Significance
Plants that provide food and housing to animals in return for defense against enemies are classic examples of mutualistic partnerships in nature. Here, we show that the evolution of such plant–animal mutualisms also can lead to a trajectory of accelerated accumulation of plant species in the lineages that participate in these cooperative interactions. We found that the evolution of plant organs (extrafloral nectaries) that facilitate mutualisms with animal defenders was repeatedly followed by increased rates of diversification across distantly related plant lineages. These results suggest that by enabling ecological interactions with animals, the convergent evolution of relatively simple glands changed the course of plant evolution toward greater protection from pests and accelerated the generation of biodiversity.
Keywords: mutualism, extrafloral nectaries, plant–insect interactions, lineage diversification rates, plant defense
Abstract
The ability of plants to form mutualistic relationships with animal defenders has long been suspected to influence their evolutionary success, both by decreasing extinction risk and by increasing opportunity for speciation through an expanded realized niche. Nonetheless, the hypothesis that defense mutualisms consistently enhance plant diversification across lineages has not been well tested due to a lack of phenotypic and phylogenetic information. Using a global analysis, we show that the >100 vascular plant families in which species have evolved extrafloral nectaries (EFNs), sugar-secreting organs that recruit arthropod mutualists, have twofold higher diversification rates than families that lack species with EFNs. Zooming in on six distantly related plant clades, trait-dependent diversification models confirmed the tendency for lineages with EFNs to display increased rates of diversification. These results were consistent across methodological approaches. Inference using reversible-jump Markov chain Monte Carlo (MCMC) to model the placement and number of rate shifts revealed that high net diversification rates in EFN clades were driven by an increased number of positive rate shifts following EFN evolution compared with sister clades, suggesting that EFNs may be indirect facilitators of diversification. Our replicated analysis indicates that defense mutualisms put lineages on a path toward increased diversification rates within and between clades, and is concordant with the hypothesis that mutualistic interactions with animals can have an impact on deep macroevolutionary patterns and enhance plant diversity.
Ever since the key innovation hypothesis was first proposed in the 1940s (1, 2), the origination of novel traits has been a popular yet controversial explanation for the exceptional disparity in species richness observed across clades in the tree of life. Despite decades of research linking traits to diversification, we have remarkably few examples of traits that have been convincingly demonstrated to spur diversification repeatedly across independent, distantly related groups. Notable exceptions include a number of ecologically important traits mediating interactions between plants and animals (3–6), suggesting that these interactions may be particularly important drivers of macroevolutionary patterns. Here, we test the hypothesis that plant defense mutualisms, a widespread and classically studied ecological interaction whereby plants provide food rewards to arthropod bodyguards in return for protection against natural enemies (7), increase the evolutionary diversification rate of the plant lineages that participate in them. The morphological traits that mediate defense mutualisms represent well-studied examples of characters hypothesized to expand a plant’s niche via interactions with mutualists and influence species success in various environmental contexts (8). Although the costs and benefits of participating in defense mutualisms are well studied (9), the hypothesis that the ecological impact of defense mutualisms leaves a predictable macroevolutionary signature, increasing lineage diversification within and among clades of plants, has only been examined in a single genus (10).
Defense mutualisms may have an impact on plant speciation and extinction rates via several mechanisms. Unlike the evolution of traits related to reproduction, which, more intuitively, could have an impact on lineage diversification (e.g., refs. 5, 11), the direct mechanism by which defense mutualisms are hypothesized to influence diversification is less obvious. One direct mechanism is a decreased incidence of damage and disease due to an enhanced defensive repertoire, which may allow for increased population sizes and, in turn, lower extinction rates (6). Additionally, by expanding the realized niche of a plant (12), defense mutualisms may broaden the range of habitats a plant can occupy (10), thereby increasing instances of allopatric speciation.
However, in addition to these direct mechanisms, the evolution of mutualistic traits may facilitate diversification indirectly. First, if niche expansion results in the successful occupation of more environments, mutualistic traits may increase the probability a lineage will encounter conditions ripe with ecological opportunity (e.g., new adaptive zones), which, in turn, will drive increases in diversification. In other words, the evolution of a trait may enable subsequent diversification via increasing exposure to new environments, some of which will harbor external drivers of radiation, such as the uplift of a mountain range or unoccupied niches. Second, the evolution of defense mutualisms may free up resources for the plant, and thereby facilitate the evolution of other innovative traits that subsequently enhance diversification. These indirect effects need not be contingent on the existence of the direct effects mentioned above, and represent a largely overlooked hypothesis concerning how traits can affect diversification (13–15).
