC was obtained by disruption of the gene encoding the subunit E2 of the cpPDC, which led to an embryo-lethal phenotype in the developing seeds of Arabidopsis (Lin et al., 2003). At the same time, alterations in the gene expression of plastid acetyl-CoA synthase (ACS), utilizing acetate to generate acetyl-CoA, did not lead to significant changes in lipid content in Arabidopsis developing seeds or leaves (Behal et al., 2002; Lin and Oliver, 2008). Recent comparative transcriptome and metabolome studies of oil and date palms further indicated the plastid carbon metabolism and the acetyl-CoA supply via PDC for FA synthesis and, ultimately, for TAG biosynthesis as important determinants of oleogenity in higher plants (Bourgis et al., 2011).
Regulation of storage lipid metabolism in photosynthetic unicellular algae is deemed to be distinct from that in the well-studied oil-storing tissues of land plants (Liu and Benning, 2013). The exposure to stress conditions, such as nitrogen starvation, arrests the cell cycle, hampers growth, and leads to a swift diversion of cellular metabolism from protein and nucleic acid synthesis to accumulation of reduced-carbon storage compounds, such as TAG, in the oleaginous species (Hu et al., 2008; Solovchenko, 2012). The flexibility of algal lipid metabolism allows the rapid remobilization of TAG reserves upon nitrogen replenishment (Khozin-Goldberg et al., 2005). Relatively little information is available on the source of acetyl-CoA in the chloroplasts of photoautotrophic oleaginous microalgae, capable of producing high amounts of the storage lipid TAG under conditions of nutrient stresses. Multiple enzymatic activities, not limited to the chloroplast, can potentially lead to acetyl-CoA production in microalgal cells other than PDC, such as ACS, acetate metabolism via acetate kinase and phosphotransacetylase, and the PDC bypass, involving pyruvate decarboxylase and aldehyde dehydrogenase, depending on the carbon and light regime (Li et al., 2014; Goodenough et al., 2014). [14C]acetate labelling studies, in various species, have demonstrated active acetate acquisition and incorporation into FA and complex lipids, implying ACS activity in microalgal cells. Moreover, the cellular content of TAG in C. reinhardtii was drastically augmented by increasing acetate concentration in the nutrient medium (Fan et al., 2012). Recent studies have shed more light on the transcriptional regulation of TAG overproduction in a starchless mutant of C. reinhardtii with an acetate boost, including the increased expression of acetate transporters and two ACSs (Goodenough et al., 2014).
There is no direct experimental evidence for the involvement of cpPDC in the supply of acetyl-CoA for de novo FA synthesis in microalgae under conditions of photoautotrophy, in general, and under conditions conducive to TAG accumulation, such as nitrogen starvation, in particular. However, much pertinent information on the abundance and patterns of gene expression of putative cpPDC-encoding genes is available from a rapidly growing number of genomic and transcriptomic studies on microalgae. Based on the transcriptional dynamics under nitrogen starvation, triggering FA and TAG biosynthesis, the transcriptional regulation of cpPDC genes is posited as an important mechanism regulating carbon flux from photoassimilates towards the biosynthesis of FA and TAG in photoautotrophic oleaginous microalgae (Rismani-Yazdi et al., 2012; Jinkerson et al., 2013; Li et al., 2014).
Remarkably, the dual role of dihydrolipoamide acetyltransferase (DLA2), an E2 subunit of the cpPDC from C. reinhardtii, in the synthesis of both acetyl-CoA and D1 protein has recently been revealed (Bohne et al., 2013). The reciprocal regulation of metabolic signals and gene expression relied on the carbon regime; in the presence of acetate, DLA2 could bind psbA mRNA favouring D1 protein synthesis and the de novo assembly of photosystem II (PSII).
Gaining further insight into the role of cpPDC in acetyl-CoA synthesis in photosynthetic microalgal cells is essential for a better understanding of the mechanisms regulating carbon flux and allocation for FAs and TAG biosynthesis in oleaginous microalgae. In the current study, C. reinhardtii recombinant lines with a decreased expression of PDC2_E1α (Cre02.g099850; http://www.phytozome.net), predicted to encode the subunit E1α of cpPDC, were generated using artificial microRNA (amiRNA) technology (Molnar et al., 2009). Transformants were used in a comparative study of the role of cpPDC in FA and TAG production in the model green microalga C. reinhardtii under photoautotrophic and mixotrophic conditions in the presence and absence of nitrogen in the nutrient medium. Significantly impaired growth was observed in the selected lines grown with a CO2 supply as the only external carbon source. Under nitrogen starvation, the deleterious effect of decreased gene expression on TAG and biomass accumulation was profound under photoautotrophy. The analysis of oxygen evolution and electron transport on the acceptor side of PSII indicated an overall decline in photosynthetic activity during photoautotrophic growth, especially under nitrogen starvation, presumably due to a sink limitation resulting from TAG biosynthesis impairment. Based on these results, we assume that cpPDC is at least one control point in the supply of acetyl-CoA for de novo FA synthesis in the plastids of C. reinhardtii under photoautotrophic conditions, and this assumption may apply to other photoautotrophic microalgae as well. In addition, this mechanism seems to play an important role in acclimation to stresses such as nitrogen limitation by channelling of the excessively fixed carbon into the biosynthesis of storage TAG.
Materials and methods
Strains and experimental conditions
A C. reinhardtii cell-wall-deficient and arginine-requiring mutant strain (CC-1618 arg7 cw15 mt–) was obtained from the Chlamydomonas Resource Center (University of Minnesota, St Paul, MN, USA; http://chlamycollection.org/). Arginine was only supplied to the original strain before transformation; all media used for the engineered strains were arginine free. The following media were used in the experiments: nitrogen-replete and nitrogen-depleted (containing 0.5mM NH4Cl, 20 times less than the full medium) Tris/acetate/phosphate (TAP: 200mM Tris, 20mM NaAc) medium, and nitrogen-replete and nitrogen-depleted Sueoka’s high-salt minimal (HSM) medium (Harris, 1989).
C. reinhardtii strain CC-1618 cells were cultured in TAP with periodic dilutions with fresh medium to maintain logarithmic growth as estimated by daily measurements of the chlorophyll (Chl) culture content (Solovchenko et al., 2013). At the onset of each experiment, cultures were diluted according to Chl content (initial Chl was 2 and 15 µg ml–1, cell number ~1×106 and 6×106 cell ml–1 for the experiments in nitrogen-replete and nitrogen-depleted medium, respectively). Before transfer to nitrogen starvation, cells were washed twice with a nitrogen-depleted medium. All experiments were performed in 250ml Erlenmeyer flasks containing 100ml (TAP) or 70ml (HSM) liquid culture in a New Brunswick Innova 43 incubator shaker (Eppendorf AG, Hamburg, Germany) at 25 °C, with CO2 supplied at 300ml min–1 under continuous illumination (75 μmol photons m–2 s–1 photosynthetically active radiation). For photoautotrophic growth in HSM medium, the flasks were sealed with AirOtop enhanced seals (Thompson Instrument Company, Oceanside, CA, USA) to provide better gas exchange.
Cultures were sampled for analyses of growth parameters and lipids at the beginning of the experiment and after 2, 4, and 6 d for the experiments in complete medium, and after 2 d when experiments were conducted under nitrogen-starvation conditions. Total Chl concentration in the cultures was determined as described by Solovchenko et al. (2013). For dry weight (DW) measurements, two types of membrane filter were set together: glass fibre filters (Sartorius Stedim Biotech, Gottingen, Germany) with a pore size of 5–8 µm and cellulose-nitrate filters with a pore size of 5 µm (Schleicher & Schuell, Dassel, Germany), which were placed on top of the glass filters. C. reinhardtii culture (5ml) were filtered through the two dried and pre-weighed filters, and filters were dried for 120min at 120 °C or until a constant weight was achieved.
Lipid extraction and FA analysis
Prior to lipid extraction, biomass samples that had been freeze dried and kept at –20 °C were treated for 10min with hot isopropanol at 80 °C to prevent lipase activity. Isopropanol was collected, and lipids were extracted from the cell pellets according to the method of Bligh and Dyer (1959) and combined with isopropanol extracts. Total lipid extract was separated into neutral (NLs) and polar (PL) lipids on Bond-Elute silica cartridges (Varian, Walnut Creek, CA, USA). TAG was isolated from NL using a solvent system of petroleum ether:diethyl ether:acetic acid (70:30:1, v/v); PL were separated into individual classes by two-dimensional thin-layer chromatography as previously described (Pal et al., 2013).
A gas chromatography (GC) analysis of FA composition and content was performed on biomass samples, aliquots of lipid extracts, and individual lipid classes isolated by thin-layer chromatography according to Pal et al. (2013). Prior to the direct transmethylation, cell pellets were dried for 45min in a Savant SPD 111 SpeedVac concentrator (Thermo Electron) at room temperature. For transmethylation, the biomass and lipid samples were incubated in dry methanol containing 2% (v/v) H2SO4 at 80 °C for 1.5h under argon atmosphere and continuous stirring. Heptadecanoic acid (C17:0; Sigma-Aldrich) was added as an internal standard. FA methyl esters were analysed on a Trace GC ultra (Thermo, Milan, Italy) equipped with a SUPELCOWAX 10 (30 m×0.32mm, 0.25 μm) capillary GC column (Sigma-Aldrich), a flame ionization detector, and a programmed temperature vaporizing injector (Pal et al., 2013).
