Significance
The stimulator of IFN genes (STING) is an important innate immune response to infection by herpes simplex virus 1 (HSV-1). STING’s impact may be deduced from the observation that HSV-1 replicates significantly better in normal immortalized cells depleted of STING. Nevertheless, published evidence shows that STING is stabilized by the virus in infected cells, and this report shows that STING is exported from infected cells in exosomes along with viral microRNAs (miRNAs) and mRNAs. Some miRNAs have been reported to suppress reactivation of latent virus. The results suggest that HSV-1 in special circumstances may control the spread of infection from cell to cell and support the hypothesis that HSV-1 controls its virulence to enable effective person-to-person transmission.
Keywords: herpes simplex viruses, exosomes, STING, microRNAs, tetraspanins
Abstract
STING (stimulator of IFN genes) activates the IFN-dependent innate immune response to infection on sensing the presence of DNA in cytosol. The quantity of STING accumulating in cultured cells varies; it is relatively high in some cell lines [e.g., HEp-2, human embryonic lung fibroblasts (HEL), and HeLa] and low in others (e.g., Vero cells). In a preceding publication we reported that STING was stable in four cell lines infected with herpes simplex virus 1 and that it was actively stabilized in at least two cell lines derived from human cancers. In this report we show that STING is exported from HEp-2 cells to Vero cells along with virions, viral mRNAs, microRNAs, and the exosome marker protein CD9. The virions and exosomes copurified. The quantity of STING and CD9 exported from one cell line to another was inoculum-size–dependent and reflected the levels of STING and CD9 accumulating in the cells in which the virus inoculum was made. The export of STING, an innate immune sensor, and of viral mRNAs whose major role may be in silencing viral genes in latently infected neurons, suggests that the virus has evolved mechanisms that curtail rather than foster the spread of infection under certain conditions.
The stimulator of IFN genes (STING) is a sensor of cytoplasmic DNA and activates immune responses to intracellular pathogens (1–3). Knockout of STING exacerbates the pathogenicity of herpes simplex virus (HSV-1) and of other pathogens in mice (2, 3). Two observations reported earlier add to the role of STING in HSV-1 infected cells. Specifically, (i) STING was readily detectable and stable in four different cell lines infected with wild-type virus (4). The stability of STING in cells infected with an HSV-1 mutant lacking the gene encoding ICP0 (infected cell protein no. 0), a key viral regulatory protein, varied. STING was stable in ΔICP0 mutant virus infected cells derived from normal tissues but was rapidly degraded in infected cells derived from human cancers (4). Implicit in this observation is that ICP0 is required to stabilize STING, although STING could also be stabilized in the absence of ICP0 by a cellular function. (ii) Depletion of STING increased wild-type virus yield 10-fold in cells derived from normal tissues but decreased the yield by the same amount in cells derived from human cancer (4). Thus, at least in cells derived from normal tissues, HSV-1 expresses ICP0 that enables the stabilization of STING even though STING is inimical to virus growth. The results of these studies suggest that HSV-1 recruits STING for a specific function, even though the persistence of STING is not beneficial to virus growth, at least in cells derived from normal tissues (4).
Here we report that STING, along with viral RNAs contained in structures that coprecipitate with exosome marker proteins, is exported from the cells in which virus is produced to uninfected cells. The quantities introduced into uninfected cells are dose-dependent and reflect the amounts produced in donor cells.
Cells continuously secrete a large number of microvesicles, macromolecular complexes, and small molecules into the extracellular space (5, 6). Of the secreted microvesicles, exosomes are 30–120 nm in diameter, containing nucleic acid and proteins, and are perceived to be carriers of this cargo between diverse locations. They are distinguished in their genesis by being budded into endosomes to form multivesicular bodies (MVBs) in the cytoplasm. The exosomes are released to extracellular fluids by fusion of these multivesicular bodies with the cell surface, resulting in secretion (7–10).
Several pathogens use the exosomes to manipulate their microenvironment (11, 12). Viruses, especially small retroviruses such as HIV, use the exosome pathway for egress and immune evasion (13–15). The hepatitis C virus uses exosomes for invasion and spread (16–18). EBV-infected cells release exosomes containing the latent membrane protein 1 to induce T-cell anergy (19–23). In the case of human cytomegalovirus the exosomes carrying viral antigens exacerbate the transplant rejection process (24). It is likely that viruses that establish long-term, latent, or chronic infections modulate exosomes to enhance their persistence (11).
Here we report that the exosomes secreted by HSV-1–infected cells deliver to uninfected cells the innate immune sensor STING along with viral RNAs. We speculate that in the long run the strategy serves the virus to fulfill its mission: to spread effectively from person to person.
Results
Accumulation of STING in Vero Cells Infected with HSV-1(F) Produced in HEp-2 Cells.
