Significance
It is clear that to address climate change, the amount of CO2 released into the atmosphere by industrial processes has to be reduced. Carbonic anhydrase regulates CO2 in nearly every single living organism and is one of the most efficient enzymes in nature. To leverage that efficiency, a β-class carbonic anhydrase was engineered using directed evolution to withstand some of the harshest conditions associated with an industrial carbon capture process. The approach laid out can be generally applied in the development of natural enzymes for their use in industrial applications.
Keywords: carbonic anhydrase, directed evolution, carbon capture
Abstract
Carbonic anhydrase (CA) is one of nature’s fastest enzymes and can dramatically improve the economics of carbon capture under demanding environments such as coal-fired power plants. The use of CA to accelerate carbon capture is limited by the enzyme’s sensitivity to the harsh process conditions. Using directed evolution, the properties of a β-class CA from Desulfovibrio vulgaris were dramatically enhanced. Iterative rounds of library design, library generation, and high-throughput screening identified highly stable CA variants that tolerate temperatures of up to 107 °C in the presence of 4.2 M alkaline amine solvent at pH >10.0. This increase in thermostability and alkali tolerance translates to a 4,000,000-fold improvement over the natural enzyme. At pilot scale, the evolved catalyst enhanced the rate of CO2 absorption 25-fold compared with the noncatalyzed reaction.
Releasing over 9 billion metric tons of CO2 worldwide each year, coal-fired power plants are the leading anthropogenic source of CO2 emission. A potential solution to reduce the release of CO2 into the atmosphere is the implementation of carbon capture and sequestration (CCS) technology on coal- and natural-gas–fired power plants. Currently, one of the most viable postcombustion CCS technology options uses an amine solvent to remove CO2 from the flue gas (1). Unfortunately, the large amount of energy required to release the CO2 and regenerate the solvent would significantly increase the cost of electricity generated (2). The ideal solvent capture process uses a solvent such as an aqueous amine with fast CO2 absorption kinetics and a low heat of desorption to minimize the solvent regeneration energy. However, a high rate of CO2 absorption kinetics for an amine solvent correlates with a higher temperature of desorption, whereas solvents with low heat of desorption tend to have much slower adsorption kinetics (3, 4).
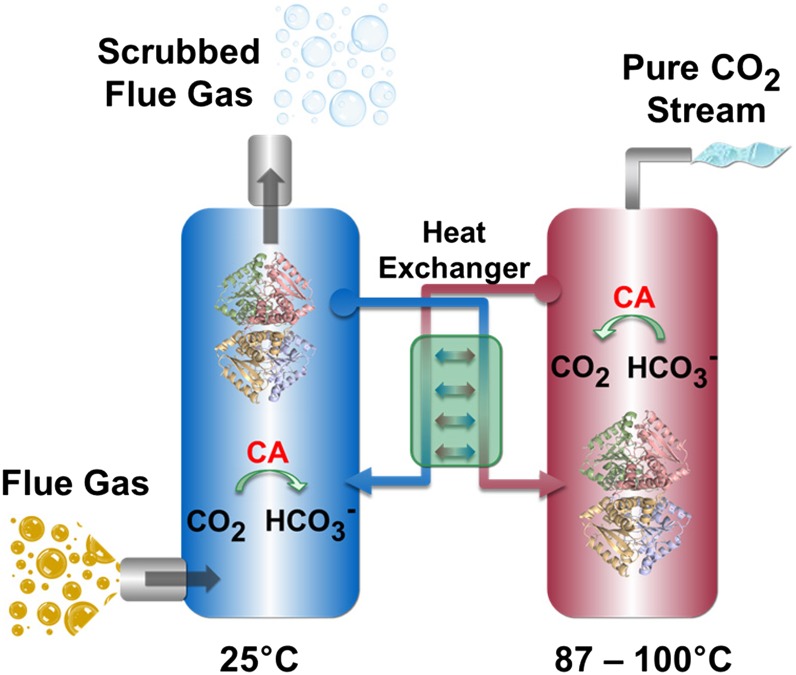
With reaction rates approaching the limits of diffusion, carbonic anhydrase (CA) is one of the fastest enzymes known. The active site of CAs can turnover CO2 and water to bicarbonate and a proton up to a million times per second, and are used by almost every living organism to maintain pH balance and transport carbon dioxide. It has been shown that CAs can be used to accelerate the capture of CO2 (5) by serving as a catalyst in alkaline capture solvents with slow absorption kinetics (6, 7). The potential improvements of CAs on CCS process economics stems from several considerations. The faster absorption and desorption kinetics allows for smaller processing equipment with reduced capital and operating costs (6). The improved kinetics also allows the use of lower temperature capture and solvent regeneration process conditions, and decreases energy losses in the capture process. Another benefit is the improved range of solvent candidates with attractive thermochemical stability (e.g., low vapor pressure, low formation of heat stable salts, etc.), but otherwise unacceptable uncatalyzed absorption kinetics. In addition, the use of a CA as an accelerant has the potential of lowering solvent replacement costs. However, implementing natural CAs in large-scale carbon capture processes has been limited by inactivation of the enzyme under the harsh alkaline conditions of an amine solvent and the elevated temperature required for CO2 desorption. To date, it has been assumed that a protein catalyst could not tolerate exposure to the high temperatures and alkaline environment of amine solvent capture and desorption process (Fig. 1).
