Significance
Staphylococcus aureus is one of the most successful and adaptable human pathogens and is a major cause of hospital-acquired infections. Here we provide insight into how S. aureus uses the catabolite control protein E (CcpE) to sense its intracellular metabolic status and to regulate its virulence-associated properties. We define a key circuit of the virulence regulatory network of S. aureus and emphasize that metabolic status may be a critical element governing the virulence of this pathogen. Understanding the role of metabolites in virulence factor expression ultimately may contribute to the development of novel strategies to combat this dreaded infectious disease.
Keywords: Staphylococcus aureus, metabolism, iron acquisition, virulence gene expression, bacterial virulence
Abstract
An effective metabolism is essential to all living organisms, including the important human pathogen Staphylococcus aureus. To establish successful infection, S. aureus must scavenge nutrients and coordinate its metabolism for proliferation. Meanwhile, it also must produce an array of virulence factors to interfere with host defenses. However, the ways in which S. aureus ties its metabolic state to its virulence regulation remain largely unknown. Here we show that citrate, the first intermediate of the tricarboxylic acid (TCA) cycle, binds to and activates the catabolite control protein E (CcpE) of S. aureus. Using structural and site-directed mutagenesis studies, we demonstrate that two arginine residues (Arg145 and Arg256) within the putative inducer-binding cavity of CcpE are important for its allosteric activation by citrate. Microarray analysis reveals that CcpE tunes the expression of 126 genes that comprise about 4.7% of the S. aureus genome. Intriguingly, although CcpE is a major positive regulator of the TCA-cycle activity, its regulon consists predominantly of genes involved in the pathogenesis of S. aureus. Moreover, inactivation of CcpE results in increased staphyloxanthin production, improved ability to acquire iron, increased resistance to whole-blood–mediated killing, and enhanced bacterial virulence in a mouse model of systemic infection. This study reveals CcpE as an important metabolic sensor that allows S. aureus to sense and adjust its metabolic state and subsequently to coordinate the expression of virulence factors and bacterial virulence.
One of the most adaptable human pathogens is Staphylococcus aureus (1, 2). Causing more deaths than AIDS in the United States (3), S. aureus owes its success largely to its antibiotic resistance and its ability to produce a wide array of virulence factors that interfere with host defenses (4, 5). Even without antibiotic resistance, S. aureus has the effective means to cause infections in almost every tissue of the human body (1). This versatility is thought to result from the remarkable capacity of this pathogen to adapt rapidly to changes in environmental conditions and to regulate the expression of a large array of virulence factors in a coordinated manner (1, 2, 4–7).
Like other living organisms, the ability of S. aureus to sense and adapt to changes in its environment is paramount to its survival. Some external signals can be sensed and responded to directly by two-component systems (8); however, other external signals that regulate the expression of virulence factors require transduction into intracellular signals, which subsequently are sensed by regulatory proteins such as transcriptional regulators CodY, CcpA, and MgrA (6). For instance, when nutrients are limited, a decrease in intracellular levels of GTP and branched-chain amino acids causes an allosteric deactivation of CodY, which leads to activated transcription of the CodY regulon that consists predominantly of genes involved in amino acid biosynthesis, transport of macromolecules, and virulence (9, 10).
To survive and replicate efficiently in the host, S. aureus has developed exquisite mechanisms for scavenging nutrients and adjusting its metabolism to maintain growth while also coping with stress (6, 11). On the other hand, S. aureus produces a wide array of virulence factors to evade host immune defenses and to derive nutrition either parasitically or destructively from the host during infections (6). Indeed, pathogen exploitation of host nutrients is one of the most fundamental aspects of host–pathogen interactions and infectious diseases (11–16). For example, in vertebrate hosts one of the first lines of defense against S. aureus infection is withholding iron to prevent the growth of S. aureus; therefore S. aureus has evolved highly efficient nutrient-retrieval strategies to counteract this nutritional deprivation (11, 17, 18). However, although it is well known that nutrients modulate intracellular metabolic status, as reflected by the concentration of key metabolic intermediates (19–22), the links between the metabolic intermediates and the expression of S. aureus virulence factors remain largely unknown (6).
Here we show that citrate, the first intermediate of the tricarboxylic acid (TCA) cycle, binds to and activates catabolite control protein E (CcpE) (23). We found that CcpE acts as a master regulator for the expression of virulence factors, central metabolism, iron acquisition, and bacterial virulence of S. aureus. This study establishes an intimate link between central metabolism and bacterial virulence in S. aureus.
Results
Deletion of ccpE Led to Enhanced Staphyloxanthin Production Mediated by the Down-Regulation of citB.
Previously, we showed that the transposon insertion in the ccpE gene (SAV0672 in S. aureus Mu50) of the S. aureus Newman strain enhances the production of staphyloxanthin (24), a virulence factor used to evade host oxidative killing (25–27). To verify further that the effect of the ccpE transposon insertion was specific to ccpE disruption and was not an artifact of transposition, we generated a ccpE-deletion mutant in the S. aureus Newman strain as described in SI Appendix, Experimental Procedures. The resulting mutant, ΔccpE, was phenotypically similar to the ccpE mutant with the transposon insertion, displaying an increase in staphyloxanthin production as compared with either the wild-type Newman strain or its complemented strain (ΔccpE/p-ccpE) (Fig. 1A and SI Appendix, Fig. S1A). When all experiments described above were repeated for the USA300 strain JE2 and its isogenic ccpE-deletion mutant (JE2-ΔccpE), similar results were observed (SI Appendix, Fig. S1 B and C), demonstrating that the effect of ccpE deletion on staphyloxanthin production is not limited to the Newman strain and that CcpE negatively controls the production of staphyloxanthin in S. aureus.
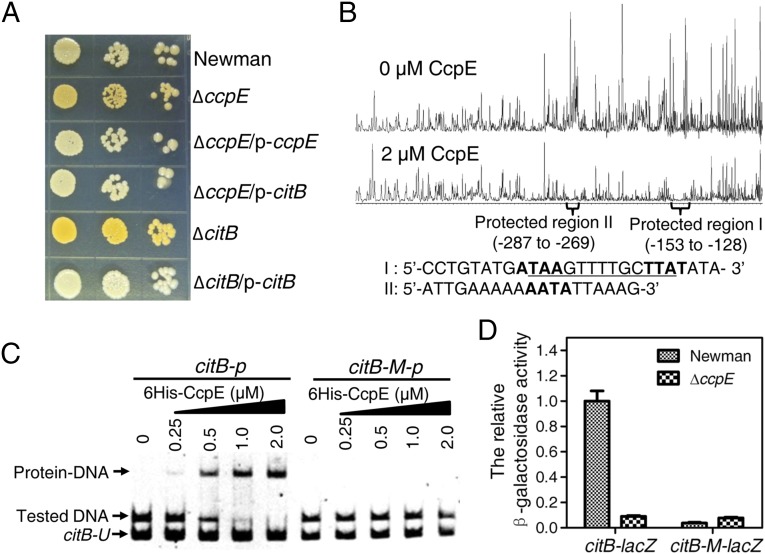
Fig. 1.
