Significance
Taste tissue regenerates continuously throughout the life span in mammals. Here, using lineage tracing and a culture system, we show that leucine-rich repeat-containing G protein-coupled receptor 5-expressing and leucine-rich repeat-containing G protein-coupled receptor 6-expressing taste stem/progenitor cells generate mature taste cells in vivo and ex vivo. Importantly, our ex vivo studies show that single-progenitor cells can generate all mature taste cell types and that differentiated taste cells form in the absence of innervation. This ex vivo model mimics the development of taste bud cells in taste papillae, recapitulates the process of taste renewal from adult taste stem cells to mature taste cells, and provides a means to study the regulation of taste cell generation and to understand the origins and cell lineage relationships within taste buds.
Keywords: Lgr5, Lgr6, taste stem cells, taste progenitor cells
Abstract
Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) and its homologs (e.g., Lgr6) mark adult stem cells in multiple tissues. Recently, we and others have shown that Lgr5 marks adult taste stem/progenitor cells in posterior tongue. However, the regenerative potential of Lgr5-expressing (Lgr5+) cells and the identity of adult taste stem/progenitor cells that regenerate taste tissue in anterior tongue remain elusive. In the present work, we describe a culture system in which single isolated Lgr5+ or Lgr6+ cells from taste tissue can generate continuously expanding 3D structures (“organoids”). Many cells within these taste organoids were cycling and positive for proliferative cell markers, cytokeratin K5 and Sox2, and incorporated 5-bromo-2’-deoxyuridine. Importantly, mature taste receptor cells that express gustducin, carbonic anhydrase 4, taste receptor type 1 member 3, nucleoside triphosphate diphosphohydrolase-2, or cytokeratin K8 were present in the taste organoids. Using calcium imaging assays, we found that cells grown out from taste organoids derived from isolated Lgr5+ cells were functional and responded to tastants in a dose-dependent manner. Genetic lineage tracing showed that Lgr6+ cells gave rise to taste bud cells in taste papillae in both anterior and posterior tongue. RT-PCR data demonstrated that Lgr5 and Lgr6 may mark the same subset of taste stem/progenitor cells both anteriorly and posteriorly. Together, our data demonstrate that functional taste cells can be generated ex vivo from single Lgr5+ or Lgr6+ cells, validating the use of this model for the study of taste cell generation.
Taste bud cells are heterogeneous and undergo constant turnover (1); however, the origins and generation of taste buds in adult mammals remain largely unclear. Based on morphological and functional characteristics, there are at least three different types of mature taste bud cells [type 1 (glial-like cells), type 2 (receptor cells, including those responsible for sensing sweet, bitter, and umami stimuli), and type 3 (presynaptic cells, including sour sensors)], and well as one type of immature taste bud cell [type 4 (basal cells that are precursors of other types of mature taste cells)] (2, 3). Mature taste bud cells are postmitotic and short-lived, with average life spans estimated at 8–12 d (4, 5), although distinct subtypes of taste bud cells may have different life spans (1, 4, 5). At present, the stem cell population and the regenerative process from adult taste stem/progenitor cells to mature taste bud cells are not well characterized.
Lgr5 (leucine-rich repeat-containing G protein-coupled receptor 5), encoded by a Wnt (wingless-type MMTV integration site family) target gene, marks adult stem/progenitor cells in taste tissue in posterior tongue that in vivo give rise to all major types of taste bud cells, as well as perigemmal cells (6, 7). Lgr5 is also known to mark actively cycling stem cells in small intestine, colon, stomach, and hair follicle, as well as quiescent stem cells in liver, pancreas, and cochlea (8). Isolated Lgr5+ adult stem cells from multiple tissues are able to generate so-called organoid structures ex vivo (9–11). For instance, Sato and colleagues (10) developed a 3D culture system to grow crypt-villus organoids from single intestinal stem cells; all differentiated cell types were found in these structures, indicating the multipotent nature of these cells. We hypothesized that Lgr5+ taste stem/progenitor cells in a 3D culture system would be capable of expanding and giving rise to taste receptor cells ex vivo. In the present study, we isolated Lgr5+ stem/progenitor cells from taste tissue and cultured them in a 3D culture system. Single Lgr5+ cells grew into organoid structures ex vivo in defined culture conditions, with the presence of both proliferating cells and differentiated mature taste cells in which taste signaling components are functionally expressed. When organoids were replated onto a 2D surface precoated with laminin and polylysine, cells grew out of the organoids and attached to the flat surface, and some cells retained the expressed taste signaling elements and responded to taste stimuli.
Lgr5 marks adult taste stem/progenitor cells in posterior tongue, which was shown using an engineered mouse model in which enhanced green fluorescent protein (EGFP) and tamoxifen-inducible Cre recombinase (CreERT2) are knocked-in to replace the coding sequence of Lgr5 and act as surrogate markers for Lgr5 (6, 7). Although Lgr5 is present in fungiform papillae in anterior tongue during embryonic stages and early life, based on the intrinsic GFP signal from the Lgr5-EGFP transgene, Lgr5-EGFP signal could not be detected in fungiform papillae cells in adult mice (6, 7). Therefore, taste stem/progenitor cells remain to be identified in fungiform papillae in anterior tongue. We hypothesized that Lgr6, an Lgr5 homolog, may mark adult taste stem/progenitor cells in anterior tongue, prompted by the finding that Lgr6 is preferentially expressed in taste tissue, but not in the surrounding epithelium devoid of taste tissue (12). Using the Lgr6-EGFP-ires-CreERT2 mouse line (13), we here show that Lgr6 is expressed in cells at the basal area of taste buds in fungiform and circumvallate papillae. By genetic lineage tracing, we show that Lgr6+ cells give rise to taste bud cells in taste papillae in both anterior and posterior tongue. RT-PCR shows that Lgr5 and Lgr6 may mark the same subset of taste stem/progenitor cells both anteriorly and posteriorly. Similar to Lgr5+ cells, isolated Lgr6+ cells can build taste organoids that generate mature taste cells.
Results
Single Isolated Lgr5+ Cells Generate Taste Organoids.
