Significance
Flowering time is one of the best studied ecologically important traits under natural or human selection for adaptation of plants to specific local environments. Photoperiodic sensitivity is a major agronomic trait that tailors vegetative and reproductive growth to local climates and is thus particularly important for crop yield and quality. This study not only identifies a major quantitative trait locus underlying photoperiod sensitivity in rice (Days to heading 7, DTH7) but also demonstrates that various haplotype combinations of DTH7 with Grain number, plant height, and heading date 7 (Ghd7) and DTH8 correlate well with the flowering time and grain yield of rice varieties under diverse cultivating conditions. Our results build a foundation for breeding of high-yield rice varieties with desired photosensitivity and optimum adaptation to the target environments.
Keywords: rice, grain yield, flowering time, photoperiod sensitivity, DTH7
Abstract
Success of modern agriculture relies heavily on breeding of crops with maximal regional adaptability and yield potentials. A major limiting factor for crop cultivation is their flowering time, which is strongly regulated by day length (photoperiod) and temperature. Here we report identification and characterization of Days to heading 7 (DTH7), a major genetic locus underlying photoperiod sensitivity and grain yield in rice. Map-based cloning reveals that DTH7 encodes a pseudo-response regulator protein and its expression is regulated by photoperiod. We show that in long days DTH7 acts downstream of the photoreceptor phytochrome B to repress the expression of Ehd1, an up-regulator of the “florigen” genes (Hd3a and RFT1), leading to delayed flowering. Further, we find that haplotype combinations of DTH7 with Grain number, plant height, and heading date 7 (Ghd7) and DTH8 correlate well with the heading date and grain yield of rice under different photoperiod conditions. Our data provide not only a macroscopic view of the genetic control of photoperiod sensitivity in rice but also a foundation for breeding of rice cultivars better adapted to the target environments using rational design.
Flowering time in plants (or heading date in crops) is a critical determinant of the distribution and regional adaptability of plants. Plants have evolved multiple genetic pathways that integrate both internal cues and extrinsic stimuli (e.g., day length and temperature) to control their flowering time to maximize their environmental fitness and survival chances (1). Among them, photoperiod response is a major genetic pathway controlling the timing of flowering in higher plants. Variation in photoperiod response has been extensively manipulated during crop domestication and improvement (2); however, the molecular mechanisms underlying photoperiod sensitivity (PS) of plants, particularly crop plants, have just began to be unraveled.
Rice (Oryza sativa L.), an important calorie source for over half of the world’s population, is a model short-day plant that exhibits robust PS. Generally its flowering is delayed when days are long and nights are short but accelerated when days are getting shorter. Cultivars with reduced PS response are characterized with early flowering and were developed to suit for growing at higher latitudes in the temperate zone (the northern limit is ∼53°N) or for two (even three) successive rounds of planting in the long growing season areas (e.g., some provinces in South China) (3, 4). By contrast, cultivars with enhanced PS (late flowering) have been developed for increased grain yield in most rice planting regions (5, 6). Thus, deciphering the molecular genetic mechanisms underlying flowering time control and regional adaptability has been a major goal of rice breeders and plant biologists.
Molecular genetic studies in the past few decades have uncovered a number of flowering-time loci (3–9). Among them, a major quantitative trait locus (QTL), Heading date 3a (Hd3a), which encodes a rice ortholog of the Arabidopsis Flowering locus T (FT), promotes floral transition under short day conditions (SDs) (9). Hd3a protein (which functions as a systemic flowering signal) is synthesized in leaves and moves through the phloem to the shoot apical meristem, where it triggers flower development (10). Recently it was found that RICE FLOWERING LOCUS T 1 (RFT1), the closest paralog of Hd3a, also functions as a mobile flowering signal that works mainly under long day conditions (LDs) (11, 12). Both of the “florigen” genes are regulated by Hd1, Early heading date 1 (Ehd1), and Days to heading 2 (DTH2) (4, 7, 13). Heading date 1 (Hd1) (counterpart of Arabidopsis CONSTANS, CO) (14) promotes Hd3a expression under SDs but suppresses it under LDs (7, 10, 11), whereas DTH2, another CO-like protein, induces flowering by promoting the expression of Hd3a and RFT1 under LDs (4). Ehd1, encoding a B-type response regulator, up-regulates both florigen gene expression under SDs and LDs (13). Further, dozens of upstream negative regulators of Ehd1 have been identified. Among them, OsphyB executes transcriptional suppression of Ehd1 expression to inhibit flowering under LDs (15, 16). Grain number, plant height, and heading date 7 (Ghd7), encoding a CO, CO-like, TOC1 (CCT) domain protein, and Days to heading 8 (DTH8), encoding a putative HAP3 subunit of the CCAAT box-binding transcription factor, act as LD-specific repressors of Ehd1 (5, 6). On the other hand, several positive Ehd1 regulators have also been cloned by isolating mutants with extreme late flowering. It was shown that Early heading date 2 (Ehd2)/Rice Indeterminate 1 (RID1)/Oryza sativa Indeterminate 1 (OsId1) (referred to as Ehd2 hereafter), encoding a Cs2/His2-type zinc finger protein; Early heading date 3 (Ehd3), encoding a putative plant homeodomain finger containing protein; and Early heading date 4 (Ehd4), encoding a CCCH-type zinc finger protein, independently promote Ehd1 expression under both SDs and LDs (3, 17, 18). Therefore, it appears that Ehd1 is a pivotal convergence point that integrates multiple signaling pathways, including “SD-activation pathway,” “LD-repression pathway,” and “LD-activation pathway,” to regulate the flowering time of rice under diverse environmental conditions (19).