CcpE negatively modulates pigment production of S. aureus by activating the expression of citB. (A) Pigmentation of S. aureus Newman strain and its derivatives grown on TSA without glucose supplement, at 37 °C for 24 h. The wild-type Newman, ΔccpE, and ΔcitB strains harbor plasmid pYJ335. (B) Electropherograms show the protection pattern of the citB promoter after digestion with DNaseI following incubation in the absence and the presence of 2 µM 6His-CcpE. There are two CcpE-protected regions (I and II) in the promoter of the citB. CcpE-protected region I harbors a box I-like sequence (shown in bold letters) [ATAA-N7-TTAT, where N is any nucleotide; the potential LysR-type transcriptional regulator (LTTR) box is underlined]. Protected region II contains a box II-like sequence (AATA, shown in bold letters), which can be found in the CcpC-binding sites of citB in B. subtilis. (C) Assessment of mutations or deletion of the protected region I on CcpE binding. EMSA assays were performed using a wild-type fragment of the citB promoter DNA (citB-p, from nucleotides −151 to +88 of the start codon of citB) or the same fragment with ATAAGTTTTGCTTAT mutated to CGCCACTTTGCTTAT (citB-M-p). Test DNA was citB-p or citB-M-p, as indicted. citB-U, a DNA fragment of the citB promoter DNA (from nucleotides −128 to +88 of the start codon of citB) containing no CcpE-protected region. (D) Effects of mutations to the protected region I on the promoter activity of citB. ATAAGTTTTGCTTAT of the citB promoter (nucleotides −767 to +78 of the start codon of citB) was mutated to CGCCACTTTGCTTAT. Bacteria were grown in TSB at 37 °C with shaking, 250 rpm of aeration, and sampled at 6 h. Values are relative to the wild-type Newman strain (set to 1). The expression level of transcriptional fusion citB-lacZ is high enough that the introduction of ccpE-null could have a detectable reducing effect. Results represent means ± SD, and data are representative of three independent experiments.
Given that the deletion of ccpE causes reduced expression of citB (the gene encoding aconitase, the second enzyme of the TCA cycle) (23) and that disruption of TCA-cycle genes causes enhanced staphyloxanthin production (24), we next sought to determine if the constitutive expression of citB could suppress the effect of ccpE deletion on the production of staphyloxanthin. As shown in Fig. 1A and SI Appendix, Fig. S1A, the introduction of p-citB (SI Appendix, Table S1) into the ΔccpE strain restored staphyloxanthin production to wild-type levels. To demonstrate further that citB is involved in the production of staphyloxanthin, we created a markerless deletion of citB in the S. aureus Newman and JE2 strains, resulting in ΔcitB and JE2-ΔcitB (SI Appendix, Table S1), respectively. Both ΔcitB and JE2-ΔcitB strains exhibited enhanced staphyloxanthin production compared with the parent strain (Fig. 1A and SI Appendix, Fig. S1). In addition, when complemented with a plasmid carrying a wild-type citB gene (p-citB), staphyloxanthin production was restored to wild-type levels in both ΔcitB and JE2-ΔcitB mutants (Fig. 1A and SI Appendix, Fig. S1). We also observed that inactivation of either ccpE or citB caused an increase in the promoter activity of the crtOPQMN operon containing staphyloxanthin biosynthesis genes (SI Appendix, Fig. S1D). Taken together, these results clearly suggest that CcpE negatively controls the production of staphyloxanthin by activating the transcription of citB.
CcpE Activates the Expression of citB in a Direct Manner.
A recent study has shown that CcpE binds to the promoter of citB and activates its expression (23). To delineate further the function of CcpE on the regulation of citB, we searched for potential CcpE-binding sites in the promoter region of citB using a dye-based DNase I footprinting analysis. As shown in Fig. 1B, in the presence of 6His-CcpE, two regions of the citB promoter apparently were protected against DNase I digestion. Protected region I extended from nucleotides −153 to −128 relative to the citB start codon, and protected region II spanned nucleotides −287 to −269. In addition, a certain degree of protection also was seen in the region between protected regions I and II. We noted that protected region I contains a box I-like sequence (ATAA-N7-TTAT, where N is any nucleotide) and that protected region II contains a box II-like sequence (AATA or TTAT), as found in the binding sites of CcpC (28), a citrate-responsive regulator in Bacillus subtilis showing 61% similarity and 35% identity to CcpE (28, 29).
Protein-sequence analysis predicted that CcpE is a LysR-type transcriptional regulator (LTTR) that binds to the LTTR box T-N11-A (where N is any nucleotide) (30). Given that the box I-like sequence contains a potential LTTR box (ATAA-N7-TTAT, shown in bold letters in Fig. 1B), we next sought to determine if the box I-like sequence is important for the binding of CcpE to the DNA fragment of the citB promoter. We performed an EMSA using an ∼0.24-kb citB-p DNA fragment (from nucleotides −151 to +88 of the start codon of citB, containing an intact box I-like sequence), a citB-U DNA fragment (from nucleotides −128 to +88 of the start codon of citB, lacking the box I-like sequence), and a citB-M-p DNA fragment in which the box I-like sequence ATAAGTTTTGCTTAT was mutated to CGCCACTTTGCTTAT). As shown in Fig. 1C, neither the citB-U nor citB-M-p DNA fragment binds to CcpE, but citB-p does, demonstrating that the box I-like sequence is critical for binding to 6His-CcpE. Moreover, the DNA fragment citB-p12 (from nucleotides −321 to +66), which contains both CcpE-protected regions I and II, showed a higher affinity for 6His-CcpE than did the citB-L-p DNA fragment (from nucleotides −194 to +88), which contains only the protected region I (SI Appendix, Fig. S2A), indicating that DNA sequence outside the protected region I also may contribute to the interaction between CcpE and the citB promoter DNA. This result is consistent with the footprinting data shown in Fig. 1B. Additionally, the dissociation constant for the binding of 6His-CcpE to a citB promoter DNA fragment is in the low micromolar range (Fig. 1C and SI Appendix, Fig. S2), indicating a relatively weak interaction between the CcpE and its target DNA sequences. The precise mechanisms of CcpE–DNA interactions require further investigation. Nonetheless, these results suggest that the box I-like sequence, which contains a potential LTTR box, is important for the citB promoter to interact with CcpE.
As is consistent with a recent study showing that CcpE is a positive regulator of citB (23), the promoter activity of citB (citB-lacZ, nucleotides −767 to +78 of the start codon) was approximately eight times lower in the ΔccpE mutant than in the wild-type strain (Fig. 1D). In addition, the mutation of the box I-like sequence in the citB promoter abolished the promoter activity of citB-M-lacZ (Fig. 1D), suggesting that an intact box I-like sequence is required for the full activity of the citB promoter. As expected, the mutant citB promoter (i.e., citB-M) did not respond to the deletion of ccpE (Fig. 1D). Taken together, these results indicate that the box I-like sequence is likely involved in the CcpE-mediated activation of the citB promoter. Based on these data, we conclude that CcpE may activate the expression of citB in a direct manner.
CcpE Is a Citrate-Responsive Regulator.
A recent study showed that citrate does not affect CcpE binding to DNA (23). However, the amino acid sequence and DNA-binding sites of CcpE (Fig. 1B) are similar to those of CcpC, a citrate-responsive regulator in B. subtilis (28, 29). In addition, deletion of ccpE causes a dramatic reduction of citB expression, resulting in the accumulation of citrate (23). These observations led us to reexamine the effect of citrate on the DNA-binding activity of CcpE using EMSA. As can be seen, although the addition of 10 mM sodium citrate increased the in vitro binding of 6His-CcpE to the citB promoter, the addition of 10 mM sodium isocitrate did not (SI Appendix, Fig. S2B). Moreover, sodium citrate enhanced CcpE binding to the citB promoter in a dose-dependent manner (Fig. 2A). These results indicate that citrate can alter the DNA-binding affinity of CcpE.
