Highlight text
This study presents the genome-wide characterization of the Populus WRKY family under biotic and abiotic stresses. Overexpression of an SA-inducible gene, PtrWRKY89, enhanced resistance to pathogens in transgenic poplar.
Key words: Pathogen, Populus, SA (salicylic acid), stress tolerance, transcription factor, WRKY.
Abstract
WRKY proteins are a large family of regulators involved in various developmental and physiological processes, especially in coping with diverse biotic and abiotic stresses. In this study, 100 putative PtrWRKY genes encoded the proteins contained in the complete WRKY domain in Populus. Phylogenetic analysis revealed that the members of this superfamily among poplar, Arabidopsis, and other species were divided into three groups with several subgroups based on the structures of the WRKY protein sequences. Various cis-acting elements related to stress and defence responses were found in the promoter regions of PtrWRKY genes by promoter analysis. High-throughput transcriptomic analyses identified that 61 of the PtrWRKY genes were induced by biotic and abiotic treatments, such as Marssonina brunnea, salicylic acid (SA), methyl jasmonate (MeJA), wounding, cold, and salinity. Among these PtrWRKY genes, transcripts of 46 selected genes were observed in different tissues, including roots, stems, and leaves. Quantitative RT-PCR analysis further confirmed the induced expression of 18 PtrWRKY genes by one or more stress treatments. The overexpression of an SA-inducible gene, PtrWRKY89, accelerated expression of PR protein genes and improved resistance to pathogens in transgenic poplar, suggesting that PtrWRKY89 is a regulator of an SA-dependent defence-signalling pathway in poplar. Taken together, our results provided significant information for improving the resistance and stress tolerance of woody plants.
Introduction
Salicylic acid (SA) is a vital hormone and signal molecule in plants, synthesized from cinnamate in a reaction catalysed by phenylalanine ammonia lyase (PAL). The bulk of SA is produced from isochorismate in plants (Chen et al., 2009b ) and has many physiological functions such as temperature resistance (Dat et al., 1998; Janda et al., 1999; Clarke et al., 2004), salt resistance (Borsani et al., 2001; Tari et al., 2002; Karlidag et al., 2009; Palma et al., 2009), drought resistance (Munné-Bosch and Peñuelas, 2003; Cho et al., 2008) and ultraviolet radiation resistance (Yalpani et al., 1994; Mahdavian et al., 2008). Moreover, SA in plant defence responses is well-established because it is essential for the onset of the hypersensitive response (HR) and systemic acquired resistance (SAR) (Malamy et al., 1990).
Induced-pathogen accumulation of SA, or treatment with SA, leads to a rapid increase in the levels of reactive oxygen species (ROS) via inhibition of the activity of SA-binding protein (SABP), which converts H2O2 to H2O and O2 (Chen et al., 1993; Baker and Orlandi, 1995; Lamb and Dixon, 1997; Klessig et al., 2000). The change in cellular redox potential results in the reduction of the NONEXPRESSOR OF PR1 (NPR1) oligomer to its active monomeric form, which is then translocated into the nucleus and enhances the binding of TGA transcription factors to SA-responsive promoter elements as a transcriptional co-activator of SA-responsive genes, such as pathogenesis-related gene (PR1) (Dong, 2004; Loake and Grant, 2007). In addition, NPR1 regulates the SA-mediated expression of WRKY70 from Arabidopsis, which encodes an activator protein of SA-dependent defence marker genes (PR1, PR2, and PR5) (Uknes et al., 1992). However, the induction of WRKY70 expression by SA in npr1 plants was only eliminated at later time points, implying that WRKY70 expression is partially NPR1-independent (Li et al., 2004). Recently, Shim et al. (2013) reported that AtMYB44 was an NPR1-independent regulatory component that directly regulated WRKY70 expression. Therefore, SA-mediated defence signalling networks require transcription factors to regulate gene expression, which provides plants with a complex control mechanism against foreign pathogen attack.
In the past 15 years, major advances in WRKY transcription factor research have been made (Rushton et al., 2010). The WRKY genes have been greatly expanded in plant genomes due to successive duplication events, resulting in large gene families that include up to 74 members in Arabidopsis (Ülker and Somssich 2004), 79 in Arabidopsis lyrata (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY), 197 in Glycine max (Schmutz et al., 2010), 93 in Sorghum bicolor, 45 in barley (Mangelsen et al. 2008), and 81 in rice (Xie et al., 2005). WRKY genes encode transcription factors characterized by the presence of one or two 60 amino-acid WRKY motifs, including a very highly conserved WRKYGQK sequence together with a zinc-finger-like motif CX4–7-CX23–28-HX1–2-(H/C) that provides binding properties to DNA (see Eulgem et al., 2000, for a review). WRKY domains (WDs) probably evolved from an ancestral group IIc-like WRKY which derived from classical C2H2 fingers (Znf) via an intermediate that was structurally close to the BED Zn-finger (Babu et al., 2006; Brand et al., 2013). The WRKY family members were classified into three groups on the basis of both the number of WDs and the features of their zinc-finger-like motif (Eulgem et al., 2000). In general, members of group I typically have two WDs, whereas most proteins with one WD belong to group II. Members of group III contain a single WD, but the pattern of the zinc-finger motif is unique. Furthermore, group II has been divided into five subgroups according to phylogenetic analysis of the WDs (Eulgem et al., 2000; Zhang and Wang, 2005). Most of the WRKY proteins bind to the conserved W-box TTGACY in the promoters of target genes related to the SA signalling pathway, such as chitinase (Yang et al., 1999; Yamamoto et al., 2004), and PR genes, such as PR10 (Despres et al., 1995; Rushton et al., 1996). W-boxes have also been found in the promoter of Arabidopsis non-expressor of PR genes 1 (NPR1) as an important component of SA-mediated induction of PR1 (Yu et al., 2001).
More recently, increasing studies have focused on elucidating roles of WRKY factors in SA-biosynthesis, SA induction, and regulation of the signalling pathway. In Arabidopsis, the wrky54wrky70 double mutant showed a remarkably high level of free SA compared to wrky70 and wrky54 single mutants, implying that WRKY70 and WRKY54 play a role as negative regulators of SA biosynthesis (Wang et al., 2006). Bioinformatics analysis and expression studies demonstrated that WRKY28 and WRKY46 were transcriptional activators of ICS1 and PBS3, which are involved in SA biosynthesis and SA-glucoside (SAG) accumulation, respectively (van Verk et al., 2011). Yu et al (2001) categorized WRKY3, WRKY4, WRKY6, WRKY7, WRKY15, WRKY21, and WRKY26 into an NPR1-independent group, and WRKY62 to an NPR1-dependent group. WRKY53 also showed normal expression at early stages but greatly decreased levels of transcripts at later times after SA treatment in the npr1 mutant. Subsequently, it was reported that overexpression of WRKY4 greatly enhanced plant susceptibility to bacterial pathogens and suppressed pathogen-induced PR1 gene expression (Lai et al., 2008). WRKY38 and WRKY62, which were induced by SA in an NPR1-dependent manner, played a negative role in basal plant defence (Kim et al., 2008). WRKY18 was dependent on NPR1 in the plant defence response, and constitutive expression of WRKY18 did not induce, but rather markedly potentiated, developmentally regulated PR gene expression (Chen and Chen, 2002). A few WRKY transcription factors have been functionally characterized in Arabidopsis and rice (Eulgem et al., 2000; Rushton et al., 2010), but our understanding of precise functions of WRKY members in the SA signalling pathway remains insufficient.
SA and jasmonic acid (JA) response pathways are one of the best studied examples of defence-related signal cross-talk in an antagonistic interaction (Pieterse et al., 2009). For example, WRKY53 mediates negative crosstalk between pathogen resistance and senescence, which is most likely to be governed by the JA and SA equilibrium in Arabidopsis (Miao and Zentgraf, 2007). WRKY70 is downstream of NPR1 and MYB44 in an SA-dependent signalling pathway. Plants overexpressing WRKY70 showed constitutive expression of PR genes and suppression of several JA responses including expression of a subset of JA- and Alternaria brassicicola-responsive genes, indicating that WRKY70 has a pivotal role in determining the balance between SA-dependent and JA-dependent defence pathways (Li et al., 2004; Li et al., 2006; Shim et al., 2013).
With the completion of the poplar genome sequence (Tuskan et al., 2006), a number of WRKY members have been found in the whole-genome sequence data from P. trichocarpa Torr. & A. Gray (http://genome.jgi-psf-org/Poptrl_l/Poptrl_l.home.html). To date, however, there are only limited elaborate studies on the functional characterization of several WRKY genes in Populus. A recent study reported the molecular cloning and functional characterization of PtWRKY23 in poplar (P. tremula × P. alba), which was induced rapidly by Melampsora infection and SA treatments, and expression patterns of PtWRKY23 and AtWRKY23 orthologues are not necessarily identical (Levée et al., 2009), indicating that WRKY in poplar has a different function from WRKY orthologues in Arabidopsis. Misexpression of PtWRKY23 in transgenic poplar plants led to increased susceptibility to Melampsora infection compared with the wild type, implying that this may be caused by deregulation of genes that disrupt redox homeostasis and cell wall metabolism. However, the role of PtWRKY23 in the SA signalling pathway is still unknown.
