Abstract
In 2014, Ebola virus (EBOV) was identified as the etiological agent of a large and still expanding outbreak of Ebola virus disease (EVD) in West Africa and a much more confined EVD outbreak in Middle Africa. Epidemiological and evolutionary analyses confirmed that all cases of both outbreaks are connected to a single introduction each of EBOV into human populations and that both outbreaks are not directly connected. Coding-complete genomic sequence analyses of isolates revealed that the two outbreaks were caused by two novel EBOV variants, and initial clinical observations suggest that neither of them should be considered strains. Here we present consensus decisions on naming for both variants (West Africa: “Makona”, Middle Africa: “Lomela”) and provide database-compatible full, shortened, and abbreviated names that are in line with recently established filovirus sub-species nomenclatures.
Keywords: Ebola, Ebola virus, ebolavirus, filovirid, Filoviridae, filovirus, genome annotation, Lomela, Lokolia, Makona, mononegavirad, Mononegavirales, mononegavirus, virus classification, virus isolate, virus nomenclature, virus strain, virus taxonomy, virus variant
1. Introduction
On 10 March 2014, a viral hemorrhagic fever (VHF) outbreak was reported among humans in Guinea, West Africa [1]. Ebola virus (EBOV), the sole member of the species Zaire ebolavirus (genus Ebolavirus, family Filoviridae, order Mononegavirales [2]), was identified as the etiological agent. Consequently, the VHF was identified as Ebola virus disease (EVD) [1,3,4]. At the time of writing, this EVD outbreak has spread from Guinea into Liberia, Nigeria, Senegal, Sierra Leone, and Mali, with individual case exportations or transport of patients to France, Germany, Norway, Spain, UK, and US. At least 15351 human infections and 5459 deaths (proportion of fatal cases ≈36%) have been recorded as of 21 November 2014, making this outbreak the largest EVD outbreak in history [5]. Through conventional Sanger [1] and next-generation sequencing [6], 102 coding-complete EBOV genome sequences have been assembled (complete genome sequences with the exception of the ultimate 3’ and 5’ untranslated regions [7]) originating from three patients from Guinea and 78 patients from Sierra Leone [1,6]. Evolutionary analyses combined with epidemiological data demonstrate that all cases are directly epidemiologically linked, tracing back to a single introduction of EBOV into the human population [1,6] as has been found for most past EVD outbreaks [8].
On 24 August 2014, another EVD outbreak was reported from Boende District, Democratic Republic of the Congo, Middle Africa [9]. A total of 66 cases and 49 deaths (proportion of fatal case ≈74%) have been recorded [5]. As in Guinea, epidemiological analyses point towards a single introduction of EBOV from its unknown natural reservoir into the human population, with subsequent spread among humans by direct person-to-person transmission [9]. Thus far, two partial L (RNA-dependent RNA polymerase) gene sequences have been deposited into GenBank, and the coding-complete sequence of one isolate has been determined [9]. Phylogenetic analysis demonstrated that the Guinea and Democratic Republic of Congo EVD outbreaks were not related. The EBOV variants causing both outbreaks were distinct from each other and from variants known from previous EVD outbreaks [1,3,4,6,9].
Next-generation sequencing techniques enable the determination of coding-complete EBOV genomes in dozens and theoretically hundreds of clinical samples in parallel in the absence of classical virus culture [6]. The rapid accumulation of sequence data challenges sequence database curators and end users when novel sequences are not uniquely named according to common standards. Here, we assign final designations to the two EBOV variants causing the 2014 Guinea and Democratic Republic of Congo EVD outbreaks and update the current GenBank sequence entries accordingly.
2. Ebola Virus Strain, Variant, and Isolate Naming
In 2013, a consortium of filovirologists and sequence database experts working at the US National Center for Biotechnology Information (NCBI) established a consistent and prospective filovirus nomenclature below the species level [10]. This nomenclature, which already has been applied to all filovirus entries in NCBI’s RefSeq database [11], is based on the template:
<virus name>(/<strain>)/<isolation host-suffix>/<country of sampling>/<year of sampling>/<genetic variant designation>-<isolate designation>. [Note: the “/” between the <virus name> and the <isolation host-suffix> field was missing in the full name template outlined in [10], but is necessary for computational purposes. It is therefore introduced here and already implemented filovirus full names [11] will be retrospectively corrected.]