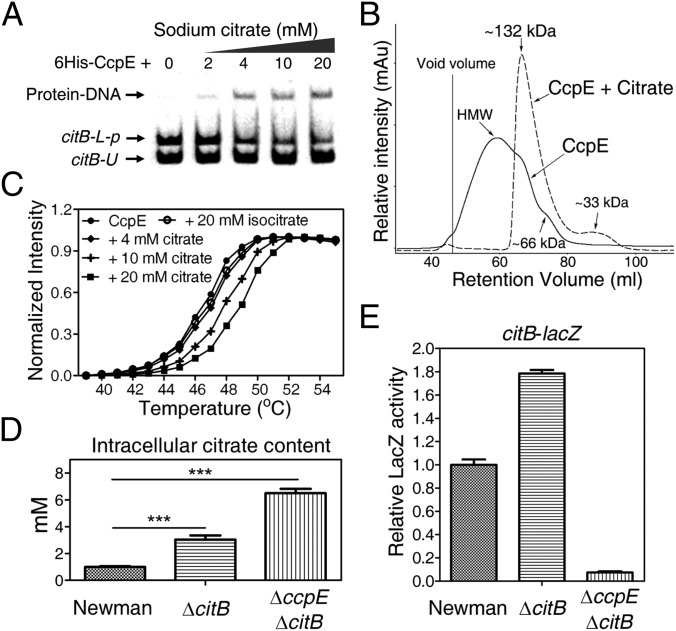
Fig. 2.
CcpE is a citrate-sensing regulator. (A) EMSA showing that sodium citrate increases the DNA-binding ability of 6His-CcpE (0.2 µM) in a dose-dependent manner. citB-L-p, a DNA fragment of the citB promoter DNA (from nucleotides −194 to +88 of the start codon of citB) containing CcpE-protected region I. citB-U, a DNA fragment of citB promoter DNA (from nucleotides −128 to +88 of the start codon of citB) containing no CcpE-protected region. (B) Gel filtration analysis of 6His-CcpE oligomerization in the absence (continuous line) and presence (dashed line) of 10 mM sodium citrate as described in SI Appendix, Experimental Procedures. HMW, high molecular weight. (C) Thermal denaturation curves for 6His-CcpE alone or in the presence of either sodium citrate or sodium isocitrate, as indicated. (D) Intracellular citrate concentrations of the S. aureus wild-type Newman, ΔcitB, and ΔccpEΔcitB strains. The intracellular citrate concentration was estimated according to the assumptions that S. aureus cell volume is 5 × 10−13 mL (57) and that 1 A600 corresponds to 2 × 108 cells/mL. (E) The expression of citB-lacZ in S. aureus wild-type Newman, ΔcitB, and ΔccpEΔcitB strains. In D and E, bacteria were grown in TSB without glucose at 37 °C with shaking, 250 rpm of aeration, and were sampled at 6 h. In E, values are relative to the wild-type Newman strain (set to 1). Results represent means ± SEM, and data are representative of three independent experiments. ***P < 0.001. The statistical difference was determined by unpaired two-tailed Student t test.
Because most LTTRs are known to be functionally active as tetramers (30), we next sought to determine if sodium citrate influences the oligomerization state of CcpE. To this end, the oligomeric state of 6His-CcpE was assessed by gel filtration experiments. As shown in Fig. 2B, the majority of 6His-CcpE was eluted as higher oligomeric forms (molecular weight [MW] >300). Meanwhile, a small percentage of tetramer species (apparent MW ∼132) was detected, and a small peak with an estimated MW of 66 (dimer species) also was observed (Fig. 2B). To our surprise, the addition of 10 mM sodium citrate shifted the equilibrium predominantly to the tetrameric species (MW ∼132) (Fig. 2B), and we also detected a small percentage of monomer species (MW 33) (Fig. 2B). These results indicate that sodium citrate could induce a dominant shift in the oligomeric state of CcpE proteins from higher oligomeric forms to tetramer species, which seem to be a biologically active form of LTTRs (30).
As a ligand binds to a protein, a shift may occur in its melting temperature (Tm) (31). We next used thermal shift assays to test the effect of sodium citrate on the thermal stability of 6His-CcpE. The ligand-induced protein-stabilization effect of 6His-CcpE protein was apparent for sodium citrate, and the protein Tm was increased with increasing concentrations of sodium citrate. As shown in Fig. 2C, the addition of 20 mM sodium citrate increased the Tm by ∼3 °C. In contrast, the addition of 20 mM sodium isocitrate had no significant effect on the Tm value of 6His-CcpE (Fig. 2C). These results suggest that citrate directly binds to CcpE.
Noting that citB deletion results in the accumulation of intracellular citrate (Fig. 2D), we next examined whether citB deletion affects the regulatory function of CcpE in vivo. We measured the regulatory activities of CcpE in the wild-type Newman strain and in a citB-deletion mutant (ΔcitB) using the citB-lacZ reporter gene as a readout. As shown in Fig. 2E, the expression level of citB-lacZ was about 1.8 times higher in the ΔcitB mutant than in the wild-type Newman strain. To examine if CcpE is required for increased expression of citB-lacZ (Fig. 2E), we measured the citB-lacZ expression in a citB and ccpE double-deletion mutant (ΔcitBΔccpE). The intracellular citrate concentration was approximately two times higher but the expression of citB-lacZ was ∼20 times lower in the ΔcitBΔccpE mutant than in the ΔcitB strain, (Fig. 2E), demonstrating that the high citrate pool alone does not increase citB expression. Therefore, the up-regulation of citB-lacZ in the ΔcitB mutant may result from the activation of CcpE by the elevated concentration of intracellular citrate (Fig. 2 A and B).
Crystal Structure of the Inducer-Binding Domain of CcpE.
Members of LTTRs have a conserved structure with an N-terminal DNA-binding domain and a C-terminal inducer-binding domain (IBD) (30). To explore the ligand-binding characteristics of CcpE and to gain structural insights into the transcriptional regulation by CcpE, we decided to crystallize CcpE. Although we could not crystallize full-length CcpE, we succeeded in crystallizing the IBD of CcpE (CcpEIBD, which comprises amino acid residues 63–288 of CcpE). The structure was solved by single-wavelength anomalous dispersion from a SeMet-substituted crystal form and was refined at 2.21-Å resolution (SI Appendix, Table S3). The final Rwork and Rfree were 20.2% and 24.3%, respectively (SI Appendix, Table S3).
As shown in Fig. 3A, the crystallized CcpEIBD is a homodimer with a head-to-tail arrangement of monomers in the asymmetric unit. In accordance with our expectations, each chain of the CcpEIBD comprises two potential regulatory domains (RD1 and RD2) by analogy with other LTTRs, and a hinge formed from the central regions of two antiparallel β-stands (β4 and β9) links these two regulatory domains of each IBD (Fig. 3A and SI Appendix, Fig. S3). These observations are highly similar to those observed for the IBDs of LTTRs (30). Because the inducer-binding cavity (IBC) of LTTRs appears to be located at the interface between RD1 and RD2 (30), we next compared the putative IBC of CcpE with that of BenM, CatM, and CysB using structure-based sequence alignment (SI Appendix, Fig. S4). In the comparison, we identified within the potential IBC of CcpE two arginine residues (Arg145 and Arg256) that frequently are involved in the formation of salt bridges with negatively charged molecules such as citrate (Fig. 3A and SI Appendix, Fig. S4). These two residues are located within the hinge region and are positioned similarly to Arg146 and Thr267 in BenM, Arg146 and Ser267 in CatM, and Thr149 and Ser268 in CysB (SI Appendix, Fig. S4). In BenM and CatM, Arg146 is a residue near the cognate inducers (SI Appendix, Fig. S4) (32). In CysB, Thr149 plays an important role in its regulatory function (33). These observations suggest that this cavity may represent the inducer-binding site of CcpE.
