Significance
The protooncogene c-Myc (Myc) is an oncogenic driver in many cancers, but is difficult to target directly with drugs. An alternative strategy is to use drugs that inhibit factors that regulate Myc expression. Notch drives Myc expression in most T-cell leukemias, but clinical trials of Notch inhibitors have been disappointing, possibly because cells emerge that express Myc in a Notch-independent fashion. Here we identify the genomic switches that regulate Myc expression in the Notch-inhibitor–sensitive and –resistant states. Our findings suggest that Notch inhibitor resistance occurs through a “switch swap” that relieves Notch dependency while increasing dependency on a different factor, bromodomain containing 4 (Brd4). These studies provide a rationale for targeting Myc in T cell leukemias with combinations of Notch and Brd4 inhibitors.
Keywords: enhancer, gene regulation, chromatin, transcription, Brd4
Abstract
Notch is needed for T-cell development and is a common oncogenic driver in T-cell acute lymphoblastic leukemia. The protooncogene c-Myc (Myc) is a critical target of Notch in normal and malignant pre-T cells, but how Notch regulates Myc is unknown. Here, we identify a distal enhancer located >1 Mb 3′ of human and murine Myc that binds Notch transcription complexes and physically interacts with the Myc proximal promoter. The Notch1 binding element in this region activates reporter genes in a Notch-dependent, cell-context–specific fashion that requires a conserved Notch complex binding site. Acute changes in Notch activation produce rapid changes in H3K27 acetylation across the entire enhancer (a region spanning >600 kb) that correlate with Myc expression. This broad Notch-influenced region comprises an enhancer region containing multiple domains, recognizable as discrete H3K27 acetylation peaks. Leukemia cells selected for resistance to Notch inhibitors express Myc despite epigenetic silencing of enhancer domains near the Notch transcription complex binding sites. Notch-independent expression of Myc in resistant cells is highly sensitive to inhibitors of bromodomain containing 4 (Brd4), a change in drug sensitivity that is accompanied by preferential association of the Myc promoter with more 3′ enhancer domains that are strongly dependent on Brd4 for function. These findings indicate that altered long-range enhancer activity can mediate resistance to targeted therapies and provide a mechanistic rationale for combined targeting of Notch and Brd4 in leukemia.
Gain-of-function Notch1 mutations occur in >50% of human T-cell acute lymphoblastic leukemia (T-ALL) and are also frequent in murine T-ALL (1). Physiologic Notch signaling occurs when a Notch ligand on one cell engages a Notch receptor on the adjacent cell, triggering changes in the extracellular juxtamembrane region of Notch that make it susceptible to cleavage by a member of the ADAM (a disintegrin and metalloprotease domain) metalloprotease family (2). This event generates a short-lived truncated form of Notch that is proteolyzed within its transmembrane region by gamma secretase, liberating the Notch intracellular domain (NICD) from the membrane. NICD then translocates to the nucleus and forms a Notch transcription complex (NTC) with the DNA-binding protein RBPJ (recombination signal binding protein for immunoglobulin kappa J) and a Mastermind-like (MAML) factor. MAML recruits p300 and other transcriptional coactivators, leading to transcription of Notch target genes. MAML also recruits proteins leading to NICD degradation (3). The activating Notch1 mutations in T-ALL lead to either ligand-independent ADAM metalloprotease cleavage and/or diminished NICD degradation (4).
Recent studies have begun to define the function of Notch in T-ALL transcriptional regulation at the genomic level. Approximately 90% of Notch/RBPJ binding sites that mediate acute changes in gene expression are found in “super-enhancers” (5), large distal regulatory switches defined by a high content of Brd4, Med1, and H3K27ac (6, 7), a histone mark associated with active chromatin and transcription that is placed by histone acetyltransferases such as p300 (8).
The protooncogene c-Myc (Myc) is a particularly important direct target of Notch in normal and malignant T cells. Notch-dependent expression of Myc is required for normal T cells to traverse early developmental checkpoints (9, 10) and for T-ALL cells to grow and survive (11–13). Moreover, retroviral expression of Myc is sufficient to rescue some Notch-addicted T-ALL cell lines from the deleterious effects of Notch inhibition (14). Initial studies showed that NTCs bound to sites within the murine Myc proximal promoter (11, 13, 15), but subsequent studies showed that murine Myc transcription required NTC dimerization (9), which is not supported by the proximal Myc promoter RBPJ binding sites.
Dimeric NTCs form on sequence-paired sites (SPSs) (16), a conserved response element consisting of two head-to-head RBPJ sites separated by a spacer of 15–17 bp. Dimerization of NTCs requires cofactors of the MAML family, which stabilize the association of the NICD ankyrin repeat domain (ANK) and RBPJ, as well as several intermolecular contacts between adjacent pairs of ANK repeats in the NTC dimer. One functionally important inter-ANK contact involves the residue R1984 (formerly denoted as R1985) (17) because the Notch1 point substitution R1984A prevents NTC dimerization on SPSs, but does not affect NTC loading on monomeric RBPJ sites. In the mouse, the R1984A mutation impairs the ability of NICD1 to stimulate Myc expression and to induce T-ALL and T-cell development (9), pointing to the existence of at least one SPS near murine Myc that is critical for NTC-dependent Myc transcription.
We now describe studies in which whole-genome approaches were used to determine how Notch regulates Myc in both human and murine T-ALL cells. In both species, the critical Notch-dependent Myc regulatory sites are found within a large enhancer region located >1 Mb 3′ of the Myc transcriptional start site (TSS) containing several discrete functional domains. The major Notch complex binding sites show differential requirements for NTC dimerization in murine and human cells, an unexpected point of divergence between murine and human T-ALL. Notably, our studies indicate that the emergence of resistance to Notch inhibitors in T-ALL cells involves altered activity and use of Myc-associated distal enhancer domains.
Results
Identification of a Notch-Dependent Distal Myc Enhancer in Murine T-ALL and Primary Pre-T Cells.