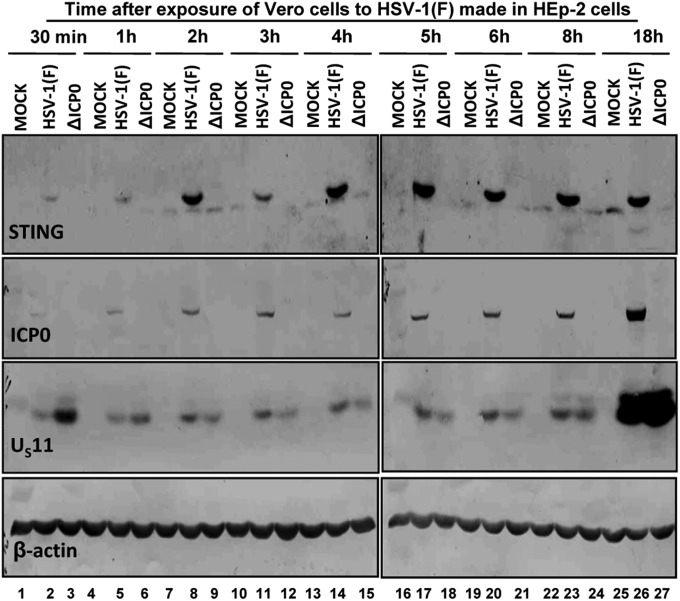
In an earlier report we showed that STING is stable in HEp-2, HeLa, HEL, and HEK-293T cells infected with wild-type HSV-1(F) virus. STING was rapidly degraded in HEp-2 and HeLa cells but not in HEL or HEK-293T cells infected with ΔICP0 mutant virus (4). The studies reported here began with the observation that Vero cells contain little endogenous STING. In contrast, STING accumulated rapidly in cells infected with wild-type virus made in HEp-2 cells but not in cells infected with ΔICP0 mutant virus. In this experiment, mock-infected or cells infected with HSV-1(F) or ΔICP0 were harvested at the times shown and then solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibodies to STING, ICP0, US11, or β-actin. As shown in Fig. 1, lanes 7 through 26, a faster migrating faint band is readily seen in both mock-infected and infected cells. A slower migrating, more intense band representing STING is seen in cells infected with HSV-1(F) but not in mock-infected cells or in cells infected with ΔICP0 mutant. This slower migrating STING was present only in cells infected with HSV-1(F), appearing as a faint band 30 and 60 min after HSV-1(F) infection (Fig. 1, lanes 2 and 5), but increasing in amount and not changing much at later times after infection. Last, the absence of the slow migrating band in ΔICP0-infected cells comes as no surprise. As noted in the introduction, STING is rapidly degraded in HEp-2 cells infected with ΔICP0 mutant viruses (4).
Fig. 1.
Accumulation of STING in Vero cells exposed to the wild-type virus made in HEp-2 cells. Vero cells seeded in 6-well plates were either mock-infected, exposed (10 pfu/cell) to HSV-1(F) virus propagated in HEp-2 cells, or to the ΔICP0 mutant virus propagated in U2OS cells. The cells were harvested at 30 min or at 1, 2, 3, 4, 5, 6, 8, or 18 h after exposure to the viruses. Approximately 60 μg of total proteins per sample were subjected to electrophoresis in a 10% (vol/vol) denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and immunoblotted with the ICP0 rabbit polyclonal antibody or with the STING, US11, and β-actin mouse monoclonal antibodies. The STING protein was visualized with ECL detection reagent, whereas the ICP0, US11, and β-actin proteins were visualized with the BCIP-NBT reagents, as described in the Materials and Methods.
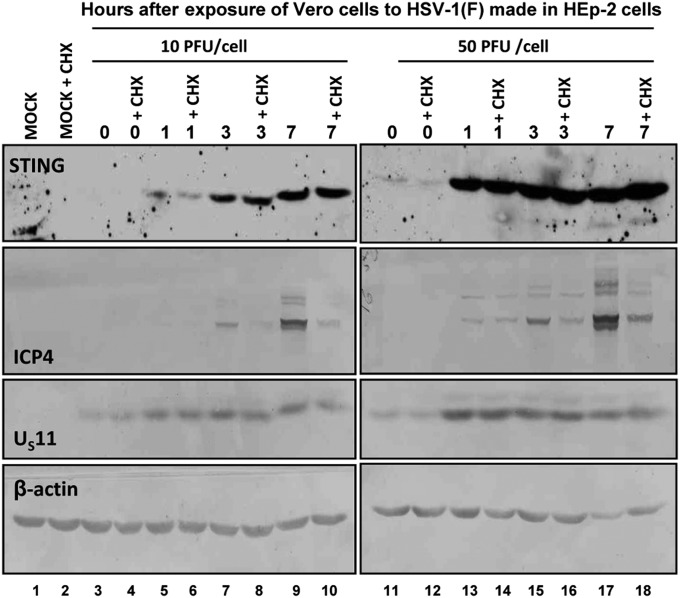
The appearance of a STING protein of slower mobility in Vero cells infected with HSV-1(F) raised questions regarding its origin. To test the hypothesis that STING is brought into Vero cells in the course of infection with HSV-1(F), Vero cells were mock-treated or exposed to cycloheximide (100 μg/mL) at the time of infection (0 h) or at 1, 3, or 7 h after exposure to 10 (Fig. 2, lanes 5–10) or 50 (Fig. 2, lanes 13–18) pfu/cell. The results shown in Fig. 2 indicate the following: (i) The amounts of STING accumulating in infected cells reflected the multiplicity of infection and (ii) cycloheximide had no effect on the accumulation of STING. The results indicate that STING was brought to the infected cells in the course of exposure of the cells to the virus.
Fig. 2.
The accumulation of STING in HSV-1(F)–infected Vero cells is independent of protein synthesis. Vero cells seeded in 6-well plates were either mock-infected or exposed to HSV-1(F) virus propagated in HEp-2 cells at 10/pfu/cell (Left) or at 50 pfu/cell (Right). Cycloheximide (100 μg/mL) was added to the cultures the time of exposure to the virus. The cells were harvested at 0, 1, 3, or 7 h after exposure to the virus. Protein analyses were done as described in Fig. 1.
Amounts of STING Accumulating in Infected Cells Reflect the Cells in Which the Virus Was Produced and the Quantity of STING Present in Recipient Cells.