The <virus name> field should contain a filovirus name as outlined in [10]. Currently, the accepted filovirus names and abbreviations are Bundibugyo virus (BDBV), Ebola virus (EBOV), Lloviu virus (LLOV), Marburg virus (MARV), Ravn virus (RAVV), Reston virus (RESTV), Sudan virus (SUDV), and Taï Forest ebolavirus (TAFV), Table 1 [2,12].
Table 1.
Summary of the current filovirus taxonomy endorsed by the 2012–2014 ICTV Filoviridae Study Group and accepted by the ICTV [2,12,13,14,15].
| Current Taxonomy and Nomenclature (Ninth ICTV Report and Updates) |
|---|
| Order Mononegavirales |
| Family Filoviridae |
| Genus Marburgvirus |
| Species Marburg marburgvirus |
| Virus 1: Marburg virus (MARV) |
| Virus 2: Ravn virus (RAVV) |
| Genus Ebolavirus |
| Species Taï Forest ebolavirus |
| Virus: Taï Forest virus (TAFV) |
| Species Reston ebolavirus |
| Virus: Reston virus (RESTV) |
| Species Sudan ebolavirus |
| Virus: Sudan virus (SUDV) |
| Species Zaire ebolavirus |
| Virus: Ebola virus (EBOV) |
| Species Bundibugyo ebolavirus |
| Virus: Bundibugyo virus (BDBV) |
| Genus Cuevavirus |
| Species Lloviu cuevavirus |
| Virus: Lloviu virus (LLOV) |
The <strain> field should contain a unique strain name in case the virus in question fulfills the criteria for being a strain (see [10]). The <isolation host-suffix> field should be provided in one word in the format “first letter of host genus name.full name of species descriptor” (e.g., “H.sapiens”) followed by suffix that denotes whether the sequence stems from an unpassaged sample (“-wt”), from virus isolated in tissue culture (“-tc”), or is a genomic fragment (“-frag). The <country of sampling> and <year of sampling> fields should contain the alpha-3 three-letter ISO 3166-1 code for the country where the virus was isolated and the year in which it was isolated, respectively. Finally, the <variant designation> and <isolate designation> fields should contain a unique variant name (i.e., a name for the virus variant that was introduced into the human population that caused an outbreak) and unique isolate name (i.e., the name for a particular representative of the variant), respectively [10]. To simplify manuscript writing, shortened and abbreviated virus designations are also defined [10]. For instance, the designations
| full: | Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Ecran |
| shortened: | EBOV/H.sap/COD/76/Yam-Ecr |
| abbreviated: | EBOV/Yam-Ecr |
specify an isolate “Ecran” of Ebola virus as a representative of the variant “Yambuku” (not a strain) that originated from a human in the Democratic Republic of the Congo in 1976 and was isolated/sequenced using tissue culture [16].