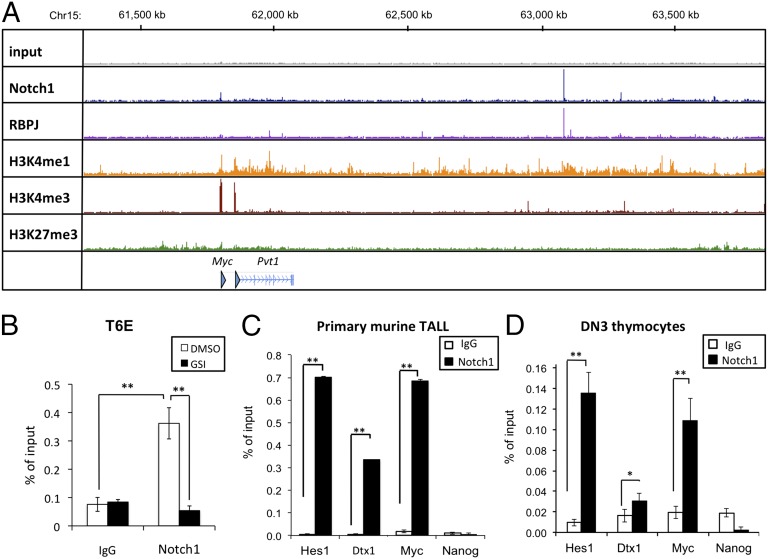
To identify dimeric NTC sites near Myc, we performed ChIP sequencing (ChIP-Seq) for Notch1 and RBPJ in the murine Notch-dependent T-ALL cell line T6E (11). Analysis of the ChIP-Seq data revealed a single prominent RBPJ/Notch1 binding site [henceforth referred to as the Notch-dependent Myc enhancer element (NDME)] located 1.27 Mb 3′ of the Myc promoter within a region with high levels of H3K4Me1, a histone mark associated with enhancers (Fig. 1A). Notch1 is depleted from the NDME site by short-term gamma-secretase inhibitor (GSI) treatment (Fig. 1B), and loading and unloading of Notch1 is associated with rapid changes in H3K27ac (Fig. S1A), features that characterize genomic Notch1 binding sites that dynamically regulate gene expression (5). Notch1 also bound the NDME in primary murine T-ALL cells (Fig. 1C) and in DN3 thymocytes (Fig. 1D), a stage of T-cell development marked by high levels of Notch1 activation (11). RBPJ/Notch1 binding was also observed at the previously identified Myc proximal promoter site (Fig. 1A), but these signals were substantially weaker than those seen at the NDME.
Fig. 1.
Identification of a site ∼1.27 Mb 3′ of the Myc gene body that binds RBPJ and Notch1 in normal and transformed murine pre-T cells. (A) ChIP-Seq read counts for Notch1, RBPJ, and H3K4me1, H3K4me3, and H3K27me3 marks in a ∼2-Mb region containing Myc. y axis, aligned reads; arrowhead, TSSs. (B) Local ChIP for Notch1 at the 3′ Myc site (NDME) in the murine T-ALL cell line T6E treated with GSI or DMSO for 4 h before harvest. Nonspecific Ig was used as a control. (C and D) Local ChIP analyses of Notch1 occupancy of the binding site 3′ of Myc (NDME) in primary murine T-ALL cells (C) and DN3 thymocytes (D). Previously characterized Notch1 binding sites in the Hes1 promoter and Dtx1 intron2 serve as positive controls; Nanog serves as a negative control. In B–D, data were obtained in triplicate in independent experiments; error bars correspond to the SEM. *P < 0.01; **P < 0.001.
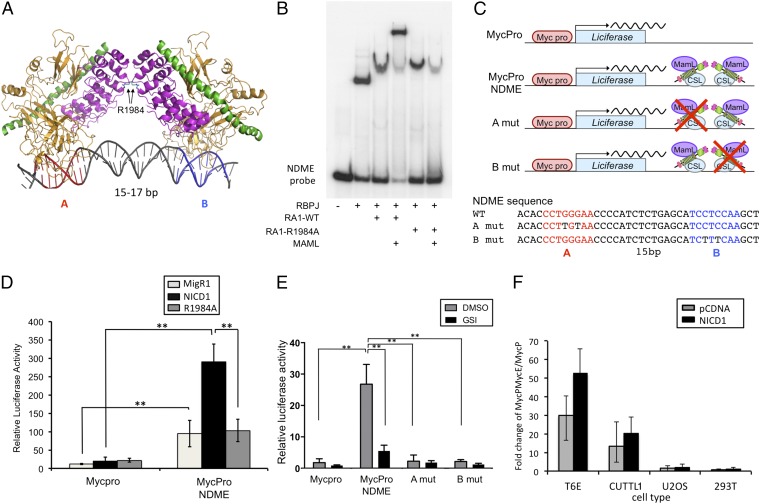
Sequence analysis of a 1-kb region centered on the RBPJ/Notch1 binding peak in the NDME revealed a single potential SPS consisting of a high-affinity RBPJ binding site (site A) oriented head-to-head with a second, lower-affinity site 15 bp away (site B) (Fig. 2A), an arrangement similar to that seen in other SPSs (16). Electrophoretic mobility shift assays (EMSAs) performed with an oligonucleotide containing the NDME site confirmed that it supported the loading of NTC dimers in the presence of the N-terminal portion of MAML1 and that a R1984A mutation in Notch1 suppressed dimerization on the NDME (Fig. 2B).
Fig. 2.
