Significance
Human lifestyles profoundly influence the communities of microorganisms that inhabit the body, that is, the microbiome; however, how the microbiomes of humans have diverged from those found within wild-living hominids is not clear. To establish how the gut microbiome has changed since the diversification of human and ape species, we characterized the microbial assemblages residing within hundreds of wild chimpanzees, bonobos, and gorillas. Changes in the composition of the microbiome accrued steadily as African apes diversified, but human microbiomes have diverged at an accelerated pace owing to a dramatic loss of ancestral microbial diversity. These results suggest that the human microbiome has undergone a substantial transformation since the human–chimpanzee split.
Keywords: microbiota, gastrointestinal tract, coevolution, Pan, Gorilla
Abstract
Humans are ecosystems containing trillions of microorganisms, but the evolutionary history of this microbiome is obscured by a lack of knowledge about microbiomes of African apes. We sequenced the gut communities of hundreds of chimpanzees, bonobos, and gorillas and developed a phylogenetic approach to reconstruct how present-day human microbiomes have diverged from those of ancestral populations. Compositional change in the microbiome was slow and clock-like during African ape diversification, but human microbiomes have deviated from the ancestral state at an accelerated rate. Relative to the microbiomes of wild apes, human microbiomes have lost ancestral microbial diversity while becoming specialized for animal-based diets. Individual wild apes cultivate more phyla, classes, orders, families, genera, and species of bacteria than do individual humans across a range of societies. These results indicate that humanity has experienced a depletion of the gut flora since diverging from Pan.
The human microbiome is shaped by host genetics, environment, and lifestyle (1–3); thus, humanity's unique evolutionary and cultural histories must have altered our associations with microorganisms (4). Despite intensive investigation of the microbiomes of humans spanning a range of geographic locations and cultures (5–7), how the composition of the microbiome has changed since humans diverged from other species, and since human populations diverged from one another, remains unclear, owing to a lack of knowledge about the microbiomes of ancestral hominid populations.
Understanding how the composition of the human microbiome has changed over evolutionary time requires the inclusion of the microbiomes of phylogenetic outgroups (i.e., the African apes) into analyses of human microbiomes. Previous comparisons of the gut microbiomes of humans and the African apes have been restricted to just a few individuals per host species (8), precluding detection of the precise compositional differences that distinguish the microbiomes of the host species. Comparing the microbiomes of populations of chimpanzees, bonobos, gorillas, and humans while considering the phylogenetic relatedness among the hosts can reveal how the composition of the microbiome has changed since the host species diversified.
Here we used a phylogenetic approach to identify the shifts in the composition of the microbiome that occurred along the lineages leading to the extant species of Homo and Pan. This analysis shows that humans across a range of cultures and geographies harbor microbiomes that are disproportionately divergent from those within wild apes. In particular, among the living hominid species, humans harbor uncharacteristically low levels of microbial diversity within their gut microbiomes.
Results
Sample Sources.
We sequenced the V4 region of 16S rDNA present in fecal samples from hundreds of wild chimpanzees from Tanzania (n = 160), wild bonobos from the Democratic Republic of the Congo (n = 70), and wild gorillas from Cameroon (n = 186). Fig. S1 shows a map of sampling locations, and Table S1 lists samples and corresponding metadata. Because methodologies were standardized across samples, we were able to compile the information obtained for these great ape populations with corresponding data from humans living urban lifestyles in the United States (n = 317) (5), rural lifestyles in Malawi (n = 114) (5), preindustrial lifestyles in the southern Amazon rainforests of Venezuela (n = 99) (5), urban lifestyles in Europe (n = 81) (6), and hunter-gatherer lifestyles in Tanzania (n = 27) (7).
Consistent Co-Occurrence Patterns Within the Core Ape Microbiome.
All humans and African ape species shared a core set of bacterial genera that were recovered from a majority of individuals from every sampled population (Fig. S2). Co-occurrence patterns among the core taxa of the ape microbiome were highly consistent across host species. For instance, the relative abundances of Prevotella and Bacteriodes were negatively correlated within each host population, whereas the relative abundances of Bacteroides were always positively correlated with those of Ruminococcus and Parabacteroides.
