Significance
We discovered that cells on micropatterned strips change from highly persistent migration into striking oscillations upon the disassembly of microtubules. The oscillation phenomenon then allowed us to apply computer modeling to understand how the positive feedback in the local-excitation–global-inhibition (LEGI) mechanism, responsible for the persistence of migration, might be converted into negative feedback to drive oscillations upon the disassembly of microtubules. Our analyses led to the conclusion that microtubules facilitate the transport of inhibitory signals and their global distribution. Depending on the relative position of excitation and inhibitory signals, the resulting feedback in the integrated control circuit may be either positive or negative. Our finding therefore provides important insights into the role of microtubules in the control circuit of cell migration.
Keywords: cell migration, cell polarity, focal adhesion, LEGI, microtubules
Abstract
Microtubules are known to play an important role in cell polarity; however, the mechanism remains unclear. Using cells migrating persistently on micropatterned strips, we found that depolymerization of microtubules caused cells to change from persistent to oscillatory migration. Mathematical modeling in the context of a local-excitation–global-inhibition control mechanism indicated that this mechanism can account for microtubule-dependent oscillation, assuming that microtubules remove inhibitory signals from the front after a delayed generation. Experiments further supported model predictions that the period of oscillation positively correlates with cell length and that oscillation may be induced by inhibiting retrograde motors. We suggest that microtubules are required not for the generation but for the maintenance of cell polarity, by mediating the global distribution of inhibitory signals. Disassembly of microtubules induces cell oscillation by allowing inhibitory signals to accumulate at the front, which stops frontal protrusion and allows the polarity to reverse.
Cell migration plays an essential role in many important physiological processes, such as embryogenesis (1, 2), wound healing (3), immune responses (4), and in engineering applications such as tissue regeneration (5). Defects in cell migration can cause such severe problems as birth defects (6), vascular disease (7), and tumor metastasis (8).
Directional cell migration requires a defined polarity, generated by an integrated network of signals, cytoskeleton, and adhesions (9–11). Morphologically, polarized cell migration is associated with the formation of distinct protrusive frontal and retractive rear regions (12, 13), which are in turn associated with polarized networks of actin filaments and microtubules together with asymmetric distribution of signaling molecules including PI3Ks, PTEN, Rac, Rho, and Cdc42 (9, 14, 15). Microtubules were proposed to play a central role in the development and/or maintenance of cell polarity, such that cells lacking an intact dynamic network of microtubules are often immobile or poorly polarized (16, 17). Polarity may be mediated by the interactions of microtubules with signaling molecules, actin cytoskeleton, and substrate adhesions (18–21), for example by differential targeting of focal adhesions for asymmetric turnover (22–25).
Despite the intensive studies in the past decades, the control circuit for cell polarization and migration is still far from clear due to its complexity. Computer modeling, especially when accompanied with simulation to generate testable predictions, is a powerful tool for building a convincing understanding of complex processes whose consequences may not be intuitively predictable (26). Perhaps the best known control model for directed cell migration is the local-excitation–global-inhibition (LEGI) mechanism, proposed initially to explain the chemotaxis of Dictyostelium discoideum (27–32). In a generic LEGI model, two antagonistic signals are released at the front, one is a localized excitation/positive signal that can self-amplify; the other is a global inhibitory/negative signal that can spread across the cell. The distribution of net signals then defines areas of protrusion or retraction. The model was later modified to account for various aspects of the directional sensing during chemotaxis (31, 33–36). In addition, it was generalized to account for stochastic persistent random walk and the associated cell shape dynamics in the absence of guidance cues, where the “front” is defined as regions with protrusions of limited duration (37). Other models have also been proposed to explain the polarity and migration behavior of cells, using pseudopodia (38) or membrane tension (39) as the feedback components. Together, these mathematical models have provided useful insights into the control and guidance of cell migration. Nevertheless, many important aspects remain unclear, such as the function of microtubules in the control circuit. A combination of pharmacological perturbations and modeling of their effects may provide important insights for understanding the control mechanism.
The present study takes advantage of the highly persistent migration of fibroblasts and epithelial cells crawling along 1D adhesive strips (12, 40), and the intriguing oscillatory movement that we discovered when these cells were treated with microtubule disassembly agents. Cells lacking an intact microtubule network are still able to migrate directionally, albeit in a transient manner such that the direction switches back and forth in an oscillatory fashion with an average period of 45–170 min depending on the cell line. By computational modeling of this phenomenon, we were able to gain insight into not only the possible roles of microtubules in the control circuit of directional cell migration, but also essential temporal relations among the components.
Results
Cultured Cells Show Highly Persistent Migration Along Micropatterned Strips.
To facilitate the study of cell polarity, we micropatterned adhesive strips 4–24 µm in width (Fig. 1A) on glass coverslips with or without fibronectin coating, using linear polyacrylamide as the blocking agent to create nonadhesive areas (41). The strips constrained cell migration along one dimension (1D) and avoided the complexity of random migration on 2D surfaces.