The murine Myc 3′ enhancer contains a SPS that drives NTC dimer-dependent transcription. (A) Schematic of the RBPJ–Notch1–MAML NTC complex on DNA containing an SPS site. The two RBPJ motifs are designated A and B, and the critical R1984 interankyrin repeat domain contact is highlighted. See ref. 16 for details. RBPJ, gold; Notch1 RAM-ANK, purple; MAML1, green. (B) Assembly of dimeric NTCs on the 3′ Myc enhancer element. EMSA was performed with oligonucleotide probes containing the putative SPS and the indicated combinations of recombinant RBPJ, Notch1 RAM-ANKs (RA1), and N-terminal MAML1 (MAML). RA1-R1984A corresponds to the dimerization-defective Notch1 mutant. (C) Schematic of the luciferase reporter gene constructs including the NDME WT and mutant sequences. Note: RBPJ is denoted by its alternative name, CSL. (D) T6E cells were transfected with Renilla luciferase control (pRLTK), the indicated Myc firefly luciferase reporter genes, and either empty vector or vectors driving expression of WT NICD1 or NICD1 with the R1984A mutation. (E) Murine T-ALL T6E cells transfected with pRL-TK and one of the reporter constructs shown in C. Cells were cultured with DMSO or 1 μM compound E for 6 h before harvest. (F) Activity of the murine Myc enhancer reporter construct (MycProNDME) in various cell lines with and without overexpression of NICD1. In D–F, cells were harvested 48 h after transfection. Data were obtained in triplicate in independent experiments; error bars correspond to the SEM. **P < 0.001.
We next tested whether the NDME enhances transcription from the murine Myc proximal promoter in an NTC dimer-dependent fashion. Luciferase reporter constructs were designed that contained either the Myc promoter alone, the Myc promoter plus the NDME, or the Myc promoter plus versions of the NDME bearing mutations in either the high-affinity (site A) or low-affinity (site B) RBPJ sites of the NDME SPS element (Fig. 2C). T6E cells were transfected with the Myc promoter or the Myc promoter/NDME reporter together with empty vector or vector encoding NICD1 or NICD1–R1984A. The NDME stimulated basal reporter gene activity in T6E cells, and this activity was increased further by expression of wild-type (WT) NICD1, but not by expression of the dimerization-defective R1984A mutant (Fig. 2D). In addition, inhibition of endogenous Notch1 with the GSI Compound E suppressed the basal activity of the NDME reporter, as did mutations in either RBPJ binding site in the NDME SPS, in line with prior data showing that loading of NTC dimers is required to activate Notch target genes with response elements containing SPSs (17) (Fig. 2E). Furthermore, dominant-negative mastermind (DN-MAML), a specific inhibitor of NTC function, had inhibitory effects on reporter activity comparable to those of GSI (Fig. S1B), consistent with the effects of GSI being mediated through Notch inhibition. In parallel studies, we noted that the NDME reporter was also active in the human T-ALL cell line CUTLL1, but not in U2OS (osteosarcoma) or HEK293T (embryonic kidney) cells (Fig. 2F). Collectively, these studies demonstrate that the NDME can stimulate NTC-dimer-dependent transcription in a cell-type–specific fashion.
The NDME Physically Associates with the Myc Promoter.
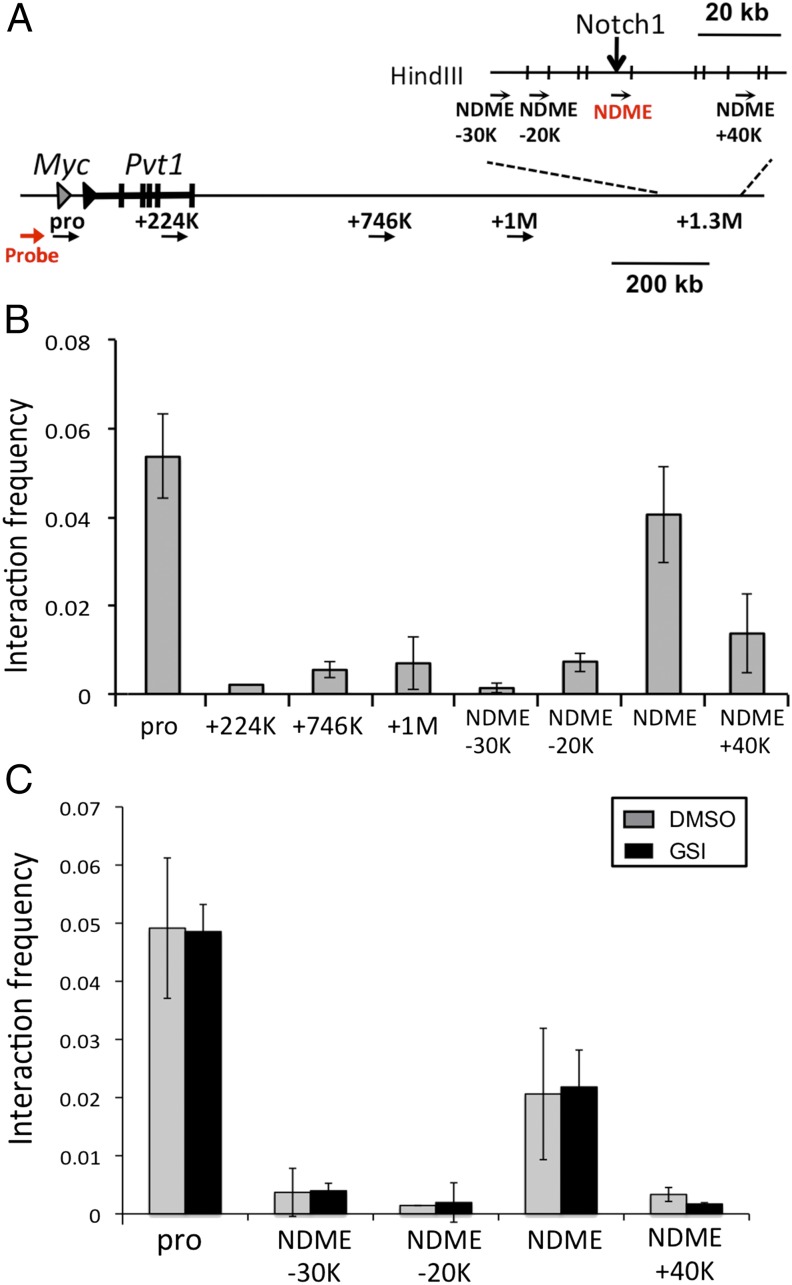
To test for chromatin looping between the NDME and the Myc promoter, chromatin conformation capture (3C) assays were carried out in T6E cells (Fig. 3A). The NDME displayed an interaction frequency with the promoter that was nearly as high as that observed with sequences immediately adjacent to the promoter, whereas minimal interaction was observed between the promoter and regions between the promoter and the NDME, or sequences 40 kb 3′ of the NDME (Fig. 3B). To determine whether Notch signaling was required to maintain the interaction between the Myc promoter and the NDME, 3C analysis was performed in T6E cells cultured in the presence or absence of GSI. Although 4-h GSI treatment depleted Notch1 from the NDME (Fig. 1B), it did not affect the promoter–enhancer interaction (Fig. 3C). Thus, interaction of the Myc promoter and NDME is not affected by acute changes in Notch1 occupancy.
