Growing up in rural China under the imposed hardships of the cultural revolution, Yigong Shi did not have the luxury of toys. Instead Shi, now the dean of the School of Life Sciences at Tsinghua University in Beijing, made elaborate models out of clay he collected from the fields. “I would stay for hours to a make a ship or train model with small parts,” he recalls. Decades later, as a structural biologist, Shi is still making models, albeit with different tools. Elected to the National Academy of Sciences in 2013, Shi studies the molecular mechanisms that govern the awakening of apoptotic enzymes in worms, fruit flies, and mammals, seeking to illuminate function by revealing structure.

Yigong Shi. Image courtesy of Tsinghua University.
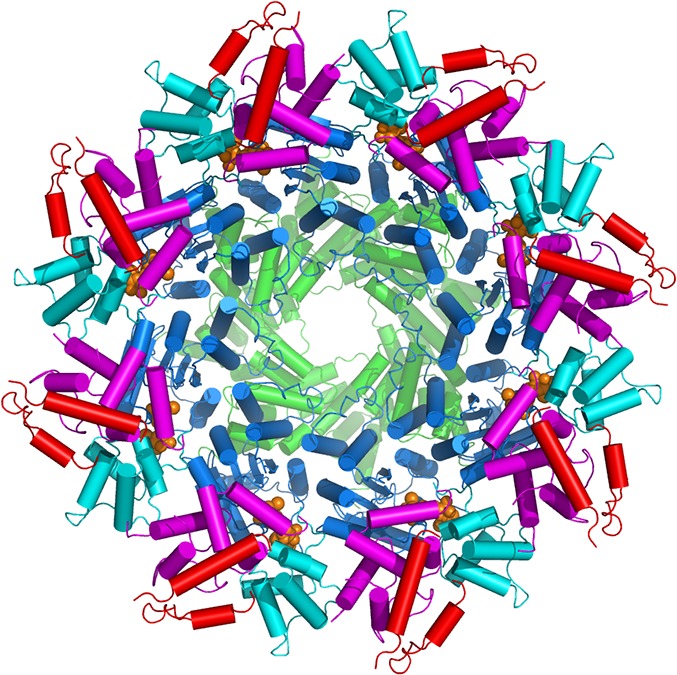
Macromolecular machine for cell death. Shown here is a 3D structure of the CED-4 apoptosome, formed by eight molecules of the CED-4 protein. Each CED-4 molecule contains five domains: caspase recruitment domain (green), nucleotide-binding domain (blue), helical domain I (cyan), winged-helix domain (magenta), and helical domain II (red).
Apoptosis, or programmed cell death, is a process akin to cellular containment and cleaning. An evolutionarily conserved cascade of enzyme activity triggers this tightly orchestrated cellular dismantling, which is especially important during embryonic development and tumor formation. In his Inaugural Article (1), Shi examines the fundamental mechanism of the final enzyme in the cascade, offering what he calls “a somewhat controversial conclusion” regarding caspase activation. He demonstrated that the enzyme caspase-9, which controls cell death triggered by intracellular stress signals, is awakened in a complex process that requires the cooperation of another protein, Apaf-1.
Working with Models
Born in 1967, Shi spent most of his early childhood in rural China. His parents were relocated to Zhumadian in 1969 as part of a government effort during the cultural revolution. The family experienced hardship, with meat rarely appearing on the dinner table. Shi enjoyed the freedom to be outdoors, however, and thinks his model making probably foreshadowed his experimental skills in the laboratory. “I have a pair of good hands,” he says. When he was still young, Shi’s parents encouraged him to pursue science as a career.
Despite his parents’ intent on a scientific future, technology did not play a big part in Shi’s upbringing. He did not use a telephone until middle school, in 1982. “When the phone rang, I didn’t even know how to pick it up.” Nevertheless, Shi focused on his schooling and discovered he had a talent for math, claiming first place in his province at a national high school mathematics contest in 1984, a feat that earned him automatic admission to one of China’s top-tier universities.