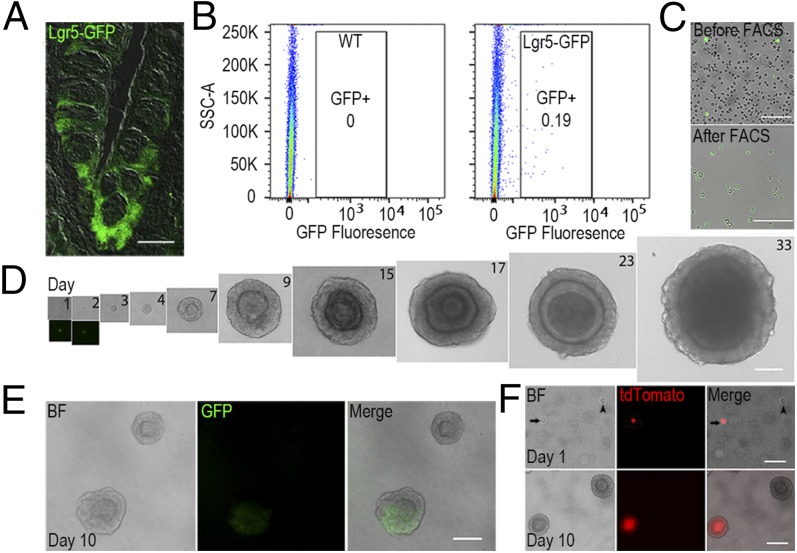
To determine whether Lgr5+ taste stem/progenitor cells are capable of expanding and generating taste cells in vitro, and to establish a taste culture system, we purified Lgr5+ taste stem/progenitor cells (Fig. 1A) from Lgr5-EGFP-ires-CreERT2 mice, using fluorescence-activated cell sorting (FACS), based on the green fluorescence signal of Lgr5-EGFP+ cells (Fig. 1 B and C). The cell sorting gates for isolating GFP-expressing cells were set such that no cells from wild-type littermate controls were isolated (Fig. 1B). All sorted cells expressed EGFP, as demonstrated by the green fluorescence signal (Fig. 1C). Single isolated Lgr5+ taste progenitor cells were embedded in Matrigel containing Clevers’ organoid culture factors for intestinal stem cells and then cultured in defined medium (14). FACS-sorted single Lgr5+ cells proliferated and formed sphere-like structures similar to those obtained from liver Lgr5+ progenitor cells (9) (Fig. 1D). The plating efficiency was about 3–10%. Cultures can be passaged at least six times and maintained for at least 2 mo (the longest time tested to date). After culturing for >3 d, most organoids showed no detectable EGFP signal; the EGFP signal decreased rapidly and became undetectable in 2–3 d. However, organoids were occasionally found to retain a strong EGFP signal: in two of 12 preparations, organoids with GFP fluorescence were seen in 43 (12%) of 374 organoids and 61 (5%) of 1201 organoids examined. Presumably this continued expression of GFP-fluorescence resulted from Lgr5-EGFP cells that were expanded from single isolated Lgr5-EGFP+ cells (Fig. 1E, Bottom).
Fig. 1.
Single isolated Lgr5+ cells generate taste-bud-like “organoids” in 3D culture. (A) Confocal images of Lgr5-EGFP+ cells (green) in a circumvallate papilla section from an Lgr5-EGFP-ires-CreERT2+/− mouse. (B) Representative results of FACS sorting of cells isolated from taste tissue from wild-type (WT) and Lgr5-EGFP-ires-CreERT2+/− mice. Boxed areas depict the gating parameters to sort only Lgr5-EGFP+ cells. (C) Single dissociated cells from digested tongue epithelium from Lgr5-EGFP-ires-CreERT2+/− mice before (Top) and after (Bottom) FACS sorting for Lgr5-EGFP+ cells. All sorted cells expressed EGFP (green). (D) Representative bright-field images of cultured organoids derived from single isolated Lgr5+ cells at indicated days in culture. At days 1 and 2, lower panels show representative fluorescence images of cultured organoids derived from Lgr5+ cells with detectable EGFP signals. (E) Representative bright-field (BF) and fluorescence images of organoids with or without the intrinsic GFP signal. (F) Representative bright-field (BF) and fluorescence images of cultured organoids derived from sorted Lgr5-EGFP+ (arrowhead) and Lgr5-EGFP+/tdTomato+ cells (arrow) at the indicated points. [Scale bars: A, C–E, 100 µm; F, 50 µm (day 1) and 100 µm (day 10).] All panels represent data from at least three independent preparations with the exception of E (two preparations).
To assess the clonality of isolated Lgr5+ cells and to determine whether organoids truly grow out from single Lgr5+ cells, as opposed to small aggregates of cells, we crossed Lgr5-EGFP-ires-CreERT2 mice with Rosa26-tdTomato mice to generate Lgr5-EGFP-ires-CreERT2+/−; Rosa26-tdTomato+/− mice in which tamoxifen-induced Cre generates expression of tdTomato fluorescence protein (red) to mark cells from the Lgr5+ lineage. Half the Lgr5-EGFP-ires-CreERT2+/−; Rosa26-tdTomato+/− mice were injected with tamoxifen 1 d before cell sorting to mark Lgr5+ progeny, using tdTomato fluorescence. Then we purified cells that were positive for EGFP (green) via FACS from both tamoxifen-treated and untreated mice and cultured them in mixture. Using this strategy, tamoxifen-induced Cre generated expression of tdTomato fluorescence protein (red) in only a fraction of single isolated Lgr5+ cells, as well as their progeny. Whole organoids grown from these isolated Lgr5+ cells were composed entirely of either tdTomato+ cells (31 and 34 organoids from two separate experiments) or tdTomato− cells (∼300 organoids each from two preparations) (Fig. 1F), never mixed populations of tdTomato+ and tdTomato− cells, indicating they are clonally expanded from single isolated Lgr5+ cells.
R-spondins have been found to be essential for culturing Lgr5+ stem cells from multiple types of tissues (15–17). To determine whether R-spondins are also critical for growth and expansion of Lgr5+ cells into taste organoids, we cultured Lgr5+ cells from taste tissue in the same medium formula without R-spondin-1 supplement. Fig. S1A shows representative fields of organoids in the presence or absence of R-spondin-1 and demonstrates that R-spondin-1 enhances the growth and expansion of organoids derived from taste Lgr5+ cells.
Organoids Derived from Single Lgr5+ Cells Contain Actively Cycling Cells.