Fig. 3.
Interaction of the 3′ Myc enhancer element and the Myc promoter. (A) Schematic of Myc locus and enhancer with superimposed primers used for chromosome conformation capture (3C) assay. (B) The 3C assay in T6E cells. Graphs show the enrichment of PCR products normalized to HindIII-digested and randomly ligated bacterial artificial chromosome (BAC) DNA spanning the region. (C) Short-term Notch inhibition does not affect chromatin looping. T6E cells were treated with DMSO or 1 μM GSI for 6 h. The 3C assay was performed on the Myc promoter and sequences 3′ of the Myc gene body. Positions of primers used in B and C are shown in A. Data were obtained in triplicate in independent experiments; error bars correspond to the SEM.
The NDME Is Conserved in Human T-ALL Cells.
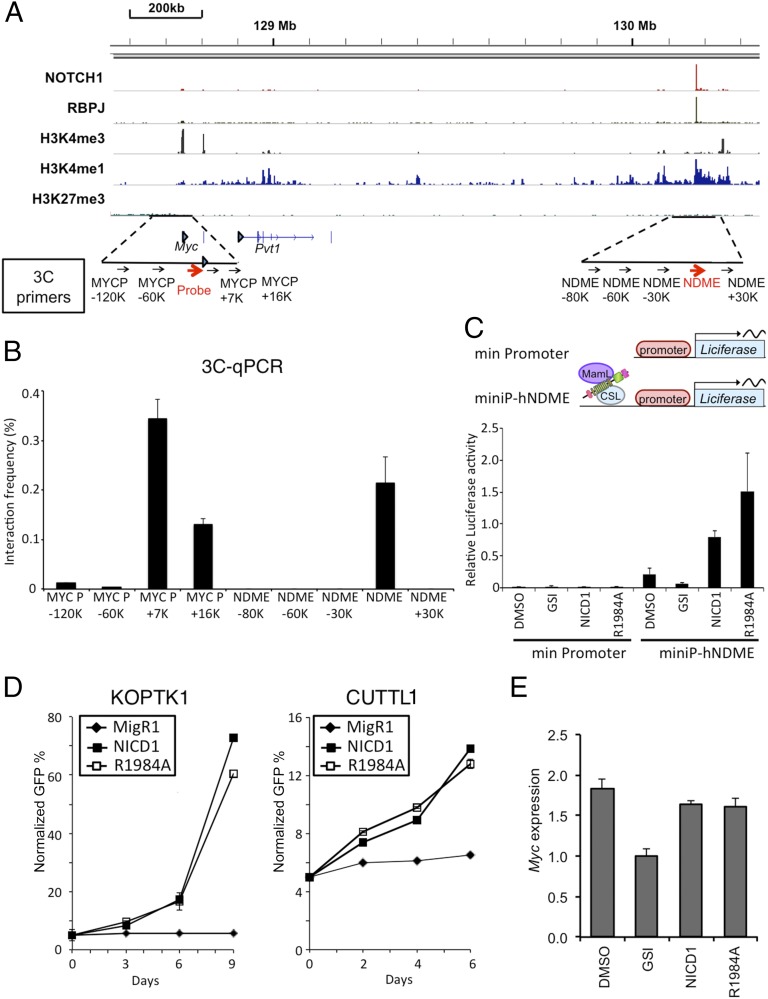
To determine how Notch signaling regulates Myc in human T-ALL cells, ChIP-Seq analysis was performed on the human T-ALL cell line CUTLL1. Analysis of the ChIP-Seq data revealed an RBPJ/Notch1 binding site associated with high levels of H3K4me1 marks located 1.43 Mb 3′ of the Myc promoter (Fig. 4A) that shares sequence homology with the murine NDME (Fig. S2). As in murine T-ALL cells, 3C assays showed that the human NDME interacted with the Myc promoter in CUTLL1 cells (Fig. 4B) and that this interaction was unaffected by short-term Notch inhibition (Fig. S3). To confirm that the putative human NDME is a Notch-responsive element, luciferase reporter gene constructs containing a minimal promoter with and without the human NDME were transfected into CUTLL1 cells (Fig. 4C). Although the promoter-only construct was unresponsive to Notch, addition of the human NDME resulted in robust reporter activity that was sensitive to Notch inhibition, showing that this element drives Notch-dependent transcription (Fig. 4C).
Fig. 4.