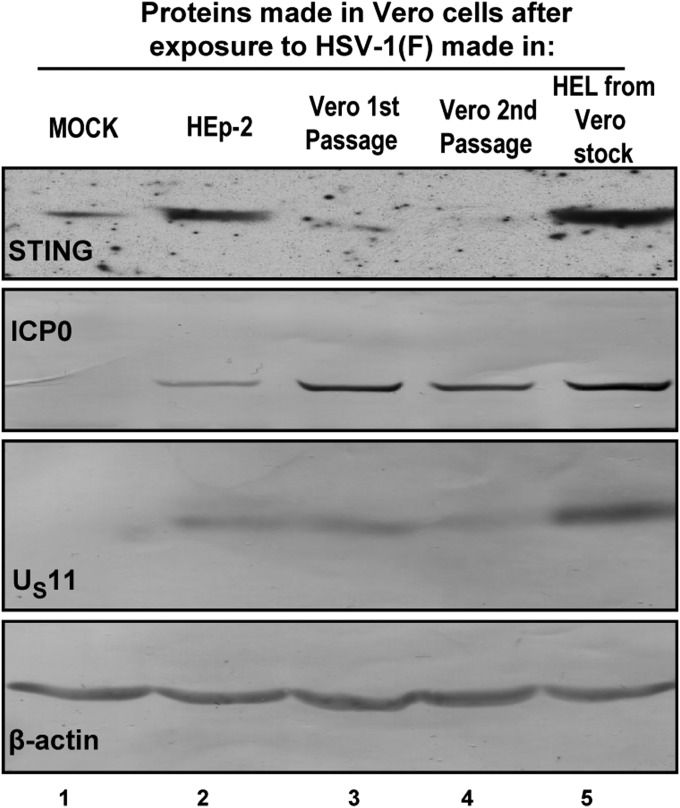
In this series of experiments, HSV-1(F) produced in HEp-2 cells was passaged serially in Vero cells. After two passages, the virus was used to infect HEp-2 cells. In each case the cells were exposed to 10 pfu of virus per cell. The cells were harvested at 6 h after exposure to the virus. The results show that the virus stock produced in HEp-2 cells introduced in Vero cells (Fig. 3, lane 2) relatively large amounts of STING. The stock produced on first and second passage in Vero cells (Fig. 3, lanes 3 and 4) introduced relatively little STING into recipient Vero cells. Finally, Vero cells infected with virus passaged first in Vero and then in HEL cells accumulated relatively large amounts of STING (Fig. 3, lane 5).
Fig. 3.
Accumulation of STING in newly infected Vero cells reflects the abundance of the protein in which the virus inoculum was made. Vero cells were either mock-infected (lane 1) or exposed to (lane 2) HSV-1(F) (10 pfu/cell) virus stock made in HEp-2 cells or to (lanes 3 and 4) virus that was propagated in HEp-2 followed by passages in Vero, or to (lane 5) virus that was propagated in Vero followed by passage in HEL cells. The cells were harvested at 6 h after exposure to the virus, and equal amounts of total proteins were analyzed as described in Fig. 1.
In Infected Cells the Patterns of Accumulation of CD9, an Exosome Marker, Reflect Those of STING.
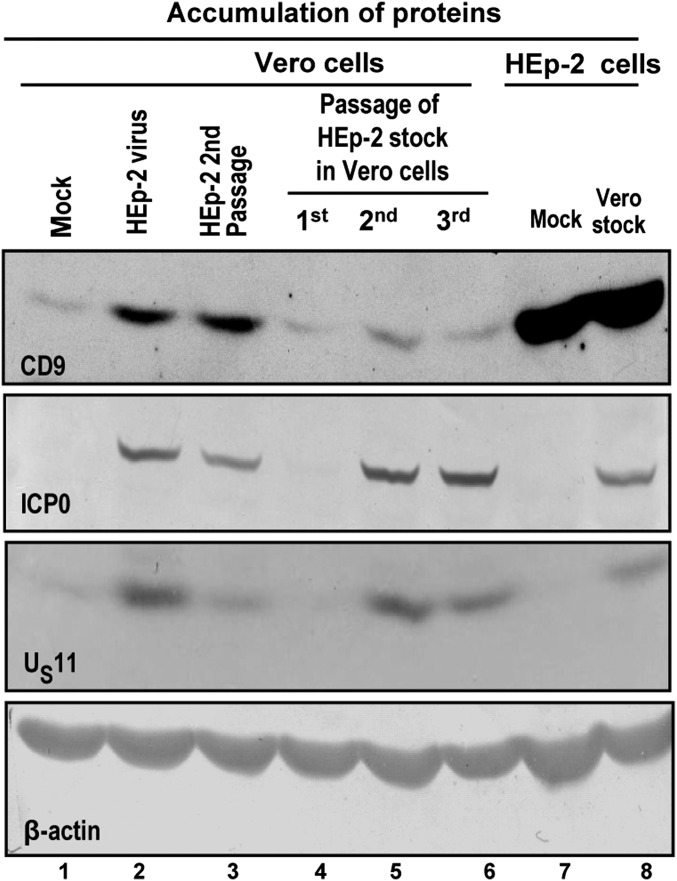
One hypothesis that could explain the data is that infected cells secrete exosomes that carry STING. Several experiments were done to test this hypothesis. In the first experiment, illustrated in Fig. 4, we examined the accumulation of tetraspanin CD9, an abundant membrane protein present in exosomes and exosome-like structures (9). Specifically, virus made in HEp-2 cells introduced relatively large amounts of CD9 into Vero cells (compare Fig. 4, lanes 1 and 2); virus passaged in HEp-2 cells again introduced large amounts of CD9 (Fig. 4, lane 3). On serial passage in Vero cells the amounts of CD9 decreased to the levels seen in mock-infected Vero cells (compare Fig. 4, lane 1 with Fig. 4, lanes 3, 4, 5, and 6). Last, the stock made in Vero cells was used to infect HEp-2 cells. The amounts of CD9 accumulating in HEp-2 cells were similar to those present in mock-infected HEp-2 cells (Fig. 4, lanes 7 and 8).
Fig. 4.