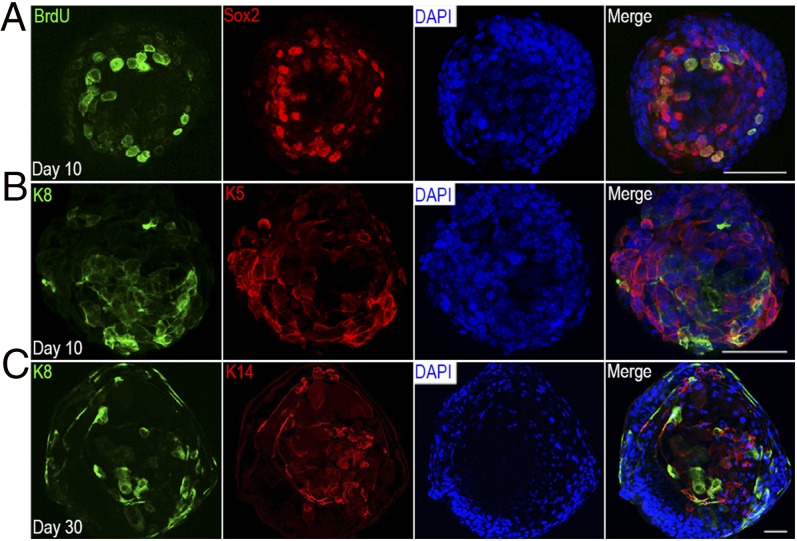
Because of the continuous expansion of organoids under our defined culture conditions, we reasoned that organoids must contain progenitor cells. To determine the proliferating capabilities of cells in cultured organoids, we performed 5-bromo-2′-deoxyuridine (BrdU) tracing. At day 10, individual organoids were examined for incorporation of BrdU after overnight incubation in BrdU-containing culture medium. As expected, numerous cells in organoids showed BrdU immunoreactivity (Fig. 2A). BrdU+ cells were typically scattered within organoid structures (Fig. 2A). Sox2 is implicated in taste cell development and taste cell renewal (18) and is a general stem cell marker (19). A subset of BrdU+ cells were also Sox2+, confirming the proliferating properties, as well as potential “stemness,” of these cells (Fig. 2A). K8 is a general marker for intragemmal taste bud cells (20). We tested cultures for the presence of K8+ cells that were distinct from proliferating cells by double immunostaining with cytokeratin K14 and K5, two additional progenitor cell markers for basal epithelial cells, including taste tissue (21). K8 immunoreactivity was detected in many organoids [23 (53%) of 43], albeit in varying numbers of cells, and these cells did not show any staining for proliferating markers such as K5 and K14 (Fig. 2 B and C), mirroring the segregation of these two cell populations in taste tissue. Organoids with detectable EGFP signal showed the presence of K5+ cells as well (Fig. S2).
Fig. 2.
Organoids derived from single Lgr5+ cells contain actively cycling cells. (A) Whole-mount immunostaining of culture day 10 organoids lacking detectable EGFP green fluorescence. Reactivity with anti-Sox2 (red) and anti-BrdU (green) antibodies shows that most actively dividing organoid cells that incorporated BrdU were also immunoreactive for the stem cell marker Sox2. (B) Whole-mount immunostaining of culture day 10 organoids with anti-K5 (basal cell marker, red) and anti-K8 (taste bud cell marker, green) antibodies. These two types of cells segregated. (C) Whole-mount immunostaining of culture day 30 organoids with anti-K14 (basal cell marker, red) and anti-K8 (taste bud cell marker, green) antibodies. These two types of cells segregated. All organoids were counterstained with DAPI provided in the mounting medium (Vector Labs) to show the cellular content and size of organoids. (Scale bars: A and B, 50 µm; C, 100 µm.) All experiments were performed in triplicate.
Intrigued by the infrequent presence (two of 12 preparations) of detectable Lgr5-EGFP signal after 2–3 d in culture in only a fraction of cultured organoids (5–11%, as detailed earlier) and by the apparent growth-promoting effects of R-spondin-1, the ligand for Lgr5, Lgr4, and Lgr6 (15–17, 22), in cultured organoids, we performed RT-PCR to determine whether Lgr5 and/or its homologs were present in cultured organoids. All three Lgr transcripts were detected in cultured organoids, despite the occasional detection of an Lgr5-EGFP signal in only a small fraction of culture organoids (Fig. S3A).
Mature Taste Cells Are Generated in Vitro from Taste Organoids.
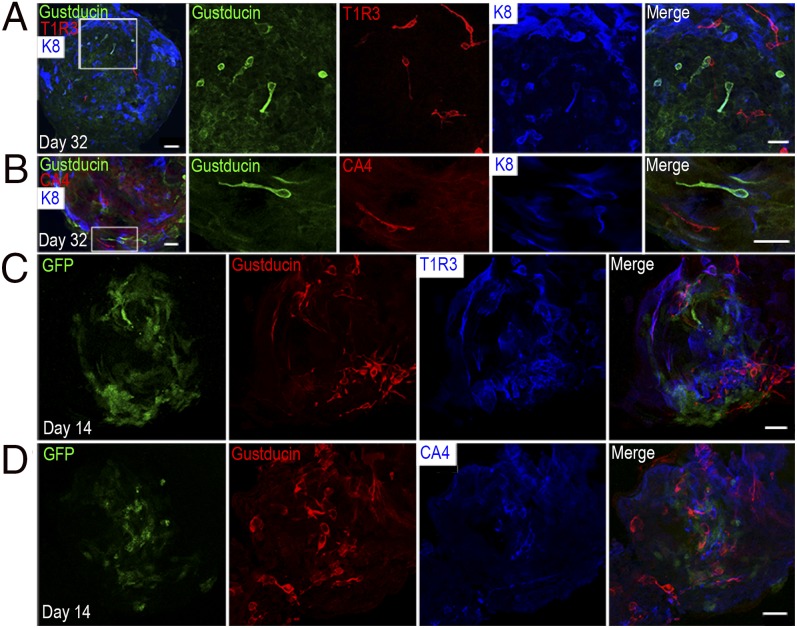
Single Lgr5+ stem cells from intestine can build organoids containing differentiated cells (10). However, in other cases, such as Lgr5+ cells isolated from pancreatic ducts, no insulin-producing cells are found in long-term organoid cultures, indicating the requirement for additional extrinsic factors or intrinsic reprogramming (11). To determine whether any mature taste receptor cells are produced in organoids derived from Lgr5+ cells, we performed double or triple immunostaining of cultured organoids, using antibodies against the taste signaling components gustducin (G protein for sweet, bitter, and umami signaling), T1R3 (taste receptor type 1 member 3, a common subunit of the sweet T1R2/T1R3 and the umami T1R1/T1R3 taste receptor) (23–25), and carbonic anhydrase 4 (CA4; a marker of sour taste cells) (26). In multiple organoids that showed no detectable EGFP signal, we identified cells immunopositive for gustducin (green) and T1R3 (red) (Fig. 3A and Table S1). Gustducin-expressing cells (mostly bitter taste cells in posterior tongue) generally did not overlap with T1R3-expressing cells (Fig. 3A), reminiscent of their segregation in the circumvallate papilla in posterior tongue (27). Of particular note, these cells displayed morphology typical of mature taste receptor cells, with a characteristic large, round nucleus and a slender cell body (Fig. 3A). Nevertheless, many K8+ cells (Fig. 3A, blue) were not immunopositive for taste receptor cell markers, suggesting many cells contain additional types of taste cells, such as type 1 nucleoside triphosphate diphosphohydrolase-2+ (NTPDase2+) cells (Fig. S3B), or cells that are differentiated but not yet committed to a particular cell type. To further assess the expression of taste signaling components in the cultured organoids, we performed RT-PCR, using taste-gene-specific primers. Transcripts for T1R1, T1R2, T1R3, gustducin, and NTPDase2 were all amplified from cDNA prepared from cultured organoids (Fig. S3C).
