Significance
Shifts in biological diversity often are associated with particular anatomical traits. Anatomical data from over 300 clades of brachiopods, molluscs, arthropods, echinoderms, and chordates show that trait-based diversification shifts are common at even fairly low (genus and species) taxonomic levels. Cambrian taxa present the lone major exception. Among post-Cambrian taxa, diversification shifts correlate strongly with elevated net extinction of primitive taxa rather than elevated net speciation of derived taxa or increased morphological disparity among derived taxa. This finding emphasizes the importance of extinction in shaping morphological and phylogenetic diversity among closely related species and genera as well as suggests another way in which Cambrian evolution was unique.
Keywords: trait-based diversification, extinction, evolvability, speciation, Cambrian
Abstract
Evolution provides many cases of apparent shifts in diversification associated with particular anatomical traits. Three general models connect these patterns to anatomical evolution: (i) elevated net extinction of taxa bearing particular traits, (ii) elevated net speciation of taxa bearing particular traits, and (iii) elevated evolvability expanding the range of anatomies available to some species. Trait-based diversification shifts predict elevated hierarchical stratigraphic compatibility (i.e., primitive→derived→highly derived sequences) among pairs of anatomical characters. The three specific models further predict (i) early loss of diversity for taxa retaining primitive conditions (elevated net extinction), (ii) increased diversification among later members of a clade (elevated net speciation), and (iii) increased disparity among later members in a clade (elevated evolvability). Analyses of 319 anatomical and stratigraphic datasets for fossil species and genera show that hierarchical stratigraphic compatibility exceeds the expectations of trait-independent diversification in the vast majority of cases, which was expected if trait-dependent diversification shifts are common. Excess hierarchical stratigraphic compatibility correlates with early loss of diversity for groups retaining primitive conditions rather than delayed bursts of diversity or disparity across entire clades. Cambrian clades (predominantly trilobites) alone fit null expectations well. However, it is not clear whether evolution was unusual among Cambrian taxa or only early trilobites. At least among post-Cambrian taxa, these results implicate models, such as competition and extinction selectivity/resistance, as major drivers of trait-based diversification shifts at the species and genus levels while contradicting the predictions of elevated net speciation and elevated evolvability models.
A basic question in evolution is whether shifts in taxonomic and/or morphologic diversification are tied to particular anatomical traits. The fossil record includes many examples of taxa possessing one set of traits losing diversity over time, whereas other taxa with different sets of traits gain diversity (1–4). Similarly, phylogenies of extant taxa often suggest that speciose subclades possessing derived traits were once much less diverse than the remainder of the clade diagnosed by primitive traits (5–7). In a different vein, morphospace studies often indicate that particular subclades diversify in regions of morphospace seemingly off limits to the remainder of the clade (8–10). Three models of trait-based diversification shifts explain these patterns. Model 1 (elevated net extinction) posits elevated extinction rates and/or decreased origination rates among taxa with primitive traits (11, 12). Model 2 (elevated net speciation) posits elevated speciation rates and/or decreased extinction rates among some taxa with derived traits (11, 13, 14). Model 3 (elevated evolvability) posits that some characters vary only among some derived taxa and not among the remainder of the clade (3, 15). These models are not mutually exclusive: elevated evolvability might elevate net speciation (models 2 and 3) (16), or elevated speciation in one part of a clade might induce elevated extinction in another part of a clade (models 1 and 2) (17). However, we do not know whether any of these three models predominates or even whether trait-based diversification shifts are the norm at low taxonomic (e.g., species and genus) levels.
Model Predictions
We can test whether traits correlate with diversification shifts on phylogenies of extant taxa (13, 14). However, accurately estimating extinction rates and recognizing lost diversity given only extant taxa are notoriously difficult (18, 19), both of which bias such tests against supporting the elevated net extinction model (20). Modifying these tests to include taxa sampled in different time intervals rather than from just the present should improve extinction rate estimates (21). Even then, error in phylogenetic reconstructions for fossil taxa is biased toward elevating early diversification rates (22). Such error biases inferred trees against supporting differential net cladogenesis and possibly, against elevated evolvability.
Trait-based diversification and trait-independent diversification make different predictions about the fossil record without reference to specific phylogenies (9, 10, 23–25). Stratigraphic patterns among compatible character pairs are one example. Character pairs are compatible if there are phylogenies that do not require parallelism or convergence for either character (26, 27). If both characters have two states, then at most, only three of four possible combinations evolve. Such pairs are stratigraphically compatible (28) if they fit one of two patterns. Suppose that we label the character states on the oldest-known species 0. Hierarchical stratigraphic compatibility (HSC) is species with 00 occurring in the oldest strata, species with 10 appearing in younger strata, and species with 11 appearing in still younger strata. HSC is consistent with a 00→10→11 sequence of evolution. Divergent stratigraphic compatibility (DSC) is species with 00 occurring in the oldest strata, with some species bearing 10 and different species bearing 01 appearing in younger strata. DSC is consistent with a sequence of evolution.
Compatible characters should represent slowly evolving characters (26, 27). Simulations confirm this expectation (29) (SI Appendix, Fig. S4). If characters change infrequently, then there usually will be several species bearing 00 (hereafter, a paraclade) (30) contemporaneous with the first species bearing 10 (31). Under trait-independent diversification, that paraclade should generate more total descendants than the sole-derived species (30) and thus, generate more opportunities for a 00→01 transition (DSC) than for a 10→11 transition (HSC). Simulations show that, given trait-independent diversification and no addition to character space, fewer than 40% of stratigraphically compatible pairs should be HSC (SI Appendix, Fig. S5). These expectations hold over a wide range of per-taxon sampling rates and evolutionary models (SI Appendix, Fig. S5) (note that the same simulations show that we should sample state pairs in correct order for 95% of compatible character pairs).