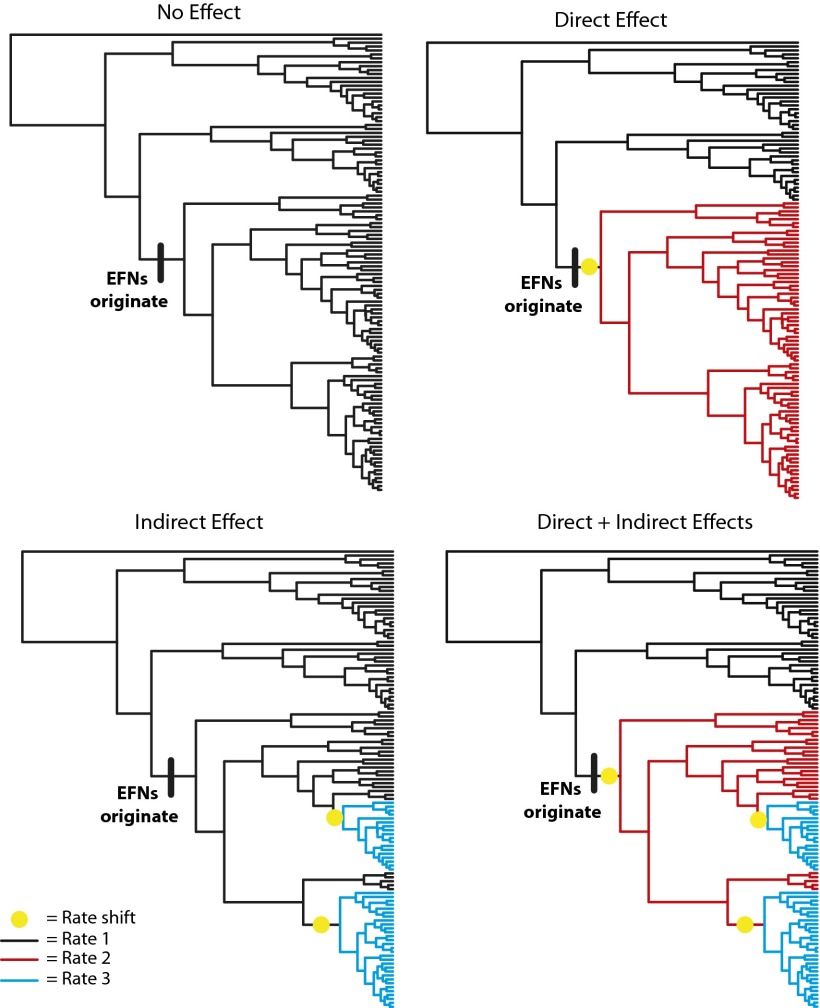
We suggest that indirect impacts of trait evolution on diversification should be reflected in a phylogenetic pattern in which the origination of a trait is followed by an increased probability of subsequent, downstream rate shifts relative to clades that lack the trait (Fig. 1). Because the indirect effect of the trait is contingent upon additional conditions (e.g., ecological opportunity, the evolution of another trait), there may be a substantial lag between the origin of the trait and rate shifts. Alternatively, a direct effect of the trait on the diversification rate is consistent with a pattern whereby a sustained rate shift occurs concomitantly with, or on the same branch as, the origin of the trait on the phylogeny (Fig. 1). Direct and indirect patterns are not mutually exclusive, and both patterns may be detectable on a single phylogeny (Fig. 1).
Fig. 1.
A conceptualization of phylogenetic patterns consistent with direct or indirect effects of EFNs (or any trait) on lineage diversification. A net change in diversification may be due to direct or indirect mechanisms. In the Upper Right, a rate shift occurs concomitantly with the origin of EFNs, consistent with a direct effect. If one or more shifts occur with some delay (Lower), this is consistent with a hypothesis that a trait has an indirect or context-dependent effect on diversification rates.
We focus on the macroevolutionary consequences of the repeated origination of extrafloral nectaries (EFNs), nectar-secreting glands found on nonfloral plant tissues that provide food for a wide array of beneficial arthropod bodyguards (16). EFNs are well studied ecologically, and their only known function is defense against herbivores and microbial pathogens by attracting natural enemies (17). Such features have evolved hundreds of times and occur in about a quarter of all vascular plant families (18). Here, we first ask whether, across all vascular plants, families containing species with EFNs are associated with higher diversification rates than families without EFNs. We then focus in on the phylogenetic history and evolution of EFNs in six distantly related plant clades to evaluate whether EFNs are linked, directly or indirectly, to increased lineage diversification rates. As such, this study represents a replicated, multiscale test of the macroevolutionary consequences of a convergently evolved and ecologically important mutualistic trait.
Results and Discussion
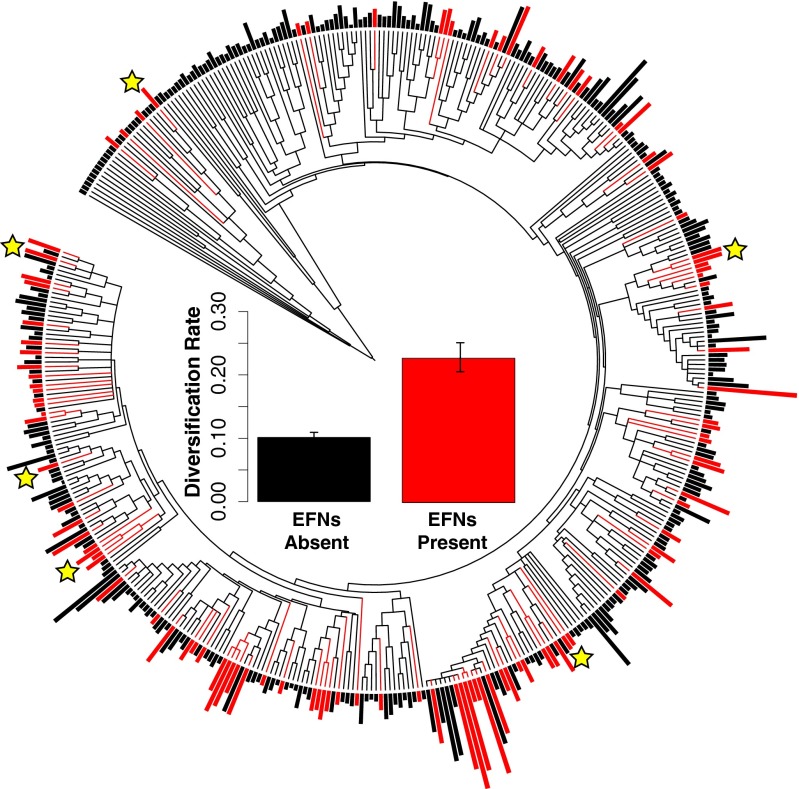
In a global analysis of vascular plant families, we combined published records of EFN occurrence (18) with fossil calibrated megatrees (19–21) to compare net diversification rates across families with and without species with EFNs. Overall, net diversification rates were more than twofold higher among the 108 families that contain instances of species with EFN compared with the ∼300 families without species with EFNs (Fig. 2 and Tables S1 and S2). Because our current knowledge likely underestimates the number of families with EFNs by ∼7% (18), we repeated this analysis with randomized inclusion of EFNs in otherwise non-EFN families and found the initial result to be robust to missing information (Fig. S1). Additionally, we found no evidence that EFN-bearing clades were older than non-EFN clades, suggesting they did not have more time to accumulate their greater number of species than clades without EFNs (Fig. S1). Finally, phylogenetic nonindependence did not confound estimates of the relationship between EFNs and species richness, because the evolution of EFNs was significantly unstructured compared with the null expectation of Brownian motion evolution [angiosperm phylogeny group megatree (APGIII): D = 0.75, P < 0.001; Zanne et al. megatree (21): D = 0.64, P = 0.006].