Fig. 3.
Mature taste-like cells are generated in cultured organoids derived from single Lgr5+ cells. (A) The first panel on the left is a whole-mount of a day 32 organoid without detectable EGFP green fluorescence immunostained with anti-gustducin (green), anti-T1R3 (red), and anti-K8 (blue) antibodies. The remainder of the images are higher magnifications of the boxed area. (B) The first panel on the left is a whole-mount of a day 32 organoid without detectable EGFP green fluorescence immunostained with anti-gustducin (green), anti-CA4 (red), and anti-K8 (blue) antibodies. The remainder of the images are higher magnifications of the boxed area. Both type 2 cells (gustducin+, green) and type 3 cells (CA4+, red) were present in the organoid and segregated. (C) Whole-mount immunostaining of day 14 organoids with anti-gustducin (red) and anti-T1R3 (blue) antibodies and intrinsic EGFP fluorescence (green). (D) Whole-mount immunostaining of day 14 organoids with anti-gustducin (red) and anti-CA4 (blue) antibodies and intrinsic EGFP fluorescence (green). (Scale bars: A and B, 100 µm; C and D, 20 µm.) The number of organoids examined for antibody staining is presented in Table S1. At least three independent experiments were performed.
To determine the lineage potential of single Lgr5+ stem/progenitor cells, whether they are multipotent and give rise to multiple taste bud cell subtypes or unipotent and committed only to one taste bud cell subtype (e.g., type 2), we stained organoids without intrinsic GFP fluorescence for markers for type 2 (gustducin, green), type 3 (CA4, red), and intragemmal (K8+, blue) taste cells (Fig. 3B). Interestingly, in a subset of organoids [18 (56%) of 32; Table S1] derived from single Lgr5+ cells, both type 2 (gustducin+) and type 3 (CA4+) taste cells were present in a nonoverlapping fashion, indicating that Lgr5+ cells are indeed multipotent.
To determine whether organoids that expressed EGFP indicative of Lgr5 expression differed in their ability to generate mature taste cells from organoids without EGFP/Lgr5, we performed immunostaining with antibodies against T1R3 (blue), gustducin (red), and CA4 (blue). All organoids with intrinsic EGFP signal (green) showed immunoreactivity to two markers among those we examined (10 of 10 for gustducin and CA4, and seven of seven for gustducin and T1R3; Fig. 3 C and D and Table S1). In contrast, only a subset of organoids without EGFP activity stained with both gustducin and CA4 markers [18 (56%) of 32; Table S1]. Thus, it would appear that organoids that continue to express Lgr5 are multipotent, whereas those without EGFP have more limited potency.
Mature Taste Cells Derived from Lgr5+ Cells Respond to Tastants.
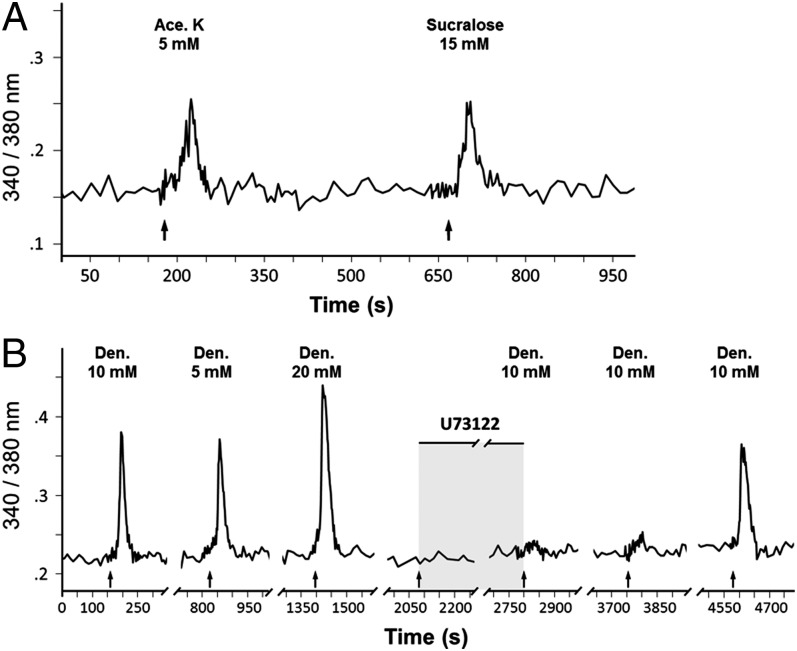
To determine whether apparent taste cells (based on immunohistochemistry and morphology) produced from progenitor cultures are functional and respond to taste stimuli, we performed calcium imaging. Because of technical difficulties in imaging or stimulating cells within 3D structures, we reseeded cultured organoids onto laminin-coated coverslips to allow cells to grow and differentiate into a 2D structure for up to 2 wk in the same taste culture medium, as described for 3D cultures. The presence of presumptive taste cells that express taste cell markers in such structures was confirmed by immunostaining and RT-PCR (Fig. S4). On the basis of immunostaining, it appeared that more gustducin+ cells than T1R3+ cells were generated under 2D culture conditions (Fig. S4 A and B). Calcium imaging of cultured cells grown on laminin-coated coverslips showed that a small