Notch1 regulates human Myc via a 3′ enhancer element. (A) ChIP-Seq read counts for Notch1, RBPJ, and H3K4me1, H3K4me3, and H3K27me3 marks in a ∼2-Mb region containing human Myc. y axis, aligned sequence tags; arrowhead, TSSs. (B) The 3C assay in the human T-ALL cell line CUTLL1. Graphs show the enrichment of PCR product normalized to HindIII-digested, randomly ligated BAC DNA spanning the region. The arrows beneath the schematic in A denote the position of primers used for 3C analysis. The Myc promoter primer and “probe” were paired with eight primers positioned in flanking sequences to detect ligated HindIII-digestion fragments. (C) The human Myc 3′ enhancer element (NDME) stimulates transcription. (Upper) Schematic of the human Myc NDME and control firefly luciferase reporter genes. A minimal TATA promoter construct was used as a negative control. Note: RBPJ is denoted by its alternative name, CSL. (Lower) CUTLL1 cells were transfected with the indicated reporter genes with and without 1 µM compound E. Luciferase assays were performed 48 h later. (D) KOPT-K1 (Left) and CUTLL1 (Right) cells transduced with either NICD1, NICD1-R1984A, or empty plasmid were cultured in the presence of 1 μM compound E beginning 24 h after transduction. The number of GFP+ transduced cells was determined at various time points and normalized to time 0 (defined as 48 h after transduction). (E) Myc mRNA expression in CUTLL1 cells transduced as in D. Except for the MigR1 empty vector control cells, 1 μM compound E was added to cells 24 h after transduction. Cells were harvested for RT-PCR analysis 48 h later. In B–E, data were obtained in triplicate in independent experiments; error bars correspond to the SEM.
Sequence alignment showed divergence in the SPS of the murine NDME and the analogous region of the human genome (Fig. S2). To address whether the human NDME required NTC dimerization for function, human CUTLL1 cells were transfected with the reporters described above and either WT NICD1 or NICD1–R1984A in the presence of GSI, which was added to inhibit endogenous Notch signaling. Unlike the murine NDME, the R1984A mutant stimulated transcription from the human NDME (Fig. 4C). In line with this observation, and in contrast to murine T-ALL cell lines (9), expression of NICD1–R1984A also rescued human Notch-addicted T-ALL lines from the effects of Notch inhibition (Fig. 4D). This phenotype correlated with stimulation of Myc expression in cells expressing either NICD1 or NICD1–R1984A (Fig. 4E). Thus, although the NDME is functionally conserved between man and mouse, it exhibits divergence in its dependence on NTC dimerization for Myc regulation.
The NDME Is Located Within a Multimodular Enhancer.
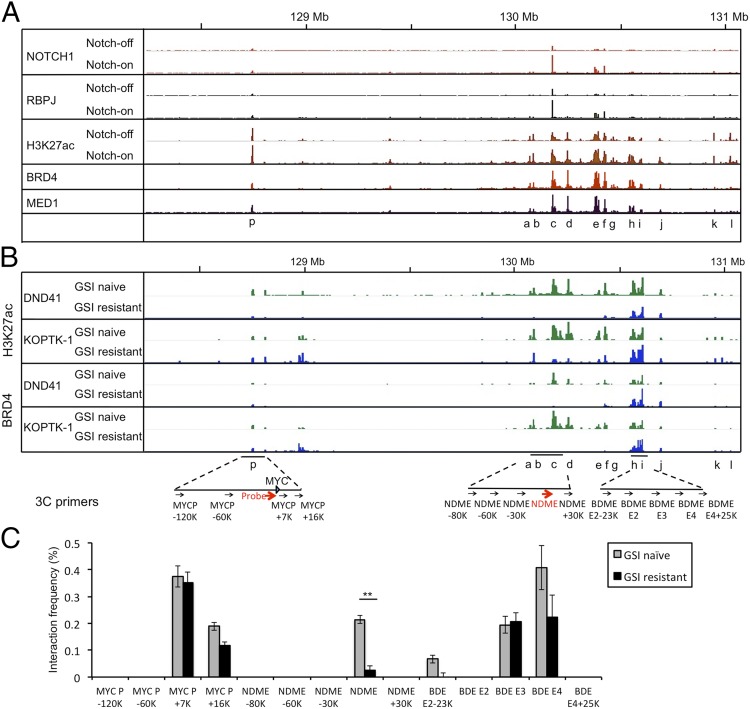
Notch activation is associated with rapid increases in H3K27ac marks across the entire breadth of enhancers containing dynamic RBPJ/Notch1 binding sites, but does not alter H3K27ac in enhancers lacking RBPJ/Notch1 binding sites (Fig. S4A; see also ref. 5). To determine the effect of acute changes in Notch activation on H3K27 acetylation in chromatin flanking the NDME, we performed CHIP-Seq for Notch1, RBPJ, and H3K27Ac in CUTLL1 cells that were (i) depleted of NICD1 for 3 d with GSI or (ii) harvested 4 h after NICD1 activation by GSI washout. Analysis of the ChIP-Seq data revealed that the RBPJ/Notch1 site in the human NDME is located within a 3′ enhancer region of >600 kb in breadth that contains multiple domains defined by discrete H3K27ac peaks associated with Brd4 and Med1 binding (Fig. 5A). The major RBPJ/Notch1 binding site in this enhancer region is in the NDME (domain c), with lower-occupancy RBPJ/Notch1 binding sites also being observed in association with H3K27ac peaks at the Myc promoter (p) and 3′ of the NDME (domains e and f). Loading of RBPJ/Notch1 was associated with a 1.5- to 2.0-fold increase in H3K27Ac signals at the Myc promoter and in multiple H3K27ac peaks (domains a–j; Fig. 5A and Fig. S4B; quantified in Fig. S4 C and D), which span a region of 635 Kb. The changes H3K27ac seen in ChIP-Seq datasets were confirmed by performing local ChIP for H3K27ac in 5′ (domain c) and 3′ (domain h) portions of the 3′ Myc enhancer region (Fig. S5A), which again revealed that Notch inhibition decreased H3K27ac across the 3′ Myc enhancer (Fig. S5 B and C). The fold change in H3K27ac upon GSI washout (Figs. S4D and S5 B and C) was comparable with the fold change in Myc expression (Fig. S6A), consistent with the finding that H3K27ac is highly sensitive to changes in NTC occupancy of Notch response elements (5) (Fig. S4A). Thus, loading of NTCs to sites 3′ of Myc causes rapid increases in active chromatin marks across the entirety of the NDME-associated 3′ enhancer that correlate with stimulation of Myc expression.