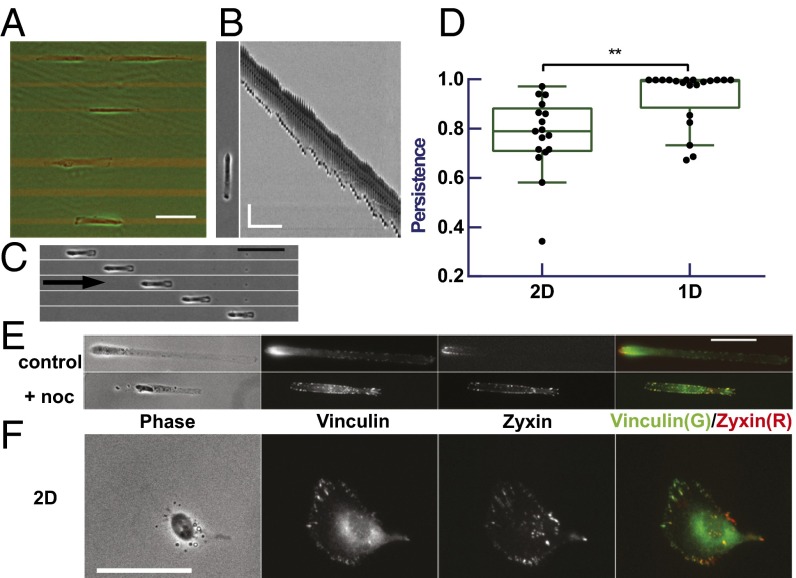
Fig. 1.
Migration of RPE-1 cells and localization of zyxin relative to vinculin. RPE-1 cells show highly persistent migration on 4- to 24-µm–wide strips (A; strips visible by red fluorescence; scale bar, 100 µm), as indicated by a typical kymograph (B; scale bar, 50 µm, 60 min) and time series (C; 60-min interval; scale bar, 100 µm; arrow indicates the direction of migration) of representative cells. Quantification of migration indicates a higher persistence on 1D strips (D; n = 20) than on 2D surface (D; n = 17, P = 0.005). On 1D strips, RFP-zyxin shows a stronger polarized localization in the rear (red) than immunostained vinculin (green) in control cells but not in nocodazole-treated cells (E). (Scale bar, 50 µm.) A similar but weaker difference is observed for cells on 2D surfaces (F). (Scale bar, 50 µm.) See also Fig. S2 and Movie S1 online.
RPE-1 cells showed highly persistent migration along 1D strips (12, 40) (Fig. 1 B and C, and Movie S1). Most cells had a measured persistence, defined as the ratio of net migration distance to total path length, of exactly 1 for a period of at least 5 h (Fig. 1D). Less than 20% of the 123 cells recorded ever changed the direction during the period of observation before the cell divided or collided with other cells (Fig. S1).
We imaged the distribution of focal adhesions in cells undergoing persistent migration, using cells expressing fluorescent zyxin or paxillin. Unexpectedly, RPE-1 cells expressing RFP-zyxin showed a much stronger polarized distribution of zyxin behind the nucleus (Fig. 1E), than other focal adhesion proteins such as paxillin (Fig. 2B), vinculin (Fig. 1E), or tensin [which like zyxin associates with focal adhesions at a late stage (42); Fig. S2A]. Zyxin also showed a preferential rear localization in RPE-1 cells undergoing persistent random walk on 2D surfaces, but the bias was not nearly as striking as in cells migrating along 1D strips (Fig. 1F). Persistent 1D migration and rear localization of zyxin were also observed in other cell lines such as NIH 3T3 fibroblasts (12) (Fig. S2 B–D). These results allowed us to use zyxin as a marker for cell polarization. The underlying mechanism for polarized zyxin localization may be related to the mechanical dependence of its binding kinetics (43, 44) and is being investigated in a separate study. Although centrosome may be considered as an alternative marker, its positioning is dependent on microtubules and its position relative to the nucleus is known to be variable particularly for cells on 1D substrate (45).
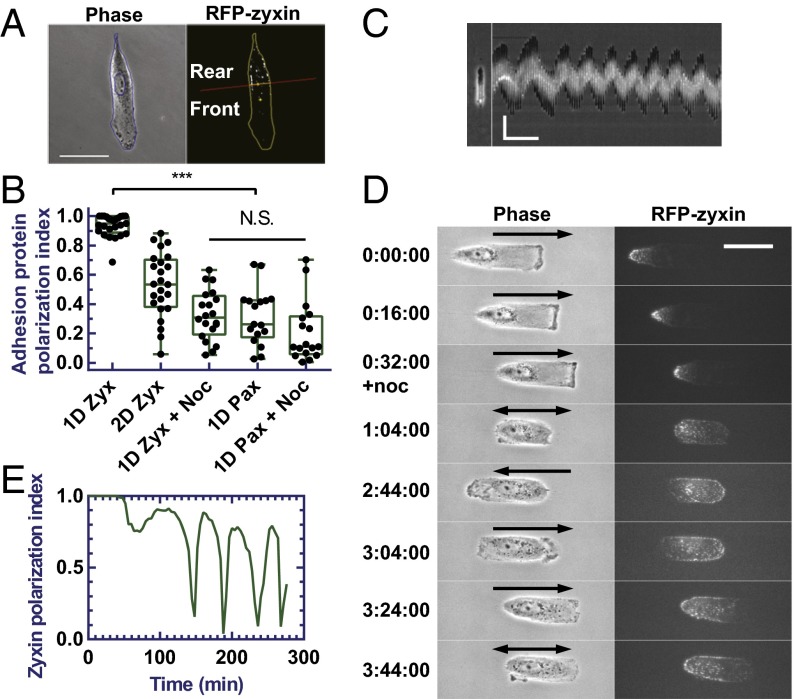
Fig. 2.