CD9 exosomal marker protein is carried with the virus inoculum to target cells. Lysates of mock-infected or infected cells (10 pfu/cell) were analyzed for the presence of ICP0, US11, and CD9 protein. In each case, β-actin served as a loading control. (lane 1) Mock-infected Vero cells; (lane 2) Vero cells infected with stock made in HEp-2 cells; (lane 3) Vero cells infected with virus stock serially passaged in HEp-2 cells; (lane 4) Vero cells infected with virus stock made in Vero cells (first Vero cell passage); (lanes 5 and 6) Vero cells infected with virus serially passaged two and three times in Vero cells; (lane 7) mock-infected HEp-2 cells; and (lane 8) HEp-2 cells infected with virus stock made in Vero cells. The CD9 protein was visualized with ECL detection reagent, whereas the ICP0, US11, and β-actin proteins were visualized with the BCIP-NBT reagents, as described in the main text.
We conclude that STING and CD9 accumulate in tandem and reflect the cells in which the virus inoculum was made. On serial passage of virus the amounts of STING and CD9 reflect the amounts present in mock-infected cells. In essence, HSV-1 enables the export but does not induce accumulation of STING or CD9 in infected cells.
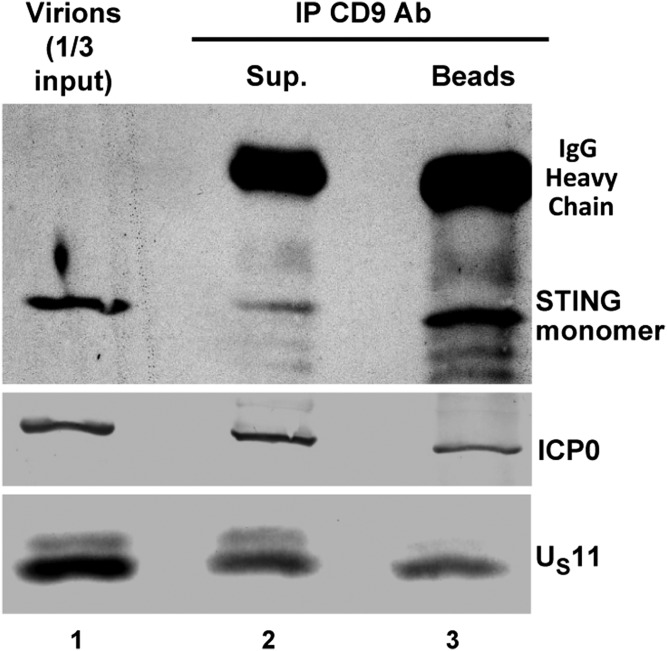
STING Coprecipitates with CD9 from HSV-1(F) Stock Purified by Centrifugation in Dextran 10 Gradients.
In this series of experiments we sought to determine the source of STING in the virus inoculum. The objective of the experiments described here was to discriminate between two hypotheses, i.e., whether STING was present in high-order structures that copurify with herpes virions or whether it was incorporated into virions. To differentiate between these hypotheses, Dextran 10 gradient-purified materials were reacted with antibody to CD9. The supernatant fluids and the precipitates were analyzed for the presence STING, ICP0, and US11 proteins. As shown in Fig. 5, the precipitates obtained with the anti-CD9 antibody were enriched with STING. The precipitates also contain ICP0 and US11 proteins, consistent with the hypothesis that exosomes could be expected to contain virion proteins present in multivesicular bodies in which they are made.
Fig. 5.
Immunoprecipitation of STING with CD9 antibody from Dextran 10 gradient-purified preparations. Dextran 10 gradient-purified fraction containing virions was diluted in 300 μL PBS and reacted with the CD9 rabbit polyclonal antibody. The immunoprecipitated proteins and the proteins in the supernatant fluid were separated in an 11% denaturing polyacrylamide gel and transferred to a nitrocellulose sheet, and the proteins were visualized as in Fig. 1.