Fig. 2.
Phylogeny of vascular plant families (APGIII) (19), with families containing species with EFNs colored red. Outer bars correspond to the age-standardized number of species [i.e., (log number of species)/(age of plant family in millions of years)]. Yellow stars mark the families of the six clades analyzed subsequently in this study. (Inset) Mean diversification rate (r) ± SE of families with and without species with EFNs calculated according to the method of Magallon and Sanderson (41) assuming no extinction. See Tables S1 and S2 for F and P statistics and calculations with additional extinction fractions for both megatrees.
Results from our global analysis are consistent with a pattern in which there is a net positive effect of this mutualistic trait on rates of species diversification across the hundreds of independent origins of EFNs. Nonetheless, these results should be interpreted with caution due to the scale of the analysis. In particular, at this broad level, it is not possible to link shifts in diversification directly with the origin and loss of EFNs. Additionally, cases of EFNs may be more likely to be reported in speciose families simply because of their relatively large size, creating a sampling effect.
To address the limitations of the global analysis and to test for direct vs. indirect evolutionary consequences of EFN evolution, we pursued analyses at a finer taxonomic scale by reconstructing the evolution of EFNs in six distantly related plant clades (yellow stars in Fig. 2): Byttneria (Malvales), Senna (Fabales), Turnera (Malpighaiales), Viburnum (Dipsacales), Polygoneae (Caryophyllales), and Pleopeltis (Polypodiales). We selected these clades because they have recently published phylogenies, are known to contain species with and without EFNs (10, 18, 22–27), and are distantly related to one another. For Senna, which was previously investigated for a link between EFN and diversification rates (10), we added recently published records on EFNs in an additional clade (28). Together, these six plant groups encompass over 350 My of evolution since diverging from a common ancestor, contain a wide variety of growth forms and life-history strategies, and occupy diverse habitats across the globe.
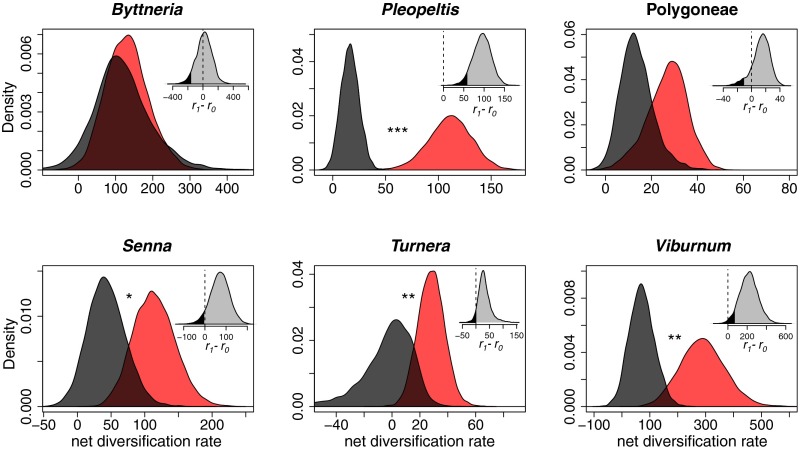
For each lineage, we first investigated whether a macroevolutionary model invoking state-dependent diversification rates [binary state speciation and extinction model (BiSSE)] (29, 30) explained the phylogenetic distribution of EFNs and diversification patterns. We found that EFNs were associated with higher mean net diversification rates (speciation rate − extinction rate) compared with lineages lacking EFNs in all six plant groups (Fig. 3). We assessed statistical significance according to the percentile of observed zero differences in state-dependent net diversification rates according to the post–burn-in Markov chain Monte Carlo (MCMC) interval. Differences in rates were below the 0.05 (one-tailed) percentile for Pleopeltis (P < 0.001), Turnera (P = 0.049), and Viburnum (P = 0.008); below 0.1 for Senna (P = 0.06); and nonsignificant for Polygoneae (P = 0.13) and Byttneria (P = 0.43) (Fig. 3, Inset). Combining the probabilities from the individual clades indicated that there was a significant overall positive association between EFNs and diversification rate (Z = 4.087, P < 0.001).
Fig. 3.
Marginal distribution of net diversification rate (speciation − extinction) parameters in EFN-present (red) and EFN-absent (black) clades from an analysis using the Bayesian implementation of BiSSE (29, 30) on MCC trees with median node heights from BEAST analyses for each lineage. *P < 0.1; **P < 0.05; ***P < 0.001. (Insets) Histograms represent the joint marginal distribution of the difference between EFN and non-EFN diversification rates, with the >0.05% probability quantile shaded dark gray and a dotted line at zero.
In simulations using the observed trees and inferred transition rates, but where EFNs evolved independently from rate shifts, we found BiSSE type 1 error rates ranging from 3 to 34% for individual topologies (Fig. S2). However, down-weighting the observed P values by the probability of seeing observed results in simulations still resulted in an overall combined probability of less than 0.001 (Z = 4.045). We also confirmed the results obtained from BiSSE using an alternative methodological approach that paired marginal ancestral state reconstructions of EFNs (31) with a recently developed reversible jump Bayesian framework for modeling diversification rates [Bayesian analysis of macroevolutionary mixtures (BAMM)] (32). Here too, we found that EFN portions of the phylogenies had higher mean net diversification rates than non-EFN portions of the phylogeny in the same four of six lineages examined (Table 1). Overall, the broad pattern across clades is consistent with hypothesis that EFNs play a role in increased plant diversification.
Table 1.