Changes to the Composition of the Microbiome During Host Evolution.
Comparisons of the gut communities of populations of ape species allowed assembly of a phylogenetic framework to determine how the composition of the microbiome has changed during hominid evolution (Fig. 1). We inferred the shifts in the relative abundances of microbial taxa along the branches of the African ape phylogeny that most parsimoniously explain the differences among the microbiomes of extant host species. A change (either increase or decrease) in relative abundance along a branch of the host phylogeny was identified if (i) there was a twofold difference between the mean relative abundance in each sampled population of the subtending lineage versus the sister lineage(s) as well as all outgroup lineages and (ii) the difference in mean relative abundance was significant based on an false discovery rate-corrected P value threshold of < 0.001. For the branch leading to humans, compositional shifts were counted only if they were identified in each sampled human population (5–7).
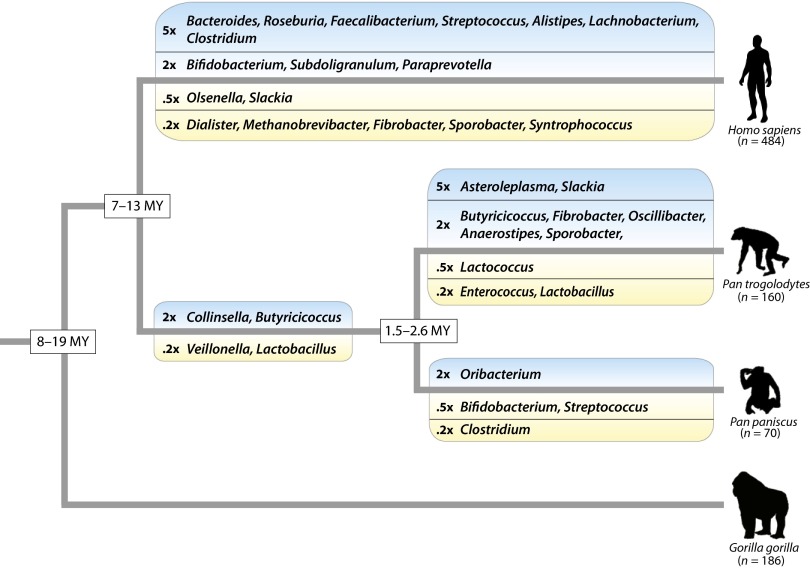
Fig. 1.
Compositional changes in the gut microbiome during African ape diversification. Shifts in relative abundances of microbial genera within the gut microbiome were inferred for each branch of the host phylogeny. Genera whose relative abundances increased or decreased are listed above or below each branch within blue-shaded or yellow-shaded boxes, respectively. Genera are grouped by degree of change in relative abundance (>5×, >2×, <0.5×, and <0.2×). Tree branches are not drawn to scale, but divergence time ranges (17, 18) are listed at each node.
We identified 35 instances in which the relative abundance of a microbial taxon shifted since the divergences of the extant species of African ape (Fig. 1), 17 of which occurred in humans since the divergence of Homo and Pan. Several of these changes in the composition of the human microbiome have functional implications for host nutrition. The relative abundance of Bacteroides, which has been positively associated with diets rich in animal fat and protein (9), has increased in relative abundance more than fivefold in humans. Conversely, the archaeon Methanobrevibacter, which promotes the degradation of complex plant polysaccharides by using the end products of fermentation for methanogenesis (10), has undergone a more than fivefold reduction within humans. Similarly, the abundance of Fibrobacter, a common plant-fermenting bacterial genus (11) of the microbiomes of wild apes, has been greatly reduced in humans.
Clock-Like Divergence of Microbiomes Among African Apes.