Increasing net extinction rates within paraclades retaining 00 pairs (model 1) reduces the chance of a 00→01 transition (and DSC) by reducing the expected descendants from paraclade members. Similarly, increasing net speciation rates for species with 10 (model 2) elevates the probability of a 10→11 transition (and HSC) by elevating the expected descendants of the species with 10. Finally, increasing the number of evolvable characters for the subclade diagnosed by 10 (model 3) introduces a suite of characters for which 10→11 (and HSC) is the only probable transition. Thus, all three models elevate expected HSC.
Models 1–3 make unique predictions about correlations between HSC and different paleontological patterns. Elevated net extinction and elevated net speciation (models 1 and 2) make distinct predictions about stratigraphic distributions of species within paraclades and whole clades, respectively. Elevated net extinction (model 1) predicts that the pooled stratigraphic distributions of species retaining primitive conditions should have lower centers of gravity than other models predict (32, 33). Elevated net speciation (model 2) predicts that the pooled stratigraphic distributions for the clade should have a higher center of gravity than other models predict.
Elevated evolvability (model 3) makes unique predictions regarding morphological diversity (disparity) relative to models 1 and 2. If fewer characters can change among early species than some derived species, then the disparity among all S/2 early species will be lower than expected given the total character space and likely rates of change (34, 35). These predictions apply to cumulative disparity (i.e., disparity among all S/2 species) rather than standing disparity (i.e., species extant halfway through a clade’s history), because extinction often greatly affects standing disparity (36) (Materials and Methods and SI Appendix, Fig. S5).
We apply stratigraphic compatibility, center of gravity, and cumulative disparity analyses to 319 published character matrices of fossil species and genera to ask three questions. (i) Are patterns consistent with trait-based diversification shifts truly common among fossil taxa at low taxonomic levels? (ii) Do these patterns vary among taxonomic groups and/or over time? (iii) Is there any general association with the expectations of elevated net extinction, elevated net speciation, or elevated evolvability?
Results
Excess HSC.
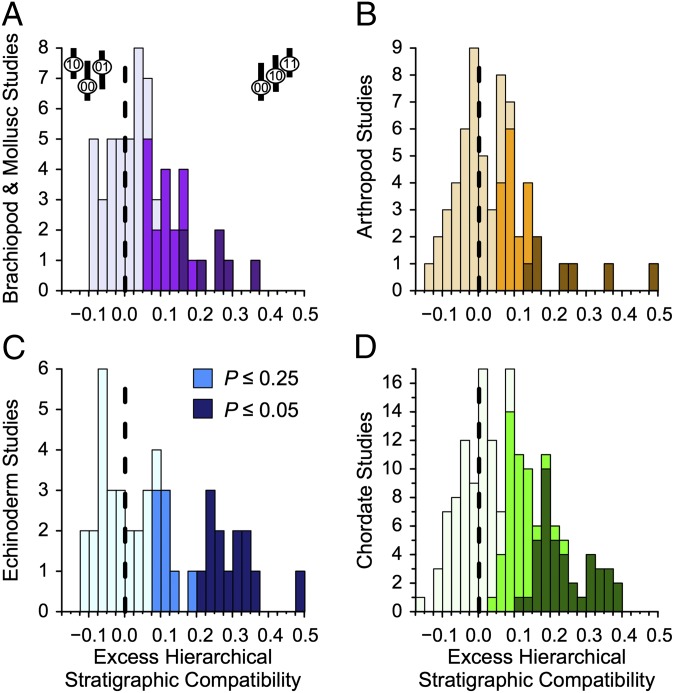
HSC exceeds expectations of trait-independent diversification in the vast majority of the clades (Fig. 1, Table 1, and SI Appendix, Table S3, results under alternative models). Only arthropods fail to have significantly more than 50% of clades with excess HSC. Major deviations are particularly common: 37–53% of clades show excess HSC deviations that 25% or fewer clades should show; 12–29% of clades show deviations that only 5% or fewer clades should show (Table 1 and SI Appendix, Fig. S7).
Fig. 1.
Deviations between observed and expected HSC for fossil (A) brachiopods and molluscs; (B) arthropods; (C) echinoderms; and (D) chordates. Positive numbers mean that 00→10→11 (upper right cartoon in A) sequences exceed Monte Carlo-generated expectations assuming continuous trait-independent diversification with empirically estimated origination, extinction, and sampling rates and simulated character evolution matching observed compatibility for each dataset. Negative numbers mean that sequences (upper left cartoon in A) exceed those same expectations. Shades correspond to the significance of the deviations.
Table 1.
Cases of excess HSC at Monte Carlo significances of P ≤ 0.05, P ≤ 0.25, and P < 0.50 assuming trait-independent diversification
| Group | N | P (HSC) ≤ 0.05 | P (HSC) ≤ 0.25 | P (HSC) < 0.50 |
| Brachiopods and molluscs | 57 | 7 (7.2 × 10−3) | 23 (3.5 × 10−3) | 39 (1.6 × 10−3) |
| Arthropods | 60 | 7 (9.8 × 10−3) | 22 (4.3 × 10−4) | 35 (0.078) |
| Echinoderms | 45 | 13 (2.3 × 10−8) | 21 (4.6 × 10−4) | 29 (0.018) |
| Chordates | 157 | 41 (1.4 × 10−18) | 83 (7.7 × 10−14) | 117 (3.0 × 10−10) |
Cases from each major group showing different levels of significance for excess HSC (measured as the proportion of Monte Carlo runs with equal or greater HSC). All cases with P ≤ 0.05 are also counted as P ≤ 0.25 and P < 0.50. Numbers in parentheses give binomial probabilities of these outcomes given expectations of 5%, 25%, and 50% of datasets. Fig. 1 describes the test.