In this study, we found that there is an expanded WRKY family with a total of 100 members in the Populus genome. A phylogenetic tree combining WRKY proteins from poplar, Arabidopsis, and other species was constructed to test their evolutionary relationships. Promoter analysis revealed that various cis-acting elements involved in stress and phytohormone responses were present in the promoter region of PtrWRKY genes. Transcriptome analysis showed that the majority (61) of the PtrWRKY genes were induced by the fungus Marssonina brunnea f.sp. multigermtubi, SA, methyl jasmonate (MeJA), wounding, and cold and salinity stresses. Furthermore, stress-response expression profiles were generated to screen candidate genes involved in signal transduction pathways initiated by SA in Populus. The function of an SA-inducible gene, PtrWRKY89, was characterized in transgenic poplar. Our results will be helpful for understanding the roles of the WRKY genes in poplar defence responses and provides valuable information for further identification of the functions of this significant gene family in Populus.
Materials and methods
Plant growth conditions and treatments
Plant material and growth conditions
Populus trichocarpa Torr. & A. Gray and P. tomentosa Carr. (clone 741) (Chinese white poplar) were grown in a greenhouse at 25°C under a 14/10h light/dark cycle.
Hormone treatments
SA (5mM in water) and MeJA [1mM in 0.1% (v/v) ethanol] were applied at the different concentrations as 5ml droplets on each plant. The treated plants were immediately covered with a transparent lid. The leaves were collected after 24h (Li et al., 2004). Additionally, the leaves applied for all stress treatments, pathogen infection, and RT-PCR analysis were excised from the second and third internodes.
Fungal inoculation
Leaves of three-month-old plants were inoculated with M. brunnea f.sp. multigermtubi and Dothiorella gregaria Sacc., respectively. Mycelial plugs (6mm) were placed on excised leaves (at least three leaves for each plant). These leaves were incubated in Petri dishes with humid filter paper in a humid chamber for 3 d (Huang et al., 2012).
Low temperature stress
The healthy, well-hydrated plants were transferred to a growth chamber at 4°C under the same light and photoperiodic conditions for 1h. After cold treatment, plants were allowed to recover at 20°C for 1h.
Wounding stress
For the wounding treatment, the young leaves of poplar plants were harvested after being punctured with sterile needles and placed at 20°C for 2h.
Salinity stress
The four-week-old seedlings were subjected to salt stress. Saline treatments had the NaCl concentrations of 100mM added to full-strength Hoagland’s solution for 2 d. The method was described previously (Yang et al., 2009a ).
Database search and sequence retrieval
The gene model IDs of the P. trichocarpa WRKY family were obtained from the DATF website (http://planttfdb.cbi.pku.edu.cn/index.php). The nucleotide and amino acid sequences of the WRKY genes were downloaded from Phytozome v9.1 website (http://www.phytozome.net/poplar). The amino acid sequences of these genes, which could not be detected in Phytozome v9.1 via ID numbers, were used as queries to perform BLAST searches against the P. trichocarpa genome database in Phytozome v9.1. After the identification of the Populus WRKY (PtrWRKY) genes, gene orthologue analysis was performed using the WRKY gene sets from Arabidopsis based on previously reported results (He et al., 2012). The catalogue of WRKY proteins in poplar is shown in Supplementary Table S1. Sequences of the AtWRKY genes were searched and downloaded from the Arabidopsis genome TAIR 9.0 website (http://www.Arabidopsis.org/index.jsp). Sequences of WRKY proteins from other species were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/) and the accession numbers are shown in Supplementary Table S2.
Phylogenetic analysis
The alignments of the amino acid sequences of full PtrWRKY proteins and WDs were performed using Clustal X 1.81 (http://www.clustal.org/) (Thompson et al., 1994, 1997). The parameters of alignment used were as follows: gap opening penalty, 10.00 (both in pairwise alignment and multiple alignment); gap extension penalty, 0.20 (both in pairwise alignment and multiple alignment); protein weight matrix, gonnet; residue-specific penalties, on; hydrophilic penalties, on; gap separation distance, 0; end-gap separation, on; use negative matrix, off; and delay divergent cutoff (%), 30. Phylogenetic trees were constructed through the neighbour-joining (NJ) method using program MEGA4.1 (http://www.megasoftware.net/mega.html) (Tamura et al., 2007). The parameters of the constructed trees were: phylogeny test and options: bootstrap (1000 replicates; random seed = 9928), gaps/missing data: complete deletion, model: amino: Poisson correction, substitutions to include: all, pattern among lineages: same (homogeneous), and rates among sites: uniform rates.
In silico analysis of regulatory elements in promoter region of PtrWRKY genes
The transcription start site was designated +1. The elements in the promoter fragments (from –1500 to +1bp) of PtrWRKY genes were found using the program PlantCARE online (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Gene expression analysis
Digital transcriptomics analysis
Digital gene expression (DGE) experiments were performed as described by Zhang et al. (2010). Briefly, total RNA was isolated from poplar leaves treated with SA andMeJA, salinity stress, low temperature, mechanical wounding, and infection with M. brunnea f.sp. multigermtubi, respectively. Approximately 1 μg of total RNA per sample was incubated with oligo (dT) beads to capture the polyadenylated RNA fraction. First-strand cDNA was synthesized using random hexamer-primer and reverse transcriptase (Invitrogen). The second-strand cDNA was synthesized using RNase H (Invitrogen) and DNA polymerase I (New England BioLabs). The cDNA libraries were then prepared according to protocols of Illumina. To summarize, one individual single-end cDNA library was constructed for each sample, and then sequenced on the Illumina GA platform for 35 cycles. The paired-end libraries were sequenced for 44–75bp.
The P. trichocarpa genome and annotated gene set were downloaded from the DOE Joint Genome Institute website (http://genome.jgi-psf.org/cgi-bin/), and 17 273 P. trichocarpa full-length cDNAs were collected from the reference genome sequence of Populus (version 2.0, Phytozome). The cDNAs were aligned to the P. trichocarpa genome and those with identities higher than 80% were retained for further analysis. After removing reads containing sequencing adapters and reads of low quality (containing Ns > 5), we aligned reads to the P. trichocarpa genome using SOAP (Li et al., 2008), allowing for a 2bp mismatch between the tag and reference transcriptome. Reads that failed to be mapped were progressively trimmed off one base from the 39-end and mapped to the genome again until a match was found (P-value ≤ 0.05). For paired-end reads, the insert size was set between paired reads at 1bp to 10kb to allow reads spanning introns of different sizes. A similar strategy was used to align reads to the non-redundant gene set, but the insert length range was restricted to 1kb for paired-end read mapping.
Semi-quantitative RT-PCR analysis
Total RNA from fresh tissues of poplar (P. trichocarpa) plants was extracted using RNA RNeasy PlantMiniKit (Qiagen, Germany) according to the manufacturer’s instructions with a modification as reported previously (Jia et al., 2010). Samples from at least three plants were pooled for analysis. The total RNA before cDNA synthesis was treated with RNase-free DNase (TaKaRA, Dalian, China), according to the manufacturer’s instructions to avoid any genomic DNA contamination. First-strand cDNA was synthesized from 2 µg RNA with RT-AMV transcriptase (TaKaRa, Dalian, China) in a total volume of 20 μl by using oligo (dT)18 at 42°C for 30min. 18S rRNA was used as an internal control. The amplification products of RT-PCR were resolved by 1% (w/v) agarose gel electrophoresis and visualized with ethidium bromide under UV light to test the expression levels of PR genes in transgenic plants.
Quantitative RT-PCR analysis
Verification of transcriptome data was conducted by checking the expression profiles of a number of the representative PtrWRKY genes in various tissues by quantitative RT-PCR (qRT-PCR) analysis. The gene-specific primers used for semi-qRT-PCR and qRT-PCR analysis are shown in Supplementary Table S3. RT-PCR analysis was based on at least two biological replicates of each sample and three technical replicates of each biological replicate.
Cloning of PtrWRKY89
The full open-reading frame of PtrWRKY89 was amplified with gene-specific primers (forward, 5′-AGTTCTTGACACCCACCACTC-3′; reverse, 5′- GGAAAATACAAAGAGGCTGC-3′; Joint Genome Institute, http://genome.jgi-psf.org/poplar/poplar.info.html) by RT-PCR with 2 μl cDNA from leaves. The PCR reaction was carried out with Pfu DNA polymerase (TaKaRa) in a total volume of 50 μl with an initial denaturing step at 94°C for 3min, 34 cycles of 94°C for 45 s, 54°C for 30 s, and 72°C for 90 s, and a final extension step at 72°C for 10min. The amplification products were cloned into the plant binary vector pCXSN, which is a zero-background TA cloning system that provides simple and high-efficiency direct cloning of PCR-amplified DNA fragments (Chen et al., 2009a ). The resulting vector p35S:PtrWRKY89, containing the PtrWRKY89 open-reading frame under the control of the cauliflower mosaic virus (CaMV) 35S promoter and the hygromycin phosphortransferase gene (Hpt) as a plant-selectable marker conferring hygromycin resistance was transferred into Agrobacterium tumefaciens EHA105 by the freeze-thaw method.
Transformation of P. tomentosa plants
A. tumefaciens strain EHA105 containing 35S:PtrWRKY89 was incubated in liquid yeast extract peptone medium supplemented with 100 mmol l–1 acetone-syringone at 18°C with constant shaking (200rpm) until the culture reached an optical density of 0.8 at OD600nm. The A. tumefaciens culture was then diluted with one volume of liquid woody plant medium (WPM) (Lloyd and McCown, 1980).