number of cells responded to sweet-tasting compounds (acesulfame-K and sucralose), suggesting the presence of functional T1R2/T1R3-expressing sweet taste cells derived from Lgr5+ cells (Fig. 4A and Fig. S5). Some cells responded to the bitter compound denatonium benzoate in a dose-dependent manner (Fig. 4B and Fig. S5). The responses to denatonium benzoate were inhibited by a brief preincubation with the phospholipase C β2 (Plcβ2) blocker U73122, and that effect was reversible after prolonged recovery (∼30 min; Fig. 4B), suggesting the responses were mediated by the Plcβ pathway, consistent with the known mechanism underlying bitter taste transduction in vivo (28). Similarly, we observed that some salts (e.g., 200 and 250 mM NaCl and 50 mM KCl) activated a subset of cells, which is indicative of the presence of salt-responsive cells (Fig. S5A). We did not find cells that responded to only 50 mM monosodium glutamate, a prototypical umami compound (Fig. S5B). However, one cell (of 153 cells tested) was tuned to multiple taste qualities and responded to monosodium glutamate, denatonium, sucralose, and acesulfame-K (Fig. S5 B and C). We also tested 50 mM citric acid for sour-responsive cells. Unexpectedly, more than 90% of cells showed responses to citric acid, presumably as a result of the presence of proton-sensitive channels in different cell types, which prevented us from specifically identifying potential sour-responsive cells (Fig. S5 B and D).
Fig. 4.
Mature taste cells derived from single Lgr5+ cells are functional. (A) Representative trace of Ca2+ responses of an organoid-derived taste cell to sucralose and acesulfame-K sweeteners. (B) Representative trace of Ca2+ responses of an organoid-derived taste cell responding to bitter denatonium benzoate in a dose-dependent fashion (5, 10, and 20 mM). The phospholipase C β2 inhibitor U73122 (10 µM) inhibited denatonium-induced calcium responses, consistent with bitter signaling pathways of taste cells in vivo. The effect of U73122 was reversible after washout for ∼30 min. Imaging experiments were performed with 2D cultures independently grown from 15 organoids (see Fig. S5 for details).
Lgr6+ Cells Are Progenitor Cells in Anterior and Posterior Tongue.
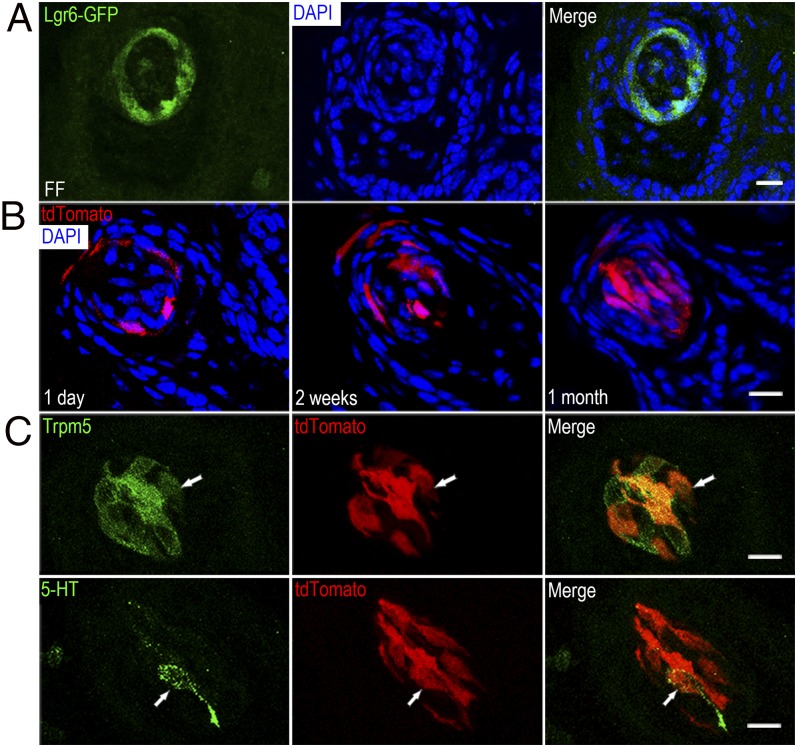
Our RT-PCR results show that in addition to Lgr5, Lgr6 can be amplified from our cultured organoids. Lgr5 marks adult taste stem/progenitor cells only in posterior tongue (6, 7). Prompted by this finding, as well as the result that Lgr6 is preferentially expressed in fungiform taste tissue but not in the surrounding epithelium devoid of taste tissue (12), we set out to determine whether Lgr6 marks adult taste stem/progenitor cells in anterior taste fields (fungiform papillae). We used heterozygous mice with one wild-type Lgr6 allele and one allele in which EGFP has been inserted into the Lgr6 gene (a knockout/knockin model) (13). Thus, EGFP serves as a surrogate marker for Lgr6 expression. EGFP green fluorescence was detected at the basal area of taste buds of the fungiform and circumvallate papillae (Fig. 5A and Fig. S6A). Occasionally, weak EGFP green fluorescence was detected in intragemmal taste cells as well. The expression pattern of Lgr6-EGFP in the circumvallate papillae resembles that of Lgr5-EGFP, which indicates that Lgr6 may also mark stem/progenitor cells in taste tissue. The low frequency of Lgr6-marked cells is likely a result of the mosaic expression of Lgr6-EGFP in taste tissue, which is known to occur as well in other tissues (13). No EGFP signal was detected in tongue epithelium devoid of taste tissue.
Fig. 5.