In this study, we report a molecular–genetic analysis of the flowering trait and PS in 91 accessions of the rice germplasm collection in four agriculture environments with a wide geographical span (from 18.3°N to 45.2°N). Map-based cloning reveals that Days to heading 7 (DTH7) is a major determinant of photosensitivity and grain yield in rice and encodes a pseudo-response regulator protein (OsPRR37). DTH7 expression is regulated by the circadian clock, and it acts downstream of OsphyB to regulate Ehd1 expression specifically under LDs. Furthermore, we demonstrate a close correlation between haplotype combinations of three major PS regulators—DTH7, Ghd7, and DTH8—with flowering time and yield in diverse environments.
Results
Rice Grain Yield Is Positively Correlated with Flowering Time in Various Natural Conditions.
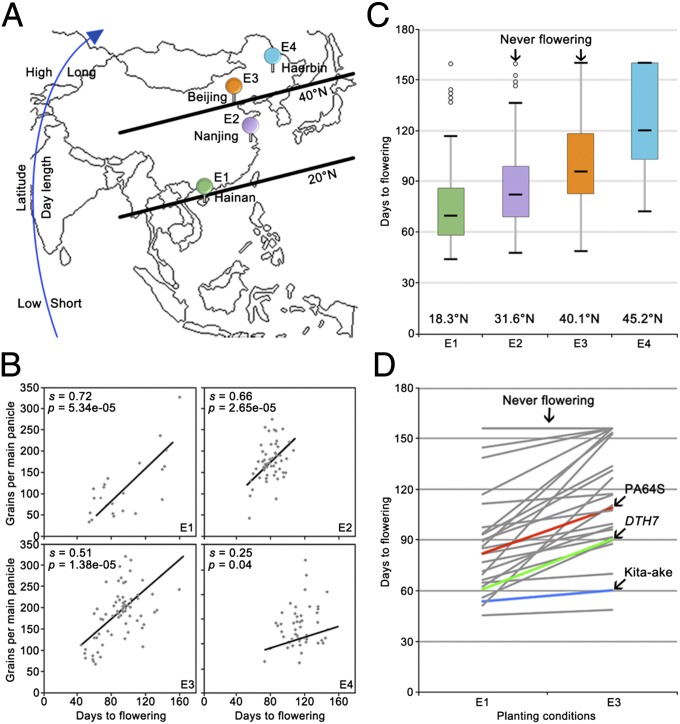
Flowering time plays an important role in regulating the biomass of crops by affecting their duration of basic vegetative growth, and thereby grain yield (1, 20). To further substantiate this notion, 91 selected cultivated accessions, representing a broad range of genetic backgrounds from around the world (4, 5), were grown under four distinct geographical locations across China (ranging from lower to higher latitudes) (Fig. 1A): E1 (Hainan, 18°30’N), E2 (Nanjing, 31°56’N), E3 (Beijing, 40°13’N), and E4 (Haerbin, 45°20’N). In general, we found that grain yield, represented by grain number per main panicle here, was positively correlated with flowering time, most evidently under long growing season areas such as E1, E2, and E3 (Fig. 1B). Despite their varying PS responses, flowering of these accessions were gradually prolonged with the increasing day length (from E1 to E4, Fig. 1 A and C), consistent with the facultative short-day plant flowering habit of rice (Fig. 1C). To better measure the PS levels in these accessions, we adopted the PS index here [PS index = (DTHE3 – DTHE1)/DTHE3; DTH, days to heading] (6). We found that accessions more sensitive to the increasing day length (with higher PS index) tend to flower later under LDs (E3) than under SDs (E1) (Fig. 1D and SI Appendix, Table S1). Therefore, these data suggest that manipulating PS could be a meaningful approach for improving the regional adaptability of rice, and thus grain yield.
Fig. 1.
Association between grain yield and flowering time in rice. (A) Geographic locations of four planting stations: E1, Hainan (18°30’N, 110°2’E); E2, Nanjing (31°56’N, 119°4’E); E3, Beijing (40°13’N, 116°13’E); and E4, Haerbin (45°20’N, 127°17’E). (B) Flowering time of 91 accessions in the core collections under E1, E2, E3, and E4 conditions. The standardized coefficient was represented by s. Student's t tests were used to generate the p values. (C) Grains per main panicle is associated with flowering time. Grains per main panicle of a partial set of accessions were record under E1 to E4 conditions. (D) Flowering time of a partial set of the core collection accessions under natural SDs (E1) and LDs (E3). Flowering time and grains per main panicle of 91 accessions were recorded under E1, E2, E3, and E4 conditions. Flowering time and grains per main panicle of each accession are presented as means ± standard deviations (n = 20).
DTH7 Is a Major Effect Locus Underlying PS and Grain Yield in Rice.