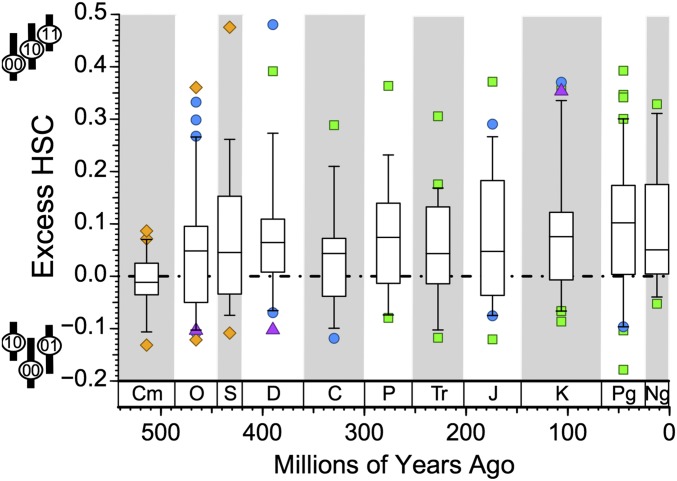
Temporally, only Cambrian clades fit null expectations (Fig. 2); excess HSC is common thereafter, with only the Carboniferous failing to show excess HSC in significantly more than 50% of clades at P ≤ 0.05. Pairwise contrasts in excess HSC between periods (SI Appendix, Table S4) show the Cambrian to be significantly different from all periods save the Carboniferous; however, only one of the remaining 45 contrasts (Ordovician vs. Paleogene) is significant at P ≤ 0.05.
Fig. 2.
Deviations between observed and expected HSC over time given budding cladogenesis. Colors denote higher taxonomic group like in Fig. 1. Binomial probabilities of deviations from an expectation of 50% excess HSC are Cambrian (Cm): P = 0.584 (11 of 22); Ordovician (O): P = 0.049 (32 of 53); Silurian (S): P = 0.025 (12 of 17); Devonian (D): P = 9.6 × 10−5 (25 of 31); Carboniferous (C): P = 0.072 (11 of 17); Permian (P): P = 0.018 (11 of 15); Triassic (Tr): P = 5.3 × 10−3 (17 of 23); Jurassic (J): P = 0.026 (18 of 27); Cretaceous (K): P = 1.1 × 10−4 (36 of 48); Paleogene (Pg): P = 2.9 × 10−5 (39 of 51); and Neogene (Ng): P = 2.9 × 10−5 (12 of 15).
Associations Between Excess HSC and Other Evolutionary Patterns.
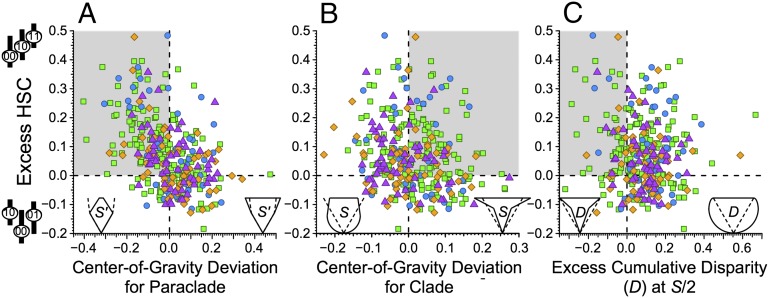
Clades with excess HSC typically have lower centers of gravity for paraclades retaining 00 pairs than expected given trait-independent diversification and origination, extinction, sampling, and character change parameters appropriate to each clade (Materials and Methods). This association (Fig. 3A) is highly significant for all clades (Kendall’s τ = −0.329, P = 1.7 × 10−18) and among brachiopod and mollusc, arthropod, echinoderm, and chordate clades separately (Table 2). The associations also are significant for Ordovician-Permian and Meso-Cenozoic clades but not Cambrian clades (Table 2). Excess HSC is also associated with whole clades having lower than expected centers of gravity. This association is much weaker than the HSC–paraclade association, and it is significant only among chordate and Meso-Cenozoic clades (Table 2). Finally, no significant associations exist between excess HSC and deviations from expected cumulative disparity (Table 2).
Fig. 3.
Associations between excess HSC and other paleontological patterns. Colors and shapes are the same as in Figs. 1 and 2. All points plot the differences between observation and expectation given continuous trait-independent diversification and no change of character space. Cartoons on the x axis idealize those deviations from the null model, with dashed lines giving expectations and solid lines giving possible patterns (D, cumulative disparity; S, richness). Gray boxes reflect predicted associations with HSC given (A) elevated net extinction, (B) elevated net speciation, and (C) elevated evolvability. (A) Observed minus expected centers of gravity for paraclades retaining 00 combinations (where 0 denotes the oldest appearing state). (B) Observed minus expected centers of gravity for whole clades. (C) Excess cumulative disparity among the first S/2 taxa in a clade of S taxa. Additional information is in Figs. 1 and 2.
Table 2.
Associations between excess HSC and other paleontological patterns
| Group | Paraclade CG | Clade CG | CD at S/2 | |||
| τ | P | τ | P | τ | P | |
| Brachiopods and molluscs | −0.234 | 0.010 | −0.135 | 0.139 | −0.068 | 0.453 |
| Arthropods | −0.307 | 5.2 × 10−4 | −0.077 | 0.386 | −0.047 | 0.592 |
| Echinoderms | −0.274 | 8.0 × 10−3 | −0.151 | 0.145 | −0.028 | 0.784 |
| Chordates | −0.367 | 8.7 × 10−12 | −0.161 | 2.8 × 10−3 | −0.048 | 0.373 |
| Cambrian | −0.074 | 0.631 | −0.017 | 0.910 | 0.052 | 0.735 |
| Paleozoic | −0.236 | 6.2 × 10−5 | −0.064 | 0.277 | −0.025 | 0.666 |
| Meso-Cenozoic | −0.398 | 4.6 × 10−14 | −0.175 | 9.3 × 10−4 | −0.058 | 0.269 |
Associations between excess HSC and deviations from expected paraclade and clade centers of gravity (CGs) and cumulative disparity (CD) halfway through clade history (S/2) broken down by taxonomic group and time. τ gives Kendall’s rank correlation statistic.