Poplar transformation methods were described previously by Jia et al. (2010). Leaves of P. tomentosa were excised from in vitro plantlets, cut into disks, and dipped in the diluted Agrobacterium culture for 8–10min. After excess liquid on the surfaces was absorbed by sterilized paper, the leaf disks were transferred to WPM medium [2.0mg l–1 zeatin, 1.0mg l–1 1-naphthalene acetic acid (NAA)]. The infected disks were co-cultivated in the dark for 2 d and then transferred to callus-inducing medium containing 2.0mg l–1 zeatin, 1.0mg l–1 NAA, 400mg l–1 cefotaxime, 9mg l–1 hygromycin, and 0.8% (w/v) agar. After 2–3 weeks of culture in the dark, these leaf disks with induced calli were subcultured on screening medium [2.0mg l–1 zeatin, 0.1mg l–1 NAA, 400mg l–1 cefotaxime, 9mg l–1 hygromycin, and 0.8% (w/v) agar] to induce adventitious buds. Regenerated shoots were transferred to rooting medium containing 0.1mg l–1 NAA, 400mg l–1 cefotaxime, and 9mg l–1 hygromycin. Transgenic plants were selected with 9mg l–1 hygromycin. Rooted plantlets were acclimatized in pots placed inside a humid chamber (16h photoperiod, 25°C, 70% relative humidity) for 2 weeks and finally transferred to the greenhouse.
Evaluation of transgenic plants for resistance against D. gregaria
To test the resistance of transgenic poplar against fungal infections, the in vivo test was performed with D. gregaria as described previously (Huang et al., 2012). Adobe Photoshop was used to calculate lesion area. Each experiment was performed with at least three replicates, and contained wild-type controls. All data were analysed by t-test at P ≤ 0.05, using the Origin 6.1 software version v6.1052 (B232) (OriginLab Corp, Northhampton, MA, USA).
Results
Identification of poplar WRKY transcription factors and phylogenetic comparison of the WRKYs from different species
WRKY proteins comprise a large family of transcription factors and have been found in various plant species (Eulgem et al., 2000; Xie et al., 2005; Rensing et al., 2008). In this study, a genome-wide analysis was carried out to identify WRKY genes in the P. trichocarpa genome using the publicly available genomic and putative full-length protein sequences, which were mainly downloaded from Phytozome v9.1 (http://www.phytozome.net/). Initially, we obtained 119 partial putative full-length protein sequences of WRKY genes in P. trichocarpa and their gene model IDs from the Plant Transcription Factor Database v2.0 (http://planttfdb.cbi.pku.edu.cn/). In an attempt to determine the reliability of these putative genes, the unique gene IDs for gene models were BLAST searched against Phytozome v6.0, resulting in a total of 105 members that were included in the WRKY superfamily identified above. Manual inspection of putative full-length protein sequences among these putative WRKY genes showed that three members contained only partial WDs whereas the other two members did not contain WDs or complete zinc finger motifs, and we removed both of them. The CDS sequences of 103 models were BLAST searched against Phytozome v9.1. Eventually we found a total of 100 members representing the unique poplar genes, and created consecutive nomenclature, designated as PtrWRKY1–PtrWRKY100 (Supplementary Table S1). These were used in the analysis.
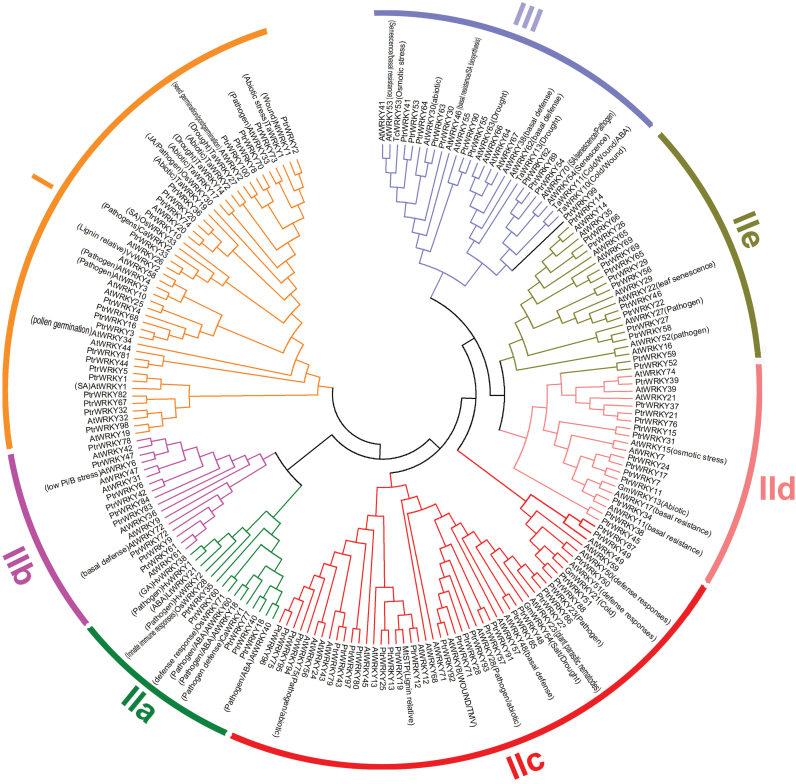
In general, transcription factor families contain a highly conserved domain or consensus motif involved in DNA binding, but the sequence similarity of other parts is relatively low in most genes (Wu et al., 2005). A conserved DNA domain is considered as an evolutionary unit whose coding sequence can be duplicated and/or undergo recombination. The most prominent structural feature of the WRKY proteins is the WD of 60 amino acid residues (Eulgem et al., 2000). All identified members of the PtrWRKY gene family contain either one or two WDs. In order to examine the phylogenetic relationships among Populus WRKY proteins, a multiple putative full-length protein sequence alignment of all putative 100 PtrWRKY proteins, 72 AtWRKYs from Arabidopsis, and 27 well-known WRKYs from other species was constructed using Clustal X (Thompson et al., 1994, 1997) and an unrooted tree was built using MEGA 4.1 (Tamura et al., 2007) by employing the NJ method. As shown in Fig. 1, this phylogram was classified into three groups (I, II, and III), based on the primary amino acid sequence. The WRKY members in group II can be further clustered into five subgroups (IIa–e) and a sole member, PtrWRKY99 (Fig. 1). Using the same method, the phylogenetic tree is also constructed based on WRKYdomainDs (Supplementary Figure S1). The members of group I contain two conserved WDs, an N-terminal WD (NT-WD) and a C-terminal WD (CT-WD), which are apparently clustered into two different clades (I-NT and I-CT), implying the retention of their divergence after duplication. The genes belonging to the NT-WDs and CT-WDs clades are assigned to IIa, IIb, and IIc, respectively, because they are obviously clustered into different clades. In addition, PtrWRKY99, which contains a C2H2 motif instead of C2HC, is clustered into subgroup III (Supplementary Figure S1).
Fig. 1.
Phylogenetic tree of WRKY proteins from poplar, Arabidopsis and other species. The complete amino acid sequences of 100 poplar and 72 Arabidopsis WRKY proteins combined with 27 WRKY proteins from other plants were aligned by ClustalW, and the NJ tree was constructed using MEGA 4.1 with 1000 bootstrap replicates. Pathogen, pathogen response; Abiotic, abiotic stress response; SA, SA response; JA, JA response; ABA, ABA response; GA, GA response; TMV, tobacco mosaic virus response. This figure is available in colour at JXB online.
Variety of cis-elements in the promoter regions of Populus WRKY genes
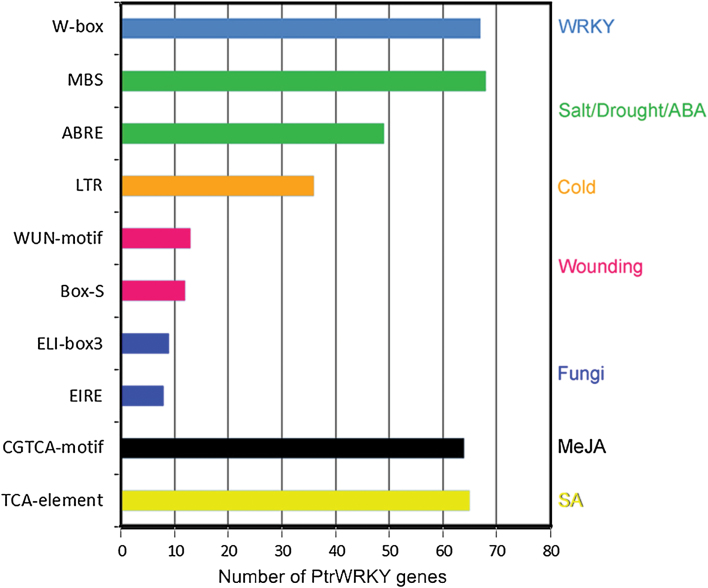
Regulatory elements of the promoter sequences are essential to temporal, spatial, and cell type-specific control of gene expression (Lescot et al., 2002). Here we searched 1500-bp upstream promoter regions of all putative Populus WRKY genes in the plant promoter database PlantCARE, and a number of cis-acting elements related to stresses as well as phytohormone responses were found. As shown in Fig. 2, SA-responsive elements (TCA-elements) and MeJA-responsive elements (CGTCA-motif) were found in the promoters of 65 and 64 PtrWRKY genes, respectively. Both of them existed in the promoter regions of 40 genes together. The conserved fungus-responsive elements, such as EIRE and ELI-box3, were present in the promoter regions of eight and nine PtrWRKYs, respectively. More than 12 promoters contained Box-S and WUN motifs involved in wounding stress. The low-temperature-responsive element (LTR) was found in 36 promoters of PtrWRKY genes. MYB-binding sites (MBSs) and ABA-responsive elements (ABREs) were abundant in PtrWRKY gene promoters and these elements were found in 68 and 49 promoters, respectively. In addition, WDs bound to W-boxes (TTGACC) with the highest affinity; the invariant TGAC core of the W-box is essential for function and WRKY binding (Brand et al., 2010; Brand et al., 2013). There were several W-boxes in the promoters of 67 PtrWRKYs, indicating that these genes might be regulated by other WRKY proteins or themselves (Supplementary Table S4). These results indicated that the Populus WRKY transcription factors are involved in the transcriptional control of the defence and stress responses.