Vector construction for silencing PDC2_E1α and Chlamydomonas transformation
Gene silencing was performed using amiRNA as described by Molnar et al. (2009). The expression vector pChlamiRNA2, containing the amiRNA precursor under the hybrid HSP70A–RBCS2 promoter and Arg7, was obtained from the Chlamydomonas Resource Center. Using the web MicroRNA designer platform (WMD3; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi), two complementary 90 nt oligonucleotides, containing 21 nt of the coding region of C. reinhardtii PDC2_E1α (Cre02.g099850, Phytozome), in forward and reverse orientations and separated by the spacer to form a loop structure, were designed (ami-PDC2_E1α-F and ami-PDC2_E1α-R; Supplementary Table S1 at JXB online). After annealing, the double-stranded oligonucleotides were ligated into the SpeI-digested pChlamiRNA2 vector using a Mighty Mix kit (Takara Bio, Otsu, Japan). The C. reinhardtii mutant strain CC-1618 arg7 cw15 mt– was transformed with the resultant pChlami2-amipdh vector using the glass-bead method (Kindle, 1990) and grown on TAP agar plates without arginine for selection. Individual colonies were detected after 1–1.5 weeks and transferred to fresh TAP plates. C. reinhardtii cells transformed with the original pChlamiRNA2 vector carrying Arg7 were used in the experiments as an empty vector (EV) control.
Selection of pdh transformants
Selection of transformants with decreased expression of C. reinhardtii PDC2_E1α (pdh lines) was based on their expected reduced ability to grow photoautotrophically. A total of 86 pdh colonies and 10 EV colonies were randomly selected and grown in 96-well culture plates in 200 µl of TAP medium. After a few days of growth, 50 µl of culture of each line was transferred to 24-well plates containing 950 µl of either TAP (mixotrophic conditions) or HSM (photoautotrophic conditions) medium. After 3 d of growth under continuous illumination (50 μmol photon m–2 s–1 photosynthetically active radiation), OD750 was measured using a microplate reader (BioTek, Winooski, VT, USA). Growth was evaluated by comparing the OD values of mutants in HSM and TAP relative to those of the EV lines. Lines exhibiting the lowest pdhOD750 HSM/TAP:EV OD750 HSM/TAP ratio were selected as potential candidates carrying the silenced PDC2_E1α.
Selected lines were examined for the presence of the vector insert by PCR using primer pair pChlamiRNA2-F/pChlamiRNA2-R (Supplementary Table S2 at JXB online). DNA from different transformant lines was isolated using the fast CHELEX-based extraction procedure (Heddad et al., 2006). A few PCR-positive clones were selected for RNA isolation and further analysis of target gene expression by real-time quantitative PCR (qPCR).
RNA isolation, cDNA synthesis, and real-time qPCR
For RNA isolation, 3ml of logarithmically growing culture of the selected pdh and EV lines was centrifuged (13 000rpm, 5min, 4 °C), and the resultant cell pellet was processed with a SV Total RNA Isolation System kit (Promega, Madison, WI, USA).
The cDNA for real-time qPCR was synthesized from 1 µg of total RNA using a Verso cDNA kit (Thermo Scientific), with a 1:3 (v/v) blend of anchored oligo(dT) and random hexamers as primers, according to the manufacturer’s recommendations.
Real-time qPCR was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) in 0.2ml 96-well low-profile skirted plates using iTAQ Universal SYBR Green Supermix (Bio-Rad). The expression of PDC2_E1α was normalized to that of RACK1 (GenBank accession no. XM_001698013) and the actin gene (GenBank accession no. XM_001699016), commonly used as housekeeping genes in Chlamydomonas. The primers and PCR programs used are listed in Supplementary Table S2. Gene expression was quantified by the ΔΔCq method in CFX Manager™ Software (Bio-Rad). Real-time qPCR procedures and analyses were performed in accordance with MIQE guidelines (Bustin et al., 2009).
Pigment analysis
Pigments were extracted and analysed using a high-performance liquid chromatography system composed of a Prostar 240 solvent-delivery module, and a Prostar 330 photodiode array detector (Varian Analytical Instruments, Walnut Creek, CA, USA) and a C18 reverse-phase column (5 µm, 250mm Lichrosphere 100; Merck, Darmstadt, Germany) (Solovchenko et al., 2013). Pigments were identified and quantified using pure pigment standards (Sigma-Aldrich, St Louis, MO, USA; Fluka, Taufkirchen, Germany).
Photosynthetic activity estimation
The photosynthetic activity of the C. reinhardtii transformants was determined by: (i) fast Chl fluorescence transients; and (ii) oxygen evolution-based photosynthesis/irradiance (P-I) curves. The induction curves of Chl fluorescence were recorded using a Fluorpen FP100s portable Pulse Amplitude Modulated Fluorometer (Photon Systems Instruments, Drasov, Czech Republic) and analysed as described elsewhere (Strasser et al., 2004; Solovchenko et al., 2013). Non-photochemical quenching (NPQ) was calculated as NPQ=Fm/Fm’ – 1 (Maxwell and Johnson, 2000). Oxygen evolution by C. reinhardtii under an actinic irradiance increasing stepwise in the range 0–800 µE m–2 s–1 was monitored with a Clark-type oxygen electrode in the temperature-regulated chamber with a built-in computer-controlled LED light source (Chlorolab III; Hansatech, King’s Lynn, Norfolk, UK).
Statistical analyses
To define whether observed differences between control and treatment cultures were significant, a two-tailed distribution Student’s t-test for two-sample unequal variance was used with an α less than 0.05. The data obtained for all replicates of two independent EV lines, 60 and 73, were combined and analysed together (designated EV).
Results
Bioinformatics survey for the Chlamydomonas PDH-E1α gene
The C. reinhardtii genome (http://www.phytozome.net/) was screened for candidate PDH subunit E1α-encoding genes for amiRNA silencing. A BLAST search was performed against an amino acid sequence of a plastid PDH subunit E1α isoform of A. thaliana (The Arabidopsis Information Resource, accession no. AT1G01090) as a query. An amino acid sequence with a high (>65%) similarity to the Arabidopsis thaliana protein and tentatively annotated as PDH E1α was found (PDC2_E1α; Cre02.g099850.t1.3). The nucleotide sequence of PDC2_E1α was selected for amiRNA silencing based on the results of the phylogenetic analysis, and a high identity of the deduced protein (60–70%) to homologous proteins from higher plants and cyanobacteria that are considered ancestors of chloroplasts (Supplementary Fig. S1 at JXB online). The BLAST search against the NCBI non-redundant protein database did not reveal a high similarity of the deduced protein to either plant or algal mitochondrial PDH E1α homologues, or to those from bacteria, which are commonly accepted as progenitors of mitochondria. Furthermore, another gene in the C. reinhardtii genome is annotated as encoding a mitochondrial PDH subunit E1α (PDC1; Cre07.g337650). A phylogenetic position similar to the putative chloroplast PDH subunit E1α was previously shown for the E2 subunit (DLA2) of the cpPDC of C. reinhardtii (Bohne et al., 2013), whose chloroplast localization was confirmed experimentally. Nevertheless, it should be noted that the protein-localization prediction servers, such as TargetP and ChloroP (http://www.cbs.dtu.dk) (Emanuelsson et al., 2000), as well as a new tool Predalgo (Tardif et al., 2012), dedicated to algae, did not predict the chloroplast localization of the protein encoded by PDC2_E1α. However, the chloroplast localization was supported by Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) and MultiLoc2-LowRes (http://abi.inf.uni-tuebingen.de/Services/MultiLoc2), the latter taking into account phylogenetic and gene ontological terms. Further experimental evidence is required to confirm the plastid localization of the protein.
Selection of the pdh silenced lines
Assuming that downregulation of the PDC2_E1α gene would impair the growth of putative transformants more strongly under photoautotrophic conditions (in HSM) than under mixotrophic conditions (in TAP), we compared the OD750HSM:OD750TAP ratio of the pdh lines with that of the control EV lines, which was set to 1. Similar results were obtained when OD680 values were recorded (not shown). Selected lines demonstrating the lowest ratio (Fig. 1A) indeed showed a 60–90% reduction in PDC2_E1α expression, as confirmed by a real-time qPCR (Fig. 1B). Three mutants (pdh21, pdh42, and pdh72) were chosen for further characterization.
Fig. 1.