Fig. 1.
Flue gas from a coal-fired power plant is piped into an absorber column (blue) where CO2 chemisorbs into an amine solvent, catalyzed by CA, and is hydrated to a proton and a bicarbonate ion. The CO2-depleted flue gas is released into the atmosphere and the HCO3−-loaded amine solvent and CA is transferred to a second column where CO2 is stripped at elevated temperatures (>87 °C), resulting in solvent regeneration. The pure CO2 stream can be compressed and stored in depositories or used in industrial processes. The regenerated solvent is returned to the absorber column to repeat the process.
Directed evolution is an efficient protein engineering strategy for the generation of very large changes in active site or surface residues to give enzyme variants capable of catalyzing new chemistries or ones which act on substrates far afield from those found in nature. Such novel enzymes have enabled efficient biocatalytic pharmaceutical manufacturing processes (8–12). In this article we describe the use of directed evolution in combination with the protein sequence activity relationships (ProSAR) algorithm (13) to create new variants of CA that function under some of the harshest industrial conditions yet applied to enzymes. We created a highly stable form of the CA from Desulfovibrio vulgaris (DvCA) that accelerates CO2 absorption at temperatures above 100 °C in alkaline solvents required for efficient carbon capture.
Results and Discussion
DvCA was selected from a set of natural CAs due to its high activity in 4.2 M N-methyldiethanolamine (MDEA). DvCA is a 243 amino acid protein with a 21 amino acid N-terminal secretion signal peptide. The mature enzyme is predicted to be a β-class CA homologous to the CA from Mycobacterium tuberculosis (MtCA), whose structure has recently been solved (14) (Fig. S1). Based on a sequence alignment of closely related homologs and the structural model, residues Cys55, Asp57, Arg59, His108, and Cys111 are predicted to be the catalytic and Zn2+ metal coordinating residues (residue numbering relative to the mature enzyme as expressed in Escherichia coli).
MDEA was selected as the solvent of choice because of its low heat of desorption for CO2 (15) and advantageous physiochemical properties. The use of MDEA in CCS has been limited by slow CO2 capture kinetics. The use of a CA was hypothesized to increase the overall rate of CO2 capture in this kinetically slow solvent while maintaining the advantage of its low heat of desorption. Although functional in 4.2 M MDEA, DvCA retained only 46% of its activity after being incubated at 40 °C for 24 h. The quest for a thermostable CA was thus focused on generating DvCA variants that tolerate temperatures of 100 °C and higher in high concentrations of MDEA.
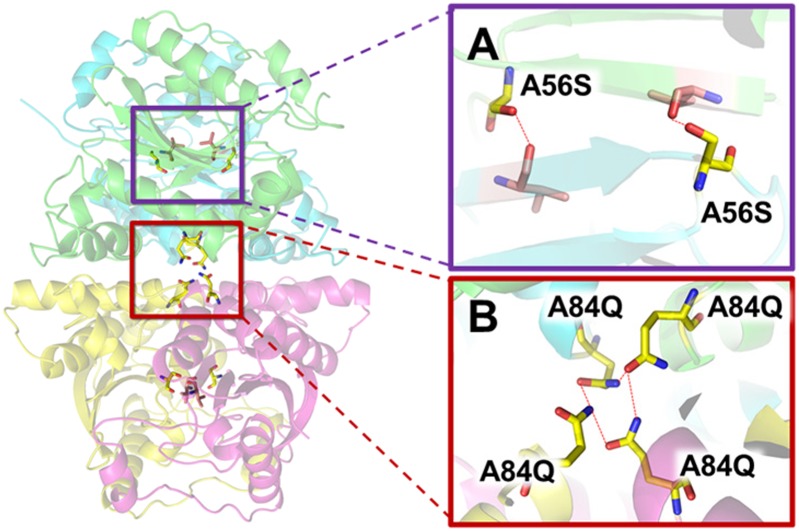
A combination of saturation mutagenesis (16–18) for effective diversity generation coupled with highly efficient recombination tools and the use of statistical analysis to maximize screening efficiency was used to improve DvCA. In nine rounds of evolution, over 27,000 DvCA variants were screened at increasingly higher temperatures in the presence of 3.0–4.2 M MDEA to identify enzymes with the desired characteristics. In the first round of evolution, saturation mutagenesis at all noncatalytic positions using a set of degenerate-codon–containing oligonucleotides was used to identify beneficial diversity. Screening of 3,780 randomly selected variants from this library provided 84 unique mutations at 44 positions that were improved over the natural enzyme when challenged at 42 °C in 3 M MDEA for 24 h (Table S1). The relatively high hit rate (2.2%) suggests that the enzyme was quite malleable and readily evolvable. The positive effect of the A56S mutation in the most active DvCA variant is thought (based on the homology model) to be a result of the formation of an additional hydrogen bond across the dimer interface and stabilization of a β-sheet in the protein core (Fig. 2A). Multiple beneficial mutations were observed at positions 30, 84, 96, 121, 139, and 147, suggesting these are hot spots for enzyme improvement.