Discussion
Our results strongly corroborate elevated net extinction (model 1), strongly contradict elevated net speciation (model 2), and are unsupportive of elevated evolvability (model 3). Before discussing the implications of these models in additional detail, we will first consider whether very different models might explain our results.
Alternative Explanations for Excess HSC.
We should sample 95% of state pairs for compatible characters in correct order, regardless of average per-taxon sampling rates (SI Appendix, Fig. S5). However, if species with derived states have vastly higher sampling rates than species with primitive states, then we could sample more state pairs out of order. We consider this an unlikely explanation for two reasons. First, such changes in preservation potential should be as apt to convert HSC to DSC as DSC to HSC. Second, it is an improbable explanation on first principles: traits, such as basic skeletal mineralogy or environmental preference, that greatly alter preservation potential rarely vary among closely related species and genera (37, 38). Instead, the vast majority of character states are variations on features with very similar preservation potentials (e.g., shapes on some region of bone or calcitic shell).
Our Monte Carlo tests use diversification models that maximize expected HSC. However, pervasive anagenesis is a very different model that also will generate copious HSC. If all species in a given dataset are morphospecies from a single anagenetically evolving lineage, then only HSC can be common: a 00→10 transition eliminates the sole (morpho-) species bearing 00. A 00→01 transition requires that the lineage first revert back to 00. Anagenesis also predicts that HSC is anagenetic: species with 00 do not occur in younger strata than the first species with 10. Anagenetic HSC is much more frequent than predicted by trait-independent diversification (SI Appendix, Fig. S8A). However, very few datasets analyzed here are good candidates for being anagenetic lineages. Most datasets include numerous clearly contemporaneous species, and reconstructed phylogenies typically imply numerous subclades within each clade. Notably, trait-independent diversification under bifurcation models that mix anagenesis and cladogenesis predicts less HSC than it does under budding models with only cladogenesis (SI Appendix, Fig. S5). Our Monte Carlo tests assume the budding model. As such, assuming no anagenesis makes our results conservative (SI Appendix, Fig. S9 and Table S3).
An evolutionary explanation for reduced durations of paraclades relative to expected paraclade durations is that turnover rates decrease over time within clades. If this happens within individual clades that we analyze, then early paraclades should have shorter durations than expected given our null model. This pattern is well-documented for the Phanerozoic as a whole (39). However, stage-to-stage variation in turnover is considerable for both metazoans (39) and larger taxonomic groups (e.g., gastropods or mammals) (40–42), which means that turnover actually varies considerably over the timespans covered by the datasets that we analyze. Moreover, individual clades often have early origination rates that are much higher than extinction rates (3, 41–44), which elevates DSC rather than HSC (SI Appendix, Fig. S5D).
Paleontologists choose species and genera for phylogenetic analyses to address particular issues, which might, in turn, bias our results. For example, workers compile many phylogenetic datasets to examine biogeographic patterns (45–47). However, biogeographic differentiation should encourage the subclade divergence and thus, should generate more DSC than null models. Other phylogenetic datasets deliberately target the oldest members of clades to unravel subclade relationships because of a concern that homoplasy among late-appearing members of subclades will confound relationships among those subclades (48–50). Deliberately targeting early members of subclades should elevate DSC. Finally, high diversification rates early in clade history also would elevate DSC (see above).
Many of the clades that we analyze actually are paraclades within larger clades. Paraclades do not affect the implications of our results. Suppose that Eocene species show high HSC and correspondingly low centers of gravity among paraclades with primitive states. The implied relationship between primitive states and elevated net extinction in the Eocene follows if the clade went extinct at the end of the Eocene or if the clade includes unanalyzed Oligocene species. Alternatively, a group might be paraphyletic relative to a contemporaneous taxon that is so different that workers have not analyzed them together. Again, subsequent evolution has no bearing on the history of character states within the paraphyletic group; moreover, if the daughter taxon is that different from its ancestors, then there probably are few character states that can be coded easily in both groups to reveal DSC. Finally, our finding that paraclades with primitive states have unusually low centers of gravity is not an artifact of paraphyly. We report the difference between expected metrics given trait-independent diversification and observed metrics; regardless of whether expected centers of gravity for paraclades are low or high (33), we find that the observed centers of gravity are too low.
General Models of Elevated Net Extinction.
We conclude that elevated net extinction of paraclades retaining primitive conditions (model 1) drives most trait-based diversification shifts at low taxonomic levels. Paleontologists have proposed several explanations for elevated net extinction, including competition (43, 51, 52) and biased survivorship over extinction pulses (53). Competition models, such as coupled logistic diversification, are particularly appealing, because they offer mechanisms for actively eliminating paraclades while not necessarily greatly increasing the overall diversity of a clade (1, 43, 52). Competition also predicts the elevated anagenetic HSC discussed above by linking elevated net extinction to the appearance of derived species (SI Appendix, Fig. S12). Competition with members of other clades could have the same effect (with or without logistic diversification) if it induces new states through mechanisms, such character displacement (54), in some lineages while elevating net extinction in paraclades. Under either case, elevated net extinction might reflect decreased origination rates rather than increased extinction rates among lineages within paraclades (32).
Extinction resistance/selectivity favoring some derived taxa (24, 53) is another plausible model. Although few of the datasets examined here span mass extinctions, many of them span extinction pulses (55). Like competition models, extinction resistance for a derived subclade predicts lower centers of gravity for many paraclades than expected without selective extinction pulses. However, extinction resistance/selectivity does not predict unusually high anagenetic HSC: The mechanism for paraclade extinction does not coincide with the appearance of derived traits. Moreover, we have empirical examples of extinction resistance associated with primitive traits (23, 56) as well as many cases in which there is no obvious selectivity at all (57). These considerations make extinction resistance/selectivity a less reliable and less powerful explanation; however, we cannot discount it entirely.
On the Viability of the Elevated Evolvability and Elevated Net Cladogenesis Models.