Fig. 2.
The number of WRKY genes containing various cis-acting elements. W-box, WRKY binding site; MBS, MYB binding site; ABRE, ABA-responsive element; LTR, cis-acting element involved in low-temperature responsiveness; WUN-motif: wound-responsive element; BOX-S, wounding responsive element; ELI-box3, elicitor-responsive element; EIRE, elicitor-responsive element; CGTCA-motif, cis-acting regulatory element involved in MeJA-responsiveness; TCA-element, cis-acting element involved in SA responsiveness. This figure is available in colour at JXB online.
Digital transcriptomics analysis of Populus WRKY genes
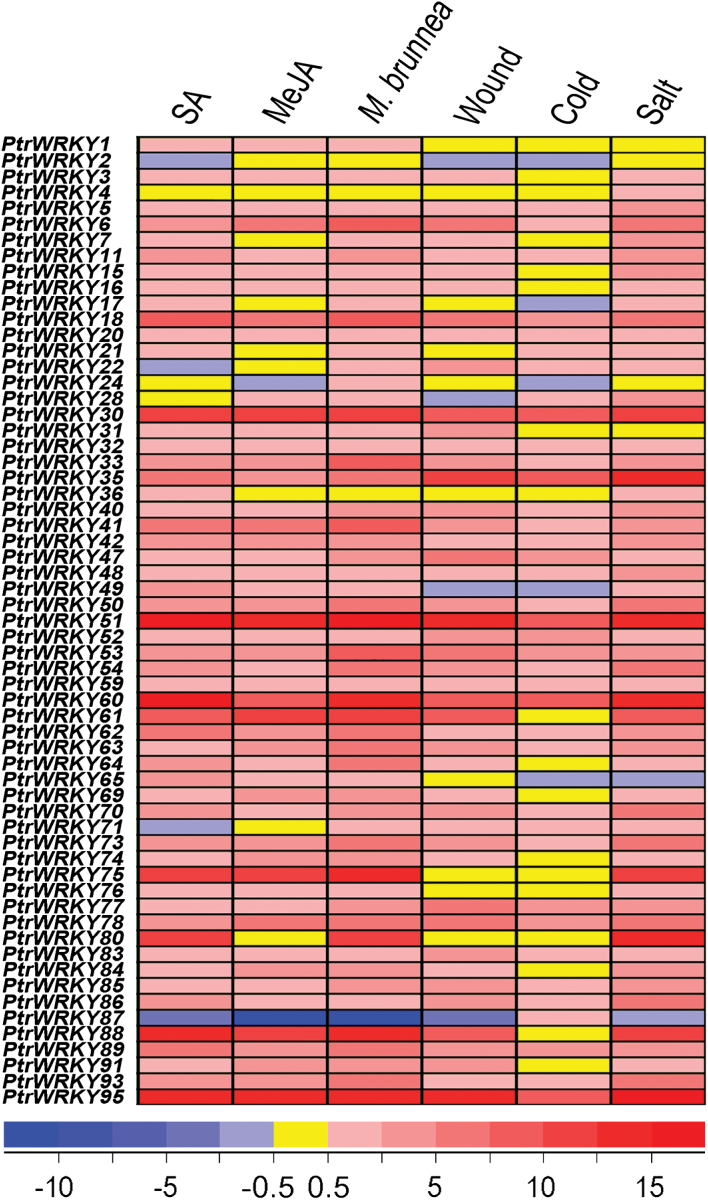
To determine the potential roles of PtrWRKY genes in plant responses to various environmental stresses, induction experiments with several treatments were conducted with wild-type poplar plants. Total RNA was isolated from leaves of control, M. brunnea-infected, SA- and MeJA-treated, wound treated, low temperature-treated, and salinity treated plants, respectively. Global transcriptomics analysis revealed that transcript abundance of 61 PtrWRKY genes changed significantly after treatments compared to control plants (Fig. 3; more detailed information is listed in Supplementary Table S5). The majority of PtrWRKY genes were induced by both SA and MeJA, except for PtrWRKY2, PtrWRKY22, PtrWRKY24, PtrWRKY71, PtrWRKY80 and PtrWRKY87. For example, PtrWRKY51 and PtrWRKY95, belonging to group IIc, were activated by both SA and MeJA treatments, resulting in an obvious increase in mRNA level. Expression of PtrWRKY80 was apparently increased in SA-treated plants, whereas no change in its expression was observed when treated with MeJA, indicating that PtrWRKY80 could be only involved in the SA signalling pathway. In contrast, PtrWRKY71 expression was downregulated significantly after SA treatment (Fig. 3). In addition, most of the PtrWRKY genes exhibited a gradual increase in expression levels in response to infection of the pathogen M. brunnea (Fig. 3 and Supplementary Table S5). Similar results were obtained in the genome-wide expression analysis of the WRKY gene superfamily in rice following SA- and MeJA-treatments, and pathogen infection (Ryu et al., 2006). Meanwhile, 61 PtrWRKY genes were induced by wounding, low temperature, or salinity, except for PtrWRKY1 (Supplementary Table S5). Transcript levels of PtrWRKY75 and PtrWRKY80 showed no change under mechanical wounding and low temperature treatments, but their mRNA levels were enhanced after salinity treatment. Expression of PtrWRKY61 and PtrWRKY88 increased after wounding and salinity treatments, but no change was detected under low temperature. In Arabidopsis, AtWRKY18, AtWRKY40, and AtWRKY60 play important roles in plant responses to both abiotic and biotic stress (Chen et al., 2010a ; Wenke et al., 2012), and their Populus orthologues, PtrWRKY18, PtrWRKY35, and PtrWRKY60, were transcriptionally upregulated by all treatments, indicating that these three PtrWRKYs are associated with responses towards biotic and abiotic stimuli. Based on the results presented here, we speculate that the functional divergence of PtrWRKY proteins plays a critical role in the responses of poplar plants to various stresses.
Fig. 3.
Digital transcriptomics analysis of PtrWRKY genes shows that 61 PtrWRKY genes are significantly sensitive to SA, MeJA, M. brunnea f.sp. multigermtubi, wounding, cold, and salinity stresses. Levels of transcript accumulation are shown. Each column represents a discreet biological sample. This figure is available in colour at JXB online.
Expression profiles of Populus WRKY genes in poplar
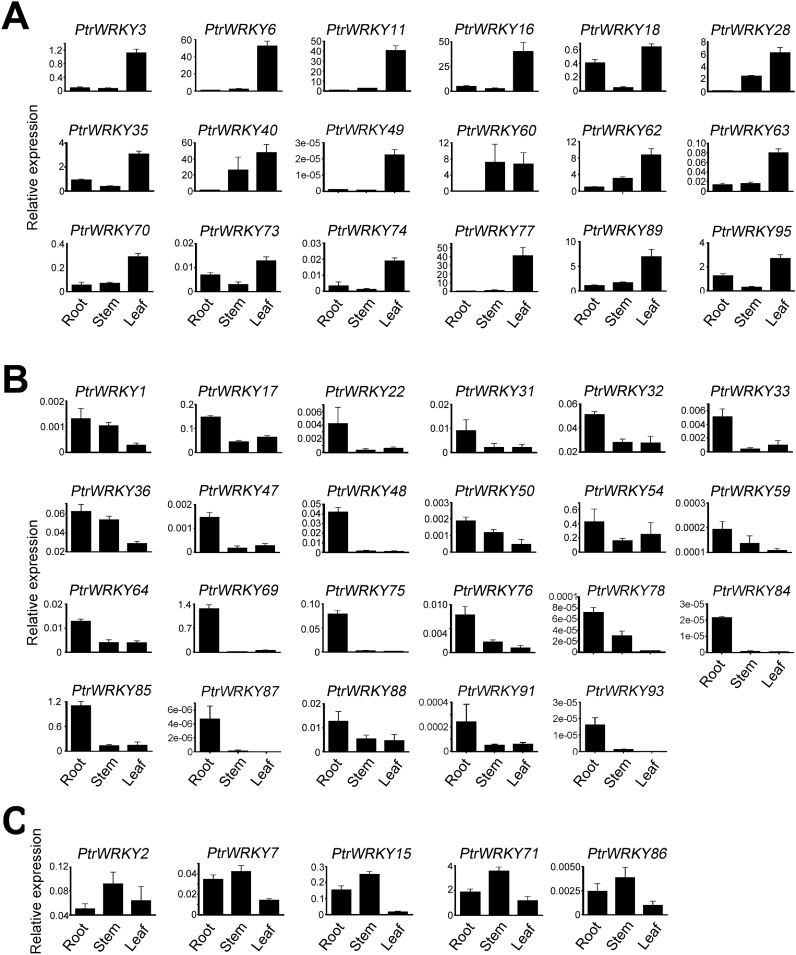
To investigate the tissue-specific expression of the Populus WRKY genes, qRT-PCR was used to determine the expression patterns of 61 PtrWRKY genes in various tissues including roots, stems, and leaves. Expression of only 46 Populus WRKY genes could be detected at the transcript level and these genes showed a diverse range of tissue-specific expression patterns in different tissues (Fig. 4). Most of Populus WRKY genes were more highly expressed in roots and leaves than in stems. Expressions of 14 PtrWRKYs, including PtrWRKY3, 6, 11, 16, 28, 35, 49, 62, 63, 70, 74, 77, 89 and 95, occur preferentially in leaves (Fig. 4A). Transcript levels of PtrWRKY40 and PtrWRKY60 were lower in roots but mRNA accumulation in stems and leaves was nearly similar. PtrWRKY18 and PtrWRKY73 showed higher expression levels in roots and leaves compare to stems. Both PtrWRKY62 and PtrWRKY89, which are orthologous genes of AtWRKY70 (Fig. 1), shared a similar expression patterns, indicating that they might play specific or redundant functional roles in poplar. For the other 23 PtrWRKY genes, high mRNA levels were observed in roots and relatively low but unambiguous expression was evident in stems and leaves (Fig. 4B). Additionally, five PtrWRKY genes, including PtrWRKY2, PtrWRKY7, PtrWRKY15, PtrWRKY71 and PtrWRKY86, were mainly expressed in stems (Fig. 4C).