Despite marked differences in lifestyle and ecology among host species, changes in microbiome the composition have accrued at a relatively clock-like rate during the diversification of the great apes (Fig. 2A). The average genus-level Bray–Curtis dissimilarity (BCD) between the gut microbiomes of each pair of wild ape species was linearly correlated with the evolutionary distance between the species (P < 10−6). Fossil and genetic evidence indicate that the divergence times for African apes range from 5 to 13 mya for the chimpanzee–human split and from 8 to 16 mya for the human–gorilla split (12, 13). Based on this range of divergence time estimates, the mean BCD between African ape species has increased at a relatively constant rate of 0.025–0.037 per million years.
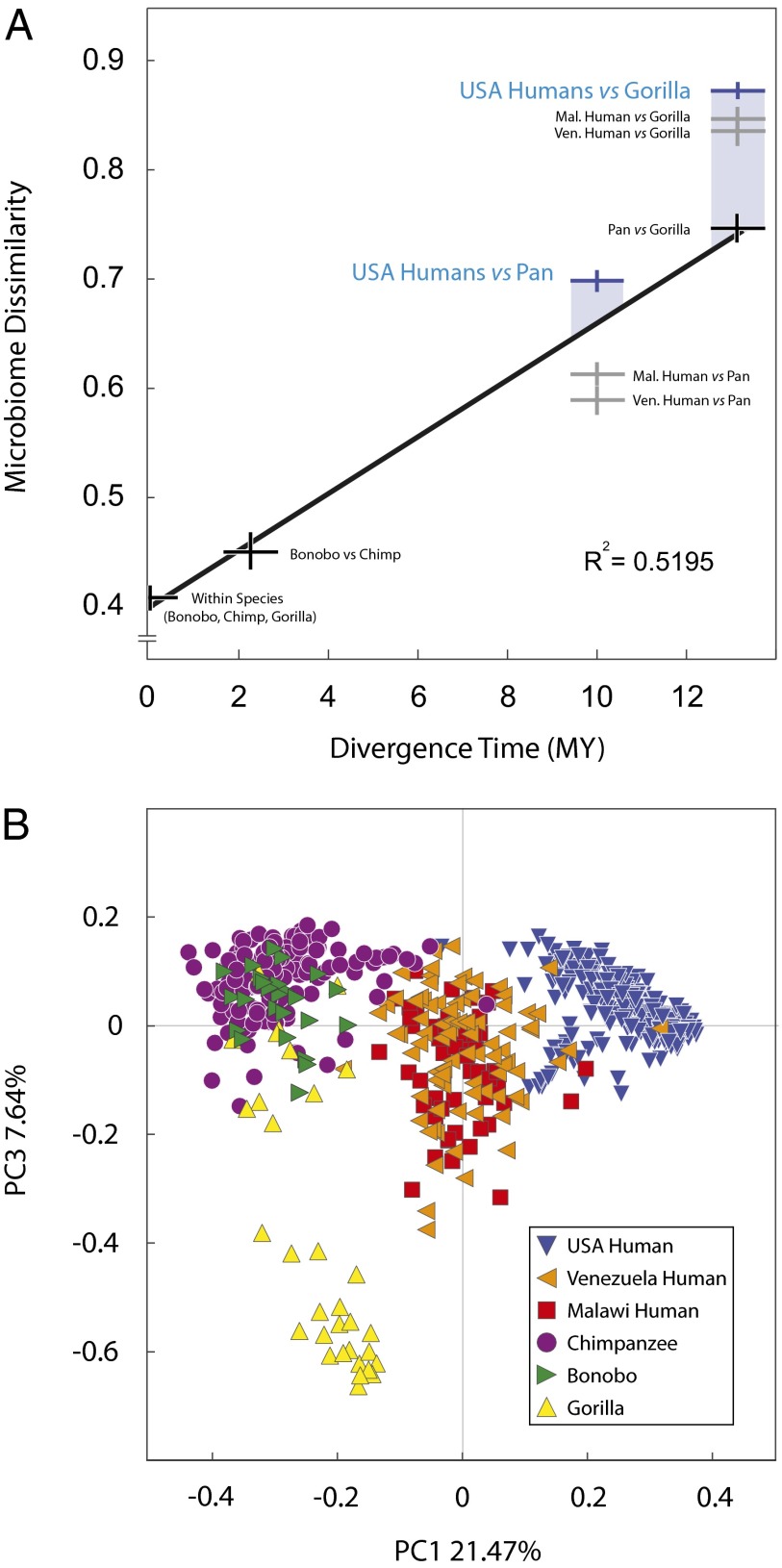
Fig. 2.