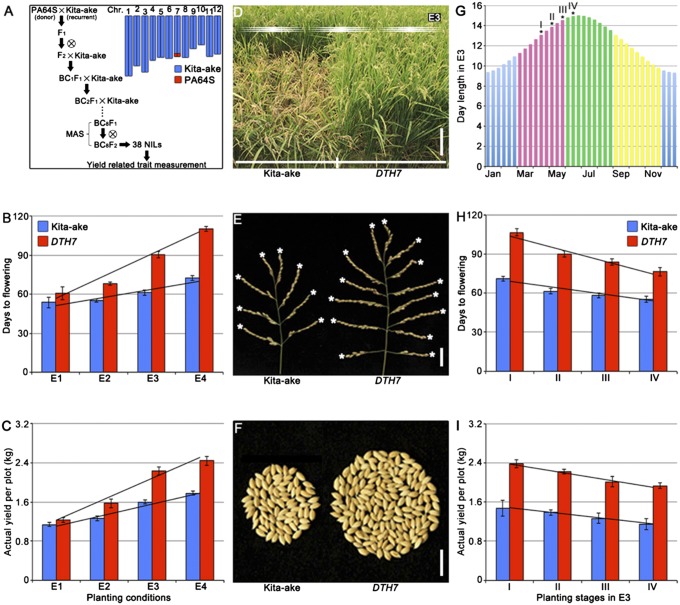
To gain a better understanding of the molecular mechanisms underlying rice grain yield and PS responses, we constructed a set of near isogenic lines (NILs) using two distinct accessions that exhibited natural variations in flowering time (Fig. 2A). PA64S (the donor parent), a parental variety of Liang–You–Pei–Jiu (a leading high-yield super hybrid cultivar widely grown in China) (21), displayed a moderate photoperiod response (PS index, 0.21; Fig. 1D), whereas Kita-ake (the recurrent parent), an early flowering variety (3), showed an impaired day-length response (PS index, 0.09; Fig. 1D). From a total of 38 NILs, one line, DTH7 (for Days to heading on chromosome 7), displayed a modest PS (PS index, 0.25; Fig. 1D) and was selected for further study.
Fig. 2.
DTH7 has a remarkable effect on photoperiod response and grain yield in rice. (A) A workflow of the NIL construction and graphical genotype of one NIL plant (BC8F2), DTH7. PA64S (donor) and Kita-ake (recurrent) were used as the parents. Blue and red bars indicate the Kita-ake chromosomes and PA64S genomic fragment, respectively. (B) Flowering time of Kita-ake and DTH7 plants under E1 to E4. (C) Grain yield per plot of Kita-ake and DTH7 plants under E1–E4. (D) Phenotypes of Kita-ake and DTH7 plants at the mature stage under E3. (Scale bar, 20 cm.) (E) Panicles of Kita-ake and DTH7 plants under E3. (Scale bar, 2 cm.) (F) Grains per panicle of Kita-ake and DTH7 plants at different planting stages under E3. (Scale bar, 2 cm.) (G) Day length in E3 during the whole year. Asterisks indicate the planting time point in E3. (H) Flowering time of Kita-ake and DTH7 plants at different planting stages under E3. (I) Grain yield per plot of Kita-ake and DTH7 plants in different planting stages under E3. Values are shown as means ± standard deviations (n = 20).
By screening a set of genome-wide InDel markers (totally 180), we found that DTH7 carries a 1.05-Mb PA64S genomic segment at the very end of chromosome 7 in the Kita-ake background (Fig. 2A). Days to flowering of DTH7 plants were prolonged more significantly than Kita-ake with elevating latitudes (from E1 to E4; Fig. 2B), indicating that DTH7 plants were more sensitive to the increasing day length than Kita-ake. For example, DTH7 plants delayed flowering time by 4.7 d under E1, but 29.3 d under E3 conditions, compared with Kita-ake (Fig. 2B). Similar results were observed under controlled SDs and LDs (Table 1). Notably, DTH7 plants showed a similar leaf emergence rate with Kita-ake under controlled conditions (SI Appendix, Fig. S1), suggesting that the differences in flowering time were not caused by alterations in growth rate. Due to the longer basic vegetative growth phase, the mature DTH7 plants were taller, producing larger panicles and more seeds than Kita-ake (Fig. 2 D–F and Table 1), leading to gradually increased grain yields when days were getting longer (Fig. 2C), despite the 1,000-grain weight being similar in Kita-ake and DTH7 plants under E1 and E3 conditions (Table 1). As another test of this notion, DTH7 and Kita-ake plants were grown at four different continuous stages (over a time course of 20 d) in a paddy field under E3 (Fig. 2G), in which day length became successively shorter as the season progressed. Strikingly, the flowering time and grain yield of DTH7 decreased more markedly than Kita-ake when days became shorter (Fig. 2 H and I). Taken together, these results suggest that DTH7 is a major QTL underlying PS and grain yield in rice.
Table 1.