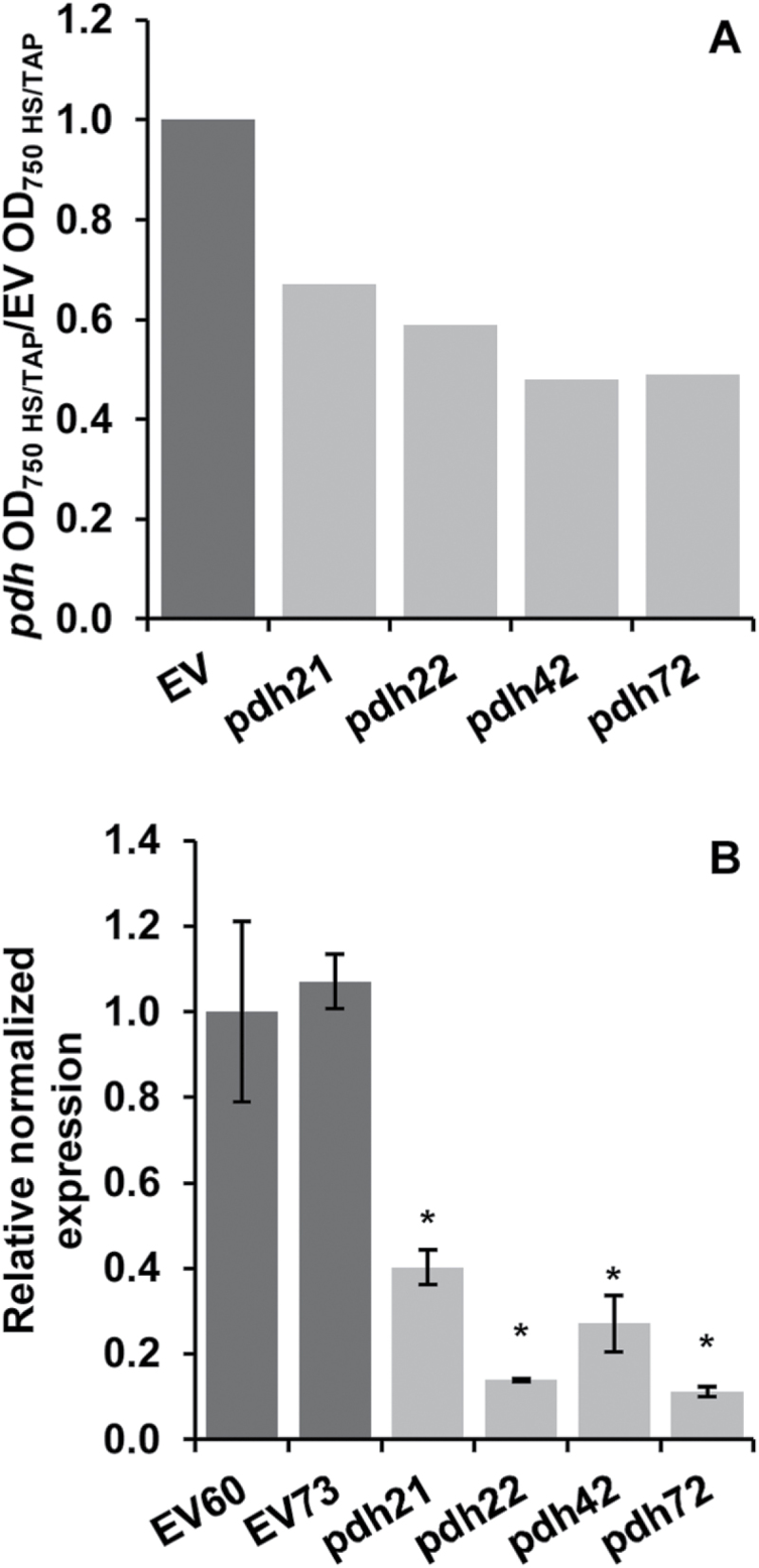
Selection of C. reinhardtii transformants. (A) OD of selected EV and pdh lines grown in HSM versus TAP medium. Dark grey bars, EV control lines; light grey bars, pdh lines. The average ratio of two different EV control lines is shown and was set to 1. (B) Expression of PDC2_E1α in the selected EV and pdh lines analysed by real-time qPCR. Statistical analysis was performed in Bio-Rad CFX Manager 3.0. Data are means±SD of three technical replicates. Asterisks indicate statistical differences between control and mutant lines at P<0.05 (Student’s t-test).
Effect of PDC2_E1α silencing on Chlamydomonas growth parameters and total FA production in an nitrogen-replete medium under photoautotrophic and mixotrophic conditions
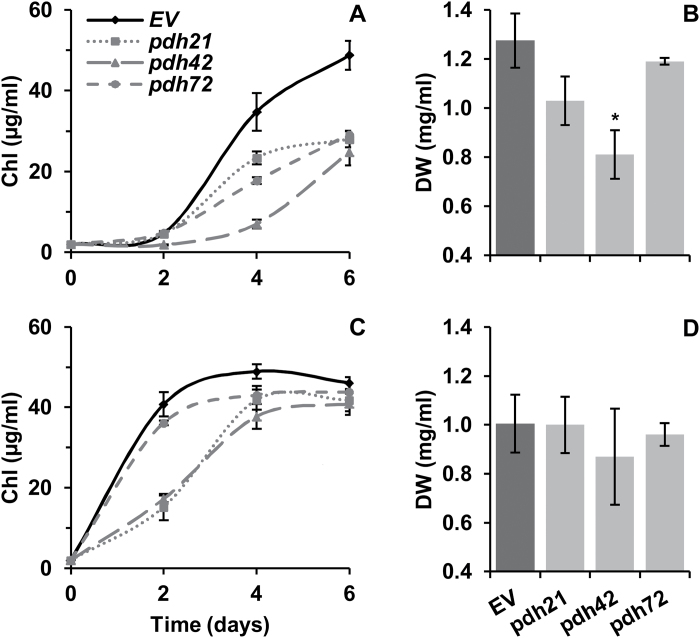
To investigate the impact of PDC2_E1α silencing on growth and total FA (TFA) production in the nitrogen-replete HSM and TAP media (designated HSM+N and TAP+N, respectively), Chlamydomonas cells were initially cultured in TAP+N and then transferred to either HSM+N or fresh TAP+N at an initial Chl concentration of 2 µg ml–1. During the duration of the experiment, lower Chl concentrations were determined in the pdh cultures in both HSM+N and TAP+N, compared with the EV lines (Fig. 2A, C). After a short initial lag, the EV cultures transferred to HSM+N resumed growth, whereas the growth of the pdh lines was substantially impaired, with a Chl content ~40–50% lower than that in the EV lines by day 6. The final biomass of the pdh cultures in HSM+N was also lower than that of the controls, but there was no significant difference in the final DW in TAP+N (Fig. 2B, D). In TAP, the growth of the pdh lines was substantially retarded within the first 2 d, particularly in lines pdh21 and pdh72, but the overall negative impact of PDC2_E1α silencing on the final Chl and DW contents was less pronounced under mixotrophic conditions (Fig. 2C).
Fig. 2.
Effect of PDC2_E1α silencing on C. reinhardtii growth in HSM (A, B) and in TAP (C, D) medium. (A, C) Chl concentration. (B, D) Biomass concentration after 6 d of growth. Values shown for the pdh lines are means±SD of two biological and two technical replicates (n=4); EV values are means of two individual lines (n=8). Asterisks indicate significant differences between the EV and pdh lines at P<0.05 (Student’s t-test).
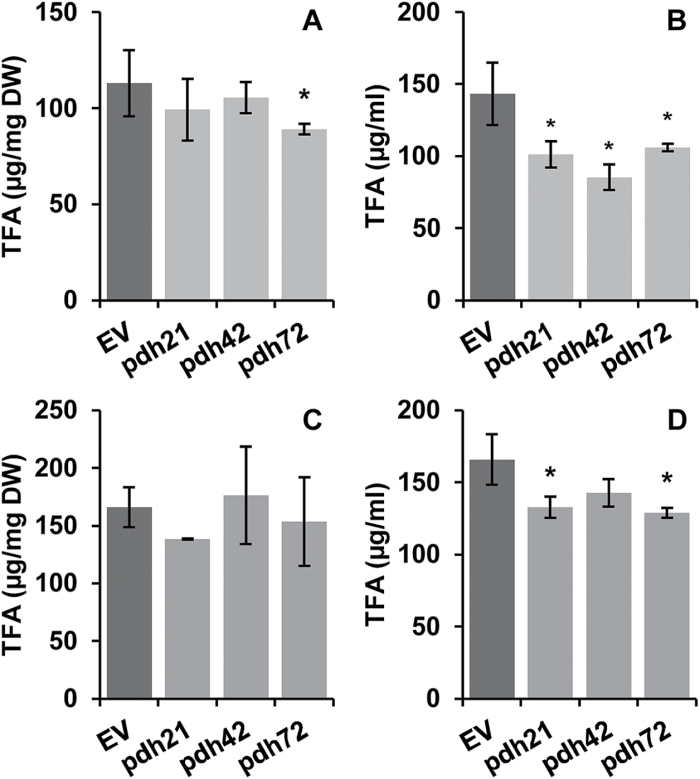
After 6 d of photoautotrophic growth, the TFA content of the biomass was about 20% lower (pdh72) or similar to that of the EVs (Fig. 3A). Notably, the difference in TFA production between EV and pdh transformants was more evident when expressed in terms of volumetric FA content (µg ml–1), which accounts for both the culture DW content and the TFA content of the biomass (Fig. 3B). Given the substantial decrease in their final DW, the pdh mutants demonstrated ~25–40% less volumetric TFA production than the control lines (Fig. 3B).
Fig. 3.
Effect of PDC2_E1α silencing on C. reinhardtii after 6 d of cultivation in HSM (A, B) and in TAP (C, D) medium. (A, C) TFA content of biomass. (B, D) Culture TFA content. Bars for the pdh lines represent means±SD of two biological and two technical replicates (n=4). EV values are means of two individual lines (n=8). Asterisks indicate significant differences between the EV and the pdh lines at P<0.05 (Student’s t-test).
A similar but less prominent effect of PDC2_E1α silencing on TFA accumulation (Fig. 3C, D) and FA composition (Supplementary Fig. S2 at JXB online) was observed under mixotrophic conditions, in TAP+N. The volumetric content of TFA in the cultures of the pdh mutants was about 15–25% lower than that in the EV line cultures (Fig. 3D).