We conclude that STING copurifies with HSV virions in separate structures, which are rich in CD9, a well-known exosome marker (9).
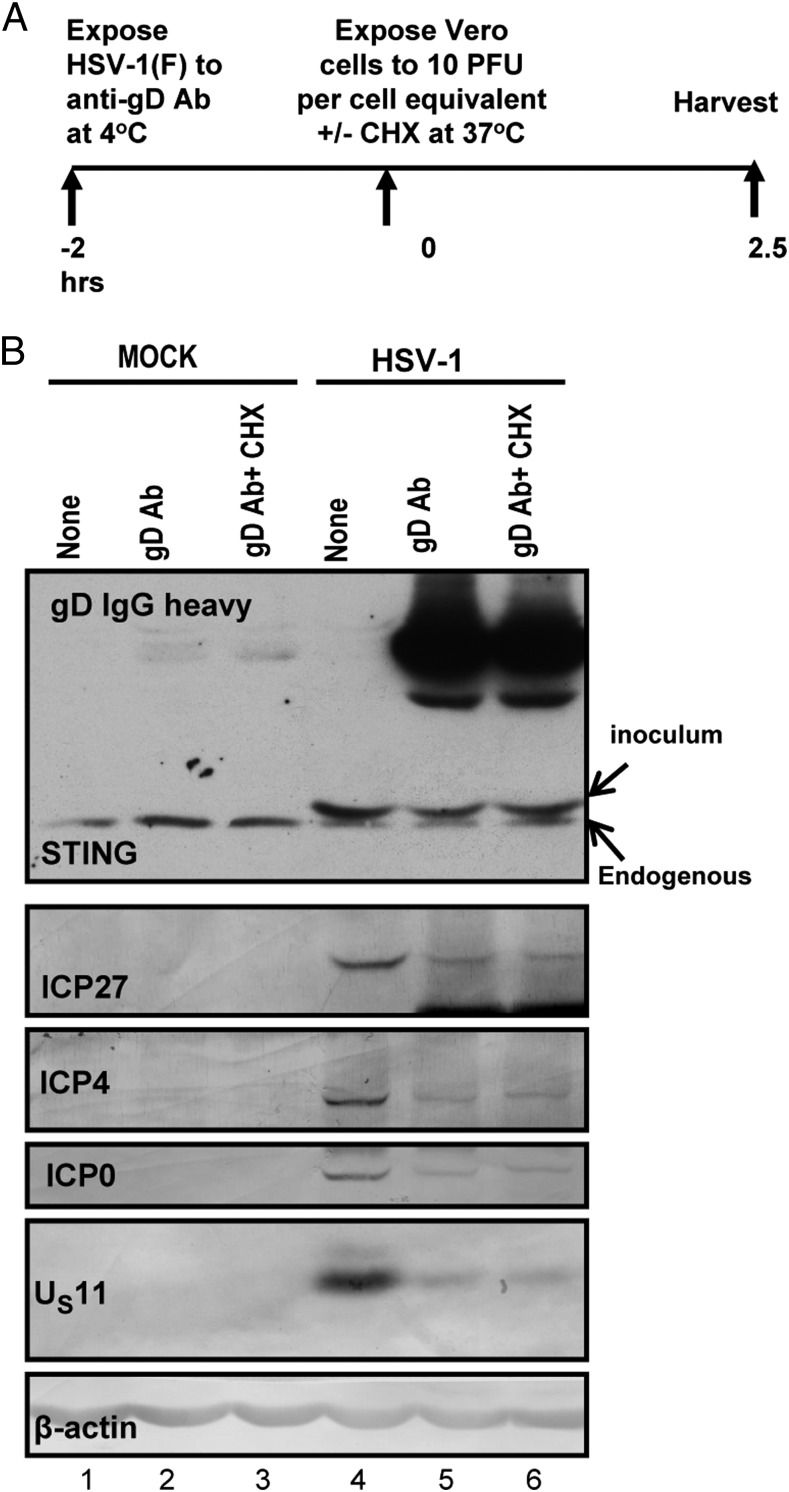
Neutralization of Virus in Extracellular Medium with Anti-Glycoprotein D Antibody Reduced the Expression of Viral Gene Products but Has Little or No Effect on Accumulation of STING.
In this series of experiments, HSV-1(F) made in HEp-2 cells was mock-treated or reacted for 2 h at 4 °C with anti-glycoprotein D (gD) antibody. HEp-2 cells were then exposed for 2.5 h alone or in the presence of cycloheximide (100 μg/mL) to 10 pfu of mock-treated or 10 pfu equivalent of neutralized virus per cell. At the end of the exposure interval the cells were harvested, solubilized, electrophoretically separated in denaturing gels, and reacted with antibody to STING, ICP27, ICP4, ICP0, US11, and β-actin. The results shown in Fig. 6 were as follows:
Fig. 6.
The neutralizing gD antibody blocks virus entry but does not affect STING entry. (A) HSV-1(F) made in HEp-2 cells was mock-treated or reacted with neutralizing anti-gD antibody for 2 h at 4 °C. Vero cells grown in 6-well plates were mock-infected or exposed to 10 pfu of virus (untreated) or the equivalent amount of neutralized virus per cell in the presence or absence of cycloheximide (100 μg/mL). The cells were harvested at 2.5 h after exposure to the virus. (B) Equal amounts of proteins were electrophoretically separated in an 11% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, and immunoblotted with the STING, ICP27, ICP4, ICP0, US11, and β-actin antibodies. The procedures were as described in Fig. 1 and in Materials and Methods.
STING formed a single band in all electrophoretically separated lysates of mock-infected cells (Fig. 6, lanes 1–3). In lysates of infected cells (Fig. 6, lanes 4–6), STING formed two bands, a fast migrating band corresponding to that seen in uninfected Vero cells and a slow migrating form corresponding in electrophoretic mobility to that seen in lysates of Vero cells shown in Fig. 1. The intensities of the STING bands in mock-treated infected cells or cells infected with neutralized virus in the presence or absence of cycloheximide were similar.
Viral gene products, i.e., ICP27, ICP4, ICP0, and US11, were present in lysates derived from cells infected with mock-treated virus (Fig. 6, lane 4). They were barely detected in cells exposed to neutralized virus alone (lane 5) or in the presence of cycloheximide (Fig. 6, lane 6).
We conclude that STING was contained in structures whose entry into cells is less amenable to neutralization by anti-gD antibodies than that of virions.
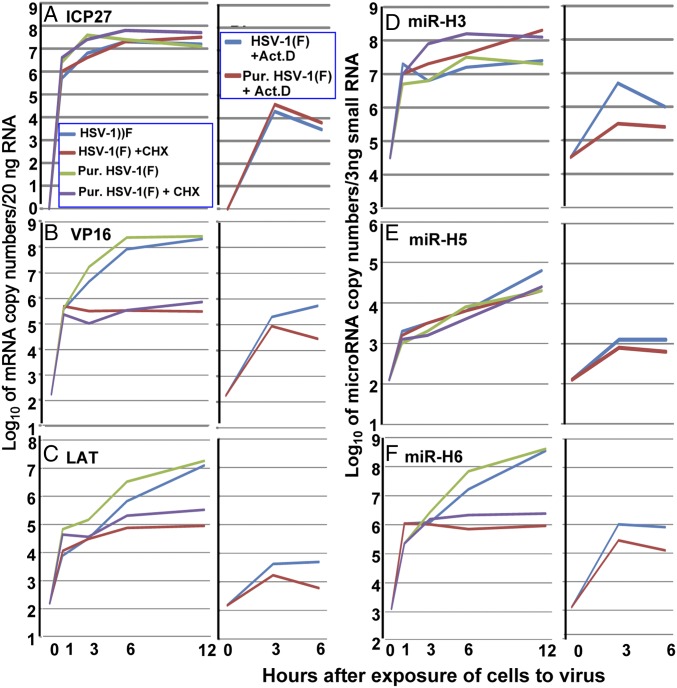
Exosomes Carry Viral microRNAs and mRNAs.