Fig. 4.
Expression analysis of 46 PtrWRKY genes using qRT-PCR. Relative quantities of PtrWRKY members in root, stem, and leaf are illustrated. (A–C) The members have highest expression levels in leaves, stems, and roots, respectively. Error bars result from three biological replicates. Poplar 18S expression was used as a control and gene-specific primers were used for qRT-PCR analysis of Populus WRKY genes.
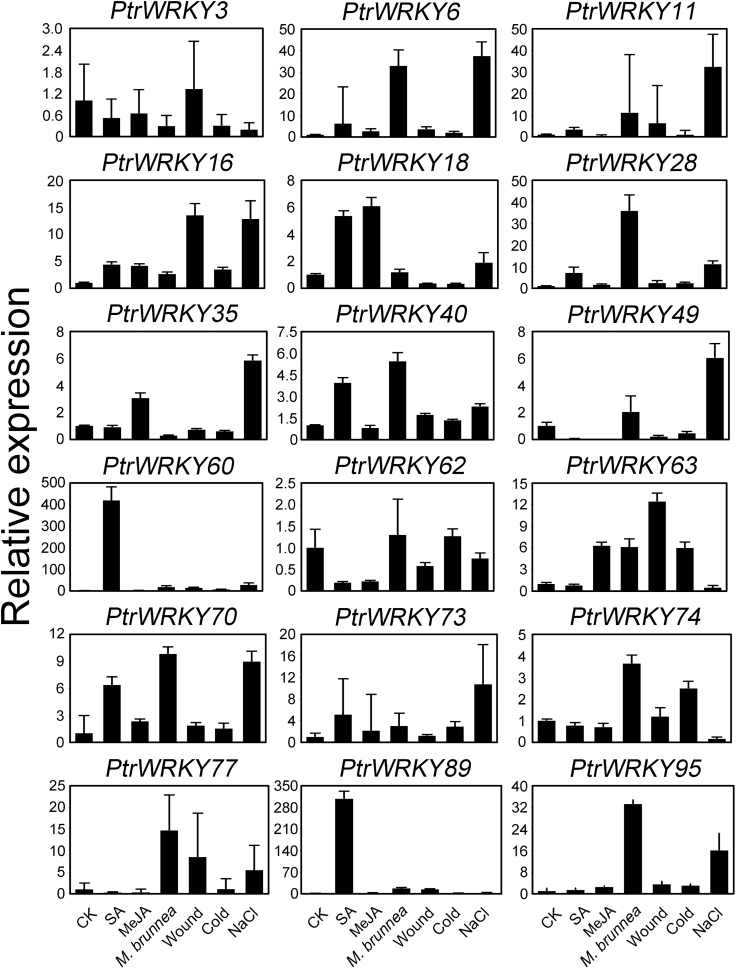
Expression patterns of 18 Populus WRKY genes in response to different treatments
To further confirm if the expression of Populus WRKY genes was induced by different biotic and abiotic stresses, 18 PtrWRKY members, whose mRNA levels were relatively high in leaves, were selected and qRT-PCRs were performed to analyse their expression patterns in response to these treatments. Overall, transcript levels of all PtrWRKY genes tested were detected to respond to at least one treatment (Fig. 5). Among them, three PtrWRKY genes were significantly induced by only one treatment. For instance, PtrWRKY28 responded to M. brunnea, whereas SA induced a remarkable increase in mRNA levels of PtrWRKY60 and PtrWRKY89. Several PtrWRKY genes, such as PtrWRKY6, PtrWRKY16, PtrWRKY18, PtrWRKY35, PtrWRKY40, PtrWRKY62 and PtrWRKY74, were able to respond to two treatments. For example, PtrWRKY18 was induced by both SA and MeJA, indicating that it might be a node of convergence for JA-mediated and SA-mediated signal transduction pathways. The expression of PtrWRKY40 was enhanced significantly after treatments with SA and M. brunnea (Fig. 5), implying that its role might be involved in pathogen resistance mediated by an SA signal.
Fig. 5.
Expression profiles of 18 Populus WRKY genes under different treatments. Leaves excised from poplar were sprayed with SA and MeJA, inoculated with M. brunnea f.sp. multigermtubi, and treated with wounding, cold, and salinity stresses. The untreated leaves were regarded as the control (CK). Leaves were collected for transcript profile analysis by qRT-PCR. Error bars result from three biological replicates. Poplar 18S expression was used as a control and gene-specific primers were used for qRT-PCR analysis of Populus WRKY genes.
Identification and sequence analysis of the PtrWRKY89 gene
As shown in Fig. 1, PtrWRKY89 has been classified into group III of the WRKY family and formed a subgroup with PtrWRKY54, PtrWRKY62, AtWRKY54 and AtWRKY70, whose domains are distinct from those of monocot species (He et al., 2012). To elucidate the role of PtrWRKY89 in SA signalling, we identified the PtrWRKY89 cDNA encoding a putative WRKY protein by RT-PCR with gene-specific primers based on the sequences deposited in Phytozome version 9.1. PtrWRKY89 appeared to be a full-lengh cDNA of 1002bp encoding a protein of 333 amino acids residues. PtrWRKY89 contains a typical WD with a C2HC-type zinc finger and putative nuclear localization signals. Interestingly, the putative full-length protein sequence of PtrWRKY89 exhibits low similarity to AtWRKY70 (48.38%) (Fig. 6), but their WDs were conserved (81.09%) (Fig. 6). Previous studies demonstrated that almost all WRKY III proteins were responsive to SA treatment (Kalde et al., 2003). Among them, AtWRKY38, AtWRKY46, AtWRKY53, AtWRKY54, AtWRKY62 and AtWRKY70 were early SA-induced, with highest expression levels 2h after SA treatment (Besseau et al., 2012). As shown in Fig, 1, PtrWRKY89 belongs to the same cluster as AtWRKY54 and AtWRKY70 characterized in Arabidopsis (Knoth et al., 2007). These results indicate that PtrWRKY89 is a potential transcriptional activator in the SA signalling pathway and is induced by SA at an early stage.
Fig. 6.
Comparison of PtrWRKY89-deduced amino acid sequence with PtrWRKY54, PtrWRKY62, AtWRKY54, and AtWRKY70 proteins. Identical amino acids are indicated by white letters on a black background, and conserved amino acids by black on a grey background. Asterisks indicate the nuclear localization signals. The WD is underlined. Putative full-length protein sequences were aligned with the DNAMAN program.
PtrWRKY89 is induced at an early stage by SA treatment
The expression pattern of PtrWRKY89 was analysed by semi-qRT-PCR at different time points when treated with SA. As shown in Fig. 7, expression of PtrWRKY89 was not induced by mock treatment. While the mRNA level of PtrWRKY89 increased 4-fold up to 2h after exposure to SA, the highest transcript level was detected from 5–8h. However, transcript accumulation decreased significantly after 24h of SA treatment. These data indicate that PtrWRKY89 is an early gene activated by the SA signal.
Fig. 7.
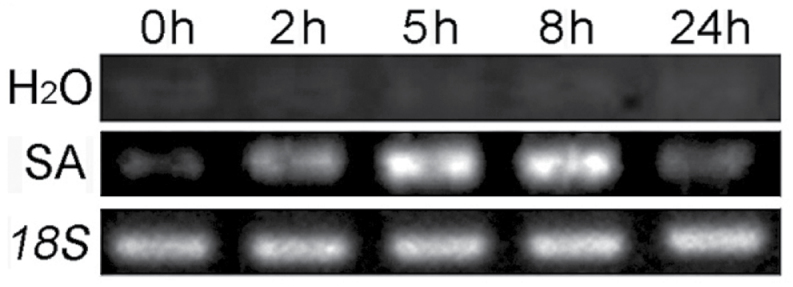
Induction profiles of PtrWRKY89. Leaves from poplar were sprayed with SA. Control plants were sprayed with H2O. Leaves were collected after 0, 2, 5, 8, and 24h for transcript profile analysis by semi-qRT-PCR. Poplar 18S expression was used as a control and gene-specific primers were used for semi-qRT-PCR analysis of PtrWRKY89.
Overexpression of PtrWRKY89 confers increased resistance to D. gregaria in transgenic poplar
To further investigate the roles of PtrWRKY89 in plant biotic stress responses, a plant binary construct containing a full-length PtrWRKY89 cDNA driven by the CaMV 35S promoter was generated and transformed into P. tomentosa by A. tumefaciens-mediated transformation. A total of 17 putative transformants with hygromycin resistance were obtained and grown in the greenhouse. No obvious phenotypic change was observed in transgenic plants when compared to wild-type plants (Supplementary Figure S3). PCR analysis using gene-specific primers was employed to confirm the presence of the transgenes in transformed plants. An expected amplification product of a 1202-bp PtrWRKY89 fragment combining a T-NOS terminal region was obtained from all transgenic lines tested, whereas no signal was detected from untransformed plants (Supplementary Figure S4), indicating the successful integration of the transgene into the poplar genome. From all independent transgenic lines containing the 35S:PtrWRKY89 construct, two lines (LC and LI) with high transcript levels of PtrWRKY89 were selected for further analysis.