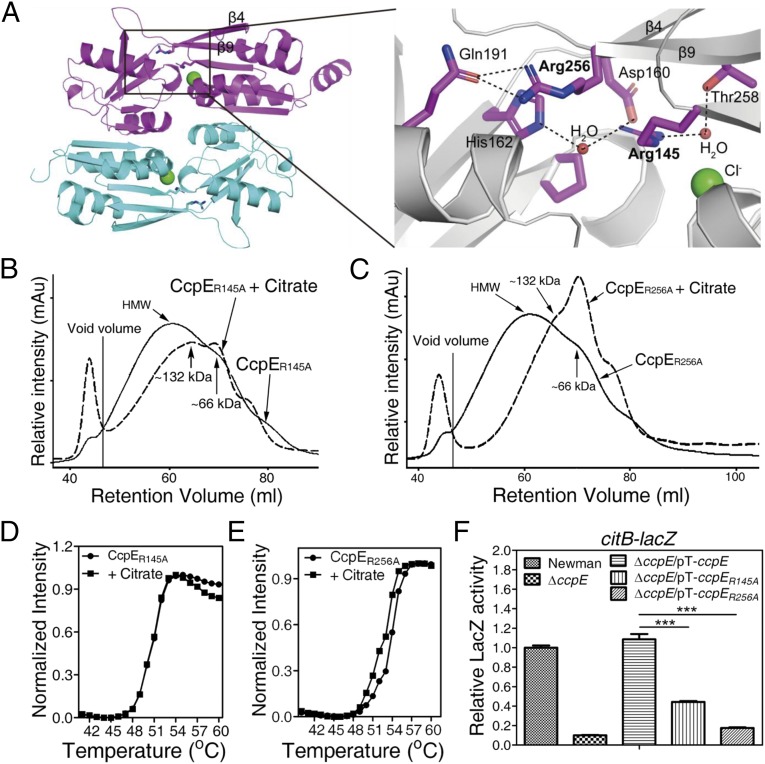
Fig. 3.
Arg145 and Arg256, located within the putative inducer-binding cavity of CcpE, are critical for the activation of CcpE by citrate. (A, Left) Overall protein folding of the inducer-binding fragment of CcpE is presented in a cartoon. One monomer is colored in magenta and the other in cyan. Each monomer possesses two domains separated by a hinge formed from the central regions of β4 and β9. (Right) A local view of the putative inducer-binding cavity of CcpE. Protein is shown in gray, amino acids in magenta sticks, water as red spheres, chloride ions as green spheres, and hydrogen bonds as black dashed lines. (B and C) Gel filtration analysis of 6His-CcpER145A (B) and 6His-CcpER256A (C) oligomerization in the absence (continuous line) and presence (dashed line) of 10 mM sodium citrate. (D and E) Thermal denaturation curves for 6His-CcpER145A (D) and 6His-CcpER256A (E) in the absence and presence of 20 mM sodium citrate. (F) Effect of the amino acid substitutions in CcpE on its ability to promote the expression of citB-lacZ in the ΔccpE mutant. ccpER145A, arginine 145 of ccpE mutated to alanine; ccpER256A, arginine 256 of ccpE mutated to alanine. The wild-type Newman and ΔccpE strains harbor the control plasmid pYJ335-Tc. S. aureus was grown in TSB at 37 °C with shaking, 250 rpm of aeration, and was sampled at 6 h. Values are relative to the wild-type Newman strain (set to 1). Results represent means ± SEM, and data are representative of three independent experiments.
Notably, in the putative IBC of CcpE, the two arginine residues (Arg145 and Arg256) formed extensive hydrogen bonds. The side chain of Arg256 comes in contact with the side chain of Gln191 through hydrogen bonding. In addition to interacting with either Thr258 or His162 via a water bridge, Arg145 comes in direct contact with Asp160 through hydrogen bonding (Fig. 3A).
Arg145 and Arg256 Are Crucial for CcpE–Citrate Interaction.
To determine the roles of Arg145 and Arg256 in the response of CcpE to citrate, we replaced either residue with Ala and, by gel filtration chromatography, assessed the oligomeric state of the resulting CcpE mutation proteins, 6His-CcpER145A and 6His-CcpER256A. As shown in Fig. 3B, the equilibrium of the 6His-CcpER145A protein was shifted to the tetrameric (MW ∼132) as well as the dimeric (MW ∼ 66) species in the presence of 10 mM sodium citrate. Moreover, the addition of 10 mM sodium citrate shifted the equilibrium of 6His-CcpER256A proteins predominantly to the dimer species (MW ∼66) (Fig. 3C), not to the tetrameric form (MW ∼132) as observed for the wild-type 6His-CcpE protein (Fig. 2B). These results indicate that both Arg145 and Arg256 are required for CcpE to evoke an appropriate response in the presence of citrate. In addition, 20 mM sodium citrate failed to increase the Tm of either the 6His-CcpER145A or the 6His-CcpER256A protein (Fig. 3 D and E), suggesting that Arg145 and Arg256 may be directly involved in CcpE binding to citrate. Intriguingly, the thermal stability of both 6His-CcpER145A and 6His-CcpER256A was increased as compared with wild-type 6His-CcpE protein (Figs. 2C and 3 D and E). The mechanisms through which these mutations increase the thermal stability of CcpE remain to be defined.
Next, we asked if the replacement of either arginine residue affects the citrate-mediated activation of CcpE in vitro. To do so, we examined the ability of sodium citrate to enhance the binding of either 6His-CcpER145A or 6His-CcpER256A to a promoter DNA fragment of citB. As shown in SI Appendix, Fig. S5, the addition of 10 mM sodium citrate failed to increase the DNA-binding ability of either 6His-CcpER145A or 6His-CcpER256A. Next, we examined the ability of ccpE, ccpER145A, or ccpER256A (SI Appendix, Table S1) to activate the expression of citB-lacZ transcriptional fusion in a ΔccpE mutant. As expected, an intact ccpE gene (pT-ccpE; SI Appendix, Table S1) was able to restore the expression of citB-lacZ in the ΔccpE mutant to a wild-type level (Fig. 3F). However, mutation of either Arg145 or Arg256 reduced the ability of ccpE to promote the expression of citB-lacZ, by 60% and 84%, respectively (Fig. 3F). In addition, although both the R145A and the R256A mutations had a significant effect on the function of CcpE (Fig. 3F), neither had an obvious effect on CcpE abundance in S. aureus (SI Appendix, Fig. S6). Therefore, it is most likely that Arg145 and Arg256 are crucial for CcpE to interact with citrate and thus may contribute to the regulatory function of CcpE.
CcpE Is a Global Regulator of Virulence Gene Expression.
To study further the roles of ccpE in the regulation of cellular functions of S. aureus, we performed microarray analysis and compared the transcriptome of the ΔccpE strain with that of the wild-type Newman strain. We identified 55 genes with transcript levels decreased twofold or more (SI Appendix, Table S4) and 71 genes with transcript levels increased twofold or more (SI Appendix, Table S5) in the ΔccpE strain as compared with the wild-type Newman strain. These 126 genes represent ∼4.7% of the total number of annotated genes in the S. aureus Newman genome. Of those 126 genes, 27% (34/126) encode hypothetical proteins of unknown functions (Fig. 4A and SI Appendix, Tables S4 and S5). The remaining genes belong to diverse functional categories, including pathogenesis, transport, metabolic process, purine biosynthesis, regulation of transcription, and translation (Fig. 4A and SI Appendix, Tables S4 and S5). To our surprise, 54% (50/92) of the genes with annotated functions are involved in the pathogenesis of S. aureus, including the genes that encode a number of superantigen-like proteins, capsular polysaccharide synthesis enzymes, fibronectin-binding proteins, and exoproteases (Fig. 4A and SI Appendix, Tables S4 and S5). These results suggest that CcpE is an important regulator of virulence gene expression in S. aureus.
Fig. 4.