Lgr6 marks taste stem/progenitor cells in fungiform papillae. (A) Representative confocal images of Lgr6+ cells in fungiform papillae. The Lgr6-EGFP transgene was detected by intrinsic fluorescence (green). Lgr6-EGFP cells were found at the base of taste buds. (B) Tamoxifen-induced Lgr6-Cre generates tdTomato activity to mark cells from the Lgr6+ lineage. After a single tamoxifen induction of Lgr6-Cre in fungiform papillae, increased numbers of taste bud cells were labeled at 2 wk (3.59 ± 0.44 per taste bud in a 10-μM section; n = 32) and 1 mo (4.66 ± 0.60; n = 38) than at 1 d (1.09 ± 0.20; n = 34) (P < 0.0001 for both comparisons). No significant difference was found between the numbers of labeled taste bud cells at 1 mo and 2 wk after tamoxifen induction (P = 0.17). (C) Lgr6+ stem/progenitor cells generate multiple subtypes of taste bud cells in fungiform papillae. Mature taste bud cells marked by Lgr6-Cre-generated tdTomato 1 mo after tamoxifen induction were stained with markers for type 2 (Trpm5) and type 3 (5-HT, serotonin) taste cells in fungiform papillae. Arrows depict tdTomato+ cells that are immunopositive for a specific taste cell marker. Three mice were used for each point of the lineage tracing experiments. (Scale bars: 40 µm.)
We next performed lineage tracing to visualize the progeny of Lgr6+ taste cells. Lgr6-EGFP-ires-CreERT2 mice were crossed with Rosa26-tdTomato mice to generate Lgr6+/−; Rosa26-tdTomato+/− mice in which tamoxifen-induced Cre generates tdTomato fluorescent protein (red) to mark cells from the Lgr6+ lineage. We examined the distribution of tdTomato+ cells at different points after a single tamoxifen injection. If the number of tdTomato+ cells increased for prolonged periods after tamoxifen induction, this would provide strong evidence that Lgr6+ cells may serve as taste progenitor cells that give rise to other types of taste cells. Indeed, at 1 d after tamoxifen induction, we observed that basal cells in the fungiform and circumvallate papillae were tdTomato+ (Fig. 5B and Fig. S6B). Occasionally, we found that intragemmal cells were tdTomato+. At 2 wk after tamoxifen induction, in both fungiform and circumvallate papillae, multiple intragemmal cells were tdTomato+. At 1 mo after tamoxifen induction in both circumvallate and fungiform papillae, the number of tdTomato+ cells within taste buds was similar to that at 2 wk (Fig. 5B and Fig. S6B). By immunostaining taste tissue from Lgr6+/−; tdTomato+/− mice 1 mo after tamoxifen induction, we found that the progeny of Lgr6+ cells included at least type 2 [transient receptor potential cation channel subfamily M member 5 (Trpm5+)] and type 3 (serotonin+) taste receptor cells (Fig. 5C and Fig. S6C).
Lgr6+ cells in posterior tongue showed a pattern of localization similar to that of Lgr5+ cells. To determine whether these two populations of cells are related, we sorted Lgr5+ cells from posterior tongue and Lgr6+ cells from posterior and from anterior tongue, generated cDNA from the different sets of sorted cells, and performed RT-PCR using intron-spanning primers to determine whether we could detect Lgr6 in Lgr5+ cells and vice versa. Interestingly, all three Lgr transcripts Lgr4–Lgr6 were detected in sorted Lgr5+ cells from posterior tongue and in sorted Lgr6+ cells from either posterior or anterior tongue (Fig. S7). In contrast, no or barely detectable Lgr5 and Lgr6 were amplified from cells negative for GFP expression sorted from Lgr5-EGFP-ires-CreERT2+/− tongue tissues (Fig. S7).
To determine whether Lgr6+ cells, similar to Lgr5+ cells, can proliferate and differentiate into taste-like cells ex vivo, we sorted Lgr6-EGFP cells on the basis of their EGFP fluorescence. Similar to Lgr5+ cells, Lgr6+ cells grew into 3D organoids (Fig. S8A), and most organoids had no intrinsic EGFP signal after being cultured for 2–3 d. However, occasionally, organoids were found to retain a strong EGFP signal: In two of eight preparations, organoids with GFP fluorescence were seen in three (3%) of 106 total organoids and 129 (9%) of 1447 organoids examined. Presumably, this continued expression of GFP resulted from Lgr6-EGFP cells that were expanded from single isolated Lgr6-EGFP+ cells (Fig. S8B). We performed immunostaining with antibodies against T1R3 (blue), gustducin (red), and CA4 (blue). Most organoids with intrinsic EGFP signal (green) showed immunoreactivity to two markers (∼80–89%: eight of 10 for gustducin and CA4 and eight of nine for gustducin and T1R3, Fig. S8 C and D and Table S1). In contrast, a smaller subset of organoids without EGFP activity stained with both markers (∼19–45%: five of 27 for gustducin and T1R3 and 13 of 29 for gustducin and CA4; Fig. S8 E and F and Table S1). Thus, it would appear that organoids that continue to express Lgr6-EGFP are more often multipotent, whereas those without EGFP may have more limited potency. Together, our lineage tracing data and cell culture data demonstrate that Lgr6 marks taste stem/progenitor cells in both anterior and posterior tongue and can generate apparent mature taste cells ex vivo.
Discussion
In this study, we used a 3D culture system to determine the regenerative capacity of Lgr5+ and Lgr6+ taste stem/progenitor cells. We found that single isolated Lgr5+ and Lgr6+ taste stem/progenitor cells can grow into 3D “organoid” structures that contain both dividing cells and mature presumptive taste cells. The presence of different types of taste cells in organoids derived from single Lgr5+ or Lgr6+ cells indicates the multipotency of such cells and sheds light on cell lineage relationships within taste papillae. Moreover, our calcium imaging studies demonstrate that some of these cells function as taste receptor cells to respond to tastants. Interestingly, similar to intestinal Lgr5+ adult stem cells that build crypt-villus structures in vitro without a mesenchymal niche (10, 14), single Lgr5+ and Lgr6+ cells can expand and differentiate into taste-like cells in vitro without a mesenchymal niche or neuronal connection, which suggests these cells are intrinsically capable of giving rise to all subtypes of taste cells.
In animal models, glossopharyngeal nerve transection leads to degeneration and subsequent regeneration of taste bud cells, which is neuronally dependent, as taste cells start to regenerate only after transected nerves reenter taste tissue (7, 29). Therefore, neuronal factors or direct neuronal contact may promote the differentiation of taste cells generated from Lgr5+ or Lgr6+ stem/progenitor cells. Given the fact that single Lgr5+ or Lgr6+ taste stem/progenitor cells can give rise to taste cells in vitro, it is likely that neuronally secreted extrinsic factors, rather than direct neuronal contact, trigger taste cell renewal from adult taste stem/progenitor cells.