Fig. 5.
Altered 3′ enhancer activity and chromatin looping in GSI-sensitive and resistant T-ALL cells. (A) Chromatin landscapes in human CUTLL1 cells (i) depleted of NICD1 by treatment with 1 μM compound E for 3 d and (ii) after 4 h of Notch activation by GSI washout. RBPJ, Notch1, and H3K27ac traces are shown in the Notch-on and -off states, whereas Brd4 and Med1 are shown only in the Notch-on state. H3K27ac peaks are seen at the Myc promoter (p) and in 3′ regions (a–l). (B) Chromatin landscapes in GSI-sensitive and -resistant human KOPT-K1 and DND-41 cells. H3K27ac and Brd4 traces are shown for each cell state. The positions of primers and probes used in the 3C analyses in C are shown below the traces. BDME, Brd4-dependent Myc enhancer. (C) The 3C analyses performed with GSI-sensitive (naïve) and GSI-resistant KOPT-K1 cells. Graphs show the enrichment of PCR products normalized to HindIII-digested, randomly ligated BAC DNA spanning the region. The 3C data were obtained in triplicate in independent experiments; error bars correspond to the SEM. **P = 0.008 (comparing GSI-naïve with GSI-resistant samples using Student t test).
GSI Resistance and Bromodomain Inhibitor Sensitivity Are Associated with Altered Function of Discrete Sets of Enhancer Domains.
Notch-addicted human T-ALL cell lines become GSI-resistant through an epigenetic mechanism characterized by Notch-independent Myc expression and increased sensitivity to Brd4 inhibitors (18) (Fig. S6B). We therefore asked whether functional changes in the Myc 3′ enhancer underlie GSI resistance. Compared with GSI-sensitive T-ALL cell lines, GSI-resistant cell lines show dramatic changes in the epigenetic landscapes of the 3′ enhancer, with a loss of H3K27ac near the NDME in domains c–f, variable loss of H3K27ac in domain g, and preservation of H3K27ac marks in domains h and i and at the Myc promoter (Fig. 5B; see Fig. S6C for blow-up of the 3′ enhancer and Fig. S4D for H3K27ac quantification within specific domains). These changes in H3K27ac are closely correlated with changes in Brd4 binding among the various enhancer domains (Fig. 5B; quantified in Fig. S4E). Of note, domains h and i correspond to a Brd4-dependent enhancer region recently characterized in acute myeloid leukemia (AML) cells (19), which we henceforth term the Brd4-dependent enhancer region (BDME). The 3C assays showed that the GSI-resistant state is associated with loss of NDME (domain c) interaction with the Myc promoter, whereas associations with the BDME (domains h and i) are maintained (Fig. 5C). Thus, GSI resistance is associated with alterations in the activity and higher-order structure of specific regions of the Myc 3′ enhancer.
Discussion
In this work, we identify a large multidomain enhancer located >1 Mb 3′ of Myc that appears to regulate Notch-dependent and -independent Myc expression in T-ALL cells. The key site mediating Notch dependence lies within a small region that we term the NDME that is conserved in position between mouse and man (20), but that shows significant divergence in RBPJ binding sites, because the murine site contains a functional paired RBPJ binding site, whereas the human site does not. Binding of Notch to chromatin in this region influences H3K27 acetylation across the entire 3′ enhancer region. Notably, T-ALL cell lines selected for resistance to Notch pathway inhibitors (18) exhibit epigenetic silencing of discrete domains within the 5′ region of the enhancer near sites of Notch binding, loss of the Myc promoter–NDME interaction, and preserved Myc promoter interaction with 3′ enhancer domains in a region termed the BDME, which was previously implicated in Brd4-dependent Myc expression in AML cells (19). These observations elucidate Notch-dependent and -independent mechanisms of Myc regulation in T-ALL cells and provide insight into GSI resistance mediated by epigenetic changes.
Our findings fill in significant gaps in our understanding of the Notch–Myc signaling axis in T-ALL cells, but also raise a number of fundamental issues that remain to be resolved. First, acute changes in Notch occupancy of the NDME lead to changes in H3K27 acetylation over the full breadth of the Myc 3′ enhancer, a region of ∼635 kb. NTC complexes recruit p300 (3), which has been implicated in the deposition of H3K27ac marks (8), but how these marks spread in a stereotypic fashion from the site of NTC recruitment to flanking chromatin is unclear. Secondly, we identified two different chromatin states in isogenic human T-ALL cells that are sensitive to the presence or absence of Notch signals, a Notch-on state dominated by Myc regulation through the NDME and a Notch-off state dominated by Myc regulation through the BDME. Single-cell cloning assays suggest that a small fraction of Notch-mutated T-ALL cells are in the GSI-resistant state before exposure of bulk cell populations to drug (18), but how individual cells shift between the GSI-sensitive and -resistant chromatin states is unknown. Third, short-term withdrawal of Notch binding did not affect looping of the NDME to the Myc promoter, whereas NDME functional activity and looping were both lost in cells selected for GSI resistance. Because the NDME remains in proximity to the Myc promoter during acute inhibition, other factors must be required to maintain transcription, as has been described for SATB1 during Th2 cell activation (21). Finally, because NTCs (22) and Brd4 (23) appear to regulate transcription by recruiting different multiprotein complexes and therefore might be expected to complement one another, it is unclear how high levels of activated Notch lower the requirement for Brd4, a conclusion drawn from both drug-sensitivity and shRNA-knockdown studies (18).