CcpE is a global regulator of virulence gene expression in S. aureus. (A) Grouping CcpE-regulated genes according to their annotated function shows that the CcpE regulon consists predominantly of genes involved in the pathogenesis of S. aureus. Numbers of genes whose expressions are down-regulated or up-regulated in the ΔccpE strain compared with the wild-type Newman strain are shown. (B) EMSA showing that 6His-CcpE binds to the promoter DNA (cap5A-p) of cap5A (NWMN_0095) but not to a DNA fragment (ccpE-O) amplified from the ccpE (NWMN_0641) gene in the absence of citrate. (C) EMSA showing that 6His-CcpE binds to the promoter DNA (SAV1168-p) of NWMN_1077 but not to a DNA fragment (citB-U) of the citB promoter (from nucleotides −128 to +88 of the start codon of citB) containing no CcpE-protected region in the absence of citrate. (D–G) The expression of transcriptional cap5A-lacZ and SAV1168-lacZ fusions in the wild-type Newman strain and its derivatives, as indicated. In F and G the wild-type Newman and ΔccpE strains harbor the control plasmid pYJ335-Tc. S. aureus was grown in TSB at 37 °C with shaking, 250 rpm of aeration, and sampled at 6 h. Values are relative to the wild-type Newman strain (set to 1). Results represent means ± SEM, and data are representative of three independent experiments. The statistical difference was determined by unpaired two-tailed Student t test (**P < 0.01, ***P < 0.001). NWMN_1077 gene of the S. aureus Newman strain is corresponding to the SAV1168 locus of S. aureus Mu50.
Consistent with the results of the reporter gene assays (Fig. 1D and SI Appendix, Fig. S1D), a sixfold decrease in citB expression (SI Appendix, Table S4) and a moderate increase (1.3- to 1.6-fold) in crtOPQMN (Gene Expression Omnibus database, accession no. GSE57260) expression was observed in the ΔccpE strain as compared with the wild-type Newman strain. In addition to the dramatic decrease in the expression level of citB, deletion of ccpE also caused a marked (more than fivefold) decrease in the expression of the SAV1609 operon (SI Appendix, Table S4), which consists of six genes including acetyl-CoA carboxylase (ACC) genes (34). ACC is a central metabolic enzyme that catalyzes the committed step in fatty acid biosynthesis, biotin-dependent conversion of acetyl-CoA to malonyl-CoA (35). Therefore, in addition to the regulation of the TCA-cycle activity, CcpE may have an important role in adjusting the level of acetyl-CoA, a key molecule in metabolism. To confirm the microarray results further, 10 genes were subjected to quantitative real-time RT-RCR (qRT-PCR) analysis. As shown in SI Appendix, Table S6, for all 10 genes, the qRT-PCR analysis showed results similar to those of microarray analysis, verifying the reliability and reproducibility of the microarray results.
CcpE Directly Controls cap5 and SAV1168 Operons.
Transcriptome analysis does not distinguish the direct target of CcpE from indirect targets. Because CcpE acts as a transcriptional regulator (Fig. 1) (23), direct targets are expected to bind to CcpE. Indeed, CcpE was able to bind to the promoter DNA of both cap5A (Fig. 4B) and SAV1168 (Fig. 4C), the two genes most highly regulated by CcpE (SI Appendix, Table S4 and S5). In addition, in agreement with the microarray data and the qRT-PCR results (SI Appendix, Tables S4–S6), deletion of ccpE resulted in an increase in cap5A-lacZ expression (Fig. 4D) but in decreased expression of SAV1168-lacZ (Fig. 4E). These results suggest that CcpE regulates the expression of these two operons in a direct manner. In S. aureus, the cap5 operon comprises 16 genes involved in the synthesis of the virulence factor capsular polysaccharide (36), whereas the predicted SAV1168 operon comprises three genes (SAV1168, SAV1167, and SAV1166) encoding superantigen-like proteins (34), which seem to play key roles in immune evasion (37).
As shown in Fig. 4 D and E, the deletion of citB in the wild-type Newman strain background causes a decrease in cap5A-lacZ expression (Fig. 4D) and an increase in SAV1168-lacZ expression (Fig. 4E). Moreover, the additional deletion of ccpE in the ΔcitB strain background offsets the effect of citB deletion on the expression of cap5A-lacZ (Fig. 4D) and SAV1168-lacZ (Fig. 4E). These results suggest that the increased concentration of intracellular citrate (Fig. 2D) augmented the function of CcpE (i.e., the repression of cap5A and activation of SAV1168). Additionally, mutation of either Arg145 or Arg256 reduced the ability of ccpE to modulate the expression of either cap5A-lacZ (Fig. 4F) or SAV1168-lacZ (Fig. 4G), indicating that these two potential citrate-binding residues are important for CcpE-dependent expression of the cap5 and SAV1168 operons.
Using a dye-based DNase I footprinting analysis, we found that two regions of the SAV1168 promoter apparently were protected against DNase I digestion by 6His-CcpE (SI Appendix, Fig. S7A). Protected region I contains a potential LTTR box (T-N11-A), and protected region II contains two box II-like sequences (AATA and TTAT) (SI Appendix, Fig. S7A). Interestingly, CcpE-protected region I of SAV1168 promoter DNA contains a nucleotide sequence (ATGATAAGTTTTGCTTAaATA) similar to that of citB promoter DNA (ATGATAAGTTTTGCTTAtATA). Mutations in this region resulted in a ninefold decrease in promoter activity of SAV1168 in the wild-type Newman strain (SAV1168-M-lacZ; SI Appendix, Fig. S7B). More importantly, the mutant promoter of SAV1168 was insensitive to the deletion of ccpE (SI Appendix, Fig. S7B), confirming the critical role of the protected region I in the CcpE-mediated regulation of SAV1168.
Additionally, like the Newman ΔccpE mutant (Figs. 1D and 4E), the JE2-ΔccpE mutant exhibits decreased expression of citB-lacZ and SAV1168-lacZ in relation to its parent strain (SI Appendix, Fig. S8), indicating a conserved function of CcpE in the S. aureus Newman and JE2 strains. Taken together, these results suggest that CcpE directly controls the expression of the cap5 and SAV1168 operons.
CcpE Regulates Iron Acquisition in S. aureus.
Because inactivation of CcpE results in the accumulation of intracellular citrate, we next examined if CcpE modulates the production of the citrate-containing siderophores, which are used by S. aureus to overcome iron limitation during infections (11, 17, 18). When bacteria were grown in RPMI medium without supplemental iron, the deletion of ccpE resulted in increased production of siderophore (Fig. 5 A and B). The introduction of p-ccpE (SI Appendix, Table S1) into the ΔccpE strain abolished the increase in siderophore production (Fig. 5B), suggesting that CcpE represses the production of siderophore in S. aureus. Moreover, consistent with the results of CAS (chrom azurol S)-based analysis (Fig. 5 A and B), the ΔccpE strain exhibited an increased growth rate in RPMI medium without supplemental iron, as compared with wild-type and the complemented strains (Fig. 5C). When the growth medium was supplemented with iron, the growth advantage of the ΔccpE strain disappeared (Fig. 5D). These results suggest that the down-regulation of CcpE may provide an advantage, allowing the S. aureus Newman strain to adapt to an iron-limiting condition. Similar results also were obtained with the S. aureus JE2 strain (SI Appendix, Fig. S9).
Fig. 5.