In our defined culture medium, we included a variety of growth factors, as well as N2 and B27. It is likely that some of these factors or functionally similar factors are normally supplied by innervating nerves or cells in mesenchymal tissue adjacent to taste epithelium to maintain taste tissue homeostasis. However, which factor or factors is key to promoting taste cell regeneration or taste cell differentiation in our cultured system merits further investigation. Prominent candidate growth factors may include brain-derived neurotrophic factor, nerve growth factor, neurotrophin-4, and R-spondins, among many others (30–34).
After 2–3 d in culture, the green fluorescent signal from Lgr5-EGFP cells or Lgr6-EGFP cells became undetectable in most organoids. One possible explanation for this is that the Lgr5-EGFP or Lgr6-EGFP transgenes are silenced in most organoids in our culture conditions, given that both Lgr5 and Lgr6 transcripts can be readily detected in cultured organoids. Another possibility is that at least two types of organoids originated from Lgr5+ or Lgr6+ cells, with distinct regeneration potentials. As reported previously, there are strong Lgr5+ cells under the trench areas of circumvallate and weak Lgr5+ cells in the basal areas of taste buds (6). Therefore, Lgr5+ cells may be heterogeneous, and we propose that the strong Lgr5+ cells are stem cells and the weak Lgr5+ cells are more committed progenitor cells. Likewise, Lgr5+- or Lgr6+-derived organoids with GFP fluorescence may have been derived from stem cells that can generate new Lgr5+ or Lgr6+ cells. In contrast, Lgr5+ or Lgr6+ organoids that lack GFP fluorescence after being cultured for a few days may have been derived from Lgr5+ or Lgr6+ committed progenitor cells incapable of self-renewal. This hypothesis is further supported by the observation that most organoids with EGFP could produce multiple subtypes of taste cells, whereas fewer organoids without detectable EGFP could do so.
Lgr5 and its homologs Lgr4 and Lgr6 interact with R-spondins to augment Wnt signaling (17–19). We noted that R-spondin-1 has a substantial growth-promoting effect on our organoid cultures. One possibility for how R-spondin works is that Lgr5, despite its low expression in organoids, may mediate R-spondin’s effect. Alternatively, R-spondin may interact with Lgr4 or Lgr6, which are Lgr5 homologs and were also expressed in our cultured organoids, to promote organoid growth. However, the exact role of the Wnt signaling pathway for taste tissue renewal during adulthood remains unclear. Nevertheless, during development, Wnt signaling plays a major role in fungiform taste papillae formation and seems to be regionally specific (35, 36).
Single Lgr5+ or Lgr6+ cells can give rise to different types of taste cells in our 3D organoid cultures, indicating that some of these cells are multipotent. It remains to be determined whether individual taste Lgr5+ or Lgr6+ cells (e.g., the strongly Lgr5+ cells from under the trench areas) are multipotent in vivo. Two contrasting models for taste cell lineages have been proposed (37): one posits existing lineage-specific stem/progenitor cells for each subtype of taste cell, and the other posits multipotent taste stem cells that can give rise to all subtypes of taste cells. However, these two models do not necessarily contradict each other: multipotent stem cells can give rise to lineage-specific progenitor cells. Our organoid culture data are consistent with both models. It is likely that some Lgr5+ or Lgr6+ cells are multipotent, such as those having EGFP activity, whereas other Lgr5+ or Lgr6+ cells are committed progenitor cells. The multipotency of taste progenitor cells is further supported by the finding that Skn-1a knockout mice have a complete loss of sweet/bitter/umami type 2 cells with a concomitant expansion of polycystic kidney disease 2-like 1 protein–expressing sour taste receptor cells, which suggests at least the binary differentiation potential of progenitor cells into type 2 and type 3 cells (38) and is consistent with the hypothesis proposed by Miura and coworkers (39, 40).
Lgr5+ cells give rise to all three major types of cells in taste tissues in posterior tongue (6, 7). Similarly, we found that Lgr6+ cells can give rise to multiple types of cells in taste tissues in both the anterior and posterior tongue, including type 2 receptor cells and type 3 presynaptic cells. Similar to Lgr5-EGFP+ cells in posterior tongue, Lgr6-EGFP+ cells sit deep in the trench area of the circumvallate papilla, as well as at the base of taste buds. In contrast to Lgr5-EGFP+ cells, Lgr6-EGFP+ cells also are detected in anterior tongue at the base of taste buds in fungiform papillae. Our lineage tracing study showed that the progeny of Lgr6+ progenitor cells can persist for 1 mo (the longest time tested to date). Although we are not certain whether Lgr6+ cells represent bona fide stem cells in fungiform papillae, they are undoubtedly progenitor cells that give rise to other types of mature taste cells both in vivo and ex vivo. Furthermore, the similar distribution pattern of Lgr5+ and Lgr6+ cells in posterior tongue; the detection of Lgr6 in Lgr5+ posterior cells and Lgr5 in Lgr6+ cells in both anterior and posterior tongue, despite no Lgr5-EGFP signal in fungiform papillae cells in adult mice, which could be a result of low expression of Lgr5 or regional silencing of Lgr5-EGFP transgene (6, 7, 12); and their similar regenerative capacity suggest they mark the same set of progenitor cells in both anterior and posterior tongue.
Materials and Methods
Genetically engineered mice (Lgr5-EGFP-ires-CreERT2 [stock # 008875], Lgr6-EGFP-ires-CreERT2 [stock # 016934], Rosa26-tdTomato [stock # 007905]) were obtained from the Jackson Laboratory. All experiments were performed under National Institutes of Health guidelines for the care and use of animals in research and approved by the Institutional Animal Care and Use Committee of the Monell Chemical Senses Center (protocol 1150). Details about lineage tracing, cell sorting, 3D organoid cultures, immunostaining, BrdU labeling, and calcium imaging are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank all members of the P.J. and R.F.M. laboratories for their input. Research reported in this publication was supported by institutional funds (P.J.) from the Monell Chemical Senses Center, as well as by NIH Grants DC010842 (to P.J.), DC000882 (to A.A.B.), DC012980 (to B.C.L.), DK081421 (to R.F.M.), and DC003055 (to R.F.M.), and an industry consortium funding Monell research (P.J., B.C.L., A.A.B., and R.F.M.). Imaging was performed at the Monell Histology and Cellular Localization Core, which is supported in part by NIH–National Institute on Deafness and Other Communication Disorders Core Grant DC011735 (to R.F.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409064111/-/DCSupplemental.