Oscillation of RPE-1 cells on 1D strips and redistribution of zyxin induced by nocodazole. Drawing shows how the fluorescent image of a cell is divided into front and rear regions, which are then used to quantify localization of focal adhesion proteins as polarization index (A). (Scale bar, 50 µm.) RFP-zyxin in control cells (n = 25) on 1D strips shows a much higher polarization index than RFP-zyxin in control cells (n = 24) on 2D surface and nocodazole-treated cells on 1D strips (n = 20), or EGFP-paxillin in control (n = 17) and nocodazole-treated (n = 17) cells on 1D strips (B; P < 0.0001). noc, nocodazole; pax, paxillin; zyx, zyxin. Cells treated with nocodazole undergo oscillatory migration as indicated by the kymograph of a representative cell (C). (Scale bar, 50 µm, 60 min.) Time-series images show the redistribution of RFP-zyxin in a representative cell migrating persistently on the strip followed by oscillations upon the treatment with nocodazole (D; migration direction indicated by arrows, double-headed arrows indicate switch of directions). (Scale bar, 50 µm.) Zyxin polarization index of the same cell shows the corresponding strong polarization before the addition of nocodazole followed by oscillations after the addition of nocodazole (E). See also Figs. S3 and S4 and Movies S1 and S2.
To quantify the polarized localization of zyxin, we separated a cell into a frontal region ahead of the nucleus and a rear region behind the nucleus, and then calculated the front and rear focal adhesion protein area density, defined as the total area occupied by a focal adhesion protein in the front or rear divided by the total area of the corresponding front or rear region (Fig. 2A). The value [1 – min (r, 1/r)], where r was the front-to-rear ratio of focal adhesion protein area densities, was then used as an index of polarization (see SI Materials and Methods for the detail). Random localization would give an index close to 0. Consistent with the qualitative observation, RFP-zyxin in cells undergoing persistent 1D migration showed an average index very close to 1, indicating a strong rear localization. In contrast to zyxin, EGFP-paxillin showed an average index of 0.3, consistent with a much more uniform distribution. RFP-zyxin in 2D cells showed an index value around 0.5, consistent with a weak rear localization (Fig. 2B).
Disassembly of Microtubules Causes Striking Cell Oscillation.
Microtubules play an important role in regulating cell polarity (18, 23), such that cells lacking an intact microtubule network typically lose their polarity (16, 24). To determine whether microtubules affect cells with a strong preestablished polarity, we treated RPE-1 cells migrating on 1D strips with 2 µM nocodazole to depolymerize microtubules.
Interestingly, treatment of nocodazole caused persistent migration to turn into oscillatory movement (Fig. 2C, Fig. S1, and Movie S1), which can last for more than 10 h until the cell entered an arrested division phase (46). Each episode of directional migration was terminated by either the appearance of protrusion at the trailing edge (Fig. S3C) or the retraction of the leading edge (Fig. S3D) to cause a reversal in direction. The period of oscillation ranged from around 20 min to more than 1 h (average, 45 ± 22 min; n = 45). NIH 3T3 cells showed a similar oscillation upon nocodazole treatment, but with a longer period of 169 ± 68 min (n = 16; Fig. S3A). Treatment of 5 µM Taxol, which is known to stabilize microtubules and cause microtubule reorganization (47), caused the cell to stop migration almost entirely, with an immobile (Fig. S4A) or slowly drifting (Fig. S4B) cell body and weak protrusions at both ends.
Zyxin, as a polarization marker, lost its strong stable rear localization in oscillating RPE-1 cells (Fig. 1E). Close observations indicated that zyxin started to spread toward the opposite end within 30 min of nocodazole treatment, while showing a concomitant decrease in overall intensity at focal adhesions that was not observed when imaging control cells over a similar period (Fig. 2D and Movie S2). Quantification of zyxin localization indicated a strong oscillation of the polarization index (Fig. 2E). The average of the polarization index decreases from 0.94 to 0.32 (Fig. 2B).