3. The 2014 Ebola Virus Variant Originating in Guinea
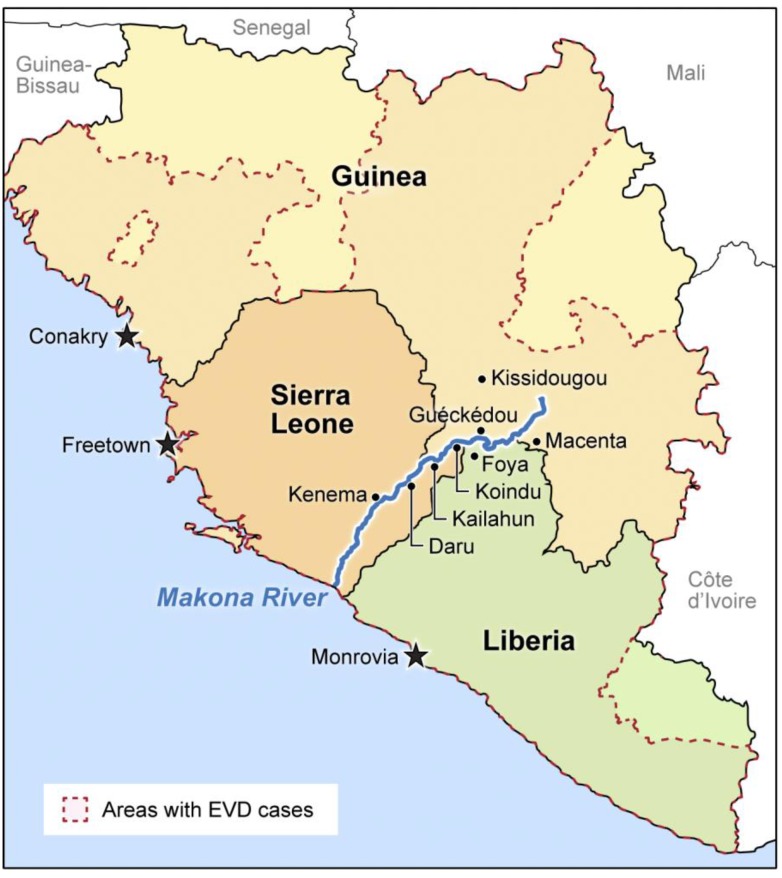
At the end of 2013, EVD broke out around Guéckédou, Kissidougou, and Macenta, Guinea [1] and consequently spread to at least five additional West African countries. Epidemiological and phylogenetic studies indicate that this large EVD outbreak was caused by a single introduction of one particular ebolavirus, Ebola virus (EBOV), into humans (Homo sapiens) from an unknown reservoir and therefore that all subsequent human cases (over 15,000 cases) are derived from one unnamed variant [1,6]. Preliminary clinical observations among EVD patients in West Africa do not contradict past descriptions of EVD [17,18,19,20,21,22], i.e., this novel unnamed EBOV variant is not a strain as defined in standardized filovirus nomenclature [10]. Here we propose the name “Makona” (IPA: [mɑ'kɔnə] or [məˈkoʊnə]; English phonetic notation: mah-kaw-nuh or muh-koh-nuh) after the Makona River close to the border between Liberia, Guinea, and Sierra Leone (Figure 1) as the variant name for this West African virus. The general name for the 2014 West African virus is therefore:
 |
Figure 1.
Location of the Makona River.
At the time of writing, 102 coding-complete genomic sequences of EBOV/Mak have been deposited into GenBank, all of which were obtained directly from clinical samples (“p0”) [1,6]. Following the rules laid out in filovirus standardized nomenclature [10], the names for these sequences therefore should contain the <suffix> “-wt”. In addition, one fragment of the L gene of one isolate of EBOV/Mak was deposited. Based on definitions described in standardized nomenclature [10], the corresponding <suffix> field should therefore be filled with “-frag”. All currently deposited sequences stem from either Guinean, Sierra Leonean, or Nigerian samples. The 3-letter country codes to be used the <country> field [10] for all countries that have thus far handled patients infected with EBOV/Mak are summarized in Table 2.
Table 2.