Rapid divergence and uniqueness of gut microbiomes of US humans. (A) Correlation between BCD among genus-level compositions of wild ape microbiomes and host divergence times. Black crosses represent comparisons within and between wild ape species, gray crosses represent comparisons between either Malawi humans or Venezuela humans and wild apes, and blue crosses represent comparisons between US humans and wild apes, with vertical lines in each cross representing 95% CIs. Shaded blue bars correspond to the differences between the observed dissimilarity in the microbiomes of US humans and wild apes and the expected dissimilarity based on divergence times. (B) Principal coordinates plot of genus-level BCDs among human and ape microbiomes. Colored shapes correspond to microbiomes of human and wild ape populations, as indicated in the key.
Despite the clock-like nature of microbiome diversification in African apes, the gut microbiomes of US humans have undergone an accelerated rate of change and are more different from those of each wild ape population than expected based on the evolutionary time separating Homo from Pan and Gorilla. Based on genus-level BCD, the microbiomes of US humans are more different from those of Malawi humans than the gut microbiomes of Malawi humans are from those of bonobos (P < 10−13). The difference between microbiomes of US humans and wild apes is particularly evident in the first two principal coordinates axes of the pairwise beta diversities among samples at both the genus (Fig. 2B) and 97% operational taxonomic unit (OTU) (Fig. S3) levels.
Changes to the Microbiome that Differentiate Human Populations.
Comparing wild ape and human microbiomes allowed us to infer the compositional changes that most parsimoniously explain the present-day variation among the microbiomes of human populations, including those that distinguish the microbiomes of US humans. We identified 23 instances in which the relative abundance of a microbial taxon shifted by at least twofold since the divergence of the sampled human populations (Fig. S4). In particular, the frequency of Bacteroides has increased more than fivefold in US humans since their separation from the other human populations.
Reduction of Ancestral Diversity Within the Human Microbiome.
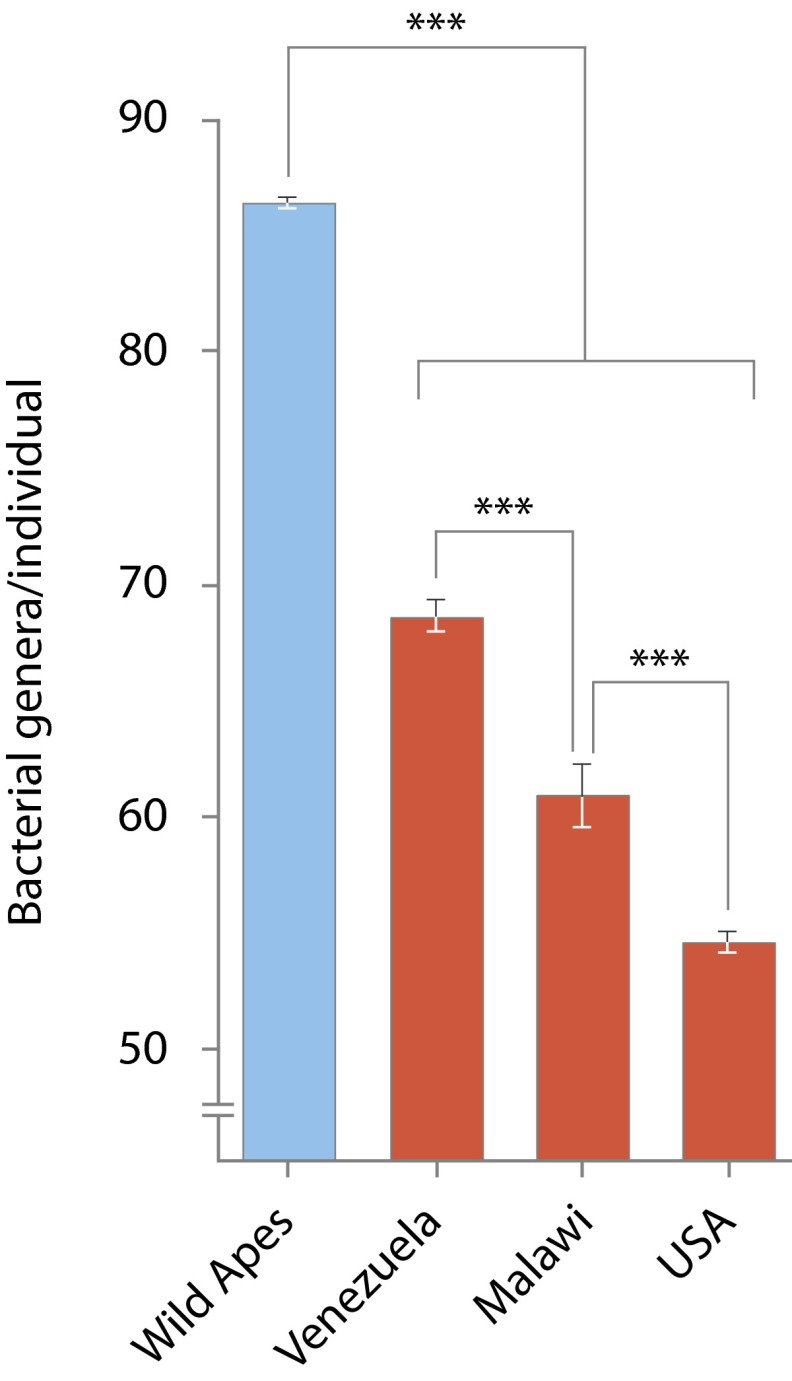
Our comparisons of V4 16S sequences from hundreds of individuals of different host species and populations allowed us to identify differences in total microbial diversity across a range of taxonomic scales. We found that humans across three continents representing a range of lifestyles harbored lower levels of microbial diversity compared with wild apes. On average, individual humans contained fewer bacterial phyla, classes, orders, families, genera, and 97% OTUs than did individual chimpanzees, bonobos, or