Phenotypes of Kita-ake, DTH7, transgene-positive and transgene-negative plants
| Genotype | Condition or location | Days to flowering | Plant height, cm | Tiller n | Panicle length, cm | Primary branches n/panicle | Secondary branches n/panicle | Grains/panicle | 1,000-grain weight, g |
| Kitaake | SD | 53.05 ± 0.91 | — | — | — | — | — | — | — |
| NIL (DTH7) | 57.63 ± 1.54 | — | — | — | — | — | — | — | |
| Kitaake | LD | 58.94 ± 0.91 | — | — | — | — | — | — | — |
| NIL (DTH7) | 79.74 ± 1.33 | — | — | — | — | — | — | — | |
| Kitaake | E1 | 53.65 ± 1.21 | 57.37 ± 2.53 | 21.65 ± 3.76 | 10.43 ± 1.28 | 4.50 ± 0.46 | 3.50 ± 0.65 | 33.85 ± 5.51 | 26.58 ± 0.21 |
| NIL (DTH7) | 60.77 ± 5.22 | 71.46 ± 2.81 | 17.88 ± 3.15 | 12.76 ± 1.23 | 5.62 ± 0.50 | 5.23 ± 1.01 | 44.73 ± 6.40 | 26.86 ± 0.22 | |
| Kitaake | E3 | 63 ± 1.3 | 72.44 ± 2.30 | 19.90 ± 2.65 | 13.97 ± 0.93 | 6.9 ± 0.85 | 11.45 ± 2.63 | 71.80 ± 9.04 | 26.93 ± 0.27 |
| NIL (DTH7) | 94 ± 1.56 | 105.27 ± 2.24 | 13.60 ± 1.67 | 18.15 ± 1.12 | 12.85 ± 0.67 | 22.40 ± 2.74 | 138.65 ± 9.53 | 27.04 ± 0.27 | |
| No. 18 (+) | E3 | 92.54 ± 2.55 | 97.07 ± 2.09 | 15.50 ± 2.14 | 17.93 ± 0.96 | 13.81 ± 1.06 | 18.85 ± 1.95 | 131.50 ± 7.65 | 27.13 ± 0.32 |
| No. 2 (+) | 92.26 ± 5.83 | 98.07 ± 2.28 | 15.43 ± 2.24 | 17.93 ± 0.67 | 13.52 ± 0.90 | 18.09 ± 1.59 | 127.87 ± 3.65 | 26.93 ± 0.43 | |
| No. 6 (+) | 96.10 ± 2.74 | 96.08 ± 1.72 | 15.22 ± 1.29 | 17.96 ± 0.67 | 14.62 ± 0.97 | 19.57 ± 3.31 | 136.76 ± 13.06 | 27.11 ± 0.21 | |
| No. 11 (–) | E3 | 65.44 ± 2.55 | 68.93 ± 3.09 | 24.50 ± 1.65 | 13.59 ± 0.70 | 6.94 ± 0.73 | 10.56 ± 3.15 | 67.94 ± 7.34 | 26.97 ± 0.27 |
| No. 12 (–) | 64.23 ± 2.25 | 70.35 ± 1.93 | 23.62 ± 1.33 | 13.87 ± 0.77 | 7.46 ± 0.90 | 10.54 ± 1.56 | 67.92 ± 4.92 | 27.01 ± 0.26 | |
| No. 16 (–) | 63.27 ± 2.75 | 75.19 ± 2.26 | 25.05 ± 1.53 | 13.58 ± 0.63 | 7.09 ± 0.81 | 10.32 ± 0.94 | 65.05 ± 8.35 | 26.95 ± 0.31 |
Agronomic traits of Kita-ake, DTH7, under natural SDs (E1) and LDs (E3), and three independent T1 transgene-positive (+)—no. 18, no. 2, and no. 6—and transgene-negative (–)—no. 11, no. 12, and no. 16—plants under E3 are shown. Days to flowering and other traits are means ± standard deviations (n = 22).
Positional Cloning of DTH7.
Genetic analysis of an F2 population derived from a cross between DTH7 and Kita-ake showed a 1:3 segregation of early- and late-flowering plants [χ2 (1:3) = 0.33 < χ20.05 = 3.84], indicating that DTH7 behaves as a single Mendelian factor. We then focused on map-based cloning of DTH7. High-resolution mapping using 4,600 early flowering F2 plants delimited DTH7 to a 114-kb region, in which 14 putative ORFs were annotated (Rice Annotation Project Database, RAP-DB; rapdb.dna.affrc.go.jp) (SI Appendix, Fig. S2). Of these, ORF6 (LOC_Os07g49460), which encodes a OsPRR37 that contains one cheY-homologous receiver (REC) domain at its N terminus and one conserved CCT domain at its C terminus, was selected as the candidate gene for DTH7 (SI Appendix, Fig. S2). The CCT domain has been shown to be involved in protein localization and probably has an important role in mediating protein–protein interaction (22–24). Interestingly, OsPRR37 has been reported to be the same allele with a major QTL, Hd2 (also known as Ghd7.1), that was firstly identified in a photoperiod-sensitive variety Nipponbare background (3, 25–27). In addition, homologous genes, such as Ppd-H1 in barley, SbPRR37 in sorghum, and Arabidopsis PRR1, 3, 5, 7, and 9 (SI Appendix, Fig. S3), have been reported as regulators of photoperiod flowering in diverse plant species (20, 28, 29). Comparison of the coding sequences of LOC_Os07g49460 in Kita-ake (designated DTH7-k) and PA64S (designated DTH7-p) identified 10 polymorphisms that lead to amino acid substitutions (SI Appendix, Fig. S2). Three of them—N214S, L462P, and P710L (in the CCT domain)—are located at the conserved positions, among their homologs (SI Appendix, Figs. S2 and S3). We speculated that these changes in conserved amino acids may affect DTH7-k function. To test this, we developed transgenic plants carrying a 16.74-kb PA64S genomic fragment containing DTH7-p in a Kita-ake background. The flowering phenotype and other agronomic traits were all restored to the DTH7 NIL level in several independent transgenic lines (T1 generation) (Table 1 and SI Appendix, Fig. S2). Thus, we concluded that ORF6 represents the PS regulator DTH7 in rice.