ISO 3166-1 alpha-3 country codes for countries with recorded cases connected to the 2014 EVD epidemic that started in Guinea [23]:
| Official short country name in English (geographical name) | Official country name in English (protocol name) | ISO 3166-1 3-letter abbreviation |
|---|---|---|
| France | French Republic | FRA |
| Germany | Federal Republic of Germany | DEU |
| Guinea | Republic of Guinea | GIN |
| LiberiaMaliNigeria | Republic of LiberiaRepublic of MaliFederal Republic of Nigeria | LBRMLINGA |
| NorwaySenegal | Kingdom of NorwayRepublic of Senegal | NORSEN |
| Sierra Leone | Republic of Sierra Leone | SLE |
| Spain | Kingdom of Spain | ESP |
| United Kingdom | United Kingdom of Great Britain and Northern Ireland | GBR |
| United States | United States of America | USA |
Three replicating isolates (C05, C07, and C15) of EBOV/Mak have been reported [1]. The 102 coding-complete genomes (including those of C05, C07, and C15) and the one fragmented L gene sequence that have been deposited into GenBank all already have assigned unique <isolate designation> descriptors. Accordingly, in all currently deposited sequences of EBOV/Mak, the definition line will be adjusted to “Zaire ebolavirus isolate Ebola virus/H.sapiens-<suffix>/<country>/2014/Makona-<isolate designation>, [coding-]complete genome, with the <suffix>, <country>, and <isolate designation> fields will be filled according to their origin. The GenBank <strain> field will be cleared throughout; the Genbank <isolate> field will be filled with “Ebola virus/H.sapiens-<suffix>/<country>/2014/Makona-<isolate designation>”, and the <organism> field will be corrected, if necessary, to “Zaire ebolavirus” (Table 3).
Table 3.
EBOV isolates from the West African EVD outbreak, 2014: Updated and/or corrected GenBank fields and final names
| GenBank Accession Number | <GenBank field> = Updated/corrected information |
|---|---|
| Final names | |
|
KJ660346 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15 | |
 | |
|
KJ660347 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C07, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C07 | |
 | |
|
KJ660348 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/GIN/2014/Makona-C05 | |
 | |
|
KM034549 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095B, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095B | |
 | |
|
KM034550 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM095 | |
 | |
|
KM034551 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM096, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM096 | |
 | |
|
KM034552 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM098, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM098 | |
 | |
|
KM034553 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3670.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3670.1 | |
 | |
|
KM034554 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.1 | |
 | |
|
KM034555 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3676.2 | |
 | |
|
KM034556 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.1 | |
 | |
|
KM034557 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3677.2 | |
 | |
|
KM034558 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3679.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3679.1 | |
 | |
|
KM034559 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3680.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3680.1 | |
 | |
|
KM034560 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3682.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3682.1 | |
 | |
|
KM034561 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3683.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3683.1 | |
 | |
|
KM034562 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-G3686.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3686.1 | |
 | |
|
KM034563 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3687.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3687.1 | |
 | |
|
KM233035 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM104, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM104 | |
 | |
|
KM233036 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM106, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM106 | |
 | |
|
KM233037 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM110, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM110 | |
 | |
|
KM233038 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM111, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM111 | |
 | |
|
KM233039 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM112, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM112 | |
 | |
|
KM233040 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM113, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM113 | |
 | |
|
KM233041 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM115, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM115 | |
 | |
|
KM233042 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM119, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM119 | |
 | |
|
KM233043 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM120, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM120 | |
 | |
|
KM233044 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM121, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM121 | |
 | |
|
KM233045 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.1 | |
 | |
|
KM233046 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.2 | |
 | |
|
KM233047 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.3 | |
 | |
|
KM233048 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-EM124.4 | |
 | |
|
KM233049 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3707, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3707 | |
 | |
|
KM233050 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.2 | |
 | |
|
KM233051 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.3 | |
 | |
|
KM233052 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3713.4 | |
 | |
|
KM233053 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3724, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3724 | |
 | |
|
KM233054 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3729, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3729 | |
 | |
|
KM233055 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3734.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3734.1 | |
 | |
|
KM233056 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.1 | |
 | |
|
KM233057 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3735.2 | |
 | |
|
KM233058 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.1 | |
 | |
|
KM233059 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.2 | |
 | |
|
KM233060 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3750.3 | |
 | |
|
KM233061 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3752, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3752 | |
 | |
|
KM233062 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3758, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3758 | |
 | |
|
KM233063 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3764, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3764 | |
 | |
|
KM233064 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3765.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3765.2 | |
 | |
|
KM233065 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.1 | |