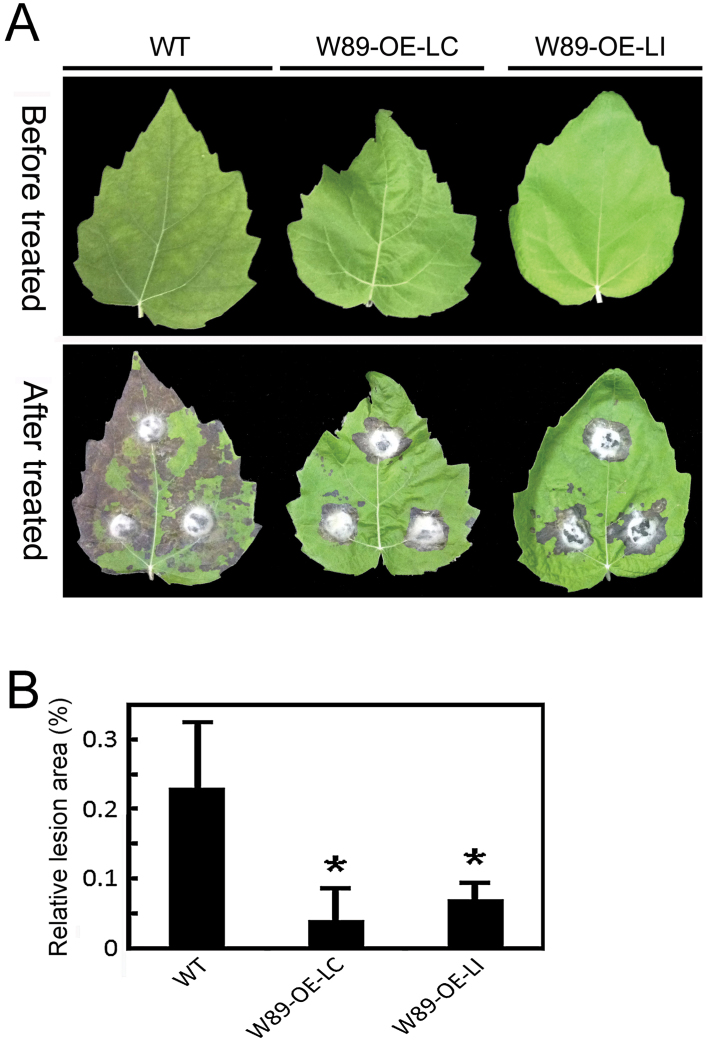
To determine the effect of PtrWRKY89 overexpression on disease resistance in poplar, leaves excised from transgenic and control lines were inoculated with agar plugs containing hyphae of D. gregaria, a hemibiotrophic fungus. Compared with the severe disease symptoms appeared on the control leaves at 3 d post inoculation (dpi), only slight necrotic lesions appeared on the leaves of the transgenic 35S:PtrWRKY89 lines tested (Fig. 8A). Quantification assays showed that the lesions were significantly (P < 0.05) smaller in 35S:PtrWRKY89 lines than in the control plants (Fig. 8B), indicating that PtrWRKY89 might act as a positive regulator of basal resistance to infection of hemibiotrophic fungal pathogens.
Fig. 8.
Resistance of transgenic poplar plants inoculated with D. gregaria. (A) The leaves from wild-type and transgenic plants before treatment and after infection with D. gregaria 3 d after inoculation were photographed. (B) Mean infected area of transgenic lines to the fungal pathogen; PtrWRKY89 confers resistance to D. gregaria in transgenic poplar plants. Values are means of three replications. Error bars indicate standard deviations. Asterisks indicate a statistically significant difference between wild-type and transgenic plants (P < 0.05 by student’s t-test). This figure is available in colour at JXB online.
Constitutive expression of the PtrWRKY89 gene in poplar resulted in the upregulation of several PR genes
Increased disease resistance in plants is often accompanied by the accumulation of elevated transcript levels of PR genes associated with the SA-mediated defence pathway (Uknes et al., 1992; Dong, 2004; Loake and Grant, 2007). Because exogenous SA-triggered expression of PtrWRKY89 and overexpression of PtrWRKY89 enhanced resistance to pathogens, we further determined whether PtrWRKY89 is directly involved in controlling expression of Populus PR genes. A genome-wide candidate gene screen found 15 putative Populus PR genes, homologues to PR1, PR2, and PR5 from Arabidopsis. Semi-qRT-PCR analysis showed that eight out of 15 PR genes, including PR1.2, PR2.3, PR2.6, and PR5s, were activated in PtrWRKY89-overexpressing lines (Fig. 9). Interestingly, the expression of other Populus PR genes was not upregulated in PtrWRKY89-overexpressing plants. In addition, the mRNA levels of NPR1.1, MYB44.1, MYB44.2, and SID2 showed no remarkable differences between wild-type and transgenic plants (Fig. 9). Similarly, AOS7 and JAZ10, involved in the JA-signalling pathway, showed no change (Fig. 9). Taken together, these results indicate that PtrWRKY89 might be one of the coactivators of SA-mediated defence marker genes in poplar.
Fig. 9.
Gene expression analyses of defence-related genes in plants overexpressing PtrWRKY89. Transcript levels of PR genes, MYB44.1, MYB44.2, NPR1.1, SID2, AOS7, and JAZ10 in wild-type and transgenic plants were elevated by semi-qRT-PCR. Poplar 18S expression was used as a control.
Discussion
Characterization of the Populus WRKY gene family
In a previous study, Zhu et al. (2007) proposed the existence of 104 WRKY family genes in the P. trichocarpa genome using the conserved WDs as the defining feature. He et al. (2012) further presented detailed information regarding the specifics of the individual Populus WRKY genes. However, in our study, only 100 members of the WRKY transcription factor superfamily were found in the P. trichocarpa genome. A new member, POPTR_0017s14630.1, was identified (Supplementary Table S1) and two members (POPTR_0003s20860.1 and POPTR_0810s00200.1) described by He et al. (2012) were removed in our database because they could not be found in the upgraded database; we also identified three members (POPTR_0014s08600.2, POPTR_0015s11130.2, and POPTR_0001s47670.2) that should be considered as alternative transcripts of POPTR_0014s08600.1, POPTR_0015s11130.1, and POPTR_0001s47670.1, respectively.
Classification and phylogenetic analysis of WRKY genes in Populus, Arabidopsis and other species
Phylogenetic comparison of the WRKY proteins has been conducted extensively in Arabidopsis, rice, canola, and poplar, and the evolutionary relationships of this gene family within and among the different species has been intensively studied (Eulgem et al., 2000; Xie et al., 2005; Yang et al., 2009b ; He et al., 2012). In Arabidopsis and rice, the WRKY members were classified into three large groups (I, II, and III) based on the number of the WD and the features of their zinc-finger-like motifs (Wu et al., 2005). However, group I contained two subgroups, Ia and Ib: members of subgroup Ia harboured two WDs; subgroup Ib contained these proteins with a single WD. Additionally, on the basis of the phylogenetic results, group II was divided into four subgroups (IIa–d) and group III divided into subgroups IIIa and IIIb (Wu et al., 2005). In Populus, three major groups were categorized as described by Wu et al. (2005). Several subgroups, such as Ia, Ib, and IIa–e, were clearly formed on the basis of the phylogenetic analysis (He et al., 2012). An alternative method also divided Arabidopsis WRKY members into three big groups (I, II, and III) (Eulgem et al., 2000). In detail, the members with two WDs with C2H2 zinc fingers belonged to group I, the members of group II encoded proteins containing a single WD and C2H2-type zinc finger, split up into five distinct subgroups (IIa–e) and the single WDs of the proteins including C2HC zinc fingers belonged to group III. However, further studies indicated that the spacing of the zinc fingers was responsible for the diversity of WRKYs (Zhang and Wang., 2005; Brand et al., 2013). For example, subgroup IIc contained C-X4-C-X22–23-HXH. Subgroup IIa, IIb, IId, and IIe shared C-X5-C-X23-HXH and group III contained the C-X4–7-C-X23-(24–30)-HXC pattern (Brand et al., 2013). According to the results reported by Eulgem et al. (2000) and Brand et al. (2013), we obtained an overall phylogenetic tree of the WRKY proteins from Arabidopsis, Populus and other species. Among the 100 putative PtrWRKY proteins, 22 members with two WDs and C2H2 zinc fingers belonged to group I. Most (68) PtrWRKY genes, which encoded proteins containing a single WD and C2H2-type zinc finger, belong to group II and there was also a sole member (PtrWRKY99) (Fig. 1 and Supplementary Figure S1). Generally, the WDs of group I and II members have the same type of zinc finger motif (C2H2), whose pattern of potential zinc ligands (C2H2) is unique among all zinc-like-motifs described (Eulgem et al., 2000). However, these WRKY members containing a C2HC motif, instead of a C2H2 pattern, were classified into group III (Eulgem et al., 2000). In Populus, 10 WRKY proteins with C2HC zinc fingers together with one WD belong to group III (Fig. 1). Interestingly, phylogenetic analyses revealed that PtrWRKY99 was clustered into subgroup III. However, this member has a typical C2H2-type zinc finger of group II (Eulgem et al., 2000). Based on the report by Brand et al. (2013), PtrWRKY99, which contained a C-X4-C-X21-HXH motif, should be assigned to group II but as a sole member (Fig. 1; Supplementary Figures S1 and S2). In addition, based on the conserved WDs, PtrWRKY49 and PtrWRKY87 with high similarity were separated from subgroup IId, IIe, and group III (Supplementary Figure S1). This finding was inconsistent with the results shown in Fig. 1, in which these two members and AtWRKY49 were clustered with subgroup IIc. In an attempt to further confirm the classification of PtrWRKY49 and PtrWRKY87, sequence alignment of their WDs was conducted. The result showed that WDs of these two members contained a C-X4-C-X23-HXH zinc finger (Supplementary Figure S2), indicating that PtrWRKY49 and PtrWRKY87 were typical members of subgroup IIc.