Deletion of ccpE results in improved ability of S. aureus to acquire iron. In all panels, the wild-type Newman and ΔccpE strains harbor the plasmid pYJ335. (A) Assessment of the siderophore production using a chrome azurol S agar diffusion assay as described in SI Appendix, Experimental Procedures. The orange halos formed around the wells correspond to the iron-chelating activity of the siderophores. (B) Siderophore levels in spent culture supernatants of the Newman strain and its derivatives, as indicated. Siderophore units were calculated as described in SI Appendix, Experimental Procedures. (C and D) Representative growth curves for S. aureus grown in iron-limited (C) and in iron-sufficient (D) medium. (E) Determination of intracellular iron content of the wild-type Newman strain and its derivatives. Bacteria were grown in RPMI medium for 24 h with shaking, 250 rpm of aeration. Then the cells were collected, prepared for, and run on atomic absorption spectroscopy. Results show iron content as a percentage of the dry weight. Values represent means ± SEM. (F) Iron release from transferrin mediated by various spent media from the wild-type Newman strain and its derivatives. A decrease in optical density signifies a release of iron from transferrin. Data are representative of three independent experiments.
To examine if CcpE is involved in iron uptake by S. aureus, we measured intracellular iron pools of the wild-type Newman, ΔccpE, and ΔccpE/p-ccpE strains using atomic absorption spectroscopy. After cultivation in RPMI medium for 12 h, the iron pool of the ΔccpE strain was about 24% larger than that of wild-type Newman strain, but the introduction of ccpE (p-ccpE; SI Appendix, Table S1) decreased the intracellular iron pool of the ΔccpE strain by ∼50% (Fig. 5E), demonstrating that CcpE negatively modulates iron uptake by S. aureus.
In addition to the increased siderophore production, the deletion of ccpE also caused a decrease in pH in the culture medium when bacteria reached the postexponential growth phase (23). Because both siderophore and acidic pH promote the release of iron from transferrin, we measured the capacity of the culture supernatants to release iron from transferrin. As shown in Fig. 5F, spent culture supernatant from the ΔccpE strain increases the rate of iron release from transferrin, but introduction of the ccpE gene completely abolished the phenotype, suggesting that inactivation of CcpE facilitates the release of iron from the host iron-sequestering protein. In sum, these results clearly suggest that CcpE regulates iron acquisition in S. aureus. This notion is substantiated further by the observation that the deletion of ccpE decreased the expression of the iron-regulated sirABC operon genes (38) in either tryptic soy broth (TSB) medium (without glucose) or RPMI medium (SI Appendix, Tables S4 and S6).
CcpE Modulates Virulence of S. aureus.
As mentioned above, the deletion of ccpE caused increased staphyloxanthin production (Fig. 1A and SI Appendix, Fig. S1), improved the ability of S. aureus to acquire iron (Fig. 5 and SI Appendix, Fig. S9), and altered the expression of a large number of genes associated with the pathogenesis of S. aureus (Fig. 4A and SI Appendix, Tables S4 and S5). Therefore we next sought to determine if ccpE contributes to the staphylococcal resistance of host innate immune responses. We mixed the wild-type strain, ΔccpE, and its complemented strain (ΔccpE/p-ccpE) with blood collected from human volunteers and measured the survival of the bacteria. As shown in Fig. 6A, the relative survival of the ΔccpE strain was increased as compared with the wild-type Newman strain (∼1.5% vs. ∼0.5%). Introduction of the ccpE gene reduced the bacterial survival to the level seen in the wild-type strain (Fig. 6A), indicating that the inactivation of CcpE increases the staphylococcal resistance to whole-blood–mediated killing.
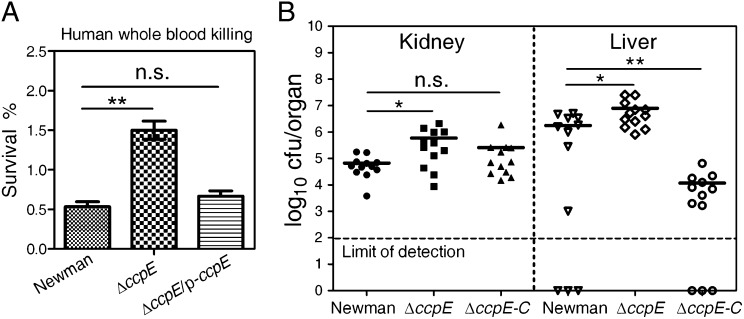
Fig. 6.
Deletion of ccpE causes increased resistance to whole-blood–mediated killing and enhanced bacterial virulence. (A) Whole-blood survival in the wild-type Newman (harboring pYJ335) and ΔccpE strains complemented with vector (pYJ335) alone or with p-ccpE. Values represent means ± SEM, and data are representative of three independent experiments. (B) Virulence of the S. aureus wild-type Newman strain (harboring pCL-lacZ) and the ΔccpE strain complemented with vector (pCL-lacZ) alone or with pCL::ccpE (ΔccpE-C) (SI Appendix, Table S1). BALB/c mice were infected by retroorbital injection of staphylococcal suspensions. Inocula of 3 × 106 cfu staphylococci per mouse were used. S. aureus colonization in murine kidney or liver was measured by tissue homogenization, dilution, and colony formation on TSA plates after 5 d of infection. Each circle, triangle, and square represents data from one experimental animal. Horizontal bars indicate observation means, and dashed lines mark limits of detection. *P < 0.05; **P < 0.01; n.s., not significant; statistical difference was determined by unpaired two-tailed Student t test.
To test further the role of ccpE in invasive staphylococcal disease, we subjected the test strains—wild type, ΔccpE, and ΔccpE-C—to a murine model of abscess formation (24, 39) and measured the bacterial survival in host organs. As shown in Fig. 6B, the lack of ccpE increased the bacterial survival in both kidney and liver. Again, the introduction of the ccpE gene lowered the bacterial survival (ΔccpE-C in Fig. 6B). Interestingly, in the liver, the bacterial load of the complemented strain (ΔccpE-C) was two-log lower than that of the wild-type Newman strain (Fig. 6B), suggesting that the appropriate expression of ccpE is important for S. aureus virulence. In sum, these results suggest that CcpE plays a key role in the pathogenesis of S. aureus in a murine model of abscess formation.
Discussion
Increasingly, metabolic potential is considered a critical element governing a pathogen’s virulence as well as its ability to survive in its host (6, 13, 16, 40–44). Here we show that the S. aureus CcpE protein captures changes in citrate levels and transforms them into various cellular responses. Intriguingly, although CcpE is a positive regulator of TCA-cycle activity, its regulon consists predominantly of virulence-associated genes. In addition to directly controlling the promoter activity of citB, CcpE also directly controls the expression of virulence-related genes such as cap5A and SAV1168. By binding to and activating CcpE, citrate appears to be a key catabolite for coordinating the S. aureus metabolic state with bacterial virulence. A model for a global regulatory role of CcpE in S. aureus is shown in Fig. 7.
Fig. 7.
Proposed role of CcpE in metabolite sensing and the information it transfers. Through the use of citrate as a key inducer, S. aureus CcpE is involved in the regulation of TCA-cycle activity, the production of staphyloxanthin, iron acquisition, virulence gene expression, and bacterial virulence, as described in the main text. CitZ, citrate synthase; CitB, aconitase; CitC, isocitrate dehydrogenase. CitB is the second enzyme of the TCA cycle and is responsible for the interconversion of citrate to isocitrate. Acetyl-CoA is the precursor for staphyloxanthin production via the mevalonate pathway (58). Lactate facilitates the release of iron from the host iron-sequestering protein transferrin (51).