Shi decided to major in biology and minor in math at Tsinghua University in Beijing. His strength lay in math, but Shi felt that biology held the future of science. After graduating in 1989 with a bachelor’s degree, Shi applied for graduate study in the United States. He left his family in 1990 for Iowa State University, but attending Johns Hopkins University’s intercampus program in molecular biophysics remained his dream. When he discovered a few months later that a waylaid letter of acceptance to Hopkins never reached him in China, Shi chose to move. “I knew Johns Hopkins would be a better choice for my education.”
Structuring His Education
During rotations at Hopkins, Shi heard Carl Pabo, then a professor in the biophysics and biophysical chemistry department at the School of Medicine, give a “beautiful lecture on zinc fingers.” Because Pabo was leaving Hopkins for the Massachusetts Institute of Technology (MIT) and not taking any new students, Shi chose instead to work with the director of the department, Jeremy Berg, who also studied zinc fingers, feeling that his strong background in math and physics made him a good fit. Shi used biochemical techniques to elucidate properties of zinc finger proteins, showing, among other things, that C2H2 zinc fingers, the classic type of this gene regulatory motif, specifically bind DNA–RNA hybrids (2). Shi, whose upbringing and culture instilled a respect for authority, credits Berg with fostering the ability to challenge the status quo, a value that Shi has held dear in his work.
After graduating with a PhD in biophysics in 1995, Shi decided that X-ray crystallography held the key to structural biology. As Shi puts it, “Questions accumulated over twenty years of biochemistry can be answered by one beautiful X-ray structure.” Shi chose Nikola Pavletich’s laboratory at Memorial Sloan Kettering Cancer Center for his postdoctoral research. Pavletich, currently the chair of the structural biology program there, had trained with Pabo at MIT, and soon Shi was learning the fine skills of crystallography, finding that he had an affinity for the precise work. “My prior experience playing with clays helped my crystallization efforts,” he says. However, he quickly chafed at the slow progress with crystallizing BRCA1, a breast cancer gene, and began to look for other proteins to target. His wife, whom he had studied with at Tsinghua and married in 1992 after reconnecting in New York City, prompted the idea of working on Smad4, a proposed human tumor suppressor protein active in a cellular signaling pathway mediated by the protein TGF-β. Listening to her describe the burgeoning field of TGF-β signaling from her own research, Shi knew that he had the opportunity to get in on the ground floor by studying proteins in this pathway. Shi eventually showed, collaborating with Memorial Sloan Kettering cancer biologist Joan Massagué, that the Smad4 structure he elucidated using heavy atom derivatives provided a suitable explanation to cancer-derived mutations (3).
Shi remembers feeling unprepared to contemplate a research laboratory of his own, but in 1997, after much urging from Pavletich, Shi applied for a position at Princeton University. Even though he had missed the deadline for applications and endured a sleepless night before the interviews, Shi received an offer that he excitedly accepted. In 1998, Shi arrived at Princeton and forged ahead with the successful TGF-β work, and his laboratory eventually characterized important elements in the Smad cascade (4).
Building a Reputation
As a newly independent scientist, Shi also needed to build a reputation. “The best way to do that is to go into a field different from that of your mentor.” Shi had followed the field of apoptosis since graduate school and had approached Pavletich about working on an apoptotic protein to no avail. Looking back, Shi is grateful that Pavletich encouraged him to pursue it once he was on his own. Shi sought insight by developing collaborations. With molecular biologist Emad Alnemri of the Kimmel Cancer Center, Shi elucidated the structure and interaction of two mammalian apoptosis proteins, Apaf-1 and caspase-9, showing how Apaf-1 recruits the caspase enzyme (5).