Our results do not support the idea that elevated evolvability (model 3) drives trait-based diversification shifts. The vast majority of clades showing excess HSC shows more disparity among early species than expected rather than less disparity. High early disparity corroborates the idea that clades rapidly exhaust available character states (34, 58). It also raises the possibly that evolvability is greatest early in clade history (25). If so, then pooling datasets to examine (say) the Carnivora as a whole might reveal associations between elevated evolvability and the founding of major clades that do not exist with the families and subfamilies examined here (16, 59).
Our results flatly contradict the idea that elevated net speciation (model 2) drives trait-based diversification shifts. The associations between clade centers of gravity and HSC actually are opposite of the model’s predictions. A corollary prediction (i.e., that major taxonomic groups with many examples of excess HSC should show rising net origination rates over time) is also incorrect. Most Cenozoic mammal clades show excess HSC (SI Appendix, Fig. S11B) without any trend in net origination rates (42). Even more damning, most Silurian-Carboniferous trilobite clades show excess HSC (SI Appendix, Fig. S11A) while showing decreasing net origination rates (40). Thus, our results are another caution that the common inference of elevated net speciation from phylogenies of extant taxa is an artifact of those trees being unable to support elevated net extinction models (18–21, 60).
Why Is the Cambrian Different?
Cambrian clades alone show neither pervasive excess HSC nor a correlation between excess HSC and low centers of gravity for paraclades. This evidence of (relatively) high divergence might reflect the radiation of clades into new ecospace, allowing for unusually high numbers of subclades to diversify (61, 62), which in turn, might generate enough DSC to cancel out excess HSC within subclades. However, major radiations in the Ordovician, Triassic, and Paleogene contradict this idea by generating frequent excess HSC (Fig. 2), despite having many plausible examples of clades radiating into “vacated” ecospace.
Nearly all Cambrian datasets represent trilobites. Thus, the Cambrian pattern might corroborate the biomere model (56), which posits that trilobites retaining primitive states selectively survived extinction pulses in the Cambrian. Such extinction would offset background loss of taxa retaining primitive states (63). Notably, post-Cambrian trilobites (and particularly, Silurian-Carboniferous trilobites) show HSC patterns comparable with other metazoans (SI Appendix, Fig. S10 and Table S4). Moreover, arthropods show a significant association between excess HSC and overly low paraclade centers of gravity, although 50 of 60 clades are trilobites (Table 2). Assessing whether this reflects something different about Cambrian trilobites or the Cambrian as a whole requires data from Cambrian molluscs, echinoderms, etc. Nevertheless, it does suggest yet another way in which Cambrian evolution was unique.
Conclusions
After the Cambrian, HSC among closely related species and genera greatly exceeds the expectations of trait-independent diversification. Our finding indicates that trait-based diversification shifts are common at low taxonomic levels. The pattern corresponds with paraphyletic groups retaining primitive conditions losing diversity faster than predicted by trait-independent diversification. Thus, elevated net extinction seems to be the primary driver of trait-based diversity shifts. Our results strongly contradict the idea that elevated net speciation within derived subclades is common, although elevated net speciation is a conclusion of many studies using phylogenies of extant species. Increased evolvability among anatomical characters also does not explain diversification shifts, although elevated evolvability might be important for the founding of the analyzed taxa. Future work should focus on assessing why we do not see clear signs of trait-based diversification shifts among Cambrian taxa and means of recognizing elevated net extinction among taxa lacking fossil records.
Materials and Methods
Datasets.
We analyze 319 published character matrices, all of which were assembled for phylogenetic analyses (SI Appendix, Tables S5 and S6). We focus on species- and genus-level datasets, because (i) we are interested in whether patterns associated with trait-based diversification shifts occur at low taxonomic levels, (ii) species- and genus-level analyzes minimize the potential for uneven species richness among taxa hiding evidence of divergence, and (iii) using species and genera instead of (say) families minimizes cases where characters used to diagnose a taxon are absent in the oldest known members of that taxon. We made exceptions for studies focusing on early members of clades that include token members of groups that diversify after the study interval of the dataset (e.g., late Eocene representatives of subfamilies that diversify in the Oligocene are included in an analysis of Eocene species). We also exclude outgroup taxa, because outgroups usually represent a small fraction of the richness in a related clade. The vast majority of our datasets lacks any extant species or genera; however, any extant taxa in a dataset are included only if they have fossil representatives.
We set polymorphic characters to states that maximized their stratigraphic compatibility. In studies including extant species, we exclude any characters not coded for extinct taxa on the assumption that they are not fossilizable characters. We also exclude characters that are invariant within the ingroup.
We derive first and last appearance data from several sources, with the original publications and the Paleobiology Database (paleobiodb.org/) being the two biggest sources. Stratigraphic ranges for extant taxa reflect the first and last fossil occurrences rather than assuming that those taxa survive to the present.
Metrics.
Our analyses measure compatibility, stratigraphic compatibility, center of gravity, and morphological disparity. Compatible characters have three of four possible combinations if the characters are binary (26, 27); if one or both characters have three or more states, then we first assess whether the pair is compatible (SI Appendix, Fig. S1), and then, we tally all binary breakdowns of the two characters with three of four possible pairs (SI Appendix, Figs. S2 and S3) (note that inapplicable and unknown conditions always are excluded from combinations). Our approach therefore treats all multistate characters as unordered, which maximizes their compatibility (27) and standardizes the inconsistent use of ordered characters among workers. We tally stratigraphic compatibility as all compatible pairs with three of four states in which species with the intermediate pair (e.g., 00 given 00, 10, and 01) do not appear last in the fossil record (28). (Note that 0 represents the first appearing state, regardless of whether those states were coded 0 in the real data.) We tally hierarchical and DSC as described in the text; in cases where species with 00 and 10 first appear in the oldest strata before species with 11, it is not clear which state for the first character appears first, and the data are consistent with both HSC and DSC. We tally such cases as one-half HSC and one-half DSC. We then use the proportion of stratigraphically compatible pairs that are HSC for comparison with Monte Carlo expectations (see below).