In this series of experiments, Vero cells were exposed to 10 pfu/cell of Dextran 10 gradient-purified virus (25) or standard inoculum prepared as described elsewhere (26). The cultures were mock-treated or exposed to actinomycin D (10 μg/mL) or cycloheximide (100 μg/mL). Samples were taken at the time of infection (0 h) and at 1, 3, 6, or 12 h after exposure of cells to the virus. The samples were analyzed for ICP27 and virion protein 16 (VP16) mRNAs and for latency-associated transcript (LAT), miR-H3, miR-H5, or miR-H6. The copy numbers were analyzed based on duplicates.
The results were as follows:
-
i)
The amounts accumulating in the presence of actinomycin D (Fig. 7) represent the minimal amounts of mRNAs, LAT, and microRNAs (miRNAs) that entered and accumulated in infected cells.
-
ii)
ICP27 and VP16 are α and γ genes, respectively (27). It would be expected that ICP27 mRNA would accumulate in the presence of cycloheximide, whereas VP16 mRNA would not. We did not detect a significant difference in the accumulation of ICP27 mRNA in the presence or absence of cycloheximide (Fig. 7A). In contrast, VP16 mRNA accumulated to at least 100-fold higher levels in the absence of cycloheximide.
-
iii)
LAT was reported to accumulate late in infection (27). The results shown in Fig. 7C indicate that optimal accumulation of LAT required prior protein synthesis and, moreover, unlike that of VP16 mRNA, the accumulation of LAT did not level off between 6 and 12 h after infection.
-
iv)
The accumulation of miR-H6 required prior protein synthesis. Moreover, the accumulation of this miRNA did not level off between 6 and 12 h after infection, suggesting that the kinetics of its synthesis are that of a late (γ) gene, in contrast to the accumulation of miR-H5. The infected cells did not show a substantive increase in miR-H3 between 1 and 12 h after infection either in the presence or absence of cycloheximide.
-
v)
Last, both the crude and Dextran 10 gradient-purified virus were made in HEp-2 cells as previously described (25, 26). The studies presented here suggest that the relative ratios of mRNAs, and miRNAs in crude virus inoculum and in the Dextran 10 gradient-purified virus, were not substantially different.
Fig. 7.
Delivery of viral mRNAs and miRNAs by crude virus preparations or Dextran 10 gradient-purified virus to Vero cells. Vero cells were exposed to 10 pfu of virus per cell alone or in the presence of cycloheximide (100 µg/mL) or actinomycin D (10 µg/mL). At indicated times after exposure to the virus the cells were harvested and analyzed for the relative amounts of ICP27 mRNA (A), VP16 mRNA (B), LAT mRNA (C), mir-H3 (D), mir-H5 (E), and mir-H6 (F). Viral mRNAs and LAT levels were normalized to 50 ng cellular RNA, which is determined by the 18S ribosomal RNA internal control, and viral miRNAs were normalized to 3 ng of small RNAs quantified by cellular miRNA lethal-7a.
Discussion
HSV-1 encodes at least 84 proteins and numerous long noncoding RNAs, stable introns, and miRNAs. Most of the proteins examined in detail to date perform multiple functions. In essence, HSV-1 encodes several hundred functions directed to one objective: to replicate and disseminate (27). Most of the functions encoded by the virus focus on suppressing innate and adaptive immune responses to infection (27). In many instances, multiple viral functions target a single innate immune response. In a colloquial sense, HSV is a control freak. In an earlier report we showed that depletion of STING in normal immortalized cells increases virus yield 10-fold (4). Hence the evidence that HSV-1 stabilizes STING in a selected number of cell lines raises the question as to how HSV-1 could possibly use this sensor of cytosolic DNA. This report adds a hitherto unrecognized dimension to the problem. In essence the studies reported here demonstrate the following:
-
i)
Vero cells contain demonstrably less STING or the exosomal marker CD9 protein than HEp-2 cells. Vero cells infected with virus stock made in HEp-2 cells acquire STING in amounts proportional to the size of the virus inoculum. Virus stocks made in Vero cells contain significantly less STING or CD9 protein than stocks made in HEp-2 or HEL cells. One hypothesis that could explain the tandem distribution of CD9 and STING is that STING is contained in exosomes.
-
ii)
Standard methods of virus purification using Dextran 10 gradients (25) did not separate STING from virions. However, immune precipitates obtained with antibody to CD9 contained STING. Similarly, exposure of virus inoculum to neutralizing anti-gD antibody had no appreciable effect on delivery of STING to recipient cells but diminished the amount of infectious virus. These results suggest that STING is delivered in exosomes.
-
iii)
Last, exosomes are known to contain RNA (5, 6, 8). Both crude and purified stocks of virus contain viral miRNAs as well as viral mRNAs. The probes used in these studies were the same as those used to detect viral mRNAs and miRNAs in ganglia harboring latent or reactivating virus (28). It is of interest to note that the data we collected indicate that the requirements and kinetics of accumulation of LAT and miRNAs are not synchronous or identical. Thus, the accumulation of LAT and miR-H6 requires prior protein synthesis and does not level off between 6 and 12 h after infection. The synthesis and accumulation of miR-H5 does not require protein synthesis but neither does it level off between 1 and 12 h after infection. The kinetics of synthesis or miR-H3 remains a puzzle. Although it was readily detected in ganglia harboring latent virus, we have no unambiguous evidence of its synthesis in Vero cells during the first 12 h of infection.
As defined by its name, STING is an activator of IFN pathway (3). At least some of the viral miRNAs target viral mRNAs (29). A central question posed by these studies is why HSV-1 would stabilize STING and export in exosomes both STING and miRNAs because the constellation of these macromolecules could impede rather than facilitate virus spread. The biology of virus infection in humans suggests one situation in which HSV would evolve a function that would limit rather than facilitate spread: In brief, following initial infection at a portal of entry into the body, HSV-1 is transported retrograde to a sensory neuron in which it establishes a latent—silent—infection. Periodically, the virus reactivates in some neurons. The reactivated virus is then transported anterograde to a site at or near the portal of entry into the body. At this site the virus is available for transmission by physical contact to another person (27). Lateral dissemination of virus from the neuron in which the virus reactivated to satellite cells and dissemination to CNS would reduce the likelihood of transmission of virus from person to person—the primary mission of HSV-1—because individuals with CNS impairments are less likely to transmit the virus (27). Sensory neurons are coated by and are intimately involved with surrounding satellite cells via vesicular traffic. Export of STING, miRNAs, and other macromolecules could significantly impact the lateral spread of infection.