 | |
|
KM233066 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.2 | |
 | |
|
KM233067 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.3 | |
 | |
|
KM233068 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.4, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3769.4 | |
 | |
|
KM233069 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.1 | |
 | |
|
KM233070 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3770.2 | |
 | |
|
KM233071 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3771, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3771 | |
 | |
|
KM233072 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3782, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3782 | |
 | |
|
KM233073 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3786, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3786 | |
 | |
|
KM233074 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3787, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3787 | |
 | |
|
KM233075 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3788, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3788 | |
 | |
|
KM233076 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3789.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3789.1 | |
 | |
|
KM233077 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3795, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3795 | |
 | |
|
KM233078 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3796, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3796 | |
 | |
|
KM233079 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3798, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3798 | |
 | |
|
KM233080 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3799, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3799 | |
 | |
|
KM233081 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3800, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3800 | |
 | |
|
KM233082 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.1 | |
 | |
|
KM233083 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3805.2 | |
 | |
|
KM233084 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3807, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3807 | |
 | |
|
KM233085 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3808, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3808 | |
 | |
|
KM233086 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3809, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3809 | |
 | |
|
KM233087 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.1 | |
 | |
|
KM233088 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3810.2 | |
 | |
|
KM233089 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3814, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3814 | |
 | |
|
KM233090 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3816, [coding-]complete genome2
= Zaire ebolavirus1 |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3816 | |
 | |
|
KM233091 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3817, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3817 | |
 | |
|
KM233092 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3818, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3818 | |
 | |
|
KM233093 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3819, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3819 | |
 | |
|
KM233094 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3820, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3820 | |
 | |
|
KM233095 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3821, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3821 | |
 | |
|
KM233096 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3822, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3822 | |
 | |
|
KM233097 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3823, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3823 | |
 | |
|
KM233098 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.1 | |
 | |
|
KM233099 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3825.2 | |
 | |
|
KM233100 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3826, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3826 | |
 | |
|
KM233101 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3827, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3827 | |
 | |
|
KM233102 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3829, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3829 | |
 | |
|
KM233103 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3831, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona- G3831 | |
 | |
|
KM233104 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3834, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3834 | |
 | |
|
KM233105 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3838, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3838 | |
 | |
|
KM233106 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3840, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3840 | |
 | |
|
KM233107 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3841, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3841 | |
 | |
|
KM233108 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3845, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3845 | |
 | |
|
KM233109 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3846, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3846 | |
 | |
|
KM233110 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3848, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3848 | |
 | |
|
KM233111 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3850, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3850 | |
 | |
|
KM233112 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3851, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3851 | |
 | |
|
KM233113 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.1 | |
 | |
|
KM233114 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3856.3 | |
 | |
|
KM233115 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3857, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-G3857 | |
 | |
|
KM233116 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.1, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.1 | |
 | |
|
KM233117 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Mak-NM042.2, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.2 | |
 | |
|
KM233118 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.3, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/SLE/2014/Makona-NM042.3 | |
 | |
|
KM251803 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-frag/NGA/2014/Makona-01072014 L gene, partial CDS3 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| <strain> = [EMPTY] | |
| </isolate> = Ebola virus/H.sapiens-frag/NGA/2014/Makona-01072014 | |
 |
1 Neither RefSeq nor GenBank currently can handle italics, which is why the species names are not italicized in the actual entry’s definition line and <organism> fields.
2 The International Nucleotide Sequence Database Collaboration (INSDC) standard currently does not offer options other than “complete” or “partial,” and, in particular, does not provide a possibility for the designation “coding-complete.” We, therefore recommend using “complete” for what is actually “coding-complete” as the word “partial” implies short sequence fragments to most users.