DTH7 Expression Exhibits a Constitutive and Diurnal Pattern.
As deduced from available microarray data (RiceXPro, ricexpro.dna.affrc.go.jp/) (30) and confirmed by real-time quantitative reverse transcription–PCR (qRT-PCR) analysis, DTH7 is expressed widely in all tissues examined, including leaf blades, leaf sheaths, roots, stems, and inflorescences (SI Appendix, Fig. S4). The highest expression of DTH7 was found in young leaf blades (SI Appendix, Fig. S4). Interestingly, the DTH7 transcript level in those tissues was about twofold higher in Kita-ake (DTH7-k) than in DTH7 (DTH7-p) plants (SI Appendix, Fig. S4). In addition, qRT-PCR analysis revealed that the DTH7 transcript exhibits a diurnal pattern in leaves with a maximum at Zeitgeber 8 under both SDs and LDs (SI Appendix, Fig. S4), indicating that DTH7 expression is modulated by the circadian clock. The diurnal expression of DTH7 greatly declined under continuous darkness (DD) (SI Appendix, Fig. S4), although the rhythm expression of several other central oscillator components such as OsELF3-1, OsLHY, OsPRR1, OsPRR73, and OsPRR95 remained unchanged in Kita-ake and DTH7 plants under both DD or continuous light conditions (31) (SI Appendix, Fig. S5). These results suggest that the diurnal waveform of DTH7 expression is regulated by light and photoperiod. To investigate the subcellular localization of the DTH7 protein, we transiently expressed the DTH7-p–GFP and DTH7-k–GFP fusion proteins in rice leaf protoplasts. Both proteins were exclusively colocalized with a nuclear marker, the OsMADS3–mCherry fusion protein (SI Appendix, Fig. S4), suggesting that DTH7 functions in the nucleus.
DTH7 Acts Downstream of phyB to Regulate Expression of Ehd1 and the Florigen Genes.
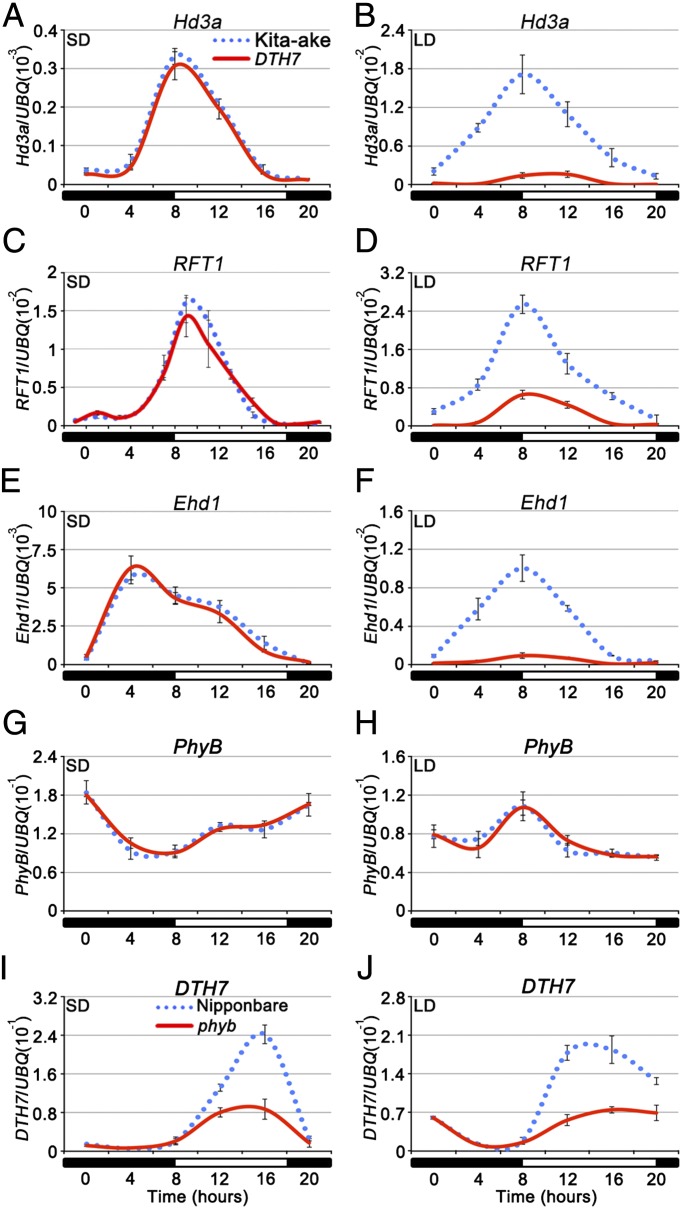
We next examined whether reduced florigen activity is responsible for the late flowering phenotype of DTH7 plants. qRT-PCR results showed that under LDs, but not SDs, Hd3a and RFT1 mRNA levels were markedly reduced in DTH7 plants (compared with Kita-ake) at all time points examined during the 24-h period (Fig. 3 A–D). In addition, the mRNA level of Ehd1, but not Hd1 or DTH2, was severely impaired in DTH7 plants in LDs, but no significant differences were observed in SDs (Fig. 3 E and F and SI Appendix, Fig. S6). Similarly, the transcript levels of Ehd1, Hd3a, and RFT1 were also greatly down-regulated in the DTH7-p transgenic plants under LDs (SI Appendix, Fig. S6). Further, in agreement with the phenotypic observations (Fig. 2 D and G), transcript levels of Ehd1, Hd3a, and RFT1, but not DTH7, gradually reduced with the increasing day length, with a critical day length of ≥14 h (SI Appendix, Fig. S7). Moreover, mRNA levels of three negative regulators (OsPhyB, Ghd7, and DTH8) and three positive regulators (Ehd2, Ehd3, and Ehd4) of Ehd1 were not significantly affected in DTH7 (Fig. 3 G and H and SI Appendix, Fig. S6). These results indicate that DTH7 functions as a negative regulator of Ehd1 and the florigen genes Hd3a and RFT1, specifically under LDs.