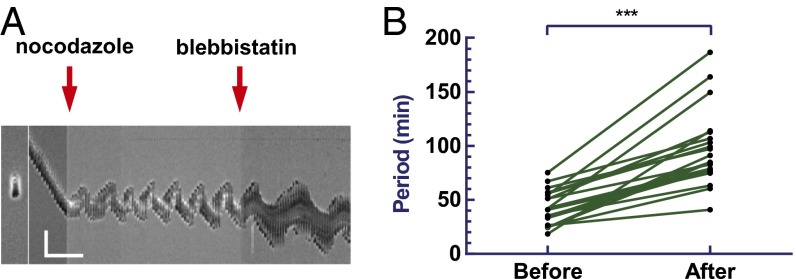
We suspect that, because the disassembly of microtubules is known to cause stabilization of focal adhesions (23–25), the resulting increase in the resistance to forward migration may eventually cause reversal of the direction of migration and drive the oscillation, by creating a negative-feedback loop. However, the ability of some oscillating cells to migrate away from the previous locations of adhesion appeared to argue against this hypothesis (Fig. S3B). However, it is difficult to rule out the possibility that a subset of focal adhesions may be responsible for such a mechanism. Also arguing against an anchorage-based mechanism, treating cells with 50 µM blebbistatin to inhibit myosin II activity and weaken focal adhesions failed to inhibit the oscillation, despite the reduction of large focal adhesions into small punctate structures (48–50) (Movie S3). Cell oscillation continued albeit with an increase in period from 43 ± 15 to 95 ± 34 min (n = 22; Fig. 3 A and B). Reversing the order of drug application did not affect this slowed oscillation (Fig. S5A). Hence, we proposed that the oscillation induced by microtubule disassembly was likely caused by the destabilization of the internal circuit that controls migration polarity, rather than the stabilization of external adhesions.
Fig. 3.
Effect of myosin II inhibition on cell oscillation. Kymograph of a representative RPE-1 cell migrating on 1D strip treated with nocodazole followed by blebbistatin (A). (Scale bar, 50 µm, 60 min.) Comparison of the oscillation before and after blebbistatin treatment indicates an increase in period upon myosin II inhibition (B; n = 22, P < 0.0001). See also Fig. S5 and Movie S3.
A Modified LEGI Model Reproduces Both Persistent Cell Migration and Nocodazole-Induced Cell Oscillation.
As described in the Introduction, there is strong evidence that directed cell migration is controlled by a LEGI-type control mechanism (29, 33). Based on the above results, we suspected that the disassembly of microtubules caused the LEGI mechanism to switch from a positive-feedback mechanism that enhances the persistence of migration, to a negative-feedback mechanism that destabilizes the persistence as the cell starts to migrate directionally. Because a key aspect of the positive feedback in LEGI mechanism is the dramatically different distributions of the excitation and inhibitory signals, we hypothesized that microtubules are required for the efficient transport of inhibitory signals away from the frontal region, such that disassembly of microtubules would cause inhibitory signals to build up in the frontal region, which may then inhibit protrusive activities soon after the cell starts to move directionally and allow the opposite end to turn into a new frontal region.
We explored this hypothesis by computer modeling of a generalized LEGI-type mechanism that takes into account the interplay between frontal excitation signals and rear inhibitory signals (37). The LEGI mechanism was used to drive both changes in cell shape and cell migration as a result of iterative changes in cell shape. This approach is able to account for a wide range of cell shapes and migration behaviors as seen on 2D surfaces in the absence of gradient sensing (37). By imposing constraints to simulate cells confined to 1D (Fig. 4A and Fig. S6), this model readily reproduced the highly persistent migration seen along the linear micropatterns (Fig. 4B and Movie S4). This may be understood intuitively if the micropattern inhibited side protrusions and dramatically decreased the probability of competing protrusions away from the existing frontal region.