Fig. 2.
A56S and A84Q mutations are shown in yellow and the hydrogen bonds as red dashed lines. Based on its closest crystallized homolog (14) the quaternary structure of DvCA is predicted to be a dimer of dimers. (A) The upper inset shows the A56S mutation hydrogen bonding to the backbone of the opposing monomer presumably stabilizing the dimer interface. The hydrogen bonds are thought to form at opposite ends of the antiparallel β-strand interaction at the core of the folded protein. (B) The lower inset depicts the hypothesized interactions between the A84Q mutations across the dimer–dimer interface. Position 84 is at the center of the tetramer interaction. It is the only amino acid that interacts with the corresponding amino acids in all four monomers.
The set of 84 beneficial mutations from round 1 was used in combinatorial libraries from rounds 2–6 and recombination of these beneficial mutations by either gene shuffling (19, 20) or simultaneous multisite targeted mutagenesis (21, 22) resulted in significant enzyme improvements (Fig. 3). Statistical analysis of the screening results using the ProSAR algorithm (13, 23) on a round 2 library containing the mutations with the largest-fold improvement from the first round revealed T30R and A84Q as two of the most beneficial mutations (Figs. S2 and S3). T30R is a mutation at a surface residue that likely increased the enzyme’s favorable interactions with the solvent, whereas the A84Q mutation is hypothesized to form an additional hydrogen bond across the dimer–dimer interface (Fig. 2B). In the context of DvCA2.0 (the best variant from round 1 that was used as the round 2 parent), 6 of the 13 mutations in the library were still predicted to be beneficial (Fig. S3). After ProSAR analysis of all of the round 2 combinatorial libraries, the pool of diversity decreased from 84 to 25 unique mutations, with many of the initially beneficial mutations dropping out in the context of the A56S mutation.
Fig. 3.
(A) Compounded fold improvement after each round of evolution. The relative fold improvement for each round is shown in red, whereas the temperature at which the half-life was determined is shown in black above each bar. (B) The round 8 parent (DvCA8.0) was exposed to 4.2 M MDEA at 50 and 60 °C for 14 wk. At regular intervals a sample was assayed for activity at 25 °C. The half-life is estimated to be 14 and 6 wk at 50 and 60 °C, respectively. This compares to a half-life of 15 min at 50 °C for the wild-type enzyme DvCA and translates to a 10,000-fold improvement in half-life of the DvCA8.0 variant. At 60 °C, the wild-type enzyme has no detectable activity.
Our strategy in subsequent rounds of evolution was to use statistical tools which allow only minimal screening of the libraries while maximizing the information obtained. When working with a library of finite size, it is common practice to screen three times the size of the library to ensure 95% coverage. In the second round of evolution, screening of over 100,000 randomly selected variants would have been required to satisfy that criterion. In our approach, fewer than 3,000 variants were screened for 1.8% coverage of the theoretical size of the library (Table 1). That was sufficient to identify an excellent hit, albeit not the best hit in the library, to serve as the parent for the next round of evolution. In addition, the use of ProSAR allowed for accurate identification of beneficial diversity for round 3 (Fig. S3).
Table 1.
Round diversity and screening coverage
| Round | Library size* | Variants screened† | Percent coverage | Beneficial mutations | Libraries |
| 1 | 4,460 | 3,780 | 57.2 | 84 | Saturation mutagenesis |
| 2 | 128,768 | 2,352 | 1.8 | 25 | Combinatorial |
| 3 | 86,144 | 2,352 | 2.7 | 22 | Combinatorial |
| 4 | 45,920 | 2,772 | 5.9 | 12 | Combinatorial |
| 5 | 4,460 | 3,948 | 58.7 | 14 | Saturation mutagenesis |
| 6 | 19,456 | 2,016 | 9.8 | 9 | Combinatorial |
| 7 | 4,460 | 3,780 | 57.2 | 20 | Saturation mutagenesis |
| 8 | 46,592 | 2,016 | 4.2 | 10 | Combinatorial |
| 9 | 69,996 | 4,200 | 5.8 | 13 | Saturation mutagenesis/combinatorial |
The library size number is a sum of the size of all of the libraries constructed and screened during that round.