Fig. 3.
DTH7 mediates repressing signaling of OsphyB to regulate Hd3a, RFT1, and Ehd1 expression under LDs. The transcript levels of Hd3a, RFT1, and Ehd1 were greatly reduced in DTH7 plants under LDs (B, D, and F), but not SDs (A, C, and E). (G and H) No significant difference of OsphyB expression level was detected in Kita-ake and DTH7 plants under both SDs and LDs. (I and J) DTH7 transcript level was decreased in osphyb mutants under both SDs and LDs. The dotted line and solid line represent Kita-ake and DTH7 plant in A–H or Nipponbare and osphyb plants in I and J, respectively. The rice Ubiquitin-1 (UBQ) gene was used as the internal control. The data are means ± standard deviations of three independent amplifications and two biological replicates.
qRT-PCR analysis also revealed that DTH7 expression was not significantly affected in NILs deficient in Ghd7, DTH8, Ehd1, Hd1, DTH2, or Hd3a (SI Appendix, Fig. S6). Strikingly, DTH7 transcript levels decreased markedly in the osphyb mutants (Fig. 3 I and J), under both SDs and LDs. These data suggest that DTH7 acts downstream of the photoreceptor phyB to regulate Ehd1 expression and flowering time in rice.
Correlation of Haplotype Combinations of DTH7, Ghd7, and DTH8 with Flowering Time Under Various Photoperiod Conditions.
We next analyzed the genetic diversity of the DTH7 coding region in our core collection. Nineteen nucleotide polymorphisms (SNPs) and two InDels were identified in the locus (SI Appendix, Fig. S8), in which 12 accessions carry the Kita-ake haplotype (Hap_2), one accession carries a mutation that leads to a premature stop codon (Hap_24), and additionally one and 14 accessions carry a 2-bp and an 8-bp deletion (Hap_3 and Hap_6), respectively; both resulted in a frame shift stop codon and premature truncated DTH7 proteins (SI Appendix, Fig. S8). Additionally, eight accessions are of the PA64S functional haplotype (Hap_7), and two accessions are of the Nipponbare haplotype (Hap_13). This result suggests that DTH7 is highly diversified in cultivated rice.
Due to the quantitative nature of the flowering trait, we thus sequenced Ghd7 (5) and DTH8 (6), the other two major genetic loci underlying flowering time, regional adaptability, and grain yield, and evaluated the combinatorial effects of DTH7 with Ghd7 and DTH8 on the local fitness of rice. In our core collection, nine and eight haplotypes were found in the coding sequences of Ghd7 and DTH8, respectively (SI Appendix, Fig. S8 and Table S1). Consistent with previous reports, we identified a MH63 haplotype (Hap_1, functional), Nipponbare haplotype (Hap_5), Kita-ake haplotype (Hap_8, nonfunctional), and ZS97 haplotype (Hap_9) at the Ghd7 locus, and an Asominori haplotype (Hap_1, functional), IR24 haplotype (Hap_7, nonfunctional), ZS97 type (Hap_4), Yangeng 15 type (Hap_2), and Longtepu B type (Hap_3) at the DTH8 locus (6) (SI Appendix, Fig. S8 and Table S1).