Diversification estimates obtained for EFN and non-EFN clades from BAMM analyses
| Whole trees | Sister clades | |||||||
| Plant group | rEFN | rnonEFN | Shifts/cladeEFN | Shifts/cladenonEFN | rEFN | rnonEFN | Shift/total timeEFN | Shift/total timenonEFN |
| Byttneria | 172.25 (53.3) | 212.40 (105.24) | — | — | — | — | — | — |
| Pleopeltis | 65.11 (29.56) | 31.5 (9.88) | 3.5 (2.45) | 3 (2.16) | 65.11 (21.18) | 46.19 (18.59) | 1.8 (0.93) | 2.63 (1.36) |
| Polygoneae | 23.66 (7.26) | 33.41 (6.83) | 2.5 (1.87) | 5.5 (3.02) | 23.66 (7.26) | 39.38 (7.73) | 2.08 (1.0) | 1.31 (0.41) |
| Senna | 113.54 (27.24) | 50.70 (28.15) | 4.6 (3.2) | 1 (1) | 113.54 (27.24) | 67.27 (39.34) | 7.02 (2.93) | 30.78 (10.28) |
| Turnera | 24.23 (6.42) | 18.22 (7.22) | — | — | — | — | — | — |
| Viburnum | 266.6 (67.92) | 114.4 (43.86) | 5 (3.32) | 2.5 (1.87) | 266.6 (67.92) | 184.42 (79.34) | 20.02 (7.35) | 30.81 (12.79) |
The mean (and SD) are reported. Sister clade comparisons were not possible for Byttneria and Turnera because of tree shape. Boldfaced numbers represent the larger of the estimates of EFN to non-EFN for each metric. EFN, EFN present; nonEFN, EFN absent; r, whole-tree net diversification rate (speciation rate − extinction rate), shifts/clade, number of rate shifts within the targeted sister clade, shift/time, number of rate shifts per total summed branch length in that targeted sister clade.
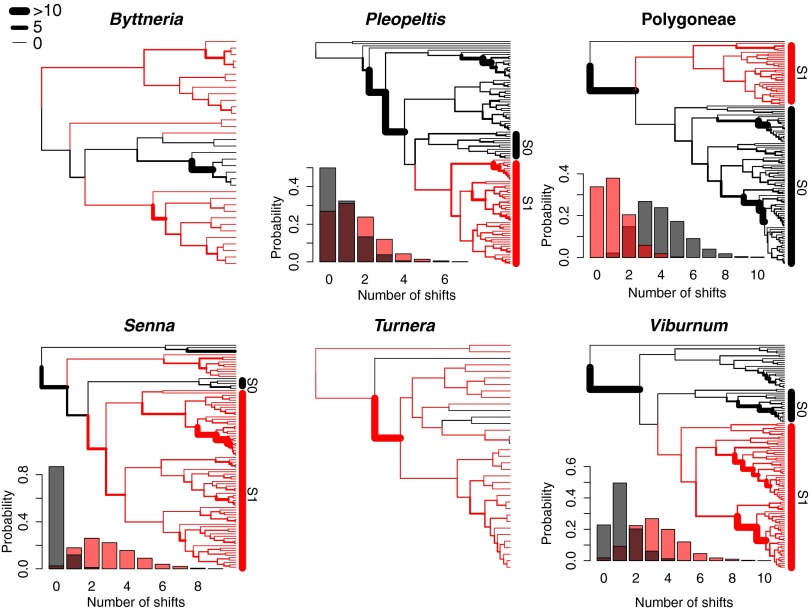
To test whether the increased rates of diversification associated with EFNs were consistent with direct or indirect effects on diversification, we used the BAMM framework (32) to model the number and placement of rate shifts on each phylogeny with respect to the marginal probability of EFN presence or absence. We found that rate shifts across our six clades were rarely placed with high confidence on the same branch as EFN transition events. Instead, the shifts that were responsible for the increased net diversification rate in EFN clades were commonly estimated to occur with some delay after the inferred origins of the trait (Fig. 4). Additionally, results were divided with respect to the hypothesis that defense mutualisms are favored by natural selection, and thus should be infrequently lost. In BiSSE analyses, rates of EFN gain were estimated as higher than rates of loss in three of the six clades (Table S3), whereas maximum likelihood (ML) estimates supported five of the six clades as having higher rates of EFN gain than loss (Table S4).
Fig. 4.
Diversification rate shifts in the EFN and non-EFN clades of six plant clades. For each group, the MCC tree is shown with branches that subtend nodes with a high marginal probability of EFNs in red and branches that subtend nodes with a high marginal probability of non-EFN in black. Branch widths are scaled to Bayes factors, representing confidence that a rate shift occurred on that branch. Bars to the right of phylogenies display the EFN (S1) and non-EFN (S0) sister clades used in sister-clade comparisons. (Insets) Histograms display the posterior distribution of the number of rate shifts in the EFN clade (red) and the sister non-EFN clade (gray).
In four groups, tree shape and the distribution of EFNs allowed for an additional comparison of the number of rate shifts that occurred in sister clades with and without EFNs. This approach allowed us to ask whether EFN clades contain more rate shifts than their non-EFN sisters while controlling for clade age. Indeed, we found that in three of the four plant groups examined, rate shifts were estimated to have occurred more frequently in EFN compared with non-EFN sister clades (Fig. 4 and Table 1). These same three groups displayed significant associations between EFNs and diversification rates in analyses with BiSSE. Sister clade comparisons of net diversification rates in the four groups mirror these results and reveal the directionality of the shifts: In three groups (Pleopeltis, Viburnum, and Senna), sister clades with EFNs had higher net diversification rates than sister clades without EFNs (Table 1). Because lineages with EFNs have greater total branch length, there is the potential for these lineages to have more shifts than lineages lacking EFNs without EFNs increasing shift density (number of shifts/total branch length) per se. Indeed, the density of rate shifts is not higher in EFN clades (Table 1). Accordingly, EFNs may be initiating a positive feedback, whereby slightly increased rates of diversification enhance the number of future rate shifts without affecting shift density. Regardless of the mechanism, EFNs are frequently associated with higher plant diversification.