Effect of PDC2_E1α silencing on Chlamydomonas biomass and total FA production in an nitrogen-depleted medium under photoautotrophic and mixotrophic conditions
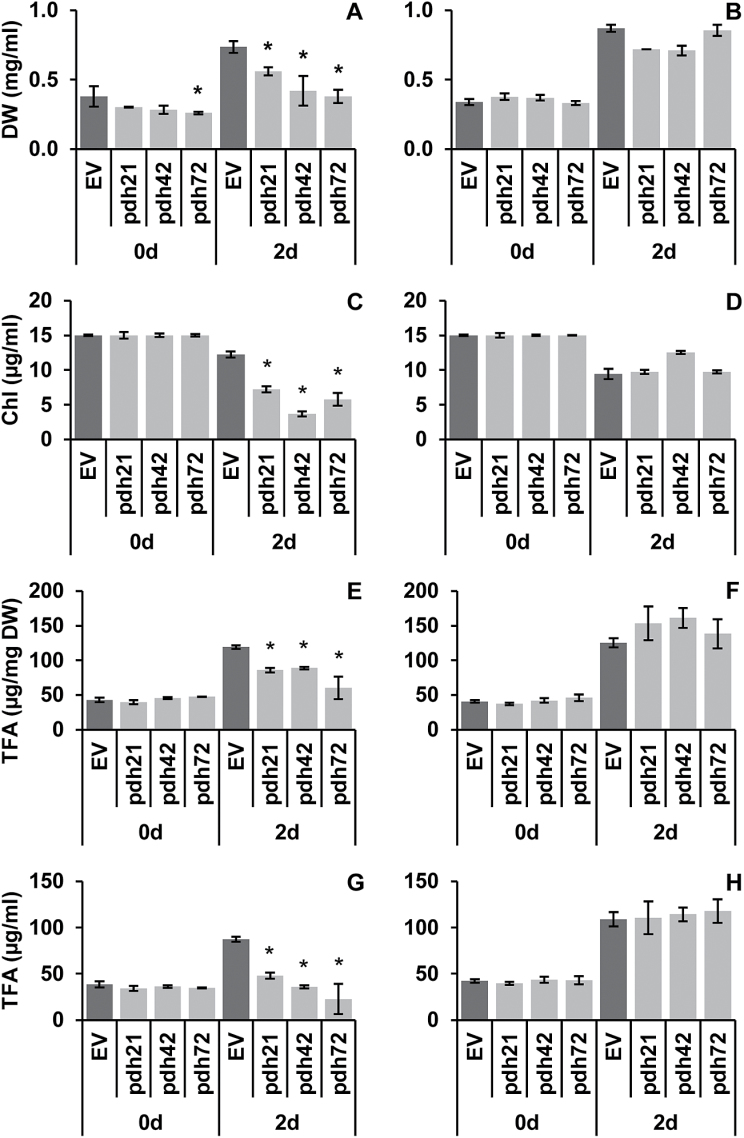
Conditions of nitrogen starvation are generally associated, in microalgal cultures, with a decline in the per cell and per culture volume contents of Chl due to gradual chloroplast dismantling (Goodson et al., 2011; Siaut et al., 2011). To avoid the strong autophagous response common to Chlamydomonas under nitrogen starvation (Perez-Perez et al., 2010), short-term experiments (lasting 48h) were performed. Three selected pdh lines demonstrated a stronger reduction in culture Chl content than the controls (Fig. 4C) cultivated in HSM-N, indicating an accelerated loss of photosynthetic pigments by the culture. The overall reduction in culture Chl content for the pdh lines was significantly more pronounced, constituting 40–70% of that in the EV cultures. Cell biomass (DW) production in the mutant cultures was also reduced, amounting to 51, 57, and 76% of the DW of the control lines for pdh72, pdh42, and pdh21, respectively (Fig. 4A). Due to a simultaneous decrease in both parameters, the Chl content of the biomass was not significantly altered compared with the EV lines, with the exception of line pdh42 (not shown). In contrast, the downregulation of PDC2_E1α did not substantially alter either the culture Chl or the DW contents of Chlamydomonas in TAP-N (Fig. 4B, D), but some decrease in biomass production was noted in the lines pdh21 and pdh42.
Fig. 4.
Effect of PDC2_E1α silencing on C. reinhardtii after 2 d of cultivation under nitrogen starvation in HSM-N (A, C, E, G) and in TAP-N (B, D, F, H) medium. (A, B) Biomass (DW) content. (C, D) Culture Chl content. (E, F) TFA content of biomass. (G, H) Culture TFA content. Values represent means±SD of n=6 for pdh lines (three biological and two technical replicates) and n=12 for EV control lines (joint results of two different control lines). Differences between controls and mutants are significant at P<0.05 (Student’s t-test).
Unlike in TAP-N, where there were no obvious differences in TFA production between the different lines (Fig. 4F, H), all pdh lines demonstrated severe impairment of TFA production in HSM-N (Fig. 4E, G). After 2 d of nitrogen starvation, the biomass content of the TFA was 25–50% lower in the pdh lines relative to the EV lines in this medium (Fig. 4E). Even more marked differences between the silenced and the control lines were observed in the volumetric content of TFA due to the cumulative effect of PDC2_E1α knockdown on biomass production and the TFA content of DW; the pdh lines produced 45–75% less TFA than the EV (Fig. 4G). Notably, TFA production was almost entirely abolished in the nitrogen-starved photoautotrophic cultures of the pdh lines 42 and 72 compared with time 0 (Fig. 4G), while it was unaltered in the mixotrophic cultures of the pdh lines in comparison with the control (Fig. 4H).
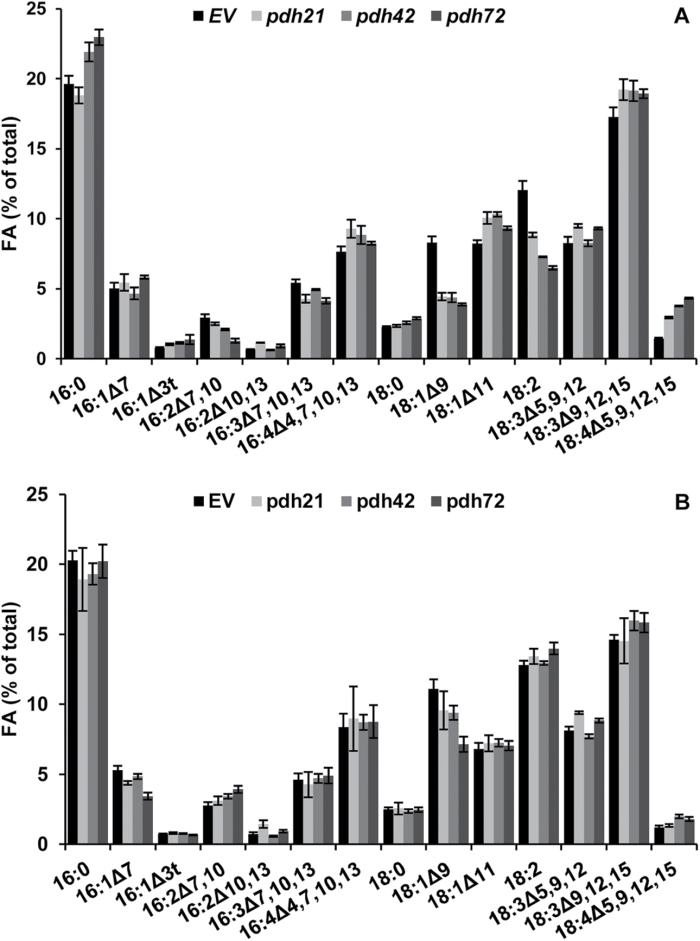
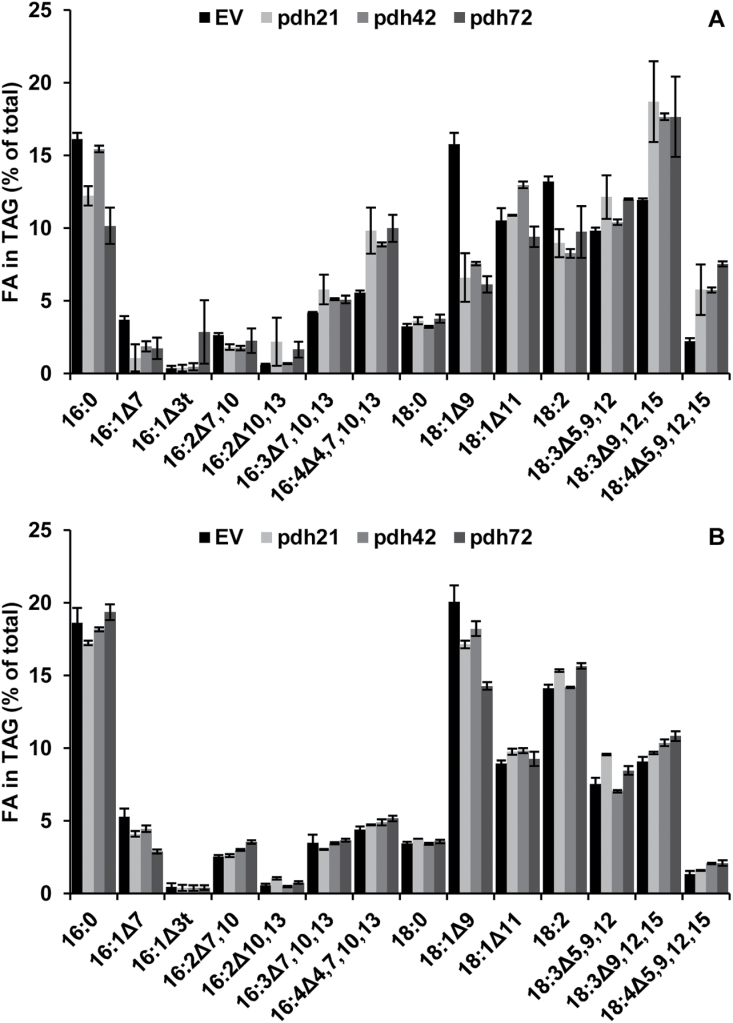
The downregulation of PDC2_E1α led to substantial changes in the FA profile of total lipids under photoautotrophic conditions, specifically to a ~50% decrease in the proportion of oleic acid (OA, 18:1Δ9) in the pdh lines relative to the EV lines after 2 d of nitrogen starvation (Fig. 5A). A decline in the relative proportions of OA, as well as linoleic acid (18:2Δ9,12), was accompanied by an increase in the proportions of the C18 polyunsaturated FA (PUFA) α-linolenic acid (ALA; 18:3∆9,12,15) and 18:4∆5,9,12,15 (Fig. 5A). Marked alterations were observed in the percentages of C16 PUFA; a decrease in the proportion of 16:3∆7,10,13 was accompanied by a concomitant increase in that of 16:4∆4,7,10,13; 16:0 increased in the pdh lines 42 and 72.