The hypothesis we are presenting is that uncontrolled replication increases virulence and diminishes person-to-person spread. HSV controls its replication. Consistent with this hypothesis is the finding reported earlier that the virulence of HSV-1 defined by virus spread and lethal infections in the mouse could be significantly increased by the insertion of a dominant-negative RE1-silencing transcription factor (REST) into the viral genome. The dominant-negative REST competed with wild-type REST to prevent the repression of viral gene expression and the establishment of latent state (30). The same effect could have been generated by mutations in REST response elements (31). In the same vein, reactivation of latent virus in neurons results from simultaneous expression of all viral genes—a process likely to significantly decrease virus yields (28). The findings reported here and earlier suggest that HSV controls its ability to replicate and spread to achieve a high rate of transmission of virus from person to person.
Last, our laboratory reported earlier on the presence of viral and cellular mRNAs in virions (32). Because it is now apparent that purified preparations of virions contain exosomes and that precipitates obtained with antibody to exosomal protein contain viral mRNAs, it is likely that the mRNAs we reported were present in exosomes rather than in virions. It should also be noted that virions have been reported to contain ICP0 and even β-actin (33–38). The question remains whether these proteins are actually packaged in virions or are present in contaminating exosomes.
Materials and Methods
Cells and Viruses.
The sources and cultivation of HEp-2 and Vero and the Telomerase-transformed human embryonic lung fibroblasts were reported elsewhere (4). HSV-1(F), a limited passage isolate, is the prototype strain used in our laboratory (39). The properties of R7910, an ICP0 null mutant virus, were reported elsewhere (40).
Purification of Virions by Dextran 10 Gradient Centrifugation and Immunoprecipitation.
The procedures were done as described elsewhere (25) with minor modifications. Briefly, HEp-2 cells (6 × 108) grown in roller bottles were infected with HSV-1(F), at 10 pfu/cell. The cells were harvested at 30 h after infection and centrifuged at 800 × g for 10 min in an Allegra 6R centrifuge equipped with a GH-3.8 rotor (Beckman Coulter, Inc.). The supernatant fluids and the infected cell pellets were collected and processed separately. Virus particles in the supernatant fluids were recovered by centrifugation at 70,000 × g for 1 h at 4 °C in an Optima LE-80K ultracentrifuge equipped with an SW28 rotor (Beckman Coulter, Inc.). The cell pellet was resuspended in 1 mM phosphate buffer and disrupted in a glass homogenizer. Extracellular and cytoplasmic fractions were layered on a 14-mL Dextran 10 gradient (1.04–1.09 g/cm3) in 1 mM phosphate buffer. The gradient was centrifuged for 1 h at 68,000 × g at 4 °C in an Optima LE-80K ultracentrifuge equipped with an SW41 rotor. After centrifugation, the virion-containing band was collected and diluted in 10 mM phosphate buffer. Purified virions were concentrated by pelleting at 75,000 × g for 2 h, as described above. The pellet was resuspended in 300 μL of 10 mM phosphate buffer and stored at −80 °C before processing.
For the immunoprecipitation assay, virions were diluted in 300 μL PBS (pH: 8), and 1 μg CD9 rabbit polyclonal antibody (Abcam) was added. The samples were incubated overnight at 4 °C on a rotating platform, and protein A beads were added for 2 h. Finally, the proteins in the supernatant fluid and those bound to beads were analyzed by immunoblotting, as described below.
RNA Isolation and Assays.
Vero cells were exposed to 10 pfu of Dextran 10 gradient-purified virus or to standard laboratory virus inoculum prepared as described elsewhere (25, 26). Infected cells were collected at 0, 1, 3, 6, and 12 h following exposure to the inoculum. Cycloheximide (100 μg/mL) or actinomycin-D (10 μg/mL) were added to the cultures at the same moment as the inoculum. RNAs depleted and enriched of small RNAs (<200 nt) were extracted by mirVana miRNA isolation kit (Ambion), according to manufacturer’s instructions. Quantifications of the LAT, VP16, and ICP27 transcripts and of the HSV-1(F) miRNAs (miR-H3, miR-H5, and miR-H6) were done as previously described (28, 41, 42).
Immunoblot Analyses.
The procedures have been described elsewhere (43). The cells were solubilized in lysis buffer and briefly sonicated (43). The protein concentration in total cell lysates was determined with the aid of Bio-Rad protein assay (Bio-Rad Laboratories). Approximately 60 μg of proteins per sample were separated in denaturing polyacrylamide gels, transferred to a nitrocellulose sheet, and reacted overnight at 4 °C with the appropriate primary antibodies, as before. The rabbit polyclonal antibody to ICP0 (40) and the mouse monoclonal antibodies to ICP4, ICP27, and US11 (Goodwin Institute for Cancer Research) were used at a 1:1,000 dilution. The mouse monoclonal antibodies to STING (R&D Systems), to β-actin (Sigma), or the rabbit polyclonal antibody to CD9 (Abcam) were used in a 1:800 dilution. Finally, protein bands were visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitrobluetetrazolium (NBT) (Denville Scientific, Inc.) or with ECL Western blotting detection reagents (Amersham Biosciences).
Neutralization Assay.
HSV-1(F) virus made in HEp-2 cells was reacted with neutralizing anti-gD mouse monoclonal antibody (Goodwin Institute for Cancer Research) for 2 h at 4 °C. Next, Vero cells grown in 6-well plates were exposed (10 pfu/cell) to mock-treated or equivalent amounts of neutralized virus in the presence or absence of cycloheximide (100 μg/mL). The cells were harvested at 2.5 h after exposure to the virus and analyzed as described above.