A recent study showed that CcpE is a major positive regulator of citB, a TCA-cycle gene, and that the DNA binding of CcpE is not affected by citrate (23). In this study we also found that CcpE binds specifically to an LTTR box of the citB promoter and activates its promoter activity (Fig. 1 B–D). However, in contrast to the previous report, we observed that citrate alters the DNA-binding affinity of CcpE (Fig. 2A and SI Appendix, Fig. S2B). The discrepancy seems to be caused by the distinct buffer composition. When we used an EMSA binding buffer similar to that described in the previous study, we also failed to observe the citrate-mediated enhancement of DNA binding by CcpE (SI Appendix, Fig. S10A). When Mg2+ was eliminated from our buffer, sodium citrate still was able to enhance the DNA-binding activity of CcpE (SI Appendix, Fig. S10B). However, when the nonionic detergent Nonidet P-40 was omitted, the citrate-mediated effect was not observed (SI Appendix, Fig. S10C), showing that the nonionic detergent plays a key role in the CcpE response to citrate. However, the mechanism by which Nonidet P-40 affects the citrate-mediated activation of CcpE requires further investigation, which may help shed light on the mode of action of CcpE.
In addition to affecting the DNA-binding activity of CcpE (Fig. 2A and SI Appendix, Fig. S2B), citrate can shift the equilibrium of CcpE proteins to a predominantly tetrameric species (Fig. 2B), which seems to be the biologically active form of LTTRs (30). Moreover, citrate is able to increase the thermal stability of CcpE (Fig. 2C), indicating that it probably binds to CcpE directly. These results also suggest that citrate likely is able to cause an allosteric activation of CcpE. Consistent with this hypothesis, we found that the deletion of citB, which leads to an accumulation of intracellular citrate (Fig. 2D), resulted in CcpE-dependent alteration of promoter activities of citB (Fig. 2E), SAV1168 (Fig. 4E), and cap5A (Fig. 4D). Structural and site-directed mutagenesis studies further revealed that Arg145 and Arg256 in the potential inducer-binding cavity (Fig. 3A and SI Appendix, Fig. S4) of CcpE are crucial for CcpE–citrate interaction and the activation of CcpE by citrate (Fig. 3 and SI Appendix, Fig. S5). Based on these data, we concluded that citrate can function as an upstream signal to determine the gene-regulatory activity of CcpE (Fig. 7). This notion is consistent with the observation that LTTRs usually require the binding of an inducer to activate transcription, and the inducers are intermediates in the degradation pathway regulated by the LTTR (30).
Inactivation of ccpE resulted in increased intracellular concentrations of citrate (23). This effect may be caused in part by the down-regulation of citB (Fig. 1D and Fig. 2D). Additionally, the increase in citrate accumulation that occurs when a ccpE-null mutation is introduced into a citB-deletion mutant (Fig. 2D) suggests that CcpE is capable of modulating intracellular citrate levels independently of aconitase (CitB). Thus, we speculate that CcpE negatively regulates a citrate-producing enzyme or positively regulates a citrate-consuming enzyme or alters a pathway that diverts acetyl-CoA from citrate production (e.g., ACC). In fact, CcpE is a positive regulator of ACC (SI Appendix, Table S4). Taken together, these findings indicate that CcpE may control the intracellular concentrations of citrate via multiple pathways.
Citrate is the first intermediate of the TCA cycle, which is highly conserved in all organisms. Citrate provides organisms with energy, reducing potential and biosynthetic intermediates (6, 45). A high level of citrate means that biosynthetic precursors are abundant and indicates a sufficient energy supply (6, 45). In organisms, intracellular concentrations of citrate are in the millimolar range (46–49). A similar intracellular level of citrate was observed for S. aureus (Fig. 2D). In addition to being an important intermediate of the TCA cycle, citrate allosterically activates ACC (50), which catalyzes the committed step in fatty acid synthesis in most organisms (35). In addition, citrate can inhibit a number of enzymes, such as phosphofructokinase, pyruvate kinase, and succinate dehydrogenase, and these inhibitions allow an immediate adjustment of glycolysis and TCA-cycle fluxes to ATP production (50). In this study we have demonstrated that citrate is able to activate CcpE, which acts as an important transcriptional regulator in S. aureus (Figs. 1 and 4). Therefore the role of citrate on cellular functions is much more profound than previously thought (Fig. 7).
Inactivation of CcpE increased the production of staphyloxanthin, and this effect likely is mediated by the down-regulation of citB (Fig. 1). In addition, we found that the ccpE-deletion mutant (ΔccpE) and the citB-deletion mutant (ΔcitB) have similar phenotypes with respect to the improved ability of S. aureus to acquire iron, increased growth rate under iron-limiting condition, and increased total intracellular iron content, as compared with the wild-type Newman strain (Fig. 5 and SI Appendix, Fig. S11). Therefore, the down-regulation of citB may contribute to the modulation of iron acquisition in S. aureus by CcpE. It has been reported that S. aureus can redirect its central metabolism to increase iron availability (51). In iron-starved S. aureus, excess lactate is produced as a result of the down-regulation of the TCA cycle and the up-regulation of the glycolytic pathway (51). The increase in lactate production in turn facilitates the release of iron from host transferrin (51). In addition, it has been reported that citrate synthase, which catalyzes the first reaction in the TCA cycle, is important for iron-regulated synthesis of staphyloferrin A, a citrate-containing siderophore (52). Therefore, given that the ability of pathogens to acquire iron in a host is an important determinant of both their virulence and the nature of the infection produced, it is likely that changes in TCA-cycle activity could provide a survival advantage for S. aureus during infection (11, 17, 18, 53, 54).
In S. aureus, TCA-cycle activity also was found to be critical for the elaboration of the capsule (55) and staphyloxanthin (24). In addition, down-regulation of the TCA cycle through aconitase (CitB) inactivation prevents the maximal expression of the virulence factors and therefore alters the interaction between S. aureus and the host (56). Because CcpE increases the expression of citB (Fig. 1), it is likely that CcpE will affect the expression of some virulence factors via citB. Moreover, CcpE is able to control the expression of the SAV1168 and cap5A operons in a direct manner (Fig. 4). Although the precise mode of action of CcpE requires further investigation, it is clear that CcpE controls important virulence-associated traits (Figs. 1A and 5) and the expression of a number of virulence-associated genes (Fig. 4 and SI Appendix, Tables S4 and S5). Importantly, the down-regulation of CcpE seems to protect the bacterium from the human whole-blood–mediated killing (Fig. 6A) and facilitates the bacterial survival in the host (Fig. 6B). Thus, the accumulation of citrate, rather than the elimination of aconitase, is likely to be the critical factor that determines the interaction between the S. aureus citB mutant and the host (56). However, this hypothesis awaits further investigation.
In summary, this study has shown that CcpE is a citrate-sensing regulator that acts as a master regulator for the virulence-associated properties of S. aureus (Fig. 7). The significance of this work extends beyond S. aureus. For instance, CcpE homologs are present in many other Gram-positive bacteria, including the important human pathogens Bacillus anthracis, Listeria monocytogenes, and Staphylococcus epidermidis. Moreover, the two arginine residues (Arg145 and Arg256) are conserved in CcpE homologs (SI Appendix, Fig. S12), indicating a conserved function. Understanding the role of metabolites in virulence factor expression ultimately may provide new insight into bacterial pathogenesis as well as therapeutic strategies to combat this dreaded infectious disease.
Experimental Procedures
SI Appendix, Table S1 lists the bacterial strains and plasmids used in this study. Unless otherwise noted, S. aureus strains were grown in TSB (Difco 286220) or on tryptic soy agar (TSA) plates without glucose supplement. Escherichia coli strains were grown in LB broth (Difco) or on LB agar plates. For plasmid maintenance, antibiotics were used at the following concentrations where appropriate: for S. aureus, erythromycin at 10 μg/mL, chloramphenicol at 10 μg/mL, and tetracyclines at 5 μg/mL; for E. coli, ampicillin at 100 μg/mL and kanamycin at 50 μg/mL. Unless otherwise noted, cultures were incubated at 37 °C in a shaker (IKA KS 4000i Control Orbital Shaker) with 250 rpm of aeration. Details of other procedures are available in SI Appendix, Experimental Procedures. Experiments with blood from human volunteers involved protocols that were reviewed, approved, and performed under regulatory supervision of Shanghai Institute of Materia Medica's Institutional Review Board. Informed consent was obtained.