Randomly selected variants screened for all of the libraries constructed during that round.
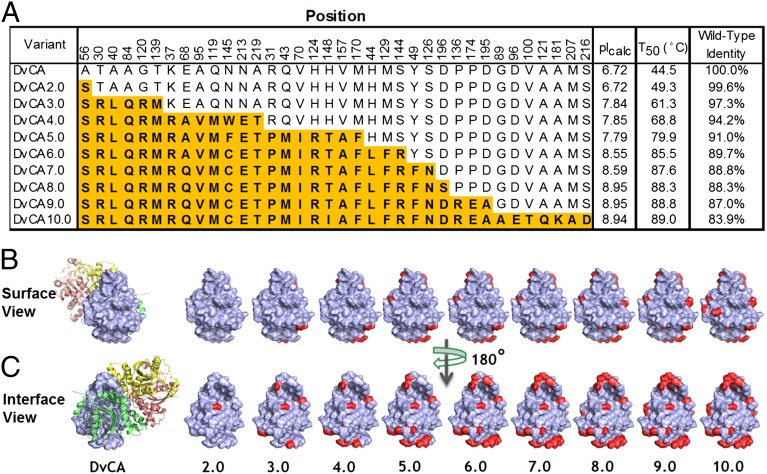
The screening procedure was adjusted to make selection pressure progressively more severe in subsequent rounds of evolution; from 24 h at 42 °C in 3 M MDEA in round 1, to the final round where the samples were challenged for 1 h at 107 °C in 4.2 M MDEA. Table 1 shows how the diversity continued to decrease in each round following the initial saturation mutagenesis round as the challenge temperature increased. The diversity identified in previous rounds was almost completely exhausted by the end of round 4. A second set of saturation mutagenesis libraries aimed at finding additional diversity was generated in round 5. After 9 rounds of evolution, a total of 36 positions representing 15% of the enzyme was mutated (Fig. 4A). Most of the mutations were located at the tetramer interface (Fig. 4 B and C). Over the course of DvCA evolution, the calculated isoelectric point (pIcalc) for the best variant of each round increased progressively from 6.7 for the natural enzyme to 8.9 for the final variant, as a result of mutations of acidic residues to basic amino acids (Table S1). This is unexpected because, based on a previous report, the enzyme’s performance was expected to decrease due to aggregation as the pIcalc approached the solvent pH of 10.0 (24).
Fig. 4.
(A) Mutations accumulated in nine rounds of evolution of DvCA are shown in orange with the corresponding residues of the wild-type enzyme shown above. The pIcalc of the enzyme increases by more than two pH units. The best round 9 variant (DvCA10) accumulated 39 mutations and shares less than 85% sequence identity with the natural sequence. (B) A surface view of one of the monomers is shown in blue with the mutations accumulated in each of the parents highlighted in red. The wild-type (DvCA) rendition also contains the ribbon diagram of the three additional monomers in red, yellow, and green. (C) Same image as in B but rotated by 180° to show the mutations at the tetramer interface.
Long-term stability studies on the round 8 parent (DvCA8.0) showed a 10,000-fold improvement in half-life compared with the native enzyme. This variant retained 40% of its activity after being challenged for 14 wk at 50 °C in 4.2 M MDEA (Fig. 3B), whereas the native enzyme had a half-life of 15 min under identical conditions, validating the evolution strategy undertaken. However, in 4.2 M MDEA and 50 °C, DvCA8.0 was shown to be twofold less active than the wild-type enzyme under the less stringent conditions of 1.0 M MDEA and 25 °C (Fig. S4). The results are consistent with previous reports that show that increased stability can come at the cost of overall activity (25).
To assess the CA-catalyzed acceleration of mass transfer of CO2 into an aqueous solution of MDEA, DvCA8.0 was used in a pilot-scale CO2 capture demonstration run at the National Carbon Capture Center (NCCC) in Wilsonville, AL. The enzyme's stability was evaluated while being cycled between the absorption temperature (25–35 °C) and the desorption temperature (87 °C) over six consecutive days. The unit was designed for continuous operation and was capable of capturing as much as 150 lb of CO2 a day at steady-state conditions (Fig. S5).