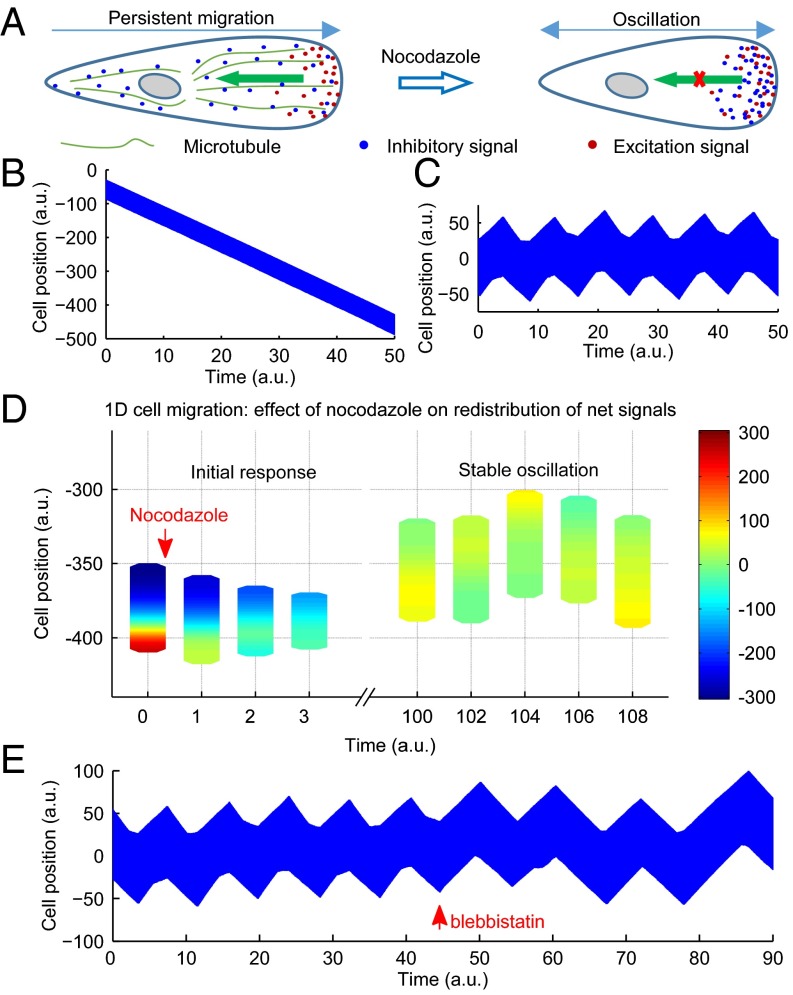
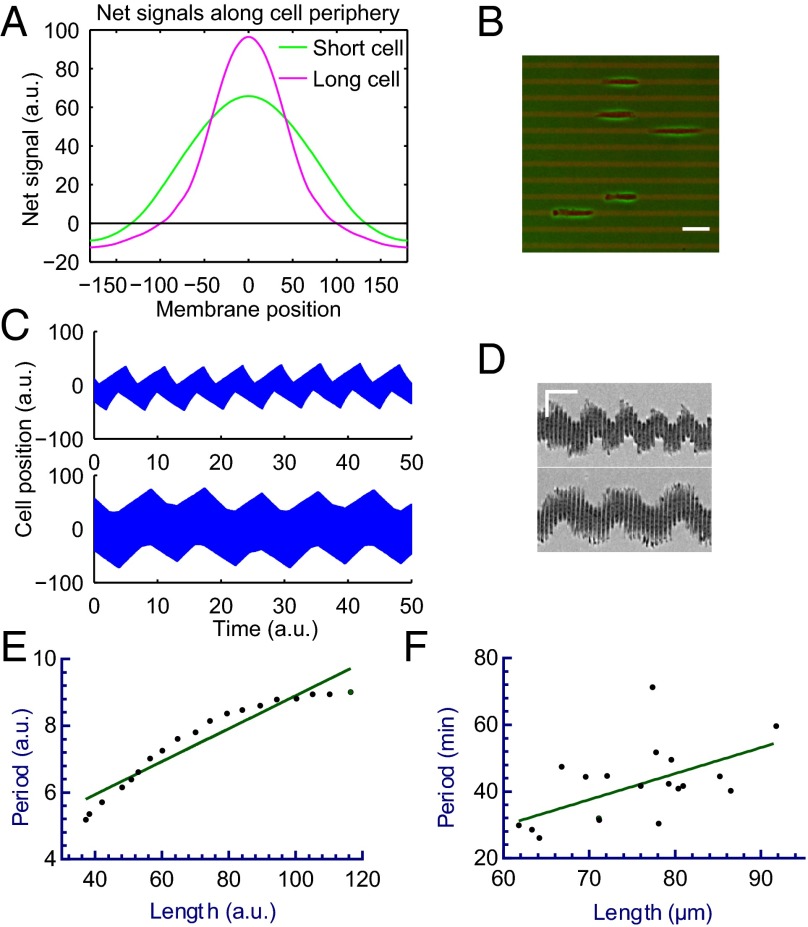
Fig. 4.
A modified LEGI model and computer simulation that reproduces both persistent cell migration and oscillations. A diagram of the modified LEGI model that involves the production of both excitation and inhibitory signals in the frontal region, followed by the transport of inhibitory signals away from the front to maintain frontal protrusions and cell polarity in control cells. In nocodazole-treated cells, the impairment of the transport and the resulting accumulation of inhibitory signals at the front act against the excitation signals and cause the head to switch into a tail (A). Computer simulation of this mechanism for cells in 1D successfully reproduces both persistent migration (kymograph in B) and oscillations (kymograph in C). Diagrams of cell location and shape, colored to show the heat map of net signals, show the dynamics of net signals before and after the treatment of nocodazole (D). In addition, the period of oscillation increases upon the impairment of the generation of inhibitory signals (by increasing the delay), which mimics the effect of blebbistatin (kymograph in E). a.u. stands for an arbitrary unit used in computer simulation. See also Fig. S6 and Movies S4–S6.
The previous simple model assumed that inhibitory signals were transported immediately and globally to maintain an instantaneous uniform distribution (37). To test the hypothesis that oscillation may be induced by impairing the propagation of these signals, we imposed a finite rate of transport away from the protrusive regions. We found that the inhibition of transport alone failed to cause oscillations, such that protrusions either never materialized given sufficiently strong inhibitory signals, or proceeded unaffected given weak inhibitory signals. However, consistent with the understanding in control theory of the importance of delay in oscillation, oscillations in cell migration were generated if inhibitory signals were generated at a limited rate or with a delay following the protrusion, which would allow protrusions to last for a limited period before inhibitory signals build up at the front to stop protrusions (Fig. 4C and Movie S5). Consistent with this notion, modeling showed that, upon the inhibition of transport in conjunction with delayed generation of inhibitory signals, the initial concentration of net positive signals at the front was followed by the propagation of net negative signals from the opposite end into the region (Fig. 4D and Movie S4). It is also noteworthy that the distribution and spreading of net negative signals resembled the behavior of zyxin in nocodazole-treated cells, suggesting that zyxin may serve as a marker of net negative signals. In addition, the effect of blebbistatin in prolonging the oscillation period may be reproduced by increasing the delay, which may be understood if these signals were mediated by myosin II-dependent inside-out signaling at focal adhesions (Fig. 4E and Movie S6).
Cell Length Affects the Regulation of Migration Behavior as Tested Mathematically and Experimentally.