Our work adds to emerging themes in an increasingly complex picture of Myc regulation by long-range enhancer elements. Cell-type–specific Myc enhancers have now been described in colorectal cancer (24, 25), T-ALL (this work), AML (19), and other hematopoietic neoplasms (7). When tested as isolated DNA fragments in reporter constructs, these elements drive transcription in a cell-context–restricted fashion, a phenomenon we observed for the NDME in T-ALL cells that is also true of the BDME in AML cells (19) and for other Myc enhancers in colorectal cancer (24, 25). This characteristic presumably stems from cell-type–specific cofactors that have yet to be identified. Moreover, the potential for coregulation of Myc by multiple enhancer inputs provides for an additional level of complexity. We observed interaction of the Myc promoter in “Notch-on” T-ALL cells with discrete enhancer domains located in the NDME and BDME, a finding similar to the cosynapsis of two Wnt-dependent enhancer elements located 5′ and 3′ of the Myc gene body with the Myc promoter in colorectal cancer cells (25). Finally, Notch-mediated Myc activation is proposed to be an important event in other human tumors besides T-ALL with Notch gain-of-function mutations—including marginal zone lymphoma (26, 27), mantle cell lymphoma (28), and breast cancer (29)—and it remains to be determined whether Notch uses the NDME in each of these varied tumor types and, if so, whether it does so together with other enhancers that interact with the Myc promoter.
These complexities may represent an opportunity for targeting Myc in cancer. It appears that Myc expression in T-ALL is regulated by at least two epigenetic states in which Myc expression is GSI-sensitive or Brd4 inhibitor-sensitive, respectively, suggesting that emergence of drug resistance to either agent may be prevented by cotreatment strategies. This cooperative regulation is supported by preclinical studies of human T-ALLs, which respond better to GSI/Brd4 inhibitor combinations than to either drug alone (18). Our findings bolster the mechanistic rationale for cotargeting of Notch and Brd4 in T-ALL and suggest that this drug combination merits study in other preclinical models of Notch-driven cancers.
Materials and Methods
Cells.
Primary murine T-ALL cells were obtained from mice that received KrasG12D bone marrow transduced with the Notch1 L1601P mutant (30); GFP-positive CD4+CD8+ T-ALL cells were isolated from the spleen by sorting. DN3 cells from Rag2−/− mice were negatively selected with biotinylated antibodies against NK1.1, CD11b, CD11c, B220, Ter119, and CD44 with streptavidin beads (Miltenyi Biotec) and confirmed to be >98% pure. Murine and human T-ALL cell lines were cultured in RPMI medium 1640 (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, 100 IU per 100 μg per mL penicillin/streptomycin, 1 mM sodium pyruvate, and 5 μM 2-mercaptoethanol. In GSI washout studies, CUTLL1 cells were treated with the GSI compound E (1 μM) (31) for various periods of time, washed, and then replated in either 1 μM GSI (washout control) or in DMSO for 4 h (washout). GSI-resistant KOPT-K1 and DND41 cells were as described (17). U2OS and 293T cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, and 100 IU per 100 μg per mL penicillin/streptomycin.
ChIP Assays.
ChIP was performed as described (32). Briefly, chromatin samples prepared from fixed cells were immunoprecipitated with rabbit IgG (no. sc-3888, Santa Cruz Biotechnology), anti-Notch1 antibodies (33), or anti-H3K27Ac antibody (no. ab4729, Abcam). Genomic DNA was quantified by real-time PCR with primers (Table S1) flanking RBPJ binding sites or H3K27ac peaks located in the NDME or BDME (in human). Positive controls for Notch1 ChIP were prepared from the Hes1 promoter and Dtx1 intron2, and ChIP for Nanog was used as an internal negative control. Enrichment was measured by using the absolute standard curve method. The DNA quantity recovered from each ChIP sample is expressed relative to input DNA.
ChIP-Seq.
ChIP was performed as described above, and libraries were constructed per the NEB NEXT kit (5, 34). High-throughput sequencing was performed by using the Illumina Genome Analyzer II. Reads were mapped to human genome hg19 or mouse genome mm9. ChIP-Seq data were processed as described (5, 34). All binding signals were normalized to reads per kilobase per million mapped reads. High-confidence peaks were defined by ChIP enrichment of greater than or equal to threefold and a false discovery rate (FDR) of <0.01. The FDR was determined by running the same peak calling analysis on a randomly selected subset of the input ChIP-Seq data. ChIP-Seq data (accession no. GSE61504) are available through the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/). Other ChIP-Seq analyses were conducted using published datasets (5, 18) (accession nos. GSE54380, GSE51800, and GSE29600).
ChIP-Seq Data Analysis.
ChIP-Seq data were aligned to either human genome build hg19 or mouse genome build mm9, and uniquely mapped, nonredundant reads were retained. Genomic wiggle traces were generated by using MACS (Version 1.4) (35), normalized by the total nonredundant read count of each sample, and displayed using IGV Browser. H3K27ac peaks were identified in T-ALL Notch-on datasets by using SICER (Version 1.1) with default parameters (36). In the Myc 3′ enhancer region, individual H3K27ac peaks were additionally filtered with a SICER island score threshold of 3,000. Comparisons of H3K27ac levels in the Notch-on/Notch-sensitive state and Notch-off/Notch-resistant state were performed by using SICER differential comparison function (36). P values were reported after Benjamini–Hochberg correction for multiple-hypothesis testing over all H3K27ac peaks in the genome.
Quantitative RT-PCR.
RNA was extracted with the QIAGEN RNeasy Mini Kit. cDNA was synthesized from RNA with the SuperScript III kit (Invitrogen). Transcripts were amplified with Sybr Green PCR Master Mix (ABI), and quantitative PCR was performed on an ABI ViiA 7 real-time PCR System. Primers are provided in Table S1.
EMSA.
EMSA was performed as described (17). Recombinant RBPJ (residues 9–435), WT or R1984A mutant Notch1 (residues 1,761–2,127), and MAML1 (residues 13–74) were expressed and purified as described (37). In the current release of the human genome, the key arginine residue at the dimerization interface of the Notch1 ankyrin repeats (formerly referred to as R1985 in prior publications) (9, 17) is R1984. Oligonucleotides used to create probes for EMSAs are listed in Table S1.