Caspase, a cysteine protease, is the cell-killing enzyme in the apoptosis cascade (6). To control a caspase is to control apoptosis, says Shi. By 2000, his laboratory had reported the structure of one of caspase’s activators known as Smac (7), and soon the team was in a race to uncover the interactions. Later that year, the team published work demonstrating the mechanism by which an N-terminal peptide in Smac binds the protein XIAP to relieve inhibition of caspase-9 (8), a result that interested cancer researchers and eventually led Shi to cofound Tetralogic Pharmaceuticals to produce anticancer compounds based on Smac mimetics.
Shi extended his apoptosis research beyond mammals. “I quickly expanded into C. elegans,” he recalls. “Then, in 2002, fruit flies. We got our hands into three important model organisms for biologists.” The Caenorhabditis elegans apoptosis pathway was simpler, but the structure work proved many times more challenging. It took a long time to express the proteins and then even more time to crystallize them. Shi recalls laboratory members abandoning the project in frustration. “We struggled with one problem after another,” he says. “At one time I felt that I would give up, so I don’t blame the students.” Eventually, graduate student Nieng Yan made headway, ultimately solving the structure of the CED-9–CED-4 complex to 2.6 Å in 2005 (9).
CED-4 is a functional homolog of the mammalian caspase activator protein Apaf-1, and Shi thought that study of its activation could provide useful insight. In 2006, in the midst of these efforts, Shi received an invitation to join the faculty at Tsinghua. The decision to accept meant turning down a hard-won, cherished offer from Howard Hughes Medical Institute and having to build a laboratory, again, from scratch without his other laboratory members.
Transition State
Shi says that he often gets asked why he went back. “I usually reply by asking, ‘why not?’” Shi acknowledges there are tradeoffs, but the gain remains clear to him. “I have helped create a strong biomedical research community and been witness to changes.” The transition, however, did not prove easy. “2008–2012 were the hardest four years of my life,” he says. In addition to rebuilding his research program from the ground up, as dean of the School of Life Sciences, he recruited more than 70 new investigators for Tsinghua, tripling the size of the biology faculty. Shi also advised the government on “how to change research culture for the better.”
By 2011, Shi felt his laboratory had eclipsed the productivity of his former laboratory at Princeton. Work on CED-4 continued, with results that showed a new mode of activation for CED-3 and potentially other caspases (10). The work suggested that specific conformations present in the protein complex, not simply a high local concentration, could lead to caspase activation (11). Shi also looked to Drosophila to investigate details of conserved and divergent mechanisms. He estimates that over the years, he and his laboratory have solved the structures of more than 40 proteins and protein complexes involved in apoptosis in worms, flies, and mammals.
Shi and his colleagues have increasingly adopted cryo-electron microscopy, a technique in which, unlike X-ray crystallography, samples can be visualized in their free form. “The biological problems remain the same. What has changed is the technology that allows us to tackle these problems. The technology has changed in fundamental ways,” he explains. Now there are “powerful synchrotrons with hard X-ray free electron laser (XFEL).” Shi expects the technology to continue to evolve, noting that within five to ten years, XFEL could “provide the means to solve structures with…little crystals.” Shi stands ready to embrace the rapidly changing technology. “I am fully ready to learn new tricks.”
Footnotes
This is a Profile of a recently elected member of the National Academy of Sciences to accompany the member’s Inaugural Article on page 16254.
References
- 1.Hu Q, et al. Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci USA. 2014;111:16254–16261. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Berg JM. Specific DNA-RNA hybrid binding by zinc finger proteins. Science. 1995;268(5208):282–284. doi: 10.1126/science.7536342. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388(6637):87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Massagué J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 5.Qin H, et al. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399(6736):549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 7.Chai J, et al. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406(6798):855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 8.Wu G, et al. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408(6815):1008–1012. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- 9.Yan N, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437(7060):831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 10.Qi S, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141(3):446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y. Caspase activation: Revisiting the induced proximity model. Cell. 2004;117(7):855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]