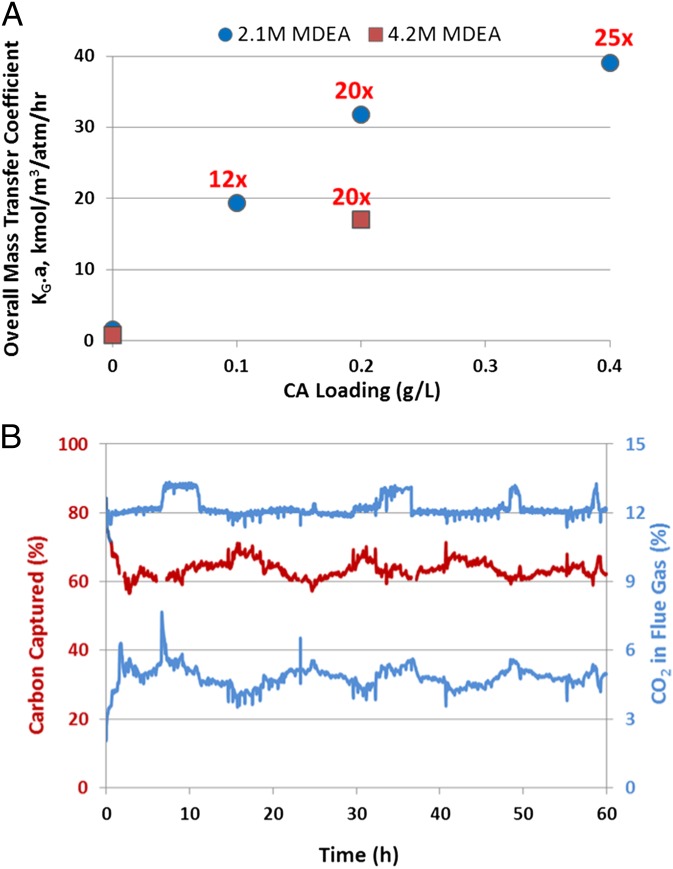
The test rig was used to determine the kinetics of CO2 absorption in MDEA using flue gas from a coal-fired power plant in the presence and absence of DvCA8.0. In a gas–liquid contacting system with CO2 absorption rates dominated by liquid-side diffusion, a fast liquid-side hydration reaction has the effect of steepening the CO2 concentration gradient near the gas–liquid interface (26). Because the rate of absorption is proportional to this gradient, this results in a net enhancement of the overall rate of CO2 capture. This effect on absorption was demonstrated and quantified by measuring the overall mass transfer coefficient KG∙a (27) of CO2 capture in the pilot-scale absorber. In the presence of 2.1 M MDEA and with 0.2 g/L of the DvCA8.0, KG∙a for the uncatalyzed and catalyzed reactions was 1.6 and 39 kmol⋅m−3⋅atm−1⋅h−1, respectively, which translates to a 25-fold improvement in the overall mass transfer coefficient (Fig. 5). Further increases in enzyme loading lead to significant but diminishing performance returns as CO2 diffusion becomes limiting.
Fig. 5.
(A) Mass transfer coefficient (KG∙a) for the carbon capture process was determined at various CA and MDEA concentrations in the test rig. The fold improvement over the no-enzyme control is shown above each data point in red. The circles indicate the results obtained at 2.1 M MDEA where the mass transfer of CO2 from the gas to the liquid phase is 25-fold greater than in the absence of enzyme. The squares show the performance of the test unit using 4.2 M MDEA. At high enzyme loadings and MDEA concentrations the absorption of CO2 becomes mass transfer limited. This is highlighted by the nonlinear correlation between KG∙a and enzyme loading at 2.1 M MDEA and by the drop in performance at 4.2 M MDEA as the solution becomes more viscous. (B) The long-term stability demonstration test was run for 60 h with DvCA8.0 in 25% MDEA solution. The enzyme was exposed to temperatures between 20 and 87 °C as it was cycled between the absorber and the desorber. The top blue trace shows the percent CO2 in the flue gas entering the absorber column, whereas the bottom blue trace shows it exiting the column. The red trace depicts the percent CO2 captured. It is clear that the CA remained active throughout the run and that there was no measurable decrease in percent carbon capture after 60 h of testing. The 60-h test spanned 5 d, operating for about 12 h per day.
Interestingly, 2.1 M MDEA and 4.2 M MDEA both show similar relative improvements in mass transfer with the addition of 0.2 g/L of enzyme. This suggests that, under the studied conditions, catalysis of CO2 hydration is insensitive to the amine concentration and results in the same enhancement factor. However, note that in both the catalyzed and uncatalyzed cases, the mass transfer coefficient for 4.2 M MDEA is about half that of 2.1 M MDEA. According to the penetration theory, the lower mass transfer coefficient at higher amine concentration seems to be accounted for by higher solvent viscosity (28).