gorillas (Fig. 3 and Figs. S5 and S6). Moreover, compared with the microbiomes of wild apes, in which many different bacterial genera coexisted at relatively low frequencies, the microbiomes of individual humans tended to be less even, with fewer dominant taxa and many low-abundance constituents (Fig. S7). The reduction of ancestral diversity within the human microbiome was evident in Venezuela and Malawi humans, but particularly pronounced in US humans, who displayed on average 30 fewer bacterial genera at a sequencing depth of 20,000 reads compared with individual chimpanzees, bonobos, or gorillas.
Fig. 3.
Diminished diversity in human gut microbiomes across populations. Mean numbers of observed bacterial genera per individual in wild apes and in human populations at a sequencing depth of 20,000 reads. Error bars correspond to 95% CIs, and asterisks denote significant differences at P < 0.001.
Discussion
Comparisons of the gut microbiomes of populations of humans, chimpanzees, bonobos, and gorillas provide insight into the evolution of hominid microbiomes. In particular, we have reconstructed how human microbiomes have changed since humans diverged from Pan by identifying the features of the microbiome shared across human populations to the exclusion of African apes. Our results demonstrate the utility of incorporating information about the phylogenetic relationships among hosts into analyses of their microbiomes.
It has been proposed that recent lifestyle changes in humans have depleted the human microbiome of microbial diversity that was present in our wild-living ancestors (4); however, this hypothesis has not been tested through comparisons of humans and closely related host species. A previous survey of two humans and 24 wild apes found that the humans contained lower levels of 99% OTU diversity than the wild apes (8), but the small sample sizes precluded both the statistical evaluation of this trend and the ability to identify bacterial taxa that are consistently not recovered from human hosts. We observed that the mean level of microbial diversity within an individual’s gut microbiome differed substantially among ape species, with the microbiomes of humans being the least diverse. This trend does not appear to be the product of any specific cultural practice and is apparent in humans regardless of whether they resided in cities in the United States, small towns in Malawi, or villages in the Amazonas or Venezuela. This observation confirms the hypothesis that the levels of microbial diversity in the human microbiomes have decreased during human evolution. Of the human populations, humans from cities in the United States harbored the lowest levels of diversity, a trend previously observed by Yatsunenko et al. (5), suggesting that microbial diversity has been reduced even further in this group.