3 CDS = coding sequence.
4. The 2014 Ebola Virus Variant Originating in the Democratic Republic of the Congo
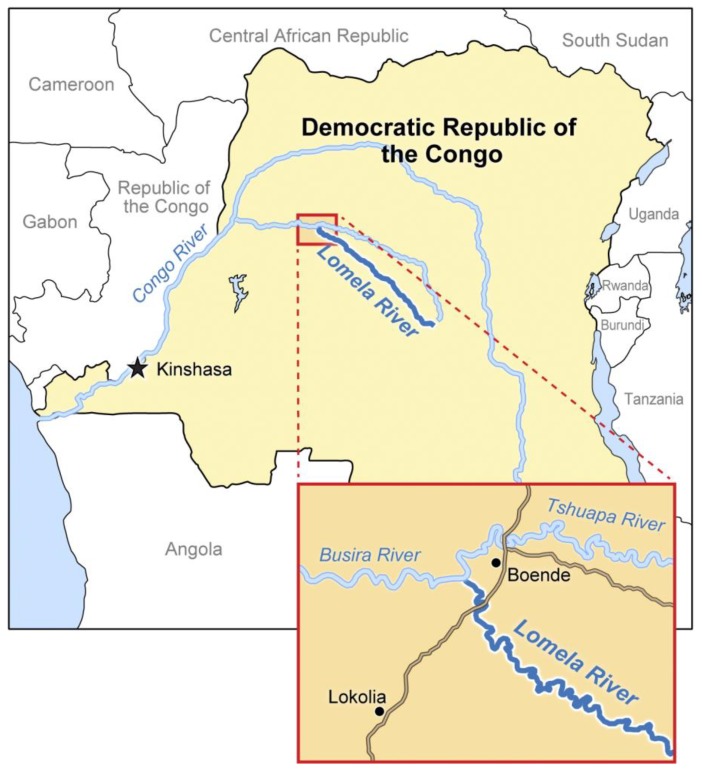
In late 2014, EVD broke out in Boende District, Democratic Republic of the Congo (3-letter country code: COD), Middle Africa. Epidemiological and phylogenetic studies indicate that this limited EVD outbreak was caused by a single introduction of one particular ebolavirus, Ebola virus (EBOV), into humans (Homo sapiens) from an unknown reservoir and therefore that all subsequent several cases (almost 70) are derived from one unnamed variant [9]. Preliminary clinical observations among EVD patients in this outbreak do not contradict past descriptions of EVD [9], i.e., this novel unnamed EBOV variant is not a strain as defined in [10]. Here we propose the name “Lomela” (IPA: [lɔ'mɛlɑ] or [lɔ'mɛlə]; English phonetic notation: law-me-lah or law-me-luh) after the Lomela River that runs through COD’s Boende District (see Figure 2) as the variant name for this Middle African virus. The general name for the 2014 Middle African virus is therefore
Figure 2.
Location of the Lomela River.
 |
Accordingly, in all currently deposited sequences of EBOV/Lom, the definition line will be adjusted to “Zaire ebolavirus isolate Ebola virus/H.sapiens-<suffix>/<COD>/2014/Lomela-<isolate designation>, with the <suffix> and <isolate designation> fields will be filled according to their origin. The GenBank <strain> field will be cleared throughout; the Genbank <isolate> field will be filled with “Ebola virus/H.sapiens-<suffix>/<COD>/2014/Lomela-<isolate designation>”, and the <organism> field will be corrected, if necessary, to “Zaire ebolavirus” (Table 4).
Table 4.
EBOV isolates from the Democratic Republic of the Congo Boende EVD outbreak, 2014: Updated and/or corrected GenBank information and final names
| GenBank Accession Number | <GenBank field> = Updated/corrected information |
|---|---|
| Final names | |
|
KM517570 |
<DEFINITION LINE> = Zaire ebolavirus1 isolate Ebola virus/H.sapiens-wt/GIN/2014/Makona-C15, [coding-]complete genome2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-frag/COD/2014/Lomela-Lokolia16 | |
 | |
|
KM517571 |
<DEFINITION LINE> = Zaire ebolavirus1 Ebola isolate virus H.sapiens-frag/COD/2014/Lomela-Lokolia17 L gene, partial CDS2 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-frag/COD/2014/Lomela-Lokolia17 | |
 | |
|
KM519951 |
<DEFINITION LINE> = Zaire ebolavirus1 Ebola isolate virus H.sapiens-wt/COD/2014/Lomela-Lokolia1, [coding-]complete genome3 |
| <SOURCE ORGANISM> = Zaire ebolavirus1 | |
| </isolate> = Ebola virus/H.sapiens-wt/COD/2014/Lomela-Lokolia1 | |
 |
1 Neither RefSeq nor GenBank currently can handle italics, which is why the species names are not italicized in the actual entry’s definition line and <organism> fields.