A phylogenetic tree combining transcription factor families from different species will not only help our understanding of the phylogenetic relationships among the members, but also allow speculation on the putative functions of the proteins based on the functional clades identified (Zhang et al., 2012). For instance, TaMYB32 was selected out from 60 isolated wheat MYB genes and demonstrated to enhance the tolerance to salt stress in transgenic Arabidopsis (Zhang et al., 2012). Because the functions of several WRKY proteins have been well characterized experimentally, phylogenetic analysis allowed the identification of several functional clades. However, it was obvious that some members had various or overlapping functions (Fig. 1). For example, AtWRKY18, AtWRKY40, and AtWRKY60 were involved in pathogen and ABA responses simultaneously (Chen et al., 2010a ), while HvWRKY38, belonging to group IIa, responded to gibberellin (GA) (Zou et al., 2008). In the clade of group III, AtWRKY53 was related to senescence and basal resistance (Miao and Zentgraf, 2007); however, TcWRKY53 was associated with the osmotic stress response (Niu et al., 2012). Additionally, PtrWRKY54, PtrWRKY62, and PtrWRKY89 could be assigned to a clade with AtWRKY54 and AtWRKY70 (Fig. 1), which seemed to have similar roles. However, PtrWRKY62 and PtrWRKY89 had different expression patterns (Fig. 5). These results imply that it is arbitrary to divide clades of the WRKY family according to their motif sequence similarity without further experiment evidence.
Populus WRKY proteins respond to biotic and abiotic stresses
Plants have had to adopt different strategies to respond to various biotic and abiotic stresses for survival in adverse environmental conditions (Ahuja et al., 2010). Increasing research has indicated that WRKY proteins play vital roles in the regulation of gene expression to deal with environmental change (Ülker and Somssich, 2004; Eulgem and Somssich, 2007; Rushton et al., 2010). In plants, response to stress needs several signalling molecules including SA, JA, abscisic acid (ABA) and ethylene (ET). The expression levels of WRKY genes changed rapidly after hormone treatments (Chen and Chen, 2002; Li et al., 2004; Lai et al., 2008; Yang et al., 2009a ; Ramiro et al., 2010) In Populus, CGTCA-motifs and TCA elements randomly distributed in the promoter regions of PtrWRKY genes (Fig. 2 and Supplementary Table 4), implying that most PtrWRKY genes were involved in SA and JA responses. PtrWRKY60 and PtrWRKY89 were significantly induced by SA (Fig. 5), indicating that they functioned as key factors in regulating specific signalling pathways. WRKY genes were also directly induced by pathogens in plants, such as AtWRKY18 (Chen and Chen, 2002), CaWRKY2 (Oh et al., 2006), AtWRKY70 (Knoth et al., 2007), PtWRKY23 (Levée et al., 2009), AtWRKY3, and AtWRKY4 (Lai et al., 2008). Many PtrWRKY genes contained EIRE and ELI-box3 elements in their promoters and 59 members were induced after inoculation of M. brunnea (Fig. 3). Additionally, WRKY transcription factors were associated with responses to abiotic stresses. For instance, TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants (Niu et al., 2012). Expression of GsWRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling (Luo et al., 2013). Overexpression of WRKY25 or WRKY33 was sufficient to increase Arabidopsis NaCl tolerance and increasing sensitivity to ABA (Jiang and Deyholos, 2009). GmWRKY13, GmWRKY21, and GmWRKY54 confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants (Zhou et al., 2008). Wounding-induced AtWRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance (Chen et al., 2010b ; Hu et al., 2013). In Populus, 10 WRKY genes, belonging to group III, were induced by varieties of stresses, such as cold, salinity, and drought, but no further analysis was performed (He et al., 2012). In this work, among the 100 PtrWRKY genes, 60 members responded differentially to at least one treatment including wounding, cold, and salt, except for PtrWRKY1 (Fig. 3), and at least eight PtrWRKY genes were significantly induced by salinity stress (Fig. 5). Overall, the results reveal that most PtrWRKY genes are involved in responses to multiple biotic and abiotic stresses, consistent with previous studies (Eulgem and Somssich, 2007).
Different expression trajectories between PtrWRKY89 and its paralogue
In planta, some genes and their paralogues had constantly redundant functions: AtWRKY18, AtWRKY40, and AtWRKY60 (Xu et al., 2006); AtWRKY11 and AtWRKY17 (Journot-Catalino et al., 2006); AtWRKY54 and AtWRKY70 (Besseau et al., 2012; Li et al., 2013); and AtWRKY3 and AtWRKY4 (Lai et al., 2008). However, expression patterns of these paralogous genes might not always be identical. In Arabidopsis, AtWRKY53 was more sensitive to SA treatment than its paralogue AtWRKY41 (Besseau et al., 2012). The synergistic effect of losing AtWRKY11 and AtWRKY17 function in untreated plants implied that they acted in a partially redundant manner as repressors of AtWRKY70, but not AtWRKY54 (Journot-Catalino et al., 2006). In addition, AtWRKY4 could be induced by B. cinerea but the expression level of AtWRKY3 was not affected (Lai et al., 2008). In Populus, we also noticed that PtrWRKY62 and PtrWRKY89 had different expression trajectories (Fig. 5). Like AtWRKY54 and AtWRKY70, PtrWRKY62 and PtrWRKY89 are paralogues but still showed sequence divergence outside the WD. Therefore, we speculate that these two members had redundant roles, but diverse functions surely exist in Populus.
Constitutive expression of PtrWRKY89 enhanced resistance to fungal pathogens
In plants, plenty of WRKY proteins played important roles in defence responses by mediating SA, JA, ethylene and ABA signalling. Ectopic expression of some WRKY genes changed the transduction of hormone signalling and then altered tolerance of transgenic plants to biotic stresses. For example, transgenic poplars overexpressing and underexpressing PtWRKY23 were both more susceptible to Melampsora infection than wild-type plants (Levée et al., 2009). In Arabidopsis, single WRKY mutants of AtWRKY18, AtWRKY40, and AtWRKY60 exhibited no or small alterations in response to the pathogens. However, wrky18/wrky40 and wrky18/wrky60 double mutants and the wrky18/wrky40/wrky60 triple mutant were substantially more resistant to P. syringae but more susceptible to B. cinerea than untransformed plants (Xu et al., 2006). Overexpression of AtWRKY70 did not affect endogenous levels of free SA in Arabidopsis (Li et al., 2004). However, AtWRKY70 was downstream of NPR1 and MYB44 in an SA-dependent signal pathway and acted as an activator of SA-induced genes such as PR1, PR2, and PR5. Meanwhile it was also a repressor of JA-responsive genes. Hence overexpression of AtWRKY70 could enhance resistance to hemibiotrophic pathogens and make plants more susceptible to necrotrophic fungal pathogens (Li et al., 2004; Shim et al., 2013). In our study, PtrWRKY89 were significantly induced by SA, indicating its potential role in SA-mediated signalling pathways (Fig. 5). Overexpression of PtrWRKY89 did not enhance the level of SID2 mRNA involved in SA synthesis (Fig. 9), indicating the stability of free SA in transgenic poplars compared with the wild type. Additionally, PtrWRKY89 was constitutively expressed in transgenic poplar plants, resulting in an increased resistance to hemibiotrophic fungal pathogen D. gregaria (Fig. 8). Moreover, expression of PR genes, as a molecular marker of the SA signalling pathway, was activated in PtrWRKY89 overexpressors (Fig. 9). These results indicate that PtrWRKY89 enhanced resistance to D. gregaria via the activation of PR genes but not improvement of SA levels. Our findings are partially consistent with a previous report in Arabidopsis WRKY70 (Li et al., 2004). In addition, we also noticed that overexpression of PtrWRKY89 did not change the levels of AOS7 and JAZ10, which were involved in the JA signalling pathway. However, it is still unknown what the precise function of PtrWRKY89 is in the JA pathway in Populus. Overall, our work provided useful information for improving the resistance and stress tolerance of woody plants and suggested that PtrWRKY89 plays a regulatory role in he SA signalling pathway to increase poplar defence.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. Common names and gene IDs of putative PtrWRKYs in different versions of Phytozome.
Supplementary Table S2. Common names of WRKYs from different species and their accession numbers in GenBank.
Supplementary Table S3. Primers for qRT-PCR and semi-qRT-PCR in this study.
Supplementary Table S4. Various cis-acting elements responsive to stresses in WRKY promoters.
Supplementary Table S5. Results of DGE analysis.
Supplementary Figure S1. Phylogenetic analyses of Populus WDs.
Supplementary Figure S2. Multiple sequence alignment of WDs using DNAMAN.
Supplementary Figure S3. Transgenic P. tomentotosa plants overexpressing PtrWRKY89.
Supplementary Figure S4. PCR analysis of transgenic poplar plants.