DvCA8.0 was evaluated for thermal stability at steady-state operation for an extended run time of 60 h as shown in Fig. 5B. Depending on the time of day and the load on the power plant, the CO2 volume percent in the flue gas varied between 12% and 13%. Steady-state operation was achieved after about 2.5 h, representing five “turnovers” of the 60 L solvent volume being circulated at 2 liters per min (LPM). Carbon capture generally stayed in the range of 60–70% with an average of 63.6%. There was no measurable trend of carbon capture performance loss in the 60-h run time. These results indicate that the enzyme is capable of withstanding the harsh solvent regeneration conditions for over 60 h of run time without any loss in activity.
As the effectiveness of enzyme evolution strategies continues to improve, the use of enzymes as catalysts for the manufacturing of everyday commodities and fine chemicals will continue to increase. The work presented here outlines a general strategy that can be readily applied to the engineering of very large changes in protein function or stability. Note that the aim of evolution is not necessarily to find the best variant in each round, but rather to quickly find beneficial diversity for recombination until activity and stability performance targets are hit. This idea, to move forward with a good enzyme and not become overburdened with an exhaustive search for the best enzyme, was key to the evolution campaign undertaken. There was a conscious effort to maximize the knowledge obtained from each library while minimizing the time spent screening it. This allowed for the evaluation of more libraries of increasingly larger size. In addition, it provided a means to move forward with more rounds of evolution in shorter period. Ultimately this strategy resulted in the successfully engineering of a CA with 4,000,000-fold compounded stability improvement over the parent enzyme.
Materials and Methods
Plasmid Construction and Expression System.
The wild-type CA amino acid sequence was obtained from the genomic sequence of Desulfovibrio vulgaris str. 'Miyazaki F' (UniProtKB ID code B8DIN9). The nucleotide sequence that codes for the mature enzyme was optimized for expression in E. coli and expressed using the pCK110900 vector (29) that contains the p15a as the origin of replication, a chloramphenicol resistance gene as the selection marker, and the lac promoter. The constructed plasmid was transformed into E. coli DH10BT by electroporation.
Library Construction.
Saturation mutagenesis libraries targeting every position of the protein were constructed by the gene splicing by overlapping extension (SOEing) technique (30). The WKG, VMA, and NDT degenerate codons were used as a pool at a ratio of 1:1.5:3 to generate fragments that coded for the 20 amino acids. The fragments were then amplified via a rescue reaction using the appropriate 3′ and 5′ flanking oligonucleotides.
Combinatorial libraries were constructed using either the QuikChange Lightning Multi Site-Directed Mutagenesis Kit from Agilent Technologies or by the SOEing technique. Multiple fragments pertaining to each mutation are created along the entire length of the gene. A second PCR is then used to assemble the fragments.
High-Throughput Library Screening.
All culturing steps, sample preparation, heat challenges, and assays were carried out in 96-well plate format. Libraries were transformed into E. coli W3110 cells and plated on Q trays with chloramphenicol (CAM). Individual colonies were picked using a Q-Pix colony picker into 96-well shallow NUNC plates containing 180 µl of LB with 1% glucose and 30 μg/mL CAM. The cells were grown overnight at 37 °C and 85% relative humidity while shaking at 200 rpm and 10 µL of the overnight culture was used to inoculate a 96–deep-well plate with 390 µl of TB and 30 μg/mL CAM in each well. The plate was shaken for 2.5 h at 250 rpm, 37 °C, and 85% humidity. Protein production was induced by the addition of 40 µl of 11 mM isopropylthio-β-galactoside and 5.5 mM ZnSO4. The plates were incubated overnight and cell pellets were harvested by centrifugation and stored at −80 °C.
Cell lysis was carried out by thawing the frozen pellets for 15 min at room temperature and then adding 200 µL of 25 mM Hepes pH 8.3, 0.5 mg/mL p-chloromercuribenzenesulfonate, and 0.25 mg/mL lysozyme to each well. The plates were sealed and shaken for 1 h at room temperature. Clarified lysates were obtained by centrifugation and 50 µL of the supernatant was mixed with 50 µL of MDEA solution to give the final challenge concentration of MDEA listed in Table S2. Plates containing lysate–MDEA mixtures were then subjected to the temperature challenge listed in Table S2. After the challenge, 10 µL of the mixture was assayed for activity in 200-μL reactions using the conditions listed in Table S2. The slope of the absorbance change at 550 nm was recorded over 10 min. The final rate was determined after subtracting the rate of the background reaction.
Enzyme Stability Confirmation.