Our results within and between clades suggest that EFNs, which are ecologically important, common, and functionally convergent traits across vascular plants, repeatedly set plant lineages on a path toward higher rates of lineage diversification. At fine phylogenetic scales, EFNs were generally associated with a higher incidence of positive, but delayed, diversification rate shifts. This result suggests that the means by which EFNs facilitate diversification may be contingent on other factors (14), such as developmental differences in EFN types, the presence or absence of other morphological traits, or the environmental conditions in which they occur. EFNs on their own may not be causally linked with immediate increased ecological opportunity; rather, they may serve as indirect innovations, facilitating subsequent diversification rate shifts. However, despite the fine-scale variation seen among the six clades studied here, we found a consistent pattern of EFNs associated with families with higher diversification across all vascular plants. Thus, although the ecological impacts of EFNs are variable in space and time (33), macroevolutionary patterns of EFNs are consistent across phylogenetic scales.
Other traits hypothesized to increase diversification rates have also shown a delayed association with rate shifts [e.g., C4 photosynthesis (34), mammary glands (35), complete metamorphism (36)], suggesting that this pattern may be widespread. However, testing causal hypotheses that link a trait with a delayed set of rate shifts can be challenging due to the possibility of interceding traits and evolutionary transitions in the same area of the phylogeny. This issue is one reason why evolutionary replication is key, because it increases our confidence that a particular trait may or may not be playing a role, allowing us to disentangle the complex ways in which traits interact with each other and the environment to influence diversification. For EFNs, our work within and between clades suggests that over deep time, these important defensive traits have enhanced diversification rates and, ultimately, the diversity of plant species.
Methods
Vascular Plant Family Analysis.
To test for a global association between defense mutualism and plant diversification, we compared net diversification rates of vascular plant families with and without species with EFNs. Families were scored as either containing or not containing accounts of at least one species with EFNs based on previous work (18). We used two published megatree phylogenies to account for phylogenetic uncertainty. The first was the APGIII megatree from the Angiosperm Phylogeny Group (19) acquired from Phylomatic (R20120829) (37) using one representative tip per family. The APGIII tree is a compilation of previously published plant phylogenies and gives the most up-to-date estimate of relationships. We time-calibrated the APGIII phylogeny by adjusting branch lengths according to fossil-based age estimates from Wikström et al. (20) using parametric rate-smoothing estimates with the program Phylocom (38). The second phylogeny was a rooted vascular plant megatree published by Zanne et al. (21), which was calibrated according to divergence time estimates from the broadly sampled molecular phylogeny of Soltis et al. (39) and 39 fossil calibration points (21). For analyses, this tree was trimmed so that each family was represented by only one tip using the drop.tip function in the R package Analyses of Phylogenetics and Evolution (APE) (40), which preserves topology and branch lengths. Genera were assigned to families according to the supplementary data in the study by Zanne et al. (21).
We calculated the net diversification rate using the method of Magallón and Sanderson (41) implemented in GEIGER (42). We calculated rates based on species richness and clade ages estimated as terminal branch lengths from both the APGIII tree and the tree published by Zanne et al. (21). We repeated calculations for four values of e, the extinction rate, expressed as a fraction of the speciation rate: 0, 0.1, 0.5, and 0.9. The number of species in each family was taken from Stevens (43). Because a range of species counts for a family was reported in some cases, we repeated all analyses across maximum and minimum species estimates. The difference in the mean net diversification rate of families with and without EFNs was then analyzed by a two-way ANOVA. Because our current knowledge likely underestimates the number of families with EFNs by 4–9% (18), we repeated this analysis 10,000 times, each time converting 10 (an additional ∼9% of the current total) randomly selected EFN-absent families to EFN-present families to simulate conservatively the discovery of new families with EFNs.
We tested for phylogenetic signal in the presence of species with EFNs in a family via the estimation of Fritz and Purvis’ D for binary traits, which is a measure of sister-clade differences in a discrete character state for a given phylogeny (44). An estimated D of 1 implies a distribution that is random with respect to the phylogeny, whereas a D of 0 implies a distribution expected under Brownian motion (44). Using the R package caper (45), we calculated D for the presence of EFNs, and to assess significance, we compared our estimate with simulated distributions of D under (i) randomly reshuffled trait values across the tips of the tree and (ii) trait evolution under Brownian motion. Each simulation included 10,000 permutations. This approach preserves the phylogenetic relationships of families, as well as the number of families assigned to each character state, although it varies the distribution of character states across the tree.
Clade-Level Analyses.
We selected six vascular plant clades for phylogenetic comparative analyses that (i) had sequence representation in GenBank and (ii) were known to contain species with and without EFNs based on descriptions in the literature: Byttneria (Malvaceae), Pleopeltis (Polypodiaceae), Polygoneae (Polygoneaceae), Senna (Fabaceae), Turnera (Passifloraceae), and Viburnum (Adoxaceae). Each of these clades represents an independent evolutionary origin of EFNs, and, together, they span a large portion of the angiosperm tree of life.
Phylogenetic Inference.
We reconstructed a distribution of time-calibrated phylogenies separately for each genus using Bayesian methods to include the highest possible number of species because the most recently published phylogenies of our clades of interest were frequently not ultrametric. Sequence availability for each group was evaluated, and sequences were obtained from the GenBank using the PhyLoTA Browser (release 1.5) (46). Molecular markers were chosen for inclusion in phylogenetic analyses if they were sampled for over 30% of the species available in the GenBank (accession numbers have been deposited in DataDryad; dx.doi.org/10.5061/dryad.17fj8). Outgroup taxa were selected based on the most recent published phylogeny or from the parent cluster in PhyLoTA based on overlapping sequence coverage with in-group taxa. Nucleotide sequences were aligned using the L-INS-I strategy in MAFFT (version 6) (47) with a gap opening penalty of 1.53 and an offset value of 0.0 using the R (48) package PHYLOCH (49). We trimmed aligned sequence ends to minimize missing data among taxa and checked alignments by hand. We used jModelTest (50) to determine appropriate substitution models for each partition based on Akaike’s information criterion (Table S5) and estimated starting parameters for Bayesian inference using the R package phanghorn (51).