Fig. 5.
Effect of PDC2_E1α silencing on FA composition of C. reinhardtii after 2 d of nitrogen starvation in HSM-N (A) and in TAP-N (B) medium. Bars show means±SD of three biological and two technical replicates (n=6). EV bars represent means of two individual lines (n=12).
Remarkably, less substantial changes in the FA profile were found between the pdh and the EV lines under mixotrophic conditions (Fig. 5B), when acetate was added to the medium along with CO2.
PDC2_E1α silencing decreases TAG production and affects the FA composition of TAG under phototrophic conditions combined with nitrogen deprivation
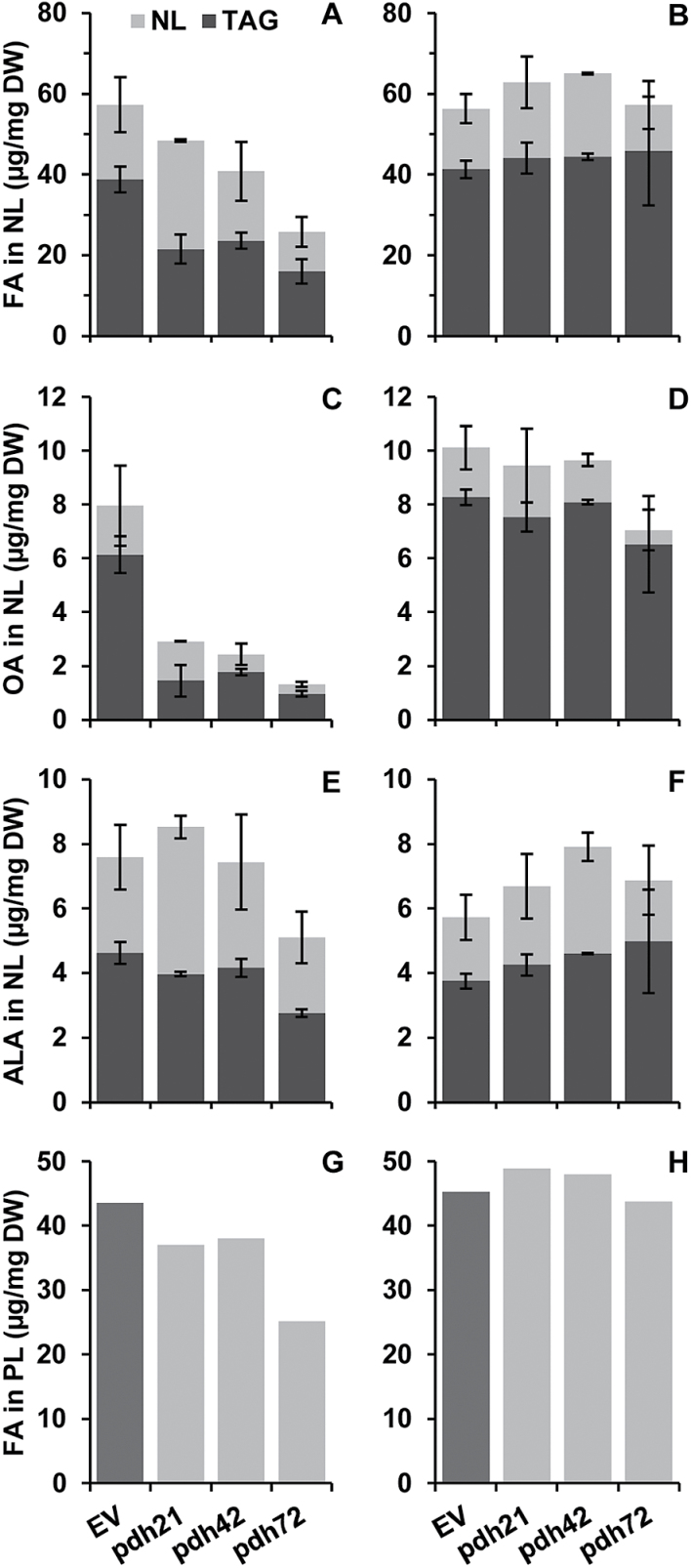
To elucidate whether the observed alterations in TFA content and FA composition were associated with alterations in the distribution of individual lipid classes and changes in their FA profiles, we further separated the total lipid extracts of the biomass samples, collected under nitrogen starvation, into NL and PL fractions to isolate TAG and individual membrane lipids, respectively. In HSM-N, the total acyl group content in the NL fraction (expressed as FA content) was significantly lower in the pdh lines than in the controls (Fig. 6A), with the most drastic decrease featured in the line pdh72. Remarkably, TAG content in the biomass of the pdh lines constituted only ~45–60% of the TAG content in the EV lines (Fig. 6A). In line with these data, microscopic observations of the cells stained with the lipophilic dye Nile red revealed a strong decrease in the number of lipid droplets in the cells of all three pdh lines (not shown).
Fig. 6.
Effect of PDC2_E1α silencing on FA distribution in NLs, TAG (A–F), and in PLs (G, H) of C. reinhardtii after 2 d of nitrogen starvation in HSM-N (A, C, E, G) and in TAP-N (B, D, F, H) medium. The contribution of TAG total FA, OA, and ALA to NLs is shown in dark grey (A)–(F). Values shown (except for PL in G and H) are the means±SD of two technical replicates from pooled samples of three biological replicates. Analysis of FA content in PL was performed from the pooled samples with two replicates. EV bars represent means of two individual lines.
The reduced TAG production in the cells of the pdh lines in HSM-N was associated with substantial alterations in the FA profile of TAG (Fig. 7A). The most drastic effect was determined on the relative percentage of 18:1∆9, which accounted for only 50–60% of that in the controls after 2 d of nitrogen starvation. Accordingly, ~60–85% and 70–85% reductions in the 18:1∆9 content (µg mg–1) allocated to NL and TAG, respectively, were found in the pdh lines compared with the controls (Fig. 6C). Similarly, the percentage of another product of de novo FA synthesis, palmitic acid (16:0), decreased in the TAG of the pdh lines 21 and 72, along with ~40–75% reductions in its content deposited in the TAG of three pdh lines (not shown). The consistent decreases in the proportions of 18:1∆9 and 18:2Δ9,12 occurred concomitantly with the increases in those of 16:4Δ4,7,10,13 and 18:3Δ9,12,15 (Fig. 7A), the latter two being the major acyl constituents of chloroplast galactolipids in Chlamydomonas (Giroud et al., 1988; Sakurai et al., 2014). Accordingly, the absolute contents of ALA (18:3Δ9,12,15) deposited in TAG did not decrease substantially in the pdh lines (Fig. 6E). These data may indicate that the flux of chloroplast-resident acyl groups into TAG (Li et al., 2012) was not altered in the pdh lines but rather the de novo TAG biosynthesis was affected that is reliant on 16:0 and 18:1∆9 production in the chloroplast. Again, the strongest impact of PDC2_E1α silencing was observed in line pdh72. In addition, a 2.5- to 3.5-fold increase was determined in the percentage of 18:4Δ5,9,12,15 in the FA profile of TAG in the pdh lines (Fig. 7A).
Fig. 7.
Effect of PDC2_E1α silencing on FA composition of TAG of C. reinhardtii after 2 d of nitrogen starvation in HSM-N (A) and in TAP-N (B) medium. Bars show the means±SD of two technical replicates from pooled samples of three biological replicates. EV bars represent means of two individual lines.
In contrast to photoautotrophic conditions, comparable FA contents in NL and TAG were determined in the mutant and control nitrogen-depleted cultures (Fig. 6B) in TAP-N; furthermore, overall alterations in the FA profile of TAG were less marked in the pdh lines (Fig. 7B). The percentage of OA in the FA profile of TAG in the pdh lines amounted to 70–90% of that found in the EV; the line pdh72 had the lowest OA content (Fig. 7B).
PDC2_E1α silencing affects the FA composition of membrane glycerolipids under phototrophic conditions combined with nitrogen deprivation
Whereas the silencing of PDC2_E1α had a crucial effect on TAG formation in HSM-N, it had a lesser effect on PL content (Fig. 6G), supporting the assumption that an alleviated acetyl-CoA supply mainly affected the induced storage lipid biosynthesis driven by nitrogen starvation. It should be noted, however, that pdh72, featuring the strongest decrease in NL, also showed the biggest decrease in PL content, implying a negative effect on structural membrane lipids.
The strongest impact of PDC2_E1α silencing was found on the relative content of the chloroplast galactolipid (MGDG), whose relative percentage of PL was markedly reduced in all pdh lines. The proportion of digalactosyldiacylglycerol (DGDG) did not seem to be substantially altered, indicating a decreased MGDG:DGDG ratio in the cells with reduced expression of PDC2_E1α (Supplementary Fig. S3 at JXB online). The percentage of the major extraplastidial betaine lipid diacylglyceryl-N,N,N-trimethylhomoserine (DGTS) in the pdh lines did not change substantially compared with the EV control.