An alternative, but less parsimonious, explanation for the differences among the observed levels of diversity within hosts of each species is that Pan and Gorilla have experienced independent increases in the levels of microbial diversity since diverging from humans. Providing an explanation for these independent increases is difficult, however, whereas cultural and ecological differences between humans and wild apes provide clear causes for the reduced microbial diversity along the human lineage. Extending sampling to wild-living populations of more distantly related primate species will provide further evaluation of these the competing hypotheses that explain the current variation in diversity levels across human and wild ape microbiomes.
Despite marked differences among the microbiomes of human and African ape species, there exists a set of bacterial taxa shared across host populations, potentially representing the ancestral core of the African ape microbiome. Moreover, co-occurrence patterns among many of these taxa are recapitulated across host species (Fig. S2). This result mirrors previous descriptions of “enterotypes” (14–16) or “community types” (17): bacterial assemblages within the gut microbiomes of humans, chimpanzees, and mice defined by differential representation of Prevotella, Bacteroides, Ruminococcus, and Parabacteroides (14–16). Thus, it is possible that these consistent co-occurrence patterns among bacterial taxa result from ecological relationships that predate the diversification of human and African ape species.
Sampling the gut microbiomes of hundreds of individuals from each host species also allows the identification of population-level differences in the mean relative abundances of scores of bacterial taxa. Analyzing these differences in a phylogenetic context provides insight into how the composition of the human gut microbiome has been reshaped since humans diverged from other species. Consistent with the known dietary shifts that occurred during human evolution (18), taxa that have been associated with the digestion of animal foodstuffs (9) have risen in relative abundance in the human gut microbiome, whereas taxa that have been associated with the digestion of plant-based diets (9) have become less prominent.
Phylogenetic comparisons of populations of host species can reveal the consistent differences between their microbiomes that arose since the host species diverged; however, the relative roles of genetic divergence and ecological/cultural divergence between host species in generating the differences between their microbiomes remain unclear. The sampling of hosts consuming similar diets in similar environments can reveal the extent to which the contents of microbiomes are attributed to innate differences between the hosts as opposed to differences between the hosts' environments or lifestyles. Some attempts have been made to compare the microbiomes of different host species that co-occur. For example, Ley et al. (19) showed that differences between the gut microbiomes of distantly related mammal taxa were maintained when hosts resided within the same zoo. Similarly, Song et al. (20) found that cohabiting dogs and humans shared more bacterial OTUs compared with hosts from separate households, but the gut microbiomes of dogs remained distinct from those of their cohabiting humans. Likewise, Moeller et al. (21) showed that sympatric chimpanzees and gorillas harbored more similar sets of bacterial species than did the gut microbiomes of allopatric chimpanzees and gorillas, but chimpanzee and gorilla microbiomes can always be differentiated, even when the host species live in sympatry (21). These results suggest that, although shared environments might lead to the exchange of some bacterial taxa, many of the differences among the microbiomes of host species are robust to environmental influences.
Conclusions
We analyzed the microbiomes of hundreds of humans and African apes in a phylogenetic framework to reconstruct how microbiomes have diverged over the course of hominid evolution. This approach, which relies on population-level microbiome data from a clade of host species for which the phylogenetic relationships are known, can be applied to interrogate the evolutionary history of the microbiomes of a diversity of host groups. Relative to the microbiomes of wild apes, human microbiomes have experienced a reduction in ancestral microbial diversity and an increase in the frequency of bacterial taxa associated with animal-based diets. The consequences of this reduction of bacterial diversity in the human gut microbiome remain unexplored; however, low levels of bacterial diversity in the microbiome have been associated with gastrointestinal disorders (22), obesity (23), and autoimmune disease (24). Understanding how recent changes in the gut microbiome have influenced human health can benefit from further study of the ancient relationships between wild apes and their resident microbial communities.