Funding
This work was supported by the National Natural Science Foundation of China (31370672, 31171620), the National Key Project for Research on Transgenic Plants (2011ZX08010-003), 100 Talents Programme of The Chinese Academy of Sciences, the Natural Science Foundation Project of CQ CSTC (CSTC2013JJB8007), the programme for New Century Excellent Talents in University (NCET-11–0700) and the Research Fund for the Doctoral Programme of Higher Education (20110182110004).
Supplementary Material
Acknowledgements
The authors thank Prof. Jingjiang Hu (Northwest A&F University, Shanxi, China) for providing M. brunnea f.sp. multigermtubi and D. gregaria, and Prof. Guoliang Wang (Hunan Agricultural University, Changsha 410128, China) for providing the plant binary vector pCXSN.
References
- Ahuja I, de Vos RC, Bones AM, Hall RD. 2010. Plant molecular stress responses face climate change. Trends in Plant Science 15, 664–674. [DOI] [PubMed] [Google Scholar]
- Babu MM, Iyer LM, Balaji S, Aravind L. 2006. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons.Nucleic Acids Research 34, 6505–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CJ, Orlandi EW. 1995. Active oxygen in plant pathogenesis. Annual Review of Phytopathology 33, 299–321. [DOI] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET. 2012. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana . Journal of Experimental Botany 63, 2667–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Valpuesta V, Botella MA. 2001. Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiology 126, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LH, Fischer NM, Harter K, Kohlbacher O, Wanke D. 2013. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Research 41, 9764–9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D. 2010. DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen Z. 2002. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiology 129, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. 2010a. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 1471–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang L, Yu D. 2010b. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis . Molecular Plant-Microbe Interactions 23, 558–565. [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009a. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Silva H, Klessig DF. 1993. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 262, 1883–1886. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. 2009b. Biosynthesis of salicylic acid in plants. Plant Signaling & Behavior 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SM, Kang BR, Han SH, Anderson AJ, Park JY, Lee YH, Cho BH, Yang KY, Ryu CM, Kim YC. 2008. 2R,3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana . Molecular Plant-Microbe Interactions 21, 1067–1075. [DOI] [PubMed] [Google Scholar]
- Clarke SM, Mur LA, Wood JE, Scott IM. 2004. Salicylic acid dependent signaling promotes basal thermo tolerance but is not essential for acquired thermo tolerance in Arabidopsis thaliana . The Plant Journal 38, 432–447. [DOI] [PubMed] [Google Scholar]
- Dat JF, Foyer CH, Scott IM. 1998. Changes in salicylic acid and antioxidants during induced thermo tolerance in mustard seedlings. Plant Physiology 118, 1455–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, Subramaniam R, Matton DP, Brisson N. 1995. The activation of the potato PR-l0a gene requires the phosphorylation of the nuclear factor PBF-1. The Plant Cell 7, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. 2004. NPR1, all things considered. Current Opinion in Plant Biology 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. 2007. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology 10, 366–371. [DOI] [PubMed] [Google Scholar]
- He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y. 2012. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa . Plant Cell Reports 31, 1199–1217. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu H, Jia Z, Fang Q, Luo K. 2012. Combined expression of antimicrobial genes (Bbchit1 and LJAMP2) in transgenic poplar enhances resistance to fungal pathogens. Tree Physiology 32, 1313–2130. [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D. 2013. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. The Plant Journal 74, 730–745. [DOI] [PubMed] [Google Scholar]
- Janda T, Szalai G, Tari I, Páldi E. 1999. Hydroponic treatment with salicylic acid decreases the effects of chilling injury in maize (Zea mays L.) plants. Planta 208, 175–180. [Google Scholar]
- Jia Z, Sun Y, Yuan L, Tian Q, Luo K. 2010. The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnology Letters 32, 1325–1332. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. 2009. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69, 91–105. [DOI] [PubMed] [Google Scholar]
- Journot-Catalino N1, Somssich IE, Roby D, Kroj T. 2006. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana . The Plant Cell 18, 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlidag H, Yildirim E, Turan M. 2009. Salicylic acid ameliorates the adverse effect of salt stress on strawberry. Scientia Agricola 66, 180–187. [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. 2008. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. The Plant Cell 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M, Barth M, Somssich IE, Lippok B. 2003. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Molecular Plant-Microbe Interactions 16, 295–305. [DOI] [PubMed] [Google Scholar]
- Klessig DF, Durner J, Noad R, et al. 2000. Nitric oxide and salicylic acid signaling in plant defence. Proceedings of the National Academy of Sciences, USA 97, 8849–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T. 2007. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica . Molecular Plant-Microbe Interactions 20, 120–128. [DOI] [PubMed] [Google Scholar]
- Lai Z, Vinod K, Zheng Z, Fan B, Chen Z. 2008. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Evolutionary Biology 8, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annual Review of Plant Physiology and Plant Molecular Biology 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levée V, Major I, Levasseur C, Tremblay L, MacKay J, Séguin A. 2009. Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytologist 184, 48–70. [DOI] [PubMed] [Google Scholar]
- Li J, Besseau S, Törönen P, Sipari N, Kollist H, Holm L, Palva ET. 2013. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis . New Phytologist 200, 457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. 2006. WRKY70 modulates the selection of signaling pathways in plant defense. The Plant Journal 46, 477–491. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. [DOI] [PubMed] [Google Scholar]
- Lloyd G, McCown B. 1980Commercially feasible micropropagation of mountain laural (Kalmla latlfolia) by use of shoot tip cultures. Combined Proceedings of the International Plant Propagators’ Society 30, 421–427. [Google Scholar]
- Loake G, Grant M. 2007. Salicylic acid in plant defence—the players and protagonists. Current Opinion in Plant Biology 10, 466–472. [DOI] [PubMed] [Google Scholar]
- Luo X, Bai X, Sun X, et al. 2013. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signaling. Journal of Experimental Botany 64, 2155–2169. [DOI] [PubMed] [Google Scholar]
- Mahdavian K, Kalantari KM, Ghorbanli M, Torkzade M. 2008. The effects of salicylic acid on pigment contents in ultraviolet radiation stressed pepper plants. Biologia Plantarum 52, 170–172. [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. 1990. Salicylic Acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Mangelsen E, Kilian J, Berendzen KW, Kolukisaoglu UH, Harter K, Jansson C, Wanke D. 2008. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. 2007. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell 19, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munné-Bosch S, Peñuelas J. 2003. Photo- and antioxidative protection, and a role for salicylic acid during drought and recovery in field-grown Phillyrea angustifolia plants. Planta 217, 758–766. [DOI] [PubMed] [Google Scholar]
- Niu CF, Wei W, Zhou QY, et al. 2012. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell and Environment 35, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Oh SK, Yi SY, Yu SH, Moon JS, Park JM, Choi D. 2006. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Molecules and Cells 22, 58–64. [PubMed] [Google Scholar]
- Palma F, Lluch C, Iribarne C, et al. 2009. Combined effect of salicylic acid and salinity on some antioxidant activities, oxidative stress and metabolite accumulation in Phaseolus vulgaris . Plant Growth Regulation 58, 307–316. [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. 2009. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Ramiro D, Jalloul A, Petitot AS, De Sá MFG, Maluf MP, Fernandez D. 2010. Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genetics & Genomes 6, 767–781. [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE. 1996. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. The EMBO Journal 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Ryu HS, Han M, Lee SK, et al. 2006. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Reports 25, 836–847. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. [DOI] [PubMed] [Google Scholar]
- Shim JS, Jung C, Lee S, Min K, Lee YW, Choi Y, Lee JS, Song JT, Kim JK, Choi YD. 2013. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. The Plant Journal 73, 483–495 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tari I, Csiszár G, Szalai F, et al. 2002. Acclimation of tomato plants to salinity stress after a salicylic acid pre-treatment. Acta Biologica Szegediensis 46, 55–56. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Difazio S, Jansson S, et al. 2006. The genome of black cottonwood Populus trichocarpa (Torr. & Gray). Science 313, 1596. [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. 1992. Acquired resistance in Arabidopsis . The Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE. 2004. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology 7, 491–498. [DOI] [PubMed] [Google Scholar]
- van Verk MC, Bol JF, Linthorst HJ. 2011. WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biology 11, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. 2006. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenke K, Wanke D, Kilian J, Berendzen K, Harter K, Piechulla B. 2012. Volatiles of two growth-inhibiting rhizobacteria commonly engage AtWRKY18 function. The Plant Journal 70, 445–459. [DOI] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J. 2005. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Research 12, 9–26. [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ. 2005. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiology 137, 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. 2006. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell 18, 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalpani N, Enyedi AJ, Leon J, Raskin I. 1994. Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 193, 372–376. [Google Scholar]
- Yamamoto S, Nakano T, Suzuki K, Shinshi H. 2004. Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochimica et Biophysica Acta 1679, 279–287. [DOI] [PubMed] [Google Scholar]
- Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. 2009a. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biology 9, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xiao XW, Zhang S, Korpelainen H, Li CY. 2009b. Salt stress responses in Populus cathayana Rehder. Plant Science 176, 669–677. [Google Scholar]
- Yang P, Chen C, Wang Z, Fan B, Chen Z. 1999. A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. The Plant Journal 18, 141–149. [Google Scholar]
- Yu D, Chen C, Chen Z. 2001. Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. The Plant Cell 7, 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Guo G, Hu X, et al. 2010. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Research 20, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, Liu X, Kong X. 2012. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. Journal of Experimental Botany 63, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang L. 2005. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology 5, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. 2008. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnology Journal 6, 486–503. [DOI] [PubMed] [Google Scholar]
- Zou X, Neuman D, Shen Q. 2008. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiology 148, 176–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, Huang M, Luo J. 2007. DPTF: a database of poplar transcription factors. Bioinformatics 23, 1307–1308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.