The individual lipid classes of Chlamydomonas possess distinct and characteristic FA signatures (Giroud et al., 1988; Sakurai et al., 2014), and they demonstrated specific alterations in the pdh lines (Supplementary Fig. S4 at JXB online). A clear trend towards more unsaturated acyl groups was observed in major lipid classes. MGDG displayed substantially higher percentages of 16:4Δ4,7,10,13 and 18:3Δ9,12,15 in all analysed pdh lines compared with the control. DGDG of the pdh lines showed a clear trend towards more unsaturated C18 FA species in comparison with the control. The chloroplast phospholipid phosphatidylglycerol featured an increased percentage of its unique FA 16:1Δ3t in the pdh lines (except for the line 42) against the background of the reduced proportion of 16:0, as well as a trend of an increasing percentage of 18:3Δ9,12,15, similar to galactolipids. The most noticeable alterations in the FA profile of DGTS accounted for the increased proportion of 16:0 and the decreased proportion of 18:2Δ9,12. Among the two Δ5 C18 PUFA, 18:3Δ5,9,12 and 18:4Δ5,9,12,15, which are particularly enriched in DGTS, a noticeable increase was documented in the proportion of 18:4Δ5,9,12,15 in all pdh lines. The sulfolipid sulfoquinovosyl diacylglycerol (SQDG) and the phospholipid phosphatidylinositol contained high proportions of 18:1Δ11. Notably, as in SQDG and phosphatidylglycerol, the proportion of 18:1Δ11 in DGTS was higher than that of 18:1Δ9, with a slight increase in the former in the silenced lines. The identity of 18:1Δ11 was confirmed by the GC mass spectrometry of 4,4-dimethyloxazoline derivatives (not shown).
Effects of PDC2_E1α silencing on carotenoid profiles and photosynthetic activity
Next, we investigated the composition of individual carotenoids in the transformed cells of C. reinhardtii. The carotenoid profiles of all of the studied transformants grown in TAP were similar (Supplementary Fig. S5 at JXB online), comprising the carotenoids typical of the wild-type C. reinhardtii (Lohr et al., 2005). Cultivation in HSM introduced no changes to the qualitative carotenoid composition regardless of the transformant line or nitrogen availability. Only a small increase in the de-epoxidation state of the violaxanthin cycle [violaxanthin, antheraxanthin, zeaxanthin (VAZ)] was detected after 2 d of nitrogen starvation; the magnitude of this increase was roughly similar in all transformant lines (Supplementary Fig. S6A at JXB online), as was the total size of the VAZ pool (Supplementary Fig. S6B).
All of the studied transformant lines (EV and pdh) grown in TAP were characterized by a uniformly high maximum quantum efficiency of PSII (estimated as Fv/Fm; Table 1), an indication of high photosynthetic activity. Cultivation in HSM brought about a small but significant decrease in Fv/Fm, which was more pronounced in the pdh lines than in the EV (Table 1). Nitrogen starvation induced an apparent and more profound decline in PSII efficiency of the pdh lines, whereas the EV lines retained Fv/Fm at a level typical of nitrogen-sufficient cultures. A similar pattern of changes was found in the slope of the linear part of the P-I curves and the maximum rate of photosynthesis (P max; Supplementary Fig. S7 at JXB online), recorded via oxygen evolution. It should be noted that, under mixotrophic conditions, P max was slightly higher in the pdh lines than in the EV. The considerable increase in F0 against the background of a decrease in Chl content (Supplementary Table S4 at JXB online), observed in the pdh lines under nitrogen starvation may suggest a certain degree of stress-induced disorders in the reaction centres of the cells.
Table 1.
Condition of photosynthetic apparatus of C. reinhardtii transformants grown in TAP+N, HSM+N for 6 d and in HSM-N for 2 dFv/Fm, maximum potential quantum yield of the PSII; M0, changes in the initial slope of the fast chlorophyll fluorescence transient (indicative of QA reduction; for the details of the calculations, see Supplementary Table S3). Values shown are the means±SD of three biological replicates. EV values represent the means of two individual lines.
| Strain | Medium | Fv/Fm | M0 |
|---|---|---|---|
| EV | TAP+N | 0.74±0.00 | 0.98±0.02 |
| HSM+N | 0.75±0.01 | 1.05±0.13 | |
| HSM-N | 0.73±0.01 | 1.36±0.05 | |
| pdh21 | TAP+N | 0.72±0.01 | 0.95±0.03 |
| HSM+N | 0.71±0.01 | 1.13±0.10 | |
| HSM-N | 0.65±0.01 | 1.65±0.04 | |
| pdh42 | TAP+N | 0.73±0.00 | 1.03±0.01 |
| HSM+N | 0.71±0.00 | 1.25±0.04 | |
| HSM-N | 0.66±0.01 | 1.76±0.04 | |
| pdh72 | TAP+N | 0.72±0.01 | 1.01±0.01 |
| HSM+N | 0.64±0.01 | 1.57±0.05 | |
| HSM-N | 0.57±0.01 | 2.10±0.05 |
To obtain additional insight into the responses of the photosynthetic apparatus of the generated C. reinhardtii transformants to cultivation in nitrogen-replete and nitrogen-depleted mineral media, the analysis of fast Chl fluorescence transients (JIP test) was employed (Supplementary Table S3 at JXB online; see also Strasser et al., 2004). Notably, little, if any, difference was found in the condition of the photosynthetic apparatus in the pdh transformant lines grown in TAP in comparison with the EV (Table 1). On the other hand, transfer into HSM+N resulted in a decrease in the maximum Chl fluorescence level, relative to F0 (Supplementary Fig. S8 at JXB online) and in an increase in the reduction of the primary acceptor, QA (Table 1), even in the dark-adapted samples. Accordingly, Stern–Volmer coefficients of NPQ in these cultures exhibited a very low response to increased irradiances (not shown). Under nitrogen starvation, these effects were much more pronounced in the pdh cells, whereas they were less conspicuous in the EV lines (Table 1 and Supplementary Table S4). The recorded changes in the Chl fluorescence parameters are evidence of the higher degree of electron carrier reduction in the chloroplast electron transport chain in the pdh cells under photoautotrophic conditions.
Discussion
The present study focused on elucidating the role of cpPDC in the supply of acetyl-CoA for de novo FA synthesis under conditions favouring TAG accumulation in the cells of photoautotrophic microalgae. As a model, we used the green microalga C. reinhardtii, which exhibits flexible responses in its central carbon and lipid metabolism to environmental perturbations, including nitrogen starvation (Merchant et al., 2012).
RNAi approaches have been widely employed in C. reinhardtii, recently also encompassing lipid metabolism (Moellering and Benning, 2010; Cerutti et al., 2011; Williams et al., 2011; Bohne et al., 2013). In the current study, C. reinhardtii mutants with reduced PDC2_E1α expression were generated and selected based on their impaired capability of photoautotrophic growth. The ability of the knockdown lines to produce TAG appeared to be severely impaired in photoautotrophic conditions when photosynthetic carbon fixation provides precursors for the synthesis of storage products. It is worth noting that RNAi-mediated deficiency in DLA2, an E2 subunit of the cpPDC from C. reinhardtii, did not cause any apparent qualitative effect on the composition of membrane glycerolipids under phototrophic nitrogen-replete conditions (Bohne et al., 2013). However, the impact of DLA2 knockdown on the storage lipid TAG accumulation under different carbon regimes was not examined.
Reduced expression of PDC2_E1α underscores the essential role of cpPDC in the carbon supply for FA synthesis from photosynthetically fixed carbon
PDH E1α downregulation did not show a strong effect on Chl, biomass, and TAG production under mixotrophic conditions, indicating that photosynthetically fixed carbon is less important in the presence of acetate. Our results firmly support the role of acetate as a source of acetyl-CoA for FA synthesis under mixotrophic conditions in Chlamydomonas in agreement with other studies (Work et al., 2010; Fan et al., 2012; Goodenough et al. 2014).
Under photoautotrophic conditions, the accelerated loss of photosynthetic pigments and reduced biomass production in the cultures of the knockdown pdh lines occurred under nitrogen starvation concomitantly with a drastic reduction in the content of TAG. Hence, the provision of acetyl-CoA through cpPDC activity to the initial and committed step of de novo FA synthesis, namely to acetyl-CoA carboxylase, seems to be indispensable for Chlamydomonas grown photoautotrophically. In addition, it should be noted that carbon precursors for FA synthesis might be provided via starch catabolism in the chloroplasts, as the synthesis and breakdown of starch occur in the same compartment as FA synthesis and precede TAG formation in Chlamydomonas (Fan et al., 2012). However, the metabolic interaction between the two major storage products is still under debate (Work et al., 2010; Siaut et al., 2011; Wase et al., 2014), and is mostly studied under conditions of mixotrophy. Regarding the origin of pyruvate in Chlamydomonas, lacking the chloroplast pyruvate kinase (Klein 1986; Terashima et al., 2011), the malic enzyme may play the role of the main pyruvate supplier for cpPDC activity. Alternatively, pyruvate can be shuttled to the chloroplast via dedicated transporters (Facchinelli and Weber, 2011; Li et al., 2014). It is noteworthy that recent ‘omic’ studies have documented an elevated level of citrate in nitrogen-starved cells of Chlamydomonas, as well as commensurate alterations in the abundances of enzymes mediating its synthesis and utilization, implying a potential role of this central metabolite as a carrier of acetyl groups for induced storage lipid accumulation (Wase et al., 2014). However, the mechanisms that may permit the bulky molecule of acetyl-CoA to enter the plastid remain elusive and warrant further investigations. We assume that conditions of nitrogen starvation lead to the partial endomembrane dismantling that might affect chloroplast membrane integrity and alter trafficking of central carbon metabolites into the plastid.