2 CDS = coding sequence
3 The International Nucleotide Sequence Database Collaboration (INSDC) standard currently does not offer options other than “complete” or “partial,” and, in particular, does not provide a possibility for the designation “coding-complete.” We, therefore recommend using “complete” for what is actually “coding-complete” as the word “partial” implies short sequence fragments to most users.
Note: “Lokolia” – IPA: [lɔ'kɔliə] or [lə'kɔliə]; English phonetic notation: law-kaw-lee-uh or luh-kaw-lee-uh.
Acknowledgments
We thank Laura Bollinger (IRF-Frederick) for carefully editing the manuscript. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services (DHHS) or of the institutions and companies affiliated with the authors. This work was funded in part through Battelle Memorial Institute’s prime contract with the US National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272200700016I. J.W. performed this work as an employee of Battelle Memorial Institute. Subcontractors to Battelle Memorial Institute who performed this work are: J.H.K., an employee of Tunnell Government Services, Inc. This research was further supported in part by the Intramural Research Program of the NIH, National Library of Medicine (Y.B., O.B., and J.R.B.).
Author Contributions
All authors were engaged in the discussion about the best possible virus variants names and were or are involved in updating/correcting current GenBank entries for isolates of these variants. The final decisions presented in the paper were reached by consensus or simple majority voting, with the understanding that all authors will apply the final decisions reached by the entire group and enforce them in their functions as authors, peer-reviewers, and/or editors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N., Soropogui B., Sow M.S., Keita S., De Clerck H., et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Jahrling P.B., Kawaoka Y., Netesov S.V., Nichol S.T., Peters C.J., Volchkov V.E., et al. Family Filoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy—Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; London, UK: 2011. pp. 665–671. [Google Scholar]
- 3.Calvignac-Spencer S., Schulze J.M., Zickmann F., Renard B.Y. Clock Rooting Further Demonstrates that Guinea 2014 EBOV is a Member of the Zaire Lineage. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.c0e035c86d721668a6ad7353f7f6fe86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudas G., Rambaut A. Phylogenetic Analysis of Guinea 2014 EBOV Ebolavirus Outbreak. PLoS Curr. 2014;6 doi: 10.1371/currents.outbreaks.84eefe5ce43ec9dc0bf0670f7b8b417d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization Global Alert and Response (GAR) Situation Reports: Ebola Response Roadmap. World Health Organization; Geneva, Switzerland: 2014. [(accessed on 23 November 2014)]. Available online: http://apps.who.int/iris/bitstream/10665/136508/1/roadmapsitrep15Oct2014.pdf?ua=1. [Google Scholar]
- 6.Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G., et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladner J.T., Beitzel B., Chain P.S., Davenport M.G., Donaldson E.F., Frieman M., Kugelman J.R., Kuhn J.H., O’Rear J., Sabeti P.C., et al. Standards for sequencing viral genomes in the era of high-throughput sequencing. MBio. 2014;5:e01360–14. doi: 10.1128/mBio.01360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn J.H. Filoviruses. A Compendium of 40 Years of Epidemiological, Clinical, and Laboratory Studies. Archives of Virology Supplementum. Volume 20. SpringerWienNewYork; Vienna, Austria: 2008. [PubMed] [Google Scholar]