Materials and Methods
Sample Collection and Species Identification.
Fresh fecal samples from gorillas and bonobos were collected from field sites in Cameroon and the Democratic Republic of the Congo, respectively. Fecal samples from chimpanzees from Gombe, Tanzania were collected immediately after being deposited. All samples were preserved in RNAlater (Ambion) and stored at –80 °C. For each sample, host species was determined by field experts and confirmed by sequencing of the host mitochondrial D-loop (25, 26).
Selection of Sample Processing, Sequencing, and Statistical Methodologies.
Previous meta-analyses have shown that systematic compositional differences in the microbiomes between Western and agrarian cultures outweigh the technical variation produced by differences among studies (e.g., DNA extraction protocol used, 16S region targeted) (27). Thus, between-species compositional differences, which are generally larger than those within a single species, also should be detectable across studies spanning a range of methodologies. Here we chose previously applied protocols, bead-beating DNA extraction and Illumina sequencing of the V4 region, to minimize technical variation and allow direct comparison of the African ape dataset with the human dataset of Yatsunenko et al. (5).
Sample Processing and Sequencing.
Total DNA was extracted from ∼400-mg aliquots using a previously described bead-beating procedure, and PCR amplifications of the V4 region of 16S rDNA were performed using 515F and 806R primers, as described previously (28). Reaction products were purified with AMPure XP Beads (Beckman Coulter). The resulting amplicons were subjected to Illumina sequencing on the MiSeq platform at the Genome Sequencing and Analysis Facility at the University of Texas at Austin.
Sequence Processing and Taxonomic Assignments.
Quality filtering and processing of sequences was performed in QIIME v1.7. Fastq files were demultiplexed with split_libraries_fastq.py using default settings. Sequences were trimmed to equal lengths of those of Yatsunenko et al. (5), and the two datasets were concatenated. Sequences were clustered into 97% OTUs through uclust_ref and assigned to the taxonomic levels (phylum, class, order, family, and genus) against the greengenes database, as done by Yatsunenko et al. (5).
Identification of a Core African Ape Microbiome.
The core microbiome of African apes and humans was defined as the bacterial genera recovered from more than two-thirds of the individuals in each population of hosts. Spearman correlations between all pairwise combinations of the relative abundances of these taxa were calculated in R for each host species. Significant correlations with an absolute value >0.3 and recapitulated across host species are shown in Fig. S2. A flowchart depicting these analyses is presented in Fig. S8.
Parsimonious Reconstruction of Microbiome Changes.
For parsimony analyses, the wild ape data produced for the present study were compared with three human microbiome datasets. All samples from infants were excluded. To reconstruct compositional shifts in the microbiome that occurred along the human lineage, we identified bacterial taxa that were overrepresented or underrepresented by at least twofold in each human population relative to each wild ape population. To reconstruct compositional shifts in the microbiome that occurred along the lineage leading to Pan, we identified bacterial taxa that were overrepresented or underrepresented by at least twofold in each Pan population relative to gorillas and to each human population. To reconstruct compositional shifts in the microbiome that occurred along the lineage leading to chimpanzees, we identified bacterial taxa that were overrepresented or underrepresented by at least twofold in chimpanzees relative to bonobos, to gorillas, and to each human population. To reconstruct compositional shifts in the microbiome that occurred along the lineage leading to bonobos, we identified bacterial taxa that were overrepresented or underrepresented by at least twofold in bonobos population relative to chimpanzees, to gorillas, and to each human population. To reconstruct compositional shifts in the microbiome that have occurred in each human population sampled by Yatsunenko et al. (5), we identified bacterial taxa that were overrepresented or underrepresented by at least twofold in one human population relative to the other two human populations, to each Pan species, and to gorillas. A flowchart depicting these analyses is presented in Fig. S8.
Estimating the Rate of Gut Microbiome Divergence During Great Ape Diversification.