References
- 1.Beidler LM, Smallman RL. Renewal of cells within taste buds. J Cell Biol. 1965;27(2):263–272. doi: 10.1083/jcb.27.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapsimali M, Barlow LA. Developing a sense of taste. Semin Cell Dev Biol. 2013;24(3):200–209. doi: 10.1016/j.semcdb.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miura H, Scott JK, Harada S, Barlow LA. Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Dev Dyn. 2014;243(10):1286–1297. doi: 10.1002/dvdy.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamamichi R, Asano-Miyoshi M, Emori Y. Taste bud contains both short-lived and long-lived cell populations. Neuroscience. 2006;141(4):2129–2138. doi: 10.1016/j.neuroscience.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 5.Perea-Martinez I, Nagai T, Chaudhari N. Functional cell types in taste buds have distinct longevities. PLoS ONE. 2013;8(1):e53399. doi: 10.1371/journal.pone.0053399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yee KK, et al. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells. 2013;31(5):992–1000. doi: 10.1002/stem.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda N, et al. Lgr5 Identifies Progenitor Cells Capable of Taste Bud Regeneration after Injury. PLoS ONE. 2013;8(6):e66314. doi: 10.1371/journal.pone.0066314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker N, Tan S, Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140(12):2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- 9.Huch M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 11.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32(20):2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hevezi P, et al. Genome-wide analysis of gene expression in primate taste buds reveals links to diverse processes. PLoS ONE. 2009;4(7):e6395. doi: 10.1371/journal.pone.0006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327(5971):1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 14.Sato T, Clevers H. Primary mouse small intestinal epithelial cell cultures. Methods Mol Biol. 2013;945:319–328. doi: 10.1007/978-1-62703-125-7_19. [DOI] [PubMed] [Google Scholar]
- 15.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2011;108(28):11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lau W, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 17.Ruffner H, et al. R-Spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS ONE. 2012;7(7):e40976. doi: 10.1371/journal.pone.0040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20(19):2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okubo T, Clark C, Hogan BL. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells. 2009;27(2):442–450. doi: 10.1634/stemcells.2008-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng P, Huang L, Wang H. Taste bud homeostasis in health, disease, and aging. Chem Senses. 2014;39(1):3–16. doi: 10.1093/chemse/bjt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: Regulator of Wnt signal strength. Genes Dev. 2014;28(4):305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Max M, et al. Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat Genet. 2001;28(1):58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 24.Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416(6877):199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- 25.Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106(3):381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 26.Chandrashekar J, et al. The taste of carbonation. Science. 2009;326(5951):443–445. doi: 10.1126/science.1174601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MR, et al. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem Biophys Res Commun. 2003;312(2):500–506. doi: 10.1016/j.bbrc.2003.10.137. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Coding of sweet, bitter, and umami tastes: Different receptor cells sharing similar signaling pathways. Cell. 2003;112(3):293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- 29.King CT, Travers SP, Rowland NE, Garcea M, Spector AC. Glossopharyngeal nerve transection eliminates quinine-stimulated fos-like immunoreactivity in the nucleus of the solitary tract: Implications for a functional topography of gustatory nerve input in rats. J Neurosci. 1999;19(8):3107–3121. doi: 10.1523/JNEUROSCI.19-08-03107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ganchrow D, Ganchrow JR, Verdin-Alcazar M, Whitehead MC. Brain-derived neurotrophic factor-, neurotrophin-3-, and tyrosine kinase receptor-like immunoreactivity in lingual taste bud fields of mature hamster. J Comp Neurol. 2003;455(1):11–24. doi: 10.1002/cne.2162. [DOI] [PubMed] [Google Scholar]
- 31.Mistretta CM, Goosens KA, Farinas I, Reichardt LF. Alterations in size, number, and morphology of gustatory papillae and taste buds in BDNF null mutant mice demonstrate neural dependence of developing taste organs. J Comp Neurol. 1999;409(1):13–24. [PMC free article] [PubMed] [Google Scholar]
- 32.Nosrat CA, Blomlöf J, ElShamy WM, Ernfors P, Olson L. Lingual deficits in BDNF and NT3 mutant mice leading to gustatory and somatosensory disturbances, respectively. Development. 1997;124(7):1333–1342. doi: 10.1242/dev.124.7.1333. [DOI] [PubMed] [Google Scholar]
- 33.Oakley B, et al. The morphogenesis of mouse vallate gustatory epithelium and taste buds requires BDNF-dependent taste neurons. Brain Res Dev Brain Res. 1998;105(1):85–96. [PubMed] [Google Scholar]
- 34.Patel AV, Huang T, Krimm RF. Lingual and palatal gustatory afferents each depend on both BDNF and NT-4, but the dependence is greater for lingual than palatal afferents. J Comp Neurol. 2010;518(16):3290–3301. doi: 10.1002/cne.22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwatsuki K, et al. Wnt signaling interacts with Shh to regulate taste papilla development. Proc Natl Acad Sci USA. 2007;104(7):2253–2258. doi: 10.1073/pnas.0607399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nat Genet. 2007;39(1):106–112. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- 37.Stone LM, Tan SS, Tam PP, Finger TE. Analysis of cell lineage relationships in taste buds. J Neurosci. 2002;22(11):4522–4529. doi: 10.1523/JNEUROSCI.22-11-04522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci. 2011;14(6):685–687. doi: 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura H, Kusakabe Y, Harada S. Cell lineage and differentiation in taste buds. Arch Histol Cytol. 2006;69(4):209–225. doi: 10.1679/aohc.69.209. [DOI] [PubMed] [Google Scholar]
- 40.Miura H, et al. Co-expression pattern of Shh with Prox1 and that of Nkx2.2 with Mash1 in mouse taste bud. Gene Expr Patterns. 2003;3(4):427–430. doi: 10.1016/s1567-133x(03)00081-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.