- 9.Maganga G.D., Kapetshi J., Berthet N., Ilunga B.K., Kabange F., Kingebeni P.M., Mondonge V., Muyembe J.J., Bertherat E., Briand S., et al. Ebola Virus Disease in the Democratic Republic of Congo. N. Engl. J. Med. 2014 doi: 10.1056/NEJMoa1411099. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn J.H., Bao Y., Bavari S., Becker S., Bradfute S., Brister J.R., Bukreyev A.A., Chandran K., Davey R.A., Dolnik O., et al. Virus nomenclature below the species level: A standardized nomenclature for natural variants of viruses assigned to the family Filoviridae. Arch. Virol. 2013;158:301–311. doi: 10.1007/s00705-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn J.H., Andersen K.G., Bao Y., Bavari S., Becker S., Bennett R.S., Bergman N.H., Blinkova O., Bradfute S., Brister J.R., et al. Filovirus RefSeq Entries: Evaluation and Selection of Filovirus Type Variants, Type Sequences, and Names. Viruses. 2014;6:3663–3682. doi: 10.3390/v6093663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T., et al. Proposal for a revised taxonomy of the family Filoviridae: Classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams M.J., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012) Arch. Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams M.J., Lefkowitz E.J., King A.M., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2014) Arch. Virol. 2014;159:2831–2841. doi: 10.1007/s00705-014-2114-3. [DOI] [PubMed] [Google Scholar]
- 15.Bukreyev A.A., Chandran K., Dolnik O., Dye J.M., Ebihara H., Leroy E.M., Muhlberger E., Netesov S.V., Patterson J.L., Paweska J.T., et al. Discussions and decisions of the 2012–2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012-June 2013. Arch. Virol. 2014;159:821–830. doi: 10.1007/s00705-013-1846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn J.H., Lofts L.L., Kugelman J.R., Smither S.J., Lever M.S., van der Groen G., Johnson K.M., Radoshitzky S.R., Bavari S., Jahrling P.B., et al. Reidentification of Ebola virus E718 and ME as Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Ecran. Genome Announc. 2014;2:e01178–14. doi: 10.1128/genomeA.01178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schieffelin J.S., Shaffer J.G., Goba A., Gbakie M., Gire S.K., Colubri A., Sealfon R.S., Kanneh L., Moigboi A., Momoh M., et al. Clinical Illness and Outcomes in Patients with Ebola in Sierra Leone. N. Engl. J. Med. 2014 doi: 10.1056/NEJMoa1411680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreuels B., Wichmann D., Emmerich P., Schmidt-Chanasit J., de Heer G., Kluge S., Sow A., Renne T., Gunther S., Lohse A.W., et al. A Case of Severe Ebola Virus Infection Complicated by Gram-Negative Septicemia. N. Engl. J. Med. 2014 doi: 10.1056/NEJMoa1411677. [DOI] [PubMed] [Google Scholar]
- 19.Briand S., Bertherat E., Cox P., Formenty P., Kieny M.P., Myhre J.K., Roth C., Shindo N., Dye C. The international Ebola emergency. N. Engl. J. Med. 2014;371:1180–1183. doi: 10.1056/NEJMp1409858. [DOI] [PubMed] [Google Scholar]
- 20.Bah E.I., Lamah M.C., Fletcher T., Jacob S.T., Brett-Major D.M., Sall A.A., Shindo N., Fischer W.A., Lamontagne F., Saliou S.M., et al. Clinical Presentation of Patients with Ebola Virus Disease in Conakry, Guinea. N. Engl. J. Med. 2014 doi: 10.1056/NEJMoa1411249. [DOI] [PubMed] [Google Scholar]
- 21.Lyon G.M., Mehta A.K., Varkey J.B., Brantly K., Plyler L., McElroy A.K., Kraft C.S., Towner J.S., Spiropoulou C., Stroher U., et al. Clinical Care of Two Patients with Ebola Virus Disease in the United States. N. Engl. J. Med. 2014 doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- 22.Barry M., Traore F.A., Sako F.B., Kpamy D.O., Bah E.I., Poncin M., Keita S., Cisse M., Toure A. Ebola outbreak in Conakry, Guinea: Epidemiological, clinical, and outcome features. Med. Mal. Infect. 2014 doi: 10.1016/j.medmal.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 23.International Organization for Standardization (ISO) Country Codes—ISO 3166. [(accessed on 24 October 2014)]. Available online: https://www.iso.org/obp/ui/#search.