We associated genus-level BCDs among the microbiomes of gorillas, chimpanzees, and bonobos with the time since their divergence across a range of divergence time estimates for the African apes. Results obtained from the recent divergence time estimates based on empirically derived generation times for wild Pan and Gorilla populations are shown in Fig. 3. To test whether human microbiomes sampled by Yatsunenko et al. (5) were more or less dissimilar from those of wild ape microbiomes than expected based on divergence times, we calculated all pairwise genus-level BCDs between human and ape populations (excluding infants), and estimated 99% confidence intervals (CIs) for the mean dissimilarities between each pair of populations. A flowchart depicting these analyses is presented (Fig. S8).
Testing for Differences in Diversity and Evenness Among Populations.
For these analyses, we compared the microbiomes of African apes with those of humans sampled by Yatsunenko et al. (5), because the datasets were produced by bead-beating DNA extraction, PCR of the V4 region of 16S rDNA, and Illumina sequencing. The Student t test was used to evaluate whether bacterial taxa per individual at a sequencing depth of 20,000 reads differed across host species and populations (excluding infants). For the genus level, we performed rarefaction analyses across a range of sequencing depths (Fig. S6). The equability of each sample was calculated at a depth of 20,000 reads, and differences in equability among host populations and species (excluding infants) were identified using the Student t test. To validate the observation that humans harbor diminished levels of microbial diversity, we calculated per-sample taxon and OTU numbers from the dataset of Ochman et al. (8), in which all samples from humans (n = 2) and wild apes (n = 24) were prepared, processed, and sequenced as a cohort using identical methods.
Supplementary Material
Acknowledgments
We thank Kim Hammond for assistance with preparation of the figures. We also thank the staff of the Prevention of AIDS in Cameroon (PRESICA) project, the Institut National de Recherches Biomédicales in Kinshasa, the World Wildlife Fund for Nature, and the Bonobo Conservation Initiative, as well as Didier Mazongo, Octavie Lunguya, Muriel Aloni, and Valentin Mbenz for field work in Cameroon and the Democratic Republic of Congo; field assistants in Gombe National Park for sample collection in Tanzania; the Cameroonian Ministries of Health, Forestry, and Wildlife, and Scientific Research and Innovation for permission to collect samples in Cameroon; the Ministries of Health and Environment and the National Ethics Committee for permission to collect samples in the Democratic Republic of Congo; the Tanzania Commission for Science and Technology; and the Tanzania Wildlife Research Institute for permission to conduct research in Gombe National Park. This work was supported by grants from the National Institutes of Health (R01 AI091595, R37 AI050529, R01 AI58715, P30 AI045008, R01 GM101209), National Science Foundation (Graduate Research Fellowship 2011119472, to A.H.M.), Agence Nationale de Recherche sur le Sida (ANRS 12125/12182/12255), and the Jane Goodall Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: All data have been deposited into the National Center for Biotechnology Information sequence read archive (accession no. SRX720662).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419136111/-/DCSupplemental.
References
- 1.Huttenhower C, et al. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 4.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnorr SL, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ochman H, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103(26):10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi Y, Shinkai T, Koike S. Ecological and physiological characterization shows that Fibrobacter succinogenes is important in rumen fiber digestion: Review. Folia Microbiol (Praha) 2008;53(3):195–200. doi: 10.1007/s12223-008-0024-z. [DOI] [PubMed] [Google Scholar]
- 12.Langergraber KE, et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci USA. 2012;109(39):15716–15721. doi: 10.1073/pnas.1211740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam M, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeller AH, et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Commun. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci USA. 2014;111(26):E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bunn HT. Archaeological evidence for meat-eating by Plio-Pleistocene hominids from Koobi Fora and Olduvai Gorge. Nature. 1981;291(5816):574–577. [Google Scholar]
- 19.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song SJ, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moeller AH, et al. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013;23(10):1715–1720. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang JY, et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197(3):435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giongo A, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460(7254):515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone CA, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23(10):1704–1714. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(1) Suppl 1:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.