Luciferase Reporter Gene Assay.
Murine Myc promoter (Mycpro) constructs containing DNA corresponding to −1,137 to +516 bp from the Myc TSS were cloned into the pGL3 vector (Promega). The murine or human Myc NDME, corresponding to positions chr15:63,087,071–63,087,595 (525 bp) and hg19 chr8:130,180,217–130,180,629 (413 bp) in the murine and human genomes, respectively, were cloned into pGL3 Mycpro or the pGL3-SV40 vector. Mutagenesis was performed with the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). T6E or CUTLL1 cells were transfected with 200 ng of the indicated reporter constructs, 50 ng of pRL-TK, and 200 ng of pcDNA–NICD1, pcDNA–NICD1–R1984A, pcDNA–DN–MAML, or empty vector with Lipofectamine LTX-plus reagent (Life Technologies). The 293T and U2OS cells were plated in triplicate and transfected with 50 ng of the indicated promoter/reporter constructs, 10 ng of pRL-TK, and 10 ng of pcDNA–NICD1 or vector control by using FuGENE 6 (Promega). For the U2OS and 293T cell assays, a Hes1 reporter was used as a positive control (33). The relative Myc enhancer activity (MycProNDME) was determined relative to the Myc promoter (MycPro) activity. Luciferase assays were performed as described (38). Firefly luciferase values were normalized with the Renilla luciferase internal control and expressed relative to activities obtained with promoter-only plasmids. The values are averages of three independent experiments; error bars show the SEM.
Chromatin Conformation Capture Assay.
Assay was performed as described (39). Briefly, cells cross-linked with 1% formaldehyde at room temperature for 10 min were quenched with glycine, lysed, and treated with HindlII followed by T4 ligase. Ligated products were quantified in triplicate by TaqMan real-time PCR. Probes and primers were designed by using Primers3 software. Control 3C template was generated by using two bacterial artificial chromosomes (BACs) that together encompass Myc and the relevant flanking regions (listed in Table S1). Equimolar amounts of the two BAC clones were digested with HindlII and ligated. PCR DNA fragments obtained from BAC 3C DNA were purified and adjusted to a concentration of 1 nM, and then used to generate PCR standard curves to which all 3C PCR products were normalized. Ct values are within the linear range over at least one order of magnitude (Figs. S7 and S8). Probe, primer sequences, and BAC clones are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Junwei Shi and Chris Vakoc (Cold Spring Harbor Laboratory) for sharing information prior to publication. The University of Pennsylvania Mouse Husbandry Core (University Laboratory Animal Resources), the Abramson Cancer Center Flow Cytometry Core (P30-CA016520), the Abramson Family Cancer Research Institute Core, the Functional Genomics Core, and the NIH/National Institute of Diabetes and Digestive and Kidney Diseases P30 Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306), contributed to this study. This work was supported by a Leukemia and Lymphoma Society fellowship (to C.Z.); Fellowships T32HD007516 and 1F31CA165813 (to W.B.); St. Baldrick's Fellowship T32HL007574 (to B.K.); National Institutes of Health Grants P01 CA119070 (to J.C.A., W.S.P., S.C.B., and X.S.L.), R01AI047833 (to W.S.P.), R01HD25147 (to S.A.L.), and ENCODE U54 HG004570 (to B.E.B.); and a Leukemia and Lymphoma Society Specialized Center of Research grant (to J.C.A. and B.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ChIP-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61504).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407079111/-/DCSupplemental.
References
- 1.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223(2):262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126(Pt 10):2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16(11):1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzoneva G, Ferrando AA. Recent advances on NOTCH signaling in T-ALL. Curr Top Microbiol Immunol. 2012;360:163–182. doi: 10.1007/82_2012_232. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci USA. 2014;111(2):705–710. doi: 10.1073/pnas.1315023111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30(2):249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, et al. Notch dimerization is required for leukemogenesis and T-cell development. Genes Dev. 2010;24(21):2395–2407. doi: 10.1101/gad.1975210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J Exp Med. 2008;205(12):2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26(21):8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110(1):278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klinakis A, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci USA. 2006;103(24):9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnett KL, et al. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat Struct Mol Biol. 2010;17(11):1312–1317. doi: 10.1038/nsmb.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci USA. 2007;104(7):2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knoechel B, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46(4):364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27(24):2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geimer Le Lay AS, et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal. 2014;7(317):ra28. doi: 10.1126/scisignal.2004545. [DOI] [PubMed] [Google Scholar]
- 21.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38(11):1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 22.Yatim A, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31(13):2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41(8):885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 25.Yochum GS, Sherrick CM, Macpartlin M, Goodman RH. A beta-catenin/TCF-coordinated chromatin loop at MYC integrates 5′ and 3′ Wnt responsive enhancers. Proc Natl Acad Sci USA. 2010;107(1):145–150. doi: 10.1073/pnas.0912294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi D, et al. The coding genome of splenic marginal zone lymphoma: Activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209(9):1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiel MJ, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553–1565. doi: 10.1084/jem.20120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kridel R, et al. Whole transcriptome sequencing reveals recurrent NOTCH1 mutations in mantle cell lymphoma. Blood. 2012;119(9):1963–1971. doi: 10.1182/blood-2011-11-391474. [DOI] [PubMed] [Google Scholar]
- 29.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelton CC, et al. Modulation of gamma-secretase specificity using small molecule allosteric inhibitors. Proc Natl Acad Sci USA. 2009;106(48):20228–20233. doi: 10.1073/pnas.0910757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23(14):1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aster JC, et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20(20):7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci USA. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25(15):1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124(5):973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 38.Bailis W, et al. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 2013;39(1):148–159. doi: 10.1016/j.immuni.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149(6):1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.