We calculate center of gravity following several prior studies (32, 33) using the stratigraphic ranges of the taxa in the dataset. We did this first for the entire clade (total clade center of gravity). For the average paraclade center of gravity within each clade, we took every HSC pair and then measured the center of gravity for the assemblage of taxa retaining the 00 condition (with 0 representing the oldest appearing states, regardless of the actual number used in the dataset). We then estimated the average center of gravity of those paraclades. (If a character pair is one-half HSC because of two states appearing in the oldest strata, then the pair is given half-weight; see above.) This average was then rescaled to the total clade center of gravity for comparisons with Monte Carlo expectations (see below).
We measure morphological disparity as the average pairwise dissimilarity among species [i.e., the (64)]. We use cumulative disparity rather than standing disparity (i.e., the average pairwise dissimilarity among all S taxa in a dataset and the average pairwise dissimilarity among the oldest S/2 taxa in that dataset). In cases where clades passed S/2 taxa partway through a stratigraphic interval, we estimate the disparity at S/2 assuming a log-linear relationship between disparity and richness (35). Suppose that a dataset with 29 species has 10 species through time 3 and 20 species through time 4 and that the average pairwise dissimilarity among the first 10 species is 0.4, whereas the average pairwise dissimilarity among the first 20 species is 0.5. Species 15 represents the halfway point. The cumulative disparity among the first 15 species is (SI Appendix, Fig. S6). We rescale for comparison with Monte Carlo expectations (see below).
Monte Carlo Analyses.
We use Monte Carlo analyses to estimate expected HSC, centers of gravity, and cumulative disparities. Unlike bootstrapping or permutation tests in similar analyses (25), Monte Carlo tests assume that some phylogeny underlies character and stratigraphic distributions. For each clade of S taxa, 1,001 phylogenies are simulated using origination and extinction rates estimated from the stratigraphic ranges of the original data until S taxa are sampled given sampling rates estimated from the same stratigraphic data. Usually, origination, extinction, and sampling are empirically estimated based on the proportions of taxa known from one, two, three, etc. intervals (65). For datasets with taxa limited to one or two intervals, we used a preliminary set of simulations to find rates maximizing the probability of observing S taxa over X intervals, with X being the number of intervals in the dataset. Origination and extinction rates are constant, which matches the null hypothesis. Also, continuous exponential diversification generates more HSC than alternative models, such as logistic diversification (SI Appendix, Fig. S5D). We simulated phylogenies under both budding cladogenesis (where species can have descendants as long as they persist) and bifurcating cladogenesis (where morphospecies disappear anagenetically on giving rise to two descendants) but present only the budding results, because budding promotes more HSC (and thus, more conservative results) than bifurcation by allowing single species to have three or more descendants instead of only two descendants (SI Appendix, Fig. S5). We simulate morphological evolution among the same numbers of characters and states as the original dataset. Change ceases when compatibility among simulated characters matches that of the original dataset (66) and thus, at a likely overall amount (SI Appendix, Fig. S4).
The Monte Carlo tests generate:
-
i)
expected HSC given continuous, trait-independent diversification over phylogeny generated under plausible rates of origination, extinction, sampling, and change;
-
ii)
expected paraclade and clade center of gravity given continuous, trait-independent diversification over phylogeny under plausible rates of origination, extinction, sampling, and change; and
-
iii)
expected cumulative disparity at S/2 over phylogeny given plausible and consistent rates of change in a single character space.
Supplementary Material
Acknowledgments
The basic analyses for this project were completed before the death of G.F.E. For comments and discussion, we thank D. H. Erwin, S. K. Lyons, F. R. McMorris, J. Marcot, and R. Warnock. We also thank the many researchers who created the original datasets. This manuscript was greatly improved by comments from two anonymous reviewers. This work is Paleobiology Database official publication 209.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.F. is a guest editor invited by the Editorial Board.
Data deposition: The datasets used in this paper have been deposited in The Paleobiology Database, www.paleobiodb.org/cgi-bin/bridge.pl?a=nexusFileSearch (reference no. 53093).
2Deceased November 24, 2011.
See Commentary on page 16240.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406304111/-/DCSupplemental.