Our model further predicted that nocodazole-induced oscillation should be affected by the length of the cell, which may be understood intuitively because increasing the length of the cell should increase the time for the signals to equilibrate across the cell, thereby prolonging the differential in net signals between the opposite ends (Fig. 5A). The period of oscillation increases as a result (Fig. 5 C and E, and Movie S7).
Fig. 5.
Model prediction and experimental verification of the relationship between cell length and oscillation period. Computed graphs show the effect of cell length on the distribution of net signals along the cell periphery, faster dissipation of signals in short cells leads to a lower peak value (A; curves show the distribution when the difference between the opposite ends is at its maximum). Experimental test using adhesive strips 10 µm in width (B; red fluorescence) yields oscillating RPE-1 cells of various length upon treatment with nocodazole (B). (Scale bar, 50 µm.) Short cells undergo fast oscillation, whereas long cells undergo slow oscillation, as indicated in the representative kymographs for both simulated cells (C) and RPE-1 cells on 1D strips (D). (Scale bar, 50 µm, 30 min.) Both simulated cells (E; n = 22, P < 0.0001) and RPE-1 cells oscillating on 10-µm–wide strips (F; n = 20, P < 0.01) show a positive relationship between cell length and oscillation period. Note simulation allows a wider range of lengths than experimentation. See also Movie S7.
To test this prediction, we measured the oscillation period of nocodazole-treated RPE-1 cells of various lengths along 1D strips 10 µm in width (Fig. 5B). Although the range of cell length was more limited in experiments than allowed in modeling, we found a similar positive correlation between the cell length and the oscillation period (Fig. 5 D and F).
Induction of Cell Oscillation upon the Inhibition of Dynein.
The notion that microtubules stabilize cell polarity by removing inhibitory signals from the frontal region pointed to possible involvement of the minus-end–directed microtubule motor, dynein (51). Inhibition of dynein-mediated rearward transport of inhibitory signals may then reproduce at least part of the behavior induced by nocodazole. To test this hypothesis, we treated RPE-1 cells with 50 µM ciliobrevin D, a small-molecular inhibitor of cytoplasmic dynein (52). A similar oscillatory behavior was observed for ciliobrevin D-treated cells as for nocodazole-treated cells, although the percentage was lower (Fig. 6, Fig. S1, and Movie S1). However, a significant fraction of ciliobrevin D-treated cells underwent random movement instead of oscillation. In addition, the oscillation appeared to be more transient in ciliobrevin D and with a longer period (Fig. S7 A and B), which may be explained if the removal of inhibitory signals from the front is only partially inhibited by the treatment of ciliobrevin D, as supported with computer modeling (Fig. S7C). The weaker effect of ciliobrevin D than nocodazole may also be understood if microtubules perform additional signaling functions such as the sequestration of a Rho GTP exchange factors (18, 53).
Fig. 6.
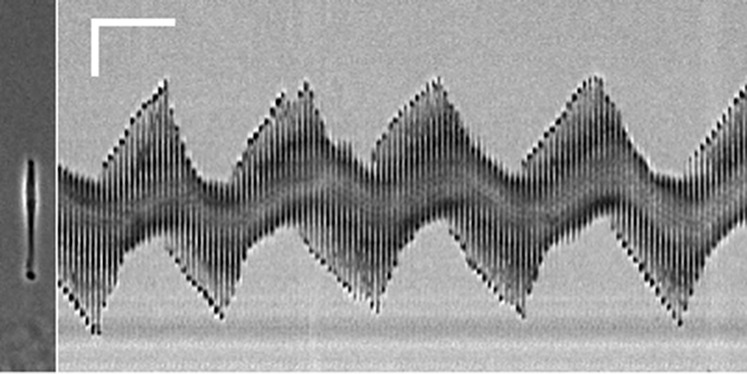
Oscillation of RPE-1 cells in 50 μM ciliobrevin D. Kymograph shows a representative RPE-1 cell treated with ciliobrevin D oscillating in a manner similar to those treated with nocodazole. (Scale bar, 50 µm, 60 min.) See also Figs. S1 and S7 and Movie S1.
Discussion
We showed that RPE-1 epithelial cells and NIH 3T3 fibroblasts migrate with a strong persistence along 1D adhesive strips and that the disassembly of microtubules turns such persistent migration into oscillations. The appearance of transient polarity in nocodazole suggested that microtubules are required not for the initial establishment but for the maintenance of polarity. Mathematical modeling further suggested that both types of behavior may be explained by a generalized LEGI-type control mechanism.