Characteristic changes in FA composition of individual glycerolipids indicate membrane lipids as the major source of FA for TAG in the pdh lines under conditions of photoautotrophy
Comparing the FA composition of TAG with that of polar lipids in the pdh lines in HSM-N, one can correlate the apparent similarity in the increased proportion of 16:4Δ4,7,10,13 and 18:3Δ9,12,15 and the decreased percentages of the less unsaturated precursors of these PUFA, in both TAG and galactolipids, specifically in MGDG. Overall alterations in the FA composition of TAG indicated a prevalence of molecular species that were assembled from diacylglycerol precursors of the chloroplast origin with a characteristic arrangement of C16 and C18 PUFA at the sn-2 and sn-1 positions of the glycerol moiety, respectively (Fan et al., 2011; Li et al., 2012). Another diagnostic feature of the TAG in the silenced lines—the decreased proportion and content of 18:1Δ9—seems to result from the suppression of the de novo FA synthesis supplying this FA for TAG assembly. Similar alterations in the FA composition of TAG were described in the pdg1 (plastid galactoglycerolipid degradation) mutant of C. reinhardtii impaired in OA channelling from the de novo synthetized MGDG (Li et al., 2012). However, in contrast to PDC2_E1α silencing, a pgd1 mutation led to drastically decreased TAG content under mixotrophic conditions (Li et al., 2012). Given that, in the TAG of Chlamydomonas, OA is predominantly localized to the sn-1/sn-3 positions (Fan et al., 2011; Li et al., 2012), it is likely that an impaired acetyl-CoA supply and reduced FA synthesis in the pdh mutants affected the flux of the de novo-produced OA onto those positions of TAG.
Notably, the mutation in PGD1 did not lead to substantial changes in the FA composition of membrane lipids under conditions of nitrogen starvation (Li et al., 2012), in contrast to the clear impact of PDC2_E1α knockdown on the FA composition of membrane lipids. The FA profile of the chloroplast lipids of the pdh lines revealed the seemingly higher level of chloroplast lipid unsaturation, and particularly of MGDG, probably retaining the FA profile of the initial non-stressed cells, which can be explained by the restricted de novo supply of FAs.
Our results are in agreement with the suggested model of cooperation of multiple pathways and cellular compartments in TAG assembly in Chlamydomonas (Fan et al., 2011; Li et al. 2012; Liu and Benning, 2013; Sakurai et al., 2014). The contribution of different pathways and compartments to the supply of acyl groups for TAG formation under nitrogen depletion seems to vary depending on the carbon regime. Notably, FA moieties characteristic of membrane lipids were prevalent in the downsized TAG of the pdh lines under conditions of phototrophy. The significance of the supply of unsaturated acyl moieties for TAG assembly from both plastidic and extraplastidial lipids has been inferred previously (Li et al., 2012; Sakurai et al., 2014), but the exact mechanisms require in-depth investigation.
Intensive membrane dismantling and increased transcript abundance of genes encoding numerous lipases and autophagy-associated genes is the characteristic Chlamydomonas response to nitrogen scarcity (Perez-Perez et al., 2010; Goodson et al., 2011; Boyle et al., 2012; Blaby et al., 2013; Goodenough et al., 2014). Notably, a stronger effect on chloroplast breakdown in the WT-12 nit – C. reinhardtii was observed under photoautotrophic conditions, while acetate addition reduced autophagy in long-term experiments (Davey et al., 2014), underscoring the alleviating effect of the organic carbon source due to its use in FA and energy production. A substantial reduction in the proportion of MGDG was documented in the wild-type C. reinhardtii strain CC1010 under photoautotrophic nitrogen-deficient conditions (Sakurai et al., 2014), whereas two other major lipids, DGDG and DGTS, sustained higher relative levels in the starved cells in line with our data on individual lipid class distribution. A reduced relative content of MGDG was documented in the pdh lines under nitrogen starvation compared with the controls in accordance with our suggestion that chloroplast lipid turnover is a major source of acyl groups for the residual TAG formation in the pdh lines under photoautotrophy. It is likely that the altered MDGD:DGDG ratio in the pdh lines that is crucial for a proper functioning of photosynthetic complexes in thylakoids could have a negative impact on the photosynthetic capacity of the knockdown lines.
TAG biosynthesis primed by cpPDC is important for nitrogen-stress acclimation under photoautotrophic conditions
Our results imply the deleterious effect of PDC2_E1α downregulation on the Chl content and photosynthetic capacity of Chlamydomonas under conditions of photoautotrophy. The impaired-growth phenotype, displaying reduced Chl culture and biomass content in the pdh mutants under nitrogen starvation, was comparable to the chlorotic phenotype observed in a pgd1 mutant of Chlamydomonas (Li et al., 2012). It was hypothesized that the reduced survival of the pgd1 mutant under nitrogen deprivation arises from cell damage by reactive oxygen species, whose level increased when photosynthesis was still functional but TAG accumulation, an important sink of the carbon excessively fixed during nitrogen starvation, was compromised (Li et al., 2012). The significance of FA synthesis, for the purposes of TAG overproduction in microalgae under conditions of an augmented carbon:nitrogen ratio, was indeed related to the prevention of photooxidative damage (Hu et al., 2008; Li et al., 2012; Solovchenko, 2012).
The reduced ability of pdh lines for TAG production might have led to an impaired sink capacity for photosynthates and hence to a decline in photosynthetic activity. According to the analysis of electron transport at the acceptor side of PSII, this impairment of photosynthetic activity could have been caused by a sustained over-reduction of electron carriers in the photosynthetic electron transport chain. It is unlikely that PDC2_E1α silencing exerted a direct detrimental effect on the photosynthetic machinery since the photosynthesis in the transformed lines appeared to be undisturbed in the TAP-grown cells in which the sink furnished by TAG biosynthesis was less impaired. It should also be emphasized that the degree of decline in TAG biosynthesis correlated well with the decline in various photosynthetic parameters. Thus, the most pronounced effects, on both TAG and the rate of photosynthesis, were recorded in the line pdh72 (Table 1, Supplementary Table S4). In contrast to the pgd1 mutant, no strong effect on TAG production was determined in the pdh lines in the presence of acetate, underscoring the essential role of cpPDC in the acetyl-CoA supply under photoautotrophic conditions. Indeed, neither pigment profile nor photosynthetic activity was affected in the pdh lines under mixotrophic conditions. It seems that when acetyl-CoA supply is not impaired, photosynthates are consumed efficiently, in the reactions of FA synthesis and consequent TAG biosynthesis, by the pdh lines to the same extent as by the EV lines, developing a phenotype that is similar to the controls.
To conclude, the results obtained in the present research suggest the dependence of de novo FA synthesis and TAG biosynthesis in photoautotrophic Chlamydomonas cells on the transcriptional activation of cpPDC. Our results strongly support a crucial role for cpPDC in supplying acetyl-CoA for de novo FA synthesis under photoautotrophic conditions; hence, the reduced expression of the E1α subunit of PDH is detrimental to storage lipid production and to the overall acclimation of the cells to nitrogen starvation. Further studies may delineate the effects of the restricted acetyl-CoA production on transcriptome reprogramming and alterations in the central carbon metabolism. In addition, further exploitation of cpPDC-encoding gene overexpression to enhance TAG production in non-model oleaginous microalgae under photoautotrophic conditions should be considered.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. 90-bp oligonucleotides used for pChlami2-amipdh vector construction for gene silencing.
Supplementary Table S2. On-colony PCR and real-time qPCR primers, and conditions of qPCR.
Supplementary Table S3. Characteristic points of OJIP and derivation of the selected JIP test parameters used in this work.
Supplementary Table S4. Characteristic parameters of chlorophyll fluorescence in the cells of pdh and EV lines of C. reinhardtii.
Supplementary Fig. S1. Phylogeny reconstruction of plastid and mitochondrial isoforms of pyruvate dehydrogenase (PDH) E1α subunit from plant, algal, and bacterial species.
Supplementary Fig. S2. Effect of PDC2_E1α silencing on FA composition of C. reinhardtii after 6 d cultivation.
Supplementary Fig. S3. Effect of PDC2_E1α silencing on relative proportion of major polar lipid classes in EV and pdh lines after 2 d of nitrogen starvation under photoautotrophic conditions.
Supplementary Fig. S4. Fatty acid composition of individual polar lipids after 2 d of nitrogen starvation under photoautotrophic conditions.
Supplementary Fig. S5. Carotenoid composition of the pdh and EV lines of C. reinhardtii cells at the onset and after 2 d of nitrogen starvation under photoautotrophic conditions.
Supplementary Fig. S6. De-epoxidation state (A) of the violaxanthin cycle and violaxanthin cycle pigment pool size (B) of the pdh and EV lines of C. reinhardtii cells at the onset and after 2 d of nitrogen starvation under photoautotrophic conditions.
Supplementary Fig. S7. Photosynthetic activity of the pdh and EV lines of C. reinhardtii cells at the onset of the experiment, and after 6 d in HSM+N and 2 d in HSM-N as manifested by P-I curves recorded via oxygen evolution.
Supplementary Fig. S8. Typical curves of chlorophyll fluorescence induction (OJIP) recorded in the initial culture of the pdh and EV lines of C. reinhardtii grown in TAP (A) and after 2 d of nitrogen starvation in HSM-N (B).
Acknowledgements
This research was financially supported by the European Commission’s Seventh Framework Program for Research and Technology Development (FP7), project GIAVAP, grant no. 266401. NS acknowledges support from the Albert Katz International School for Desert Studies and the Kreitman School of Advanced Graduate Studies at Ben-Gurion University. We would like to thank Prof. Ivo Feussner and Dr. J. Popko for the GC mass spectrometry analysis of 4,4-dimethyloxazoline derivatives of FAs.