For each of the six clades, we estimated the joint posterior distribution of topologies and relative node divergence times using three independent Bayesian MCMC searches in Bayesian Evolutionary Analysis Sampling Trees (BEAST) (52). Each marker was partitioned with its own unlinked, previously estimated substitution model. We used one uncorrelated exponential relaxed clock model to estimate node heights for all of the partitions. For each clade, three MCMC searches were run for 1 million generations sampled every 10,000 generations using a random starting tree. Trees were rooted by constraining the in-group to be monophyletic. Convergence of each Bayesian run was assessed by plotting the log-likelihood of sampled trees and parameters using Tracer (version 1.5) (53). The first 25% of sampled trees were removed from each run as a burn-in. A maximum clade credibility (MCC) tree was identified from the combined output of the three MCMC runs using LogCombiner (54) and TreeAnnotator (55).
Character State Assignment.
The presence or absence of EFNs was coded as a discrete, binary character state. The distribution of EFNs within each clade was evaluated using previous publication records (10, 18, 22–27) and herbarium specimens from the Bailey Hortorium of Cornell University, the herbaria of the Yale Peabody Museum of Natural History, and digitized specimens in the JSTOR Global Plants database (plants.jstor.org). The world list of plants with EFNs database can be accessed at www.extrafloralnectaries.org (56).
Lineage Diversification Analyses.
We evaluated whether net diversification rates in each clade were dependent on EFN state using BiSSE (29) and BAMM (32). Outgroup taxa and multiple individuals per species were pruned from the MCC tree for each analysis so that resulting phylogenies contained only one sample per species.
We used the BiSSE (29) state-dependent speciation and extinction model to estimate net diversification (speciation − extinction) rates in lineages with and without EFNs. We implemented the Bayesian MCMC BiSSE algorithm in the diversitree package in R (31). Because some species are missing from our phylogenies for each of our six groups, we accounted for missing taxa in these analyses by including information on the proportions of taxa (included and missing) assigned to each character state (30) using species descriptions and clade estimates from previous publications (10, 23–26, 57). In cases where the character state of missing species was unknown, we assumed the proportions of character states in our known samples were representative (Table S6).
To test whether EFN and non-EFN lineages had different diversification rates, we used Bayesian MCMC BiSSE analyses on the MCC tree for each clade. We used exponential priors for all parameters estimated using the starting.point.bisse function in diversitree, with a mean of twice the state-independent net diversification rate. Initial models were fit using parameter starting points estimated from a constant-rate birth–death model. We assumed a Markov (Mk) model of evolution for the trait in all cases. First, a primary MCMC was run for each clade for 1,000 generations with an arbitrary tuning parameter of 0.1. We used the posterior parameter distributions of these initial MCMC runs to estimate tuning parameters of the final MCMC analyses, which each ran for 10,000 generations. Significance was assessed according to the credible set of the differences between state-dependent net diversification rates. Finally, to gain insight into the probability of seeing our results if EFNs were evolving independent of rate shift location, we conducted ML-BiSSE analyses across the same topologies using simulated trait data. EFN tip states were simulated as discrete characters 100 times for each topology using an Mk2 model with parameter estimates fit using the observed trait data with the fitDiscrete function in the R package GEIGER (41). Simulations with fewer than five species in each state were rejected to condition on having more than a small number of species in one state. Root states for simulations were determined using a random draw from a binomial distribution with a probability of successfully drawing a state proportional to the marginal probability of that state at the root in the ancestral reconstruction using the asr.bisse function in diversitree (31). We used a weighted Z-test (58) to combine probabilities from the six BiSSE analyses, both with and without weights equal to the inverse of the probability of seeing our observed P value in simulations (to account for type 1 error rate).
To examine (i) whether lineages with EFNs contained more rate shifts than lineages without EFNs, (ii) whether EFN lineages had a higher density of rate shifts than non-EFN clades, and (iii) where rate shifts occurred on phylogenies in relation to EFN origination or loss, we used BAMM (32) and the package BAMMtools (59). For each clade, we performed three BAMM runs on the MCC phylogeny from the log-combined BEAST analyses to avoid getting stuck in local optima. Each BAMM analysis was run for 1 million MCMC generations, sampling parameters every 50,000 generations. We accounted for incomplete sampling in each clade according to diversity estimates from publications (10, 23–26, 57). We ran Bayesian MEDUSA-like models, where the rates of speciation and extinction were constant within shift regimes by setting the updateRateLambdaShift and lambdaShift0 parameters to 0. We computed tree appropriate rate priors using the setBAMMpriors function in BAMMtools, and used a flattened PoissonRatePrior of 0.1 and a minimum clade size for rate shifts (minCladeSizeforShift) of 2. We assessed convergence of the three BAMM runs for each clade by ensuring the effective sample sizes of log-likelihoods, number of processes, and evolutionary rate parameters were greater than 500 using the CODA library (60).
We assigned the presence or absence of EFNs to clades according to the probability of each state at internal nodes using the asr.marginal function in diversitree (31), which performs marginal reconstructions of ancestral states for each node. To account for potentially misleading effects of trait-associated diversification rates, we reconstructed ancestral states of EFNs under the BiSSE model, which was fit for each clade using the using the make.bisse and find.mle functions, with starting parameter guesses of the mean parameter estimates from MCMC BiSSE analyses. We then asked whether portions of the tree in the EFN state have a higher net diversification rate than branches in the non-EFN state. Diversification rates were calculated using the getcladerate function in BAMMtools on BAMMobjects pruned to include only EFN or only non-EFN taxa using the subtreeBAMM function. This approach results in some internal branches being included in both EFN and non-EFN rate estimates; however, this double inclusion is conservative with respect to the hypothesis.