E.coli cultures overexpressing carbonic anhydrase variants were grown, and the crude lysates were lyophilized to prepare enzyme powders for testing. To measure the activity and stability of the carbonic anhydrase, the enzyme was added to the MDEA solution and incubated at the specified incubation temperature. Periodically, the solution was removed from the incubator and adjusted to the assay temperature and assayed for activity using a stirred cell reactor (SCR) as described elsewhere (31). Briefly, 100 mL of enzyme plus MDEA solution was added to a 500-mL (total reactor volume) SCR. The reactor was then evacuated until the pressure reached the vapor pressure of the solvent. After the temperature and pressure reached equilibrium, the reactor was pressurized with carbon dioxide gas to about 10 pounds per square inch gauge (psig), after which the flow of CO2 was stopped. The pressure drop with time (corresponding to the absorption of CO2 into the solvent) was recorded, along with the temperature, and these data were used to calculate the overall rate constant for absorption (kOV) using the equations described elsewhere (31). After the assay was complete, the solution was returned to the incubation temperature until the next assay was performed.
Carbon Capture Pilot Test.
The pilot test unit was custom-built for transport and installation at the NCCC in Wilsonville, AL. NCCC is a US Department of Energy sponsored user facility to test CO2 capture technologies under commercially representative conditions with coal-derived flue gas. The pilot-scale test rig consisted of two main process modules––a small absorption column to treat an industrial flue gas stream and a heated desorption vessel to drive off absorbed CO2 and regenerate the solvent. The absorber was composed of two inline 3.15-m columns, 150 mm in diameter and packed with 16-mm pall rings. Liquid redistributors were installed for every 1 m of packing height. Because of enhanced mass transfer in the desorber, desorption is accomplished in a stirred vessel where mixing is enhanced by desorbing CO2. This is an inexpensive design and a deviation from the conventional design consisting of a packed column. Solvent flow rates were varied from 1 to 5 LPM and flue-gas flow rates from 30 to 500 standard liters per min (SLPM). Lean solvent temperature ranged from 20 to 40 °C and desorber temperature was varied between 87 and 100 °C. Nominal conditions for steady-state experiments were 2 LPM of solvent, 180 SLPM of flue gas, a 25 °C absorber temperature, and 87 °C desorber temperature. All enzyme–solvent mixtures were prepared using municipal tap water with no further treatment. The flow rate, temperature, pressure, and compositions of the flue gas entering and leaving the absorber were measured and used for real-time mass balance calculations of percent carbon capture. The cooling system for the column’s solvent temperature became burdened during daytime temperatures of ∼35 °C, which resulted in the lean solvent feed temperature ranging from 20 to ∼30 °C between early morning and afternoon. The 60-h test spanned 5 d, operating for about 12 h per day. The test was initiated with unloaded 25 wt % MDEA containing 0.2 g/L of active DvCA8.0. The overall mass transfer coefficient KG.a was estimated by the methods outlined below.
For CO2 absorption in MDEA, the convective mass transfer takes the form
| [1] |
where NCO2 is the flux of CO2 into the solvent, yCO2 represents the mole fraction of CO2 in the flue gas or exerted by the vapor pressure of the solvent, and P is the total pressure.
Mass balance across a differential volume in the column gives
| [2] |
where dz represents a differential in the packing height, a is the specific surface area of the packing material (e.g., m2/m3), and G′ is the total molar flow rate inert components of the flue gas (mostly N2 and O2).
Some judicious simplifications yield the following expression for estimating Kga:
| [3] |
where y1 is CO2 mole fraction entering the section of absorber column and y2 is the CO2 mole fraction leaving the section, R is the ideal gas constant, and T is the temperature.
Supplementary Material
Acknowledgments
We thank Frank Morton and Thomas Carter at the NCCC for their help in setting up the test unit. In addition, we acknowledge Dr. Gjalt Huisman and Dr. Scott Baskerville for critically reviewing the manuscript. This program is funded in part by the Advanced Research Projects Agency–Energy, US Department of Energy, under Award DE-AR0000071.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411461111/-/DCSupplemental.
References
- 1.Rochelle GT. Amine scrubbing for CO2 capture. Science. 2009;325(5948):1652–1654. doi: 10.1126/science.1176731. [DOI] [PubMed] [Google Scholar]
- 2.Lin LC, et al. In silico screening of carbon-capture materials. Nat Mater. 2012;11(7):633–641. doi: 10.1038/nmat3336. [DOI] [PubMed] [Google Scholar]
- 3.Chu S, Majumdar A. Opportunities and challenges for a sustainable energy future. Nature. 2012;488(7411):294–303. doi: 10.1038/nature11475. [DOI] [PubMed] [Google Scholar]
- 4.Oexmann J, Kather A. Minimising the regeneration heat duty of post-combustion CO2 capture by wet chemical absorption: The misguided focus on low heat of absorption solvents. Int J Greenh Gas Control. 2010;4(1):36–43. [Google Scholar]
- 5.Blais R, Rogers P. 2003. Process and apparatus for the treatment of carbon dioxide with carbonic anhydrase. US Patent 09/424,852:US6524843.