Acknowledgments
These studies were aided by a grant from the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14(1):19–26. doi: 10.1038/ni.2491. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa H, Barber GN. The STING pathway and regulation of innate immune signaling in response to DNA pathogens. Cell Mol Life Sci. 2011;68(7):1157–1165. doi: 10.1007/s00018-010-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalamvoki M, Roizman B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci USA. 2014;111(5):E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosaka N, et al. Trash or treasure: Extracellular microRNAs and cell-to-cell communication. Front Genet. 2013;4:173. doi: 10.3389/fgene.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bang C, Thum T. Exosomes: New players in cell-cell communication. Int J Biochem Cell Biol. 2012;44(11):2060–2064. doi: 10.1016/j.biocel.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int J Biochem Cell Biol. 2012;44(1):11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3(5):321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 10.van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: A common pathway for a specialized function. J Biochem. 2006;140(1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 11.Fleming A, et al. The carrying pigeons of the cell: Exosomes and their role in infectious diseases caused by human pathogens. Pathog Dis. 2014;71(2):109–120. doi: 10.1111/2049-632X.12135. [DOI] [PubMed] [Google Scholar]
- 12.Hwang I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol Cells. 2013;36(2):105–111. doi: 10.1007/s10059-013-0154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izquierdo-Useros N, et al. HIV and mature dendritic cells: Trojan exosomes riding the Trojan horse? PLoS Pathog. 2010;6(3):e1000740. doi: 10.1371/journal.ppat.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Izquierdo-Useros N, Puertas MC, Borràs FE, Blanco J, Martinez-Picado J. Exosomes and retroviruses: The chicken or the egg? Cell Microbiol. 2011;13(1):10–17. doi: 10.1111/j.1462-5822.2010.01542.x. [DOI] [PubMed] [Google Scholar]
- 15.Pelchen-Matthews A, Raposo G, Marsh M. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 2004;12(7):310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Cosset FL, Dreux M. HCV transmission by hepatic exosomes establishes a productive infection. J Hepatol. 2014;60(3):674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Greenhill C. Hepatitis: New route of HCV transmission. Nat Rev Gastroenterol Hepatol. 2013;10(9):504. doi: 10.1038/nrgastro.2013.148. [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110(32):13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middeldorp JM, Pegtel DM. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin Cancer Biol. 2008;18(6):388–396. doi: 10.1016/j.semcancer.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Canitano A, Venturi G, Borghi M, Ammendolia MG, Fais S. Exosomes released in vitro from Epstein-Barr virus (EBV)-infected cells contain EBV-encoded latent phase mRNAs. Cancer Lett. 2013;337(2):193–199. doi: 10.1016/j.canlet.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Keryer-Bibens C, Pioche-Durieu C, Villemant C, Souquere S, Nishi N, Hirashima M, et al. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer. 2006;6:283. doi: 10.1186/1471-2407-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meckes DG, Jr, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107(47):20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij FJ, et al. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 2011;30(11):2115–2129. doi: 10.1038/emboj.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker JD, Maier CL, Pober JS. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J Immunol. 2009;182(3):1548–1559. doi: 10.4049/jimmunol.182.3.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spear PG, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpes virion. J Virol. 1972;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roizman B, Spear PG. Preparation of herpes simplex virus of high titer. J Virol. 1968;2(1):83–84. doi: 10.1128/jvi.2.1.83-84.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, et al., editors. Fields' Virology. 6th Ed. Wolters Kluwer/Lippincott–Williams and Wilkins; New York: 2013. pp. 1823–1897. [Google Scholar]
- 28.Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci USA. 2011;108(46):18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores O, et al. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol. 2013;87(12):6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du T, Zhou G, Khan S, Gu H, Roizman B. Disruption of HDAC/CoREST/REST repressor by dnREST reduces genome silencing and increases virulence of herpes simplex virus. Proc Natl Acad Sci USA. 2010;107(36):15904–15909. doi: 10.1073/pnas.1010741107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou G, Du T, Roizman B. HSV carrying WT REST establishes latency but reactivates only if the synthesis of REST is suppressed. Proc Natl Acad Sci USA. 2013;110(6):E498–E506. doi: 10.1073/pnas.1222497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sciortino MT, Suzuki M, Taddeo B, Roizman B. RNAs extracted from herpes simplex virus 1 virions: Apparent selectivity of viral but not cellular RNAs packaged in virions. J Virol. 2001;75(17):8105–8116. doi: 10.1128/JVI.75.17.8105-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Rio T, DeCoste CJ, Enquist LW. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J Virol. 2005;79(13):8614–8619. doi: 10.1128/JVI.79.13.8614-8619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maringer K, Elliott G. Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle. J Virol. 2010;84(9):4682–4696. doi: 10.1128/JVI.00126-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedlackova L, Rice SA. Herpes simplex virus type 1 immediate-early protein ICP27 is required for efficient incorporation of ICP0 and ICP4 into virions. J Virol. 2008;82(1):268–277. doi: 10.1128/JVI.01588-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao F, Courtney RJ. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66(5):2709–2716. doi: 10.1128/jvi.66.5.2709-2716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loret S, Lippé R. Biochemical analysis of infected cell polypeptide (ICP)0, ICP4, UL7 and UL23 incorporated into extracellular herpes simplex virus type 1 virions. J Gen Virol. 2012;93(Pt 3):624–634. doi: 10.1099/vir.0.039776-0. [DOI] [PubMed] [Google Scholar]
- 38.Stegen C, et al. Analysis of virion-incorporated host proteins required for herpes simplex virus type 1 infection through a RNA interference screen. PLoS ONE. 2013;8(1):e53276. doi: 10.1371/journal.pone.0053276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 40.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71(10):7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci USA. 2010;107(41):17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]