Glossary
Abbreviations:
- ALA
α-linolenic acid
- ACS
acetyl-CoA synthase
- amiRNA
artificial microRNA
- Chl
chlorophyll
- cp
chloroplast
- DGDG
digalactosyldiacylglycerol
- DGTS
diacylglyceryl-N,N,N-trimethylhomoserine
- DLA2
dihydrolipoamide acetyltransferase
- DW
dry weight
- EV
empty vector
- FA
fatty acid
- GC
gas chromatography
- HSM
high-salt minimal media
- MGDG
monogalactosyldiacylglycerol
- NL
neutral lipids
- NPQ
non-photochemical quenching
- OA
oleic acid
- PDC
pyruvate dehydrogenase complex
- PDH
pyruvate dehydrogenase
- PEP
phosphoenolpyruvate
- P-I
photosynthesis/irradiance
- PL
polar lipids
- PSII
photosystem II
- PUFA
polyunsaturated fatty acid
- qPCR
quantitative PCR
- SQDG
sulfoquinovosyl diacylglycerol
- TAG
triacylglycerol
- TAP
Tris-acetate-phosphate medium
- TFA
total fatty acids
- VAZ
violaxanthin, antheraxanthin, zeaxanthin.
References
- Behal RH, Lin M, Back S, Oliver DJ. 2002. Role of acetyl-coenzyme A synthetase in leaves of Arabidopsis thaliana . Archives of Biochemistry and Biophysics 402, 259–267. [DOI] [PubMed] [Google Scholar]
- Blaby IK, Glaesener AG, Mettler T, et al. 2013. Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25, 4305–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Bohne AV, Schwarz C, Schottkowsk M, Lidschreiber M, Piotrowski M, Zerges W, Nickelsen J. 2013. Reciprocal regulation of protein synthesis and carbon metabolism for thylakoid membrane biogenesis. PLoS Biology 11, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgis F, Kilaru A, Cao X, Ngando-Ebongue GF, Drira N, Ohlrogge JB, Arondel V. 2011. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proceedings of the National Academy of Sciences, USA 108, 12527–12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, et al. 2012. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. Journal of Biological Chemistry 287, 15811–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Ma X, Msanne J, Repas T. 2011. RNA-Mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryotic Cell 10, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT. 2013. Commentary: Why don’t plant leaves get fat? Plant Science 207, 128–134. [DOI] [PubMed] [Google Scholar]
- Davey MP, Horst I, Duong GH, Tomsett E, Litvinenko AC, Howe CJ, Smith AG. 2014. Triacylglyceride production and autophagous responses in Chlamydomonas reinhardtii depend on resource allocation and carbon source. Eukaryotic Cell 3, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne GJ. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Facchinelli F, Weber APM. 2011. The metabolite transporters of the plastid envelope: an update. Frontiers in Plant Science 2, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Andre C, Xu C. 2011. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii . FEBS Letters 585, 1985–1991. [DOI] [PubMed] [Google Scholar]
- Fan J, Yan C, Andre C, Shanklin J, Schwender J, Xu C. 2012. Oil accumulation is controlled by carbon precursor supply for fatty acid synthesis in Chlamydomonas reinhardtii . Plant and Cell Physiology 53:1380–1390. [DOI] [PubMed] [Google Scholar]
- Giroud C, Gerber A, Eichenberger W. 1988. Lipids of Chlamydomonas reinhardtii. Analysis of molecular species and intracellular site(s) of biosynthesis. Plant and Cell Physiology 29, 587–595. [Google Scholar]
- Goodenough U, Blaby I, Casero D, et al. 2014. The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii . Eukaryotic Cell 13, 591–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson C, Roth R, Wang ZT, Goodenough U. 2011. Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryotic Cell 10, 1592–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. 1989. The Chlamydomonas sourcebook . San Diego, CA: Academic Press. [Google Scholar]
- Heddad M, Willig A, Delessert C, Rahire M, Rochaix JD. 2006. DNA analysis. In: EMBO Practical Course. Molecular Genetics of Chlamydomonas. Laboratory protocols . http://www.chlamy.org/methods.html. [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant Journal 54, 621–639. [DOI] [PubMed] [Google Scholar]
- Jinkerson RE, Radakovits R, Posewitz MC. 2013. Genomic insights from the oleaginous model alga Nannochloropsis gaditana . Bioengineered 4, 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Behal RH, Back SL, Nikolau BJ, Wurtele ES, Oliver DJ. 2000. The role of pyruvate dehydrogenase and acetyl-coenzyme A synthetase in fatty acid synthesis in developing Arabidopsis seeds. Plant Physiology 123, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khozin-Goldberg I, Shrestha P, Cohen Z. 2005. Mobilization of arachidonyl moieties from triacylglycerols into chloroplastic lipids following recovery from nitrogen starvation of the microalga Parietochloris incisa . Biochimica et Biophysica Acta 1738, 63–71. [DOI] [PubMed] [Google Scholar]
- Kindle KL. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii . Proceedings of the National Academy of Sciences, USA 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U. 1986. Compartmentation of glycolysis and of the oxidative pentose-phosphate pathway in Chlamydomonas reinhardii . Planta 167, 81–86. [DOI] [PubMed] [Google Scholar]
- Li J, Han D, Wang D, Ning K, et al. 2014. Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae Plant Cell 26, 1645–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C. 2012. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii . Plant Cell 24, 4670–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Behal R, Oliver DJ. 2003. Disruption of plE2, the gene for the E2 subunit of the plastid pyruvate dehydrogenase complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Molecular Biology 52, 865–872. [DOI] [PubMed] [Google Scholar]
- Lin M, Oliver DJ. 2008. The role of acetyl-coenzyme A synthetase in Arabidopsis. Plant Physiology 147, 1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Benning C. 2013. Lipid metabolism in microalgae distinguishes itself. Current Opinion in Biotechnology 24, 300–309. [DOI] [PubMed] [Google Scholar]
- Lohr M, Im CS, Grossman AR. 2005. Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii . Plant Physiology 138, 490–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson G. 2000. Chlorophyll fluorescence—a practical guide. Journal of Experimental Botany 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J. 2012. TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Current Opinion in Biotechnology 23, 352–363. [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii . Eukaryotic Cell 9, 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. 2009. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii . The Plant Journal 58, 165–174. [DOI] [PubMed] [Google Scholar]
- Pal D, Khozin-Goldberg I, Didi-Cohen S, Solovchenko A, Batushansky A, Kaye Y, Sikron N, Samani T, Fait A, Boussiba S. 2013. Growth, lipid production and metabolic adjustments in the euryhaline eustigmatophyte Nannochloropsis oceanica CCALA 804 in response to osmotic downshift. Applied Microbiology and Biotechnology 97, 8291–8306. [DOI] [PubMed] [Google Scholar]
- Perez-Perez ME, Florencio FJ, Crespo JL. 2010. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii . Plant Physiology 152, 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid EE, Thompson P, Lyttle CR, Dennis DT. 1977. Pyruvate dehydrogenase complex from higher plant mitochondria and proplastids. Plant Physiology 59, 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismani-Yazdi H, Haznedaroglu BZ, Hsin C, Peccia J. 2012. Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnology for Biofuels 5, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Moriyama T, Sato N. 2014. Detailed identification of fatty acid isomers sheds light on the probable precursors of triacylglycerol accumulation in photoautotrophically grown Chlamydomonas reinhardtii . Eukaryotic Cell 13, 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Cuiné S, Cagnon C, Fessler B, et al. 2011. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnology 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SR, Abbriano RM, Hildebrand M. 2012. Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Research 1, 2–16. [Google Scholar]
- Solovchenko A. 2012. Physiological role of neutral lipid accumulation in eukaryotic microalgae under stresses. Russian Journal of Plant Physiology 59, 167–176. [Google Scholar]
- Solovchenko A, Solovchenko O, Khozin-Goldberg I, Didi-Cohen S, Pal D, Cohen Z, Boussiba S. 2013. Probing the effects of high-light stress on pigment and lipid metabolism in nitrogen-starving microalgae by measuring chlorophyll fluorescence transients: studies with a Δ5 desaturase mutant of Parietochloris incisa (Chlorophyta, Trebouxiophyceae). Algal Research 2, 175–182. [Google Scholar]
- Strasser R, Tsimilli-Michael M, Srivastava A. 2004. Analysis of the chlorophyll a fluorescence transient. In: Papageorgiou G, Govindjee, eds. Chlorophyll a fluorescence: a signature of photosynthesis . Heidelberg: Springer, 321–362. [Google Scholar]
- Tardif M, Atteia A, Specht M, et al. 2012. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Molecular Biology and Evolution 29, 3625–3639. [DOI] [PubMed] [Google Scholar]
- Terashima M, Specht M, Hippler M. 2011. The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Current Genetics 57, 151–168. [DOI] [PubMed] [Google Scholar]
- Wase N, Black PN, Stanley BA, DiRusso CC. 2014. Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. Journal of Proteome Research 13, 1373–1396. [DOI] [PubMed] [Google Scholar]
- Williams JA, Dijk K, Sarka N. 2011. Selectable and inheritable gene silencing through RNA interference in the unicellular alga Chlamydomonas reinhardtii . In: Williams JA, ed Strain engineering . New York: Humana Press, 457–476. [DOI] [PubMed] [Google Scholar]
- Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LM, Dismukes GC, Posewitz MC. 2010. Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryotic Cell 9, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.