Next, we investigated the relationship between the different haplotype combinations of DTH7, Ghd7, and DTH8 and flowering times of the core collection across E1 to E4 conditions. These accessions were clustered into four groups: group I carries three nonfunctional loci, group II carries two nonfunctional loci, group III carries one nonfunctional locus, and group IV carries three functional loci (Fig. 4A and SI Appendix, Table S1). As expected, flowering time among group I, group II, group III, and group IV was gradually prolonged under these four conditions (Fig. 4A), whereas accessions carrying two functional loci (group III) flowered much later than accessions of group I and II when days were getting longer, and some accessions of group III never flowered under E4 (Fig. 4A and SI Appendix, Table S1). Strikingly, flowering of accessions carrying all three functional loci (group IV) was further delayed than other groups, and many accessions could not flower under E3 and E4 conditions (Fig. 4A and SI Appendix, Table S1). These data suggest that rice flowering is controlled to a great extent by the combinatorial effects of the haplotype combinations of DTH7, Ghd7, or DTH8 under various agricultural environments, especially under long photoperiod conditions. In support of this observation, variance analysis (ANOVA analysis) showed that functional differences at the DTH7, Ghd7, or DTH8 individual locus could partially explain the phenotypic variations and their contributions became increasingly bigger with the prolonged photoperiods (Fig. 4B). For example, although functional differences at the DTH7 locus explained 20.1%, 28.2%, 42.2%, and 41.2% of the flowering time variations at E1, E2, E3, and E4, respectively [bigger than that of Ghd7 (explained 14.7%, 21.3%, 34.5%, and 40.5%) and DTH8 (explained 6.9%, 11.8%, 17.5%, and 19.4%)], haplotype combinations of these three loci could explain 21.3%, 32.3%, 45.6%, and 52.3% of the flowering time variations at E1, E2, E3, and E4, respectively (Fig. 4B). These observations together suggest that selection of the haplotype combinations of these three loci (and together with other flowering regulators) could improve the local fitness of rice, especially under LDs. Thus, developing molecular markers for the various haplotypes of DTH7, Ghd7, and DTH8 could facilitate molecular breeding of rice cultivars suitable for different environment conditions with desired PS responses.
Fig. 4.
Association analysis of haplotype combinations of DTH7, Ghd7, and DTH8 with flowering time. (A) Association analysis of various combinations of functional and nonfunctional haplotypes of DTH7, Ghd7, and DTH8 with flowering time under E1, E2, E3, and E4 conditions. (B) Variance analysis of the contributions of DTH7, Ghd7, or DTH8 individual locus and their combined effects on flowering time in the core collection. Flowering time of each accession is presented as means ± standard deviations (n = 20).
Discussion
In this study, we demonstrated that DTH7 is a major regulator of PS and grain yield in rice. We show that an NIL (in Kita-ake background) containing DTH7-p (from PA64S) was more sensitive to increasing day length than Kita-ake (carries DTH7-k), and accordingly its heading date was more delayed in higher geographical regions. The longer basic vegetative growth phase allows the mature DTH7 plants to grow much more taller, produce larger panicles and more seeds than Kita-ake, and as a result, increase grain yield under LDs (Fig. 2). Map-based cloning revealed that DTH7 encodes an OsPRR37 that is most closely related to the gene products encoded by barley Ppd-H1, sorghum SbPRR37, and Arabidopsis AtPRR7. DTH7 expression is regulated by the circadian clock under both LDs and SDs, but it acts as a LD-specific suppressor of Ehd1 transcription. Our data suggest that DTH7 activity is gradually enhanced with the increased day length, causing more significant delay in rice flowering. However, how DTH7 activity is regulated by day length remains to be elucidated in future studies.
It is notable that despite a number of flowering-related genes being cloned and characterized as important targets of natural or artificial selection for adaptation of rice to specific natural conditions, the genetic interplay and regulatory relationship among these genes/loci still remain largely unknown. In this study, we performed association analysis of flowering time and different haplotype combinations of DTH7 with two other major flowering time QTL loci, Ghd7 and DTH8, in the core germplasm collection, which contains 91 accessions collected around the world under four geographical conditions across the major rice cultivation areas in China. Our variance analysis showed that each of these three loci contributes significantly to the long photoperiod response and that haplotype combinations of DTH7, Ghd7, and DTH8 could explain most of the phenotype variations and how their contributions became bigger with the prolonged photoperiods from E1 to E4 conditions (Fig. 4). In addition, our data suggest that these genes also play a minor but important role in regulating flowering under SDs, as accessions carrying all three functional loci flowered later than accessions that lost the three loci under SDs. Future efforts to comprehensively analyze the respective contribution and allelic combinations of DTH7, Ghd7, and DTH8 with other genetic loci controlling flowering should allow us to develop a platform for rational design of rice cultivars best adapted to different environmental conditions with maximized grain yield potential and fitness. For example, accessions clustered into group I could be selected for planting in higher latitude conditions, such as E4, to secure a harvest before cold weather approaches, whereas allelic combinations similar to group IV should be excluded when improving and creating new varieties with longer vegetative growth in target environments like E4.
Methods
Plant Material and Growth Conditions.
A DTH7 NIL plant was isolated from a set of NILs in a backcrossing program using two rice parents, Kita-ake (the recurrent parent) and PA64S (the donor parent). F2 population was used for genetic analysis and positional cloning of DTH7. A set of 91 O. sativa rice accessions collected from 17 countries was used in this study (SI Appendix, Table S1). All plants were grown in a paddy field in Hainan (18°30’N, 110°2’E), Nanjing (31°56’N, 119°4’E), Beijing (40°13’N, 116°13’E), and Haerbin (45°20’N, 127°17’E), respectively.
Map-Based Cloning.
A DTH7 NIL plant was crossed with Kita-ake, and then the resultant F1 plants were self-crossed to generate a F2 mapping population. Two DNA pools were generated from 15 F2 late-flowering and 15 early-flowering plants, respectively, were used for rough mapping. We used 4,600 early-flowering plants derived from the F2 population for fine mapping.
Vector Construction and Plant Transformation.