Supplementary Material
Acknowledgments
We thank Taeok Bae (Indiana University School of Medicine Northwest) for helpful discussions and S. F. Reichard for editing the manuscript. This work was supported financially by National Natural Science Foundation of China Grants 21472207 (to L.L.), 31270126 (to L.L.), and 91313303 (to C.-G.Y.), Shanghai Committee of Science and Technology Grants 12JC1410200 (to L.L.) and 12ZR1453200 (to F.C.), the National Science and Technology Major Project “Key New Drug Creation and Manufacturing Program” Grants 2014ZX09507009-015 (to L.L.) and 2013ZX09507-004 (to C.-G.Y.), and the Hundred Talents Program of the Chinese Academy of Sciences (L.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4QBA). The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE57260).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411077111/-/DCSupplemental.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119(9):2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis. 2007;13(12):1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40(1):1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 5.Bronner S, Monteil H, Prévost G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol Rev. 2004;28(2):183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73(2):233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 8.Beier D, Gross R. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol. 2006;9(2):143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Stenz L, et al. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol Med Microbiol. 2011;62(2):123–139. doi: 10.1111/j.1574-695X.2011.00812.x. [DOI] [PubMed] [Google Scholar]
- 10.Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8(2):203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6(8):e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19(7):341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8(6):401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs TM, Eisenreich W, Heesemann J, Goebel W. Metabolic adaptation of human pathogenic and related nonpathogenic bacteria to extra- and intracellular habitats. FEMS Microbiol Rev. 2012;36(2):435–462. doi: 10.1111/j.1574-6976.2011.00301.x. [DOI] [PubMed] [Google Scholar]
- 15.Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4(12):e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown SA, Palmer KL, Whiteley M. Revisiting the host as a growth medium. Nat Rev Microbiol. 2008;6(9):657–666. doi: 10.1038/nrmicro1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beasley FC, Heinrichs DE. Siderophore-mediated iron acquisition in the staphylococci. J Inorg Biochem. 2010;104(3):282–288. doi: 10.1016/j.jinorgbio.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12(5):327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 20.Rabinowitz JD, Silhavy TJ. Systems biology: Metabolite turns master regulator. Nature. 2013;500(7462):283–284. doi: 10.1038/nature12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: Fine-tuning metabolism. Cell Metab. 2014;20(1):26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol Cell. 2013;49(3):379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann T, et al. Catabolite control protein E (CcpE) is a LysR-type transcriptional regulator of tricarboxylic acid cycle activity in Staphylococcus aureus. J Biol Chem. 2013;288(50):36116–36128. doi: 10.1074/jbc.M113.516302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan L, Cheng A, Dunman PM, Missiakas D, He C. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J Bacteriol. 2010;192(12):3068–3077. doi: 10.1128/JB.00928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu GY, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu CI, et al. A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence. Science. 2008;319(5868):1391–1394. doi: 10.1126/science.1153018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clauditz A, Resch A, Wieland KP, Peschel A, Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74(8):4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jourlin-Castelli C, Mani N, Nakano MM, Sonenshein AL. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J Mol Biol. 2000;295(4):865–878. doi: 10.1006/jmbi.1999.3420. [DOI] [PubMed] [Google Scholar]
- 29.Mittal M, et al. Dual role of CcpC protein in regulation of aconitase gene expression in Listeria monocytogenes and Bacillus subtilis. Microbiology. 2013;159(Pt 1):68–76. doi: 10.1099/mic.0.063388-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154(Pt 12):3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 31.Pantoliano MW, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6(6):429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 32.Clark T, Haddad S, Neidle E, Momany C. Crystallization of the effector-binding domains of BenM and CatM, LysR-type transcriptional regulators from Acinetobacter sp. ADP1. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 1):105–108. doi: 10.1107/s0907444903021589. [DOI] [PubMed] [Google Scholar]
- 33.Colyer TE, Kredich NM. In vitro characterization of constitutive CysB proteins from Salmonella typhimurium. Mol Microbiol. 1996;21(2):247–256. doi: 10.1046/j.1365-2958.1996.6301347.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Trawick JD, Yamamoto R, Zamudio C. Genome-wide operon prediction in Staphylococcus aureus. Nucleic Acids Res. 2004;32(12):3689–3702. doi: 10.1093/nar/gkh694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polyak SW, Abell AD, Wilce MC, Zhang L, Booker GW. Structure, function and selective inhibition of bacterial acetyl-coa carboxylase. Appl Microbiol Biotechnol. 2012;93(3):983–992. doi: 10.1007/s00253-011-3796-z. [DOI] [PubMed] [Google Scholar]
- 36.O’Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17(1):218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fraser JD, Proft T. The bacterial superantigen and superantigen-like proteins. Immunol Rev. 2008;225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 38.Dale SE, Sebulsky MT, Heinrichs DE. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J Bacteriol. 2004;186(24):8356–8362. doi: 10.1128/JB.186.24.8356-8362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun F, et al. Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci USA. 2012;109(38):15461–15466. doi: 10.1073/pnas.1205952109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song C, et al. Metabolic reconstruction identifies strain-specific regulation of virulence in Toxoplasma gondii. Mol Syst Biol. 2013;9:708. doi: 10.1038/msb.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘Nutritional virulence’ as an emerging paradigm. Cell Microbiol. 2013;15(6):882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 42.Poncet S, et al. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol. 2009;16:88–102. doi: 10.1159/000219374. [DOI] [PubMed] [Google Scholar]
- 43.Görke B, Stülke J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6(8):613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 44.Barbier T, Nicolas C, Letesson JJ. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 2011;585(19):2929–2934. doi: 10.1016/j.febslet.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Meléndez-Hevia E, Waddell TG, Cascante M. The puzzle of the Krebs citric acid cycle: Assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. J Mol Evol. 1996;43(3):293–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]
- 46.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nurmohamed S, et al. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli. J Biol Chem. 2011;286(16):14315–14323. doi: 10.1074/jbc.M110.200741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Promper C, Schneider R, Weiss H. The role of the proton-pumping and alternative respiratory chain NADH:ubiquinone oxidoreductases in overflow catabolism of Aspergillus niger. Eur J Biochem. 1993;216(1):223–230. doi: 10.1111/j.1432-1033.1993.tb18136.x. [DOI] [PubMed] [Google Scholar]
- 50.Icard P, Poulain L, Lincet H. Understanding the central role of citrate in the metabolism of cancer cells. Biochim Biophys Acta. 2012;1825(1):111–116. doi: 10.1016/j.bbcan.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Friedman DB, et al. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006;2(8):e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheldon JR, Marolda CL, Heinrichs DE. TCA cycle activity in Staphylococcus aureus is essential for iron-regulated synthesis of staphyloferrin A, but not staphyloferrin B: The benefit of a second citrate synthase. Mol Microbiol. 2014;92(4):824–839. doi: 10.1111/mmi.12593. [DOI] [PubMed] [Google Scholar]
- 53.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 54.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2(12):946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 55.Sadykov MR, et al. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. J Bacteriol. 2010;192(5):1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somerville GA, et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun. 2002;70(11):6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vilhelmsson O, Miller KJ. Humectant permeability influences growth and compatible solute uptake by Staphylococcus aureus subjected to osmotic stress. J Food Prot. 2002;65(6):1008–1015. doi: 10.4315/0362-028x-65.6.1008. [DOI] [PubMed] [Google Scholar]
- 58.Pelz A, et al. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem. 2005;280(37):32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.