Oscillation is a highly informative phenomenon particularly amendable to mathematical modeling, due to its requirement of a negative-feedback loop and definable conditions that control the period and other characteristics. Therefore, detailed analysis of oscillation can often unveil important aspects of the underlying control mechanism. Although the segregation of excitation and inhibitory signals plays a critical role in creating a positive feedback essential for the traditional LEGI mechanism (29, 33), we demonstrated that an easy way to create a negative-feedback circuit from a LEGI mechanism would be to slow down the propagation of inhibitory signals away from the front. Oscillation would take place as long as there is a finite period for the inhibitory signals to build up. This would then allow directional migration to last for a limited period before inhibitory signals reach a level to overcome the excitation signals and cause a switch in direction. The existence of such buildup of inhibitory signals may involve either a delay or a slow rate in their generation following protrusion, as was suggested by a transient peak of various activities at the front, such as Ras, upon the stimulation of cell migration (54, 55). On 2D surfaces, we would expect similar oscillations around the cell perimeter upon the disassembly of microtubules. However, the lack of synchronization of oscillations around the perimeter would cause the cell as a whole to undergo only short-range random walks, as reported in the literature (56, 57). We would expect similar oscillations for cells migrating in confined 3D environments such as narrow channels. Indeed, there was a report that several kinds of cancer cells lost migration directionality upon the treatment with colchicine (58). The lack of obvious oscillations in the study may reflect either a very slow oscillation as seen in some NIH 3T3 cells (Fig. S3A), or a slightly different state of the migration mechanism due to the involvement of chemotaxis and/or lateral compression of the nucleus, which is known to cause cell migration to favor a blebbing-dependent mechanism (58, 59).
Based on the above analysis, we suggest that a main function of microtubules is to facilitate the transport of inhibitory signals, which may include the PTEN phosphatase in Dictyostelium (29, 60, 61), a tentative heterotrimeric G protein (31), or zyxin as indicated in this study, away from the front as soon as they are generated by the protrusive activities. In contrast, microtubules may not play as much a role in the generation and frontal localization of excitation signals, which may include the small GTPase Rac (62), Cdc42 (63), or PI3 kinase (61). This explanation appears most compatible with the well-known function of microtubules in mediating intracellular transport. Consistent with this idea, the inhibition of dynein, which drives microtubule-mediated retrograde transport, is known to inhibit cell polarity and migration (64). In addition, treatment of cells in 1D with a dynein inhibitor ciliobrevin D induced oscillatory migration in the present experiments as did the disassembly of microtubules. Supported by modeling, the less consistent induction of oscillation by ciliobrevin D than nocodazole may be explained by a weaker inhibition of the transport of inhibitory signals than the disassembly of microtubules (Fig. S7C). Other signaling functions of microtubules, such as the regulation of Rho-GEF (18, 53), may provide additional mechanisms for suppressing the effect of inhibitory signals at the front.
Our model further revealed a number of ways to affect the period of nocodazole-induced oscillations. An intuitive way to lengthen the period would be to increase the length of the cell, which should prolong the time required for inhibitory signals to build up at the front before they reach a level to stop protrusions. This prediction was supported both mathematically and experimentally—we confirmed that longer cells oscillate at a longer period. In addition, because myosin II is required for controlling cell length (12), this length-dependent mechanism may also play a role in the increase of oscillation period upon blebbistatin treatment. However, length dependence alone cannot explain the increase in oscillation period of cells treated with both blebbistatin and nocodazole (Fig. S5B). Most likely, the reduction of retraction rate (12), and the effect on myosin II-mediated inside-out signaling also play a role. For example, blebbistatin may decrease the rate of the generation of inhibitory signals, which, according to our modeling, has a striking effect on oscillation period by prolonging the buildup of inhibitory signals at the front (Fig. 4E).
To our knowledge, there has been only one complex model that may be appropriate for addressing cellular oscillations (65, 66). However, the main purpose of the study was to explore the range of possible cell behaviors as a result of the interplay between shape, mechanics, and polarity, with oscillation demonstrated as one of the possible behaviors without considering the trigger or underlying control circuit. In addition, its prediction that cells must contract before repolarization is inconsistent with our observations, which indicated that the switch of migration direction may be initiated by a slowdown of migration and subsequent extension at the opposite end (hence an extension of cell length; Fig. S3C), retraction of the leading edge (hence contraction of the cell length; Fig. S3D), or both frontal retraction and rear extension roughly simultaneously (Fig. S3 C and D). Our model generated consistent results (Fig. S3 E and F) and supported the notion that the front and rear of a cell are driven by independent stochastic events, coupled only loosely to each other through the distribution of signals.
A previous report showed that nocodazole treatment caused detachment of fibroblasts from linear strips and that cells treated with both nocodazole and low dose of blebbistatin showed random membrane activities in all directions (67). However, the study was done with very narrow strips 1.5 µm in width, where the area for cell adhesion was small compared with the cell-spreading area such that most of the cell body dangled above the strips. The resulted weak adhesion was likely responsible for the detachment of cells upon the contraction induced by nocodazole. Even when detachment was avoided by blebbistatin treatment, the small protrusions allowed by the limited adhesion area combined with compromised global inhibition caused by nocodazole may allow random membrane activities to appear all along the length.
Consistent with a previous report (58), we found Taxol treatment almost brought the cell to a complete halt, with only small protrusions extending at both ends (17) (Fig. S4). Thus, unlike nocodazole, which affects the maintenance of polarity, Taxol appears to inhibit both the development and maintenance of cell polarity. The more severe effect of Taxol than nocodazole may be understood because Taxol is known to not only suppress microtubule dynamics but also cause an extensive reorganization of microtubules, keeping them from reaching the leading edge (47). Taxol-stabilized microtubules may also be defective in their interactions with other structures such as focal adhesions to affect the production of excitation and/or inhibitory signals. In addition, although nocodazole allows an anterior end to turn into a retracting posterior end during oscillation, Taxol-induced stabilization of microtubules has been shown to block cell retraction (57), which may be sufficient to prevent oscillatory migration.