We visualized the probability of rate shifts on branches of the tree by scaling the edge widths of each plotted phylogeny according to the Bayes factor associated with that branch using the bayesFactorBranches function. This method corrects for differences in branch length or biases introduced by the prior on the number of diversification rates. We estimated the number of shifts in sister clades with and without EFNs for groups that had sister groups with and without EFNs that included at least two species each. We used the subtreeBAMM function to extract sister clades from the original BAMM objects. The shift density for each extracted sister subclade was calculated by dividing each sample from the subclade’s posterior by the sum of branch lengths for that subclade.
Data Availability.
Phylogenies, character states, GenBank accession numbers, and R scripts are deposited in DataDryad (dx.doi.org/10.5061/dryad.17fj8).
Supplementary Material
Acknowledgments
We thank María Mercedes Arbo, Ana Maria Gonzalez, Elizabeth Otto, Harald Schneider, Matthew Jebb, and Tanja Schuster for help with EFN distributions within clades. We thank Gideon Bradburd, Michael Donoghue, Monica Geber, Harry Greene, Marc Johnson, Irby Lovette, Luke Mahler, Brigitte Marazzi, Nicholas Mason, Sally Otto, Dan Rabosky, Michael Sanderson, Catherine Wagner, and three anonymous reviewers for providing discussion or comments that improved this project. A.A.A. was supported by Grant 1118783 of the Division of Environmental Biology of the National Science Foundation (NSF) and by the John Templeton Foundation. M.G.W. was supported by the Society for the Study of Evolution’s Rosemary Grant Award and by the NSF (Graduate Research Fellowship and Doctoral Dissertation Improvement Grant).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The phylogenies, character matrices, GenBank accession numbers, and R scripts reported in this paper have been deposited in DataDryad, dx.doi.org/10.5061/dryad.17fj8.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413253111/-/DCSupplemental.
References
- 1.Miller AH. Some ecologic and morphologic considerations in the evolution of higher taxonomic categories. Ornithologie als Biologische Wissenschaft 1949 , eds Mayr E, Schüz E (Universitätsverlag Winter, Heidelberg), pp 84–88. [Google Scholar]
- 2.Simpson GG. Tempo and Mode in Evolution. Columbia Univ Press; New York: 1944. [Google Scholar]
- 3.Hodges SA. Floral nectar spurs and diversification. Int J Plant Sci. 1997;158(Suppl 6):S81–S88. [Google Scholar]
- 4.Lengyel S, Gove AD, Latimer AM, Majer JD, Dunn RR. Ants sow the seeds of global diversification in flowering plants. PLoS ONE. 2009;4(5):e5480. doi: 10.1371/journal.pone.0005480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proc Biol Sci. 2004;271(1539):603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell BD, Dussourd DE, Mitter C. Escalation of plant defense: Do latex and resin canals spur plant diversification? Am Nat. 1991;138(4):881–900. [Google Scholar]
- 7.Janzen DH. Coevolution of mutualism between ants and acacias in Central America. Evolution. 1966;20(3):249–275. doi: 10.1111/j.1558-5646.1966.tb03364.x. [DOI] [PubMed] [Google Scholar]
- 8.Boucher DH. The Biology of Mutualism: Ecology and Evolution. Oxford Univ Press; New York: 1985. [Google Scholar]
- 9.Heil M, McKey D. Protective ant-plant interactions as model systems in ecological and evolutionary research. Annu Rev Ecol Evol Syst. 2003;34:425–453. [Google Scholar]
- 10.Marazzi B, Sanderson MJ. Large-scale patterns of diversification in the widespread legume genus Senna and the evolutionary role of extrafloral nectaries. Evolution. 2010;64(12):3570–3592. doi: 10.1111/j.1558-5646.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- 11.Barraclough TG, Harvey PH, Nee S. Sexual selection and taxonomic diversity in passerine birds. Proc R Soc Lond B Biol Sci. 1995;259(1355):211–215. [Google Scholar]
- 12.Bruno JF, Stachowicz JJ, Bertness MD. Inclusion of facilitation into ecological theory. Trends Ecol Evol. 2003;18(3):119–125. [Google Scholar]
- 13.de Queiroz A. Contingent predictability in evolution: Key traits and diversification. Syst Biol. 2002;51(6):917–929. [PubMed] [Google Scholar]
- 14.Donoghue MJ. Key innovations, convergence, and success: Macroevolutionary lessons from plant phylogeny. Paleobiology. 2005;31(2) Suppl:77–93. [Google Scholar]
- 15.Wainwright PC, et al. The evolution of pharyngognathy: A phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Syst Biol. 2012;61(6):1001–1027. doi: 10.1093/sysbio/sys060. [DOI] [PubMed] [Google Scholar]
- 16.Koptur S. Insect Plant Interactions. Vol IV. CRC; Boca Raton, FL: 1992. Extrafloral nectary-mediated interactions between insects and plants; pp. 81–129. [Google Scholar]
- 17.Bentley BL. Extrafloral nectaries and protection by pugnacious bodyguards. Annu Rev Ecol Syst. 1977;8:407–427. [Google Scholar]
- 18.Weber MG, Keeler KH. The phylogenetic distribution of extrafloral nectaries in plants. Ann Bot (Lond) 2013;111(6):1251–1261. doi: 10.1093/aob/mcs225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angiosperm Phylogeny Group An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APGIII. Bot J Linn Soc. 2009;161(2):105–121. [Google Scholar]
- 20.Wikström N, Savolainen V, Chase MW. Evolution of the angiosperms: Calibrating the family tree. Proc Biol Sci. 2001;268(1482):2211–2220. doi: 10.1098/rspb.2001.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506(7486):89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 22.Marazzi B, Endress PK, Queiroz LP, Conti E. Phylogenetic relationships within Senna (Leguminosae, Cassiinae) based on three chloroplast DNA regions: Patterns in the evolution of floral symmetry and extrafloral nectaries. Am J Bot. 2006;93(2):288–303. doi: 10.3732/ajb.93.2.288. [DOI] [PubMed] [Google Scholar]