For the genetic complementation test, the 16.7-kb genomic DNA fragment containing DTH7-p (including a 3.3-kb promoter region, 11.2-kb coding region, and 2.2-kb downstream sequence) was amplified using PrimeSTAR GXL DNA Polymerase (Takara) and then cloned into the binary vector pCAMBIA1305.1 using In-Fusion Advantage PCR Cloning Kits (Clontech). The resultant plasmid was transformed into the Agrobacterium tumefaciens strain EHA105 and then introduced into Kita-ake via agrobacteria-mediated transformation.
Subcellular Localization of DTH7 Protein.
The GFP coding region was fused in frame into the DTH7 C terminus and controlled by the 35S CaMV promoter. The DTH7-p–GFP or DTH7-k–GFP fusion protein and the nucleus marker OsMADS3–mCherry fusion protein were transiently coexpressed in rice leaf protoplasts using polyethylene glycol (PEG) treatment (32). Fluorescence was observed using a Leica TCS-SP4 confocal microscope.
Real-Time qRT-PCR.
For real-time qRT-PCR, total RNA was extracted from various tissues using the RNeasy Plant Mini Kit (QIAGEN). The first-strand cDNA was synthesized using the QuantiTect Reverse Transcription Kit (QIAGEN). PCR was performed using gene-specific primers and SYBR Premix ExTaq reagent (Takara) with an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) according to the manufacturer’s instructions. PCR was performed in triplicate for each sample from two independent biological replicates. The rice Ubiquitin-1 gene was used as the internal control. Plants were grown in controlled growth chambers (Conviron) under SDs (9 h light at 30 °C/15 h dark at 25 °C) or LDs (15 h light at 30 °C/9 h dark at 25 °C) with a relative humidity of 70%. The light intensity was set at 800 μmol·m−2·s−1.
Primers.
The primers used in this study are listed in SI Appendix, Table S2.
Accession Codes.
The following GenBank accession codes were used: full-length DTH7-p CDS, KF977867; DTH7-p genomic DNA, KF977869; full-length DTH7-k CDS, KF815740; and DTH7-k genomic DNA, KF977868.
Supplementary Material
Acknowledgments
We thank Dr. Masahiro Yano (The National Institute of Agrobiological Sciences) for providing NILs (Hd1 and Hd3a) and mutants (ehd2 and ehd3), Dr. Atsushi Yoshimura (Kyushu University) for a mutant (osphyb), Dr. Qifa Zhang (Huazhong Agricultural University) for an NIL (Ghd7), and George Coupland (Max Planck Institute for Plant Breeding Research), Amaury de Montaigu (Max Planck Institute for Plant Breeding Research), Song Ge (Institute of Botany, Chinese Academy of Sciences), and Jiankang Wang (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for critical reading and comments on the manuscript. We acknowledge funding from the 863 Program of China Grants 2011AA10A101 and 2012AA100101, National Natural Science Foundation of China Grants 31000534 and 31300276, and Jiangsu Cultivar Development Program Grant BE2012303 (to J.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418204111/-/DCSupplemental.
References
- 1.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 2.Xing Y, Zhang Q. Genetic and molecular bases of rice yield. Annu Rev Plant Biol. 2010;61:421–442. doi: 10.1146/annurev-arplant-042809-112209. [DOI] [PubMed] [Google Scholar]
- 3.Gao H, et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013;9(2):e1003281. doi: 10.1371/journal.pgen.1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA. 2013;110(8):2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 6.Wei X, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153(4):1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yano M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y, Shomura A, Sasaki T, Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc Natl Acad Sci USA. 2001;98(14):7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima S, et al. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43(10):1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- 10.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 11.Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135(4):767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- 12.Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136(20):3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- 13.Doi K, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18(8):926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80(6):847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. Plant J. 2010;63(1):18–30. doi: 10.1111/j.1365-313X.2010.04226.x. [DOI] [PubMed] [Google Scholar]
- 16.Takano M, et al. Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc Natl Acad Sci USA. 2009;106(34):14705–14710. doi: 10.1073/pnas.0907378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsubara K, et al. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 2008;148(3):1425–1435. doi: 10.1104/pp.108.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsubara K, et al. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011;66(4):603–612. doi: 10.1111/j.1365-313X.2011.04517.x. [DOI] [PubMed] [Google Scholar]
- 19.Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14(1):45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Murphy RL, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA. 2011;108(39):16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z-Y, et al. Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc Natl Acad Sci USA. 2013;110(35):14492–14497. doi: 10.1073/pnas.1306579110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazaro A, Valverde F, Piñeiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24(3):982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L-J, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20(2):292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenkel S, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin H, Yamamoto T, Sasaki T, Yano M. Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor Appl Genet. 2000;101(7):1021–1028. [Google Scholar]
- 26.Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci Biotechnol Biochem. 2005;69(2):410–414. doi: 10.1271/bbb.69.410. [DOI] [PubMed] [Google Scholar]
- 27.Yan W, et al. Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 2013;23(7):969–971. doi: 10.1038/cr.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22(3):594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310(5750):1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 30.Sato Y, et al. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41(Database issue) D1:D1206–D1213. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, et al. OsELF3-1, an ortholog of Arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PLoS ONE. 2012;7(8):e43705. doi: 10.1371/journal.pone.0043705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bart R, Chern M, Park C-J, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2(1):13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.