References
- 1.Lidgard S, McKinney FK, Taylor PD. Competition, clade replacement, and a history of cyclostome and cheilostome bryozoan diversity. Paleobiology. 1993;19(3):352–371. [Google Scholar]
- 2.Roy K. The roles of mass extinction and biotic interaction in large-scale replacements: A reexamination using the fossil record of stromboidean gastropods. Paleobiology. 1996;22(3):436–452. [Google Scholar]
- 3.Eble GJ. Contrasting evolutionary flexibility in sister groups: Disparity and diversity in Mesozoic atelostomate echinoids. Paleobiology. 2000;26(1):56–79. [Google Scholar]
- 4.Lupia R, Lidgard S, Crane PR. Comparing palynological abundance and diversity: Implications for biotic replacement during the Cretaceous angiosperm radiation. Paleobiology. 1999;25(3):305–340. [Google Scholar]
- 5.Sanderson MJ, Donoghue MJ. Shifts in diversification rate with the origin of angiosperms. Science. 1994;264(5165):1590–1593. doi: 10.1126/science.264.5165.1590. [DOI] [PubMed] [Google Scholar]
- 6.Purvis A, Nee S, Harvey PH. Macroevolutionary inferences from primate phylogeny. Proc Biol Sci. 1995;260(1359):329–333. doi: 10.1098/rspb.1995.0100. [DOI] [PubMed] [Google Scholar]
- 7.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106(32):13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foote M. Contributions of individual taxa to overall morphological disparity. Paleobiology. 1993;19(4):403–419. [Google Scholar]
- 9.Stockmeyer Lofgren A, Plotnick RE, Wagner PJ. Morphological diversity of Carboniferous arthropods and insights on disparity patterns of the Phanerozoic. Paleobiology. 2003;29(3):350–369. [Google Scholar]
- 10.Wagner PJ, Ruta M, Coates MI. Evolutionary patterns in early tetrapods. II. Differing constraints on available character space among clades. Proc Biol Sci. 2006;273(1598):2113–2118. doi: 10.1098/rspb.2006.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanley SM. A theory of evolution above the species level. Proc Natl Acad Sci USA. 1975;72(2):646–650. doi: 10.1073/pnas.72.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenzweig ML, McCord RD. Incumbent replacement: Evidence for long-term evolutionary progress. Paleobiology. 1991;17(3):202–213. [Google Scholar]
- 13.Slowinski JB, Guyer C. Testing whether certain traits have caused amplified diversification: An improved method based on a model of random speciation and extinction. Am Nat. 1993;142(6):1019–1024. doi: 10.1086/285586. [DOI] [PubMed] [Google Scholar]
- 14.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56(5):701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 15.Wagner GP, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50(3):967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 16.Rabosky DL, et al. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat Commun. 2013;4:1958. doi: 10.1038/ncomms2958. [DOI] [PubMed] [Google Scholar]
- 17.Simpson C. Species selection and driven mechanisms jointly generate a large-scale morphological trend in monobathrid crinoids. Paleobiology. 2010;36(3):481–496. [Google Scholar]
- 18.Quental TB, Marshall CR. Extinction during evolutionary radiations: Reconciling the fossil record with molecular phylogenies. Evolution. 2009;63(12):3158–3167. doi: 10.1111/j.1558-5646.2009.00794.x. [DOI] [PubMed] [Google Scholar]
- 19.Rabosky DL. Extinction rates should not be estimated from molecular phylogenies. Evolution. 2010;64(6):1816–1824. doi: 10.1111/j.1558-5646.2009.00926.x. [DOI] [PubMed] [Google Scholar]
- 20.Freckleton RP, Phillimore AB, Pagel M. Relating traits to diversification: A simple test. Am Nat. 2008;172(1):102–115. doi: 10.1086/588076. [DOI] [PubMed] [Google Scholar]
- 21.Liow LH, Quental TB, Marshall CR. When can decreasing diversification rates be detected with molecular phylogenies and the fossil record? Syst Biol. 2010;59(6):646–659. doi: 10.1093/sysbio/syq052. [DOI] [PubMed] [Google Scholar]
- 22.Wagner PJ. The quality of the fossil record and the accuracy of phylogenetic inferences about sampling and diversity. Syst Biol. 2000;49(1):65–86. doi: 10.1080/10635150050207393. [DOI] [PubMed] [Google Scholar]
- 23.Liow LH. A test of Simpson’s “rule of the survival of the relatively unspecialized” using fossil crinoids. Am Nat. 2004;164(4):431–443. doi: 10.1086/423673. [DOI] [PubMed] [Google Scholar]
- 24.Liow LH. Do deviants live longer? Morphology and longevity in trachyleberidid ostracodes. Paleobiology. 2006;32(1):55–69. [Google Scholar]
- 25.Hughes M, Gerber S, Wills MA. Clades reach highest morphological disparity early in their evolution. Proc Natl Acad Sci USA. 2013;110(34):13875–13879. doi: 10.1073/pnas.1302642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Quesne WJ. A method of selection of characters in numerical taxonomy. Syst Zool. 1969;18(2):201–205. [Google Scholar]
- 27.Estabrook GF, Johnson CS, Jr, McMorris FR. An idealized concept of the true cladistic character. Math Biosci. 1975;23(2):263–272. [Google Scholar]
- 28.Estabrook GF, McMorris FR. The compatibility of stratigraphic and comparative constraints on estimates of ancestor–descendant relations. Syst Biodivers. 2006;4(2):9–17. [Google Scholar]
- 29.O'Keefe FR, Wagner PJ. Inferring and testing hypotheses of correlated character evolution using character compatibility. Syst Biol. 2001;50(5):657–675. doi: 10.1080/106351501753328794. [DOI] [PubMed] [Google Scholar]
- 30.Raup DM. Mathematical models of cladogenesis. Paleobiology. 1985;11(1):42–52. [Google Scholar]
- 31.Estabrook GF. Does common equal primitive? Syst Bot. 1977;2(1):16–42. [Google Scholar]
- 32.Gilinsky NL, Bambach RK. Asymmetrical patterns of origination and extinction in higher taxa. Paleobiology. 1987;13(4):427–445. [Google Scholar]
- 33.Uhen MD. An evaluation of clade-shape statistics using simulations and extinct families of mammals. Paleobiology. 1996;22(1):8–22. [Google Scholar]
- 34.Foote M. Morphological diversification of Paleozoic crinoids. Paleobiology. 1995;21(3):273–299. [Google Scholar]