- 23.Mercedes Arbo M, Espert SM. Morphology, phylogeny and biogeography of Turnera L. (Turneraceae) Taxon. 2009;58(2):457–467. [Google Scholar]
- 24.Otto EM, Janssen T, Kreier H-P, Schneider H. New insights into the phylogeny of Pleopeltis and related Neotropical genera (Polypodiaceae, Polypodiopsida) Mol Phylogenet Evol. 2009;53(1):190–201. doi: 10.1016/j.ympev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Schuster TM, Reveal JL, Kron KA. Phylogeny of Polygoneae (Polygonaceae: Polygonoideae) Taxon. 2011;60(6):1653–1666. [Google Scholar]
- 26.Whitlock BA, Hale AM. The phylogeny of Ayenia, Byttneria, and Rayleya (Malvaceae s. l.) and its implications for the evolution of growth forms. Syst Botany. 2011;36(1):129–136. [Google Scholar]
- 27.Weber MG, Clement WL, Donoghue MJ, Agrawal AA. Phylogenetic and experimental tests of interactions among mutualistic plant defense traits in Viburnum (Adoxaceae) Am Nat. 2012;180(4):450–463. doi: 10.1086/667584. [DOI] [PubMed] [Google Scholar]
- 28.Marazzi B, Conti E, Sanderson MJ, McMahon MM, Bronstein JL. Diversity and evolution of a trait mediating ant-plant interactions: Insights from extrafloral nectaries in Senna (Leguminosae) Ann Bot (Lond) 2013;111(6):1263–1275. doi: 10.1093/aob/mcs226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maddison WP, Midford PE, Otto SP. Estimating a binary character’s effect on speciation and extinction. Syst Biol. 2007;56(5):701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 30.FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst Biol. 2009;58(6):595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- 31.FitzJohn RG. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3(6):1084–1092. [Google Scholar]
- 32.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE. 2014;9(2):e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bronstein JL, Alarcón R, Geber M. The evolution of plant-insect mutualisms. New Phytol. 2006;172(3):412–428. doi: 10.1111/j.1469-8137.2006.01864.x. [DOI] [PubMed] [Google Scholar]
- 34.Spriggs EL, Christin P-A, Edwards EJ. C4 photosynthesis promoted species diversification during the Miocene grassland expansion. PLoS ONE. 2014;9(5):e97722. doi: 10.1371/journal.pone.0097722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 36.Nel A, Roques P, Nel P, Prokop J, Steyer JS. The earliest holometabolous insect from the Carboniferous: A “crucial” innovation with delayed success (Insecta Protomeropina Protomeropidae) Annales de la Société Entomologique de France. 2007;43(3):349–355. [Google Scholar]
- 37.Webb CO, Donoghue MJ. Phylomatic: Tree assembly for applied phylogenetics. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 38.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24(18):2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 39.Soltis DE, et al. Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot. 2011;98(4):704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- 40.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 41.Magallón S, Sanderson MJ. 2001 Absolute diversification rates in angiosperm clades. Evolution 55(9):1762–1780, and erratum (2006) 60(11):2411. [Google Scholar]
- 42.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24(1):129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- 43.Stevens P. 2001. Angiosperm Phylogeny Website, Version 12, July 2012 (Missouri Botanical Garden, University of Missouri, St. Louis, MO). Available at www.mobot.org/MOBOT/research/APweb/. Accessed September 3, 2014.
- 44.Fritz SA, Purvis A. Selectivity in mammalian extinction risk and threat types: A new measure of phylogenetic signal strength in binary traits. Conserv Biol. 2010;24(4):1042–1051. doi: 10.1111/j.1523-1739.2010.01455.x. [DOI] [PubMed] [Google Scholar]
- 45.Orme CDL, Freckleton RP, Thomas GH, Petzold T, Fritz SA. caper: Comparative analyses of phylogenetics and evolution in R. 2013 R package version 0.5.2. Available at R-Forge.R-project.org/projects/caper/. Accessed December 4, 2013.
- 46.Sanderson MJ, Boss D, Chen D, Cranston KA, Wehe A. The PhyLoTA Browser: Processing GenBank for molecular phylogenetics research. Syst Biol. 2008;57(3):335–346. doi: 10.1080/10635150802158688. [DOI] [PubMed] [Google Scholar]
- 47.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 48.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2012. [Google Scholar]
- 49.Heibl C. PHYLOCH: R language tree plotting tools and interfaces to diverse phylogenetic software packages. 2008 Available at www.christophheibl.de/Rpackages.html. Accessed November 1, 2013.
- 50.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 51.Schliep KP. phangorn: Phylogenetic analysis in R. Bioinformatics. 2011;27(4):592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rambaut A, Drummond AJ. Tracer v1.5. 2007 Available at beast.bio.ed.ac.uk/Tracer. Accessed December 1, 2009.
- 54.Rambaut A, Drummond A. 2012. LogCombiner v1. 7.5 MCMC Output Combiner. Available at beast.bio.ed.ac.uk. Accessed April 15, 2013. [Google Scholar]
- 55.Rambaut A, Drummond A. TreeAnnotator, version 1.7.5. 2012 Available at beast.bio.ed.ac.uk/TreeAnnotator. Accessed April 15, 2013.
- 56.Keeler KH. 2014. World list of plants with extrafloral nectaries. Available at www.extrafloralnectaries.org. Accessed October 14, 2014.
- 57.Clement WL, Donoghue MJ. Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evol Biol. 2012;12(1):73. doi: 10.1186/1471-2148-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18(5):1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 59.Rabosky DL, et al. BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol. 2014;5(7):701–707. [Google Scholar]
- 60.Plummer M, Best N, Cowles K, Vines K. CODA: Convergence diagnosis and output analysis for MCMC. R News. 2006;6(1):7–11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phylogenies, character states, GenBank accession numbers, and R scripts are deposited in DataDryad (dx.doi.org/10.5061/dryad.17fj8).