- 6.Chu S. Carbon capture and sequestration. Science. 2009;325(5948):1599. doi: 10.1126/science.1181637. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Lu H, Lu Y. Enhanced stability and chemical resistance of a new nanoscale biocatalyst for accelerating CO2 absorption into a carbonate solution. Environ Sci Technol. 2013;47(23):13882–13888. doi: 10.1021/es4031744. [DOI] [PubMed] [Google Scholar]
- 8.Bommarius AS, Blum JK, Abrahamson MJ. Status of protein engineering for biocatalysts: How to design an industrially useful biocatalyst. Curr Opin Chem Biol. 2011;15(2):194–200. doi: 10.1016/j.cbpa.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Liszka MJ, Clark ME, Schneider E, Clark DS. Nature versus nurture: Developing enzymes that function under extreme conditions. Annu Rev Chem Biomol Eng. 2012;3:77–102. doi: 10.1146/annurev-chembioeng-061010-114239. [DOI] [PubMed] [Google Scholar]
- 10.Savile CK, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329(5989):305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 11.Bornscheuer UT, et al. Engineering the third wave of biocatalysis. Nature. 2012;485(7397):185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 12.Blomberg R, et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503(7476):418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 13.Fox RJ, et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25(3):338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 14.Covarrubias AS, Bergfors T, Jones TA, Högbom M. Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J Biol Chem. 2006;281(8):4993–4999. doi: 10.1074/jbc.M510756200. [DOI] [PubMed] [Google Scholar]
- 15.Kim I, Svendsen HF. Comparative study of the heats of absorption of post-combustion CO2 absorbents. Int J Greenh Gas Control. 2011;5(3):390–395. [Google Scholar]
- 16.Brissos V, Eggert T, Cabral JM, Jaeger KE. Improving activity and stability of cutinase towards the anionic detergent AOT by complete saturation mutagenesis. Protein Eng Des Sel. 2008;21(6):387–393. doi: 10.1093/protein/gzn014. [DOI] [PubMed] [Google Scholar]
- 17.Parikh MR, Matsumura I. Site-saturation mutagenesis is more efficient than DNA shuffling for the directed evolution of beta-fucosidase from beta-galactosidase. J Mol Biol. 2005;352(3):621–628. doi: 10.1016/j.jmb.2005.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reetz MT, Soni P, Fernández L, Gumulya Y, Carballeira JD. Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem Commun (Camb) 2010;46(45):8657–8658. doi: 10.1039/c0cc02657c. [DOI] [PubMed] [Google Scholar]
- 19.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370(6488):389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 20.Fox FJ, Giver L. Principles of enzyme optimization for the rapid creation of industrial biocatalysts. In: Yeh W, Yang H, McCarthy JR, editors. Enzyme Technologies: Metagenomics, Evolution, Biocatalysis, and Biosynthesis. Wiley; Hoboken, NJ: 2010. pp. 99–123. [Google Scholar]
- 21.Hogrefe HH, Cline J, Youngblood GL, Allen RM. Creating randomized amino acid libraries with the QuikChange Multi Site-Directed Mutagenesis Kit. Biotechniques. 2002;33(5):1158–1165. doi: 10.2144/02335pf01. [DOI] [PubMed] [Google Scholar]
- 22.Kim YG, Maas S. Multiple site mutagenesis with high targeting efficiency in one cloning step. Biotechniques. 2000;28(2):196–198. doi: 10.2144/00282bm01. [DOI] [PubMed] [Google Scholar]
- 23.Fox R. Directed molecular evolution by machine learning and the influence of nonlinear interactions. J Theor Biol. 2005;234(2):187–199. doi: 10.1016/j.jtbi.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129(33):10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meiering EM, Serrano L, Fersht AR. Effect of active site residues in barnase on activity and stability. J Mol Biol. 1992;225(3):585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 26.Aboudheir A, Tontiwachwuthikul P, Chakma A, Idem R. Kinetics of the reactive absorption of carbon dioxide in high CO2-loaded, concentrated aqueous monoethanolamine solutions. Chem Eng Sci. 2003;58(23-24):5195–5210. [Google Scholar]
- 27.Perry RH, Green DW. Perry's Chemical Engineers' Handbook. McGraw-Hill; New York: 2008. [Google Scholar]
- 28.Danckwerts PV. 1970. Gas-Liquid Reactions. McGraw-Hill Chemical Engineering Series (McGraw-Hill, New York), p 276.
- 29.Davis SC, Huisman GW, Jenne SJ, Krebber A, Newman LM. 2009. Ketoreductase polypeptides and related polynucleotides.US Patent US7629157.
- 30.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8(5):528–535. [PubMed] [Google Scholar]
- 31.Kucka L, Richter J, Kenig EY, Górak A. Determination of gas-liquid reaction kinetics with a stirred cell reactor. Separ Purif Tech. 2003;31(2):163–175. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.