- 35.Foote M. Models of morphologic diversification. In: Jablonski D, Erwin DH, Lipps JH, editors. Evolutionary Paleobiology: Essays in Honor of James W Valentine. Univ of Chicago Press; Chicago: 1996. pp. 62–86. [Google Scholar]
- 36.Foote M. Discordance and concordance between morphological and taxonomic diversity. Paleobiology. 1993;19(2):185–204. [Google Scholar]
- 37.Jablonski D, Sepkoski JJ, Jr, Bottjer DJ, Sheehan PM. Onshore-offshore patterns in the evolution of phanerozoic shelf communities. Science. 1983;222(4628):1123–1125. doi: 10.1126/science.222.4628.1123. [DOI] [PubMed] [Google Scholar]
- 38.Bottjer DJ, Jablonski D. Paleoenvironmental patterns in the evolution of post-Paleozoic benthic marine invertebrates. Palaios. 1988;3(6):540–560. [Google Scholar]
- 39.Alroy J. Colloquium paper: Dynamics of origination and extinction in the marine fossil record. Proc Natl Acad Sci USA. 2008;105(Suppl 1):11536–11542. doi: 10.1073/pnas.0802597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webster M. A Cambrian peak in morphological variation within trilobite species. Science. 2007;317(5837):499–502. doi: 10.1126/science.1142964. [DOI] [PubMed] [Google Scholar]
- 41.Wagner PJ. Diversification among early Paleozoic gastropods - contrasting taxonomic and phylogenetic descriptions. Paleobiology. 1995;21(4):410–439. [Google Scholar]
- 42.Alroy J. Constant extinction, constrained diversification, and uncoordinated stasis in North American mammals. Palaeogeogr Palaeoclimatol Palaeoecol. 1996;127(1/4):285–311. [Google Scholar]
- 43.Miller AI, Sepkoski JJ., Jr Modeling bivalve diversification: The effect of interaction on a macroevolutionary system. Paleobiology. 1988;14(4):364–369. doi: 10.1017/s0094837300012100. [DOI] [PubMed] [Google Scholar]
- 44.Brayard A, et al. Good genes and good luck: Ammonoid diversity and the end-Permian mass extinction. Science. 2009;325(5944):1118–1121. doi: 10.1126/science.1174638. [DOI] [PubMed] [Google Scholar]
- 45.Lieberman BS. Systematics and biogeography of the “Metacryphaeus group” Calmoniidae (Trilobita, Devonian), with comments on adaptive radiations and the geological history of the Malvinokaffric Realm. J Paleontol. 1993;67(4):549–570. [Google Scholar]
- 46.Rode AL, Lieberman BS. Phylogenetic and biogeographic analysis of Devonian phyllocarid crustaceans. J Paleontol. 2002;76(2):271–286. [Google Scholar]
- 47.Mihlbachler MC. Species taxonomy, phylogeny, and biogeography of the Brontotheriidae (Mammalia: Perissodactyla) Bull Am Mus Nat Hist. 2008;311(1):1–475. [Google Scholar]
- 48.Froehlich DJ. Phylogenetic systematics of basal perissodactyls. J Vertebr Paleontol. 1999;19(1):140–159. [Google Scholar]
- 49.Smith AB. Probing the cassiduloid origins of clypeasteroid echinoids using stratigraphically restricted parsimony analysis. Paleobiology. 2001;27(2):392–404. [Google Scholar]
- 50.Wesley-Hunt GD, Flynn JJ. Phylogeny of the Carnivora: Basal relationships among the carnivoramorphans, and assessment of the position of “Miacoidea” relative to crown-clade Carnivora. J Syst Palaeontol. 2005;3(1):1–28. [Google Scholar]
- 51.Stanley SM. Macroevolution: Pattern and Process. Elsevier; Amsterdam: 1979. [Google Scholar]
- 52.Sepkoski JJ, Jr, McKinney FK, Lidgard S. Competitive displacement among post-Paleozoic cyclostome and cheilostome bryozoans. Paleobiology. 2000;26(1):7–18. doi: 10.1666/0094-8373(2000)026<0007:cdappc>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 53.Benton MJ. On the nonprevalence of competitive displacement in the evolution of tetrapods. In: Jablonski D, Erwin DH, Lipps JH, editors. Evolutionary Paleobiology: Essays in Honor of James W Valentine. Univ of Chicago Press; Chicago: 1996. pp. 185–210. [Google Scholar]
- 54.Schluter D. Ecological character displacement in adaptive radiation. Am Nat. 2000;156(Suppl):S4–S16. [Google Scholar]
- 55.Foote M. Pulsed origination and extinction in the marine realm. Paleobiology. 2005;31(1):6–20. [Google Scholar]
- 56.Palmer AR. Biomere: A new kind of biostratigraphic unit. J Paleontol. 1965;39(1):149–153. [Google Scholar]
- 57.Bapst DW, Bullock PC, Melchin MJ, Sheets HD, Mitchell CE. Graptoloid diversity and disparity became decoupled during the Ordovician mass extinction. Proc Natl Acad Sci USA. 2012;109(9):3428–3433. doi: 10.1073/pnas.1113870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner PJ. Exhaustion of morphologic character states among fossil taxa. Evolution. 2000;54(2):365–386. doi: 10.1111/j.0014-3820.2000.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 59.Erwin DH. Disparity: Morphological pattern and developmental context. Palaeontology. 2007;50(1):57–73. [Google Scholar]
- 60.Quental TB, Marshall CR. The molecular phylogenetic signature of clades in decline. PLoS ONE. 2011;6(10):e25780. doi: 10.1371/journal.pone.0025780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foote M. Ecological controls on the evolutionary recovery of Post-Paleozoic crinoids. Science. 1996;274(5292):1492–1495. doi: 10.1126/science.274.5292.1492. [DOI] [PubMed] [Google Scholar]
- 62.Erwin DH. Novelties that change carrying capacity. J Exp Zoolog B Mol Dev Evol. 2012;318(6):460–465. doi: 10.1002/jez.b.21429. [DOI] [PubMed] [Google Scholar]
- 63.Jablonski D. Background and mass extinctions: The alternation of macroevolutionary regimes. Science. 1986;231(4734):129–133. doi: 10.1126/science.231.4734.129. [DOI] [PubMed] [Google Scholar]
- 64.Foote M. Paleozoic record of morphologica diversity in blastozoan echinoderms. Proc Natl Acad Sci USA. 1992;89(16):7325–7329. doi: 10.1073/pnas.89.16.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foote M, Raup DM. Fossil preservation and the stratigraphic ranges of taxa. Paleobiology. 1996;22(2):121–140. doi: 10.1017/s0094837300016134. [DOI] [PubMed] [Google Scholar]
- 66.Wagner PJ. Modelling rate distributions using character compatibility: Implications for morphological evolution among fossil invertebrates. Biol Lett. 2012;8(1):143–146. doi: 10.1098/rsbl.2011.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.