In summary, we have discovered an oscillation phenomenon using cells migrating on micropatterned substrates that, through a combination of experimental and modeling approaches, has yielded important insights into the control circuit for cell migration. The main hypothesis supported by this study is that microtubules and associated motors are responsible for the efficient global distribution of inhibitory signals to support a LEGI-type control mechanism, although microtubules may play additional roles such as the regulation of small GTPase activities. Although our mathematical model likely represents a simplification of the actual mechanism, it has provided important insights into the control mechanism and is capable of explaining a range of cell behaviors and generating testable predictions.
Materials and Methods
Cell culture, plasmids, reagents, micropatterning, microscopy, and statistics are described in detail in SI Materials and Methods. Manual or automated image analysis was carried out using ImageJ, Matlab, and a custom software (SI Materials and Methods). Computer simulations of the modified LEGI model were carried out in Matlab with details provided in SI Materials and Methods and Table S1.
Supplementary Material
Acknowledgments
This work was supported by Grant GM-32476 from the National Institutes of Health (to Y.-L.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410533111/-/DCSupplemental.
References
- 1.Detrich HW, 3rd, et al. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA. 1995;92(23):10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiery JP, Duband JL, Tucker GC. Cell migration in the vertebrate embryo: Role of cell adhesion and tissue environment in pattern formation. Annu Rev Cell Biol. 1985;1:91–113. doi: 10.1146/annurev.cb.01.110185.000515. [DOI] [PubMed] [Google Scholar]
- 3.Krawczyk WS. A pattern of epidermal cell migration during wound healing. J Cell Biol. 1971;49(2):247–263. doi: 10.1083/jcb.49.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: Present and future therapeutic targets. Nat Immunol. 2005;6(12):1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 5.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 6.Finnell RH, Waes JG, Eudy JD, Rosenquist TH. Molecular basis of environmentally induced birth defects. Annu Rev Pharmacol Toxicol. 2002;42:181–208. doi: 10.1146/annurev.pharmtox.42.083001.110955. [DOI] [PubMed] [Google Scholar]
- 7.Charo IF, Taubman MB. Chemokines in the pathogenesis of vascular disease. Circ Res. 2004;95(9):858–866. doi: 10.1161/01.RES.0000146672.10582.17. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Weinberg RA. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 10.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 11.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10(8):538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo WH, Wang YL. A three-component mechanism for fibroblast migration with a contractile cell body that couples a myosin II-independent propulsive anterior to a myosin II-dependent resistive tail. Mol Biol Cell. 2012;23(9):1657–1663. doi: 10.1091/mbc.E11-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheetz MP, Felsenfeld D, Galbraith CG, Choquet D. Cell migration as a five-step cycle. Biochem Soc Symp. 1999;65:233–243. [PubMed] [Google Scholar]
- 14.Goehring NW, Grill SW. Cell polarity: Mechanochemical patterning. Trends Cell Biol. 2013;23(2):72–80. doi: 10.1016/j.tcb.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman RD. The role of three cytoplasmic fibers in BHK-21 cell motility. I. Microtubules and the effects of colchicine. J Cell Biol. 1971;51(3):752–762. doi: 10.1083/jcb.51.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci USA. 1980;77(3):1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etienne-Manneville S. Microtubules in cell migration. Annu Rev Cell Dev Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15(2):76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez OC, et al. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5(7):599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 21.Waterman-Storer CM, Salmon E. Positive feedback interactions between microtubule and actin dynamics during cell motility. Curr Opin Cell Biol. 1999;11(1):61–67. doi: 10.1016/s0955-0674(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 22.Broussard JA, Webb DJ, Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr Opin Cell Biol. 2008;20(1):85–90. doi: 10.1016/j.ceb.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Small JV, Kaverina I. Microtubules meet substrate adhesions to arrange cell polarity. Curr Opin Cell Biol. 2003;15(1):40–47. doi: 10.1016/s0955-0674(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 24.Kaverina I, et al. Enforced polarisation and locomotion of fibroblasts lacking microtubules. Curr Biol. 2000;10(12):739–742. doi: 10.1016/s0960-9822(00)00544-3. [DOI] [PubMed] [Google Scholar]
- 25.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7(6):581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 26.Danuser G, Allard J, Mogilner A. Mathematical modeling of eukaryotic cell migration: Insights beyond experiments. Annu Rev Cell Dev Biol. 2013;29:501–528. doi: 10.1146/annurev-cellbio-101512-122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284(5415):765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 28.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 29.Levchenko A, Iglesias PA. Models of eukaryotic gradient sensing: Application to chemotaxis of amoebae and neutrophils. Biophys J. 2002;82(1):50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher PR. Pseudopodium activation and inhibition signals in chemotaxis by Dictyostelium discoideum amoebae. Semin Cell Biol. 1990;1(2):87–97. [PubMed] [Google Scholar]
- 31.Levine H, Kessler DA, Rappel W-J. Directional sensing in eukaryotic chemotaxis: A balanced inactivation model. Proc Natl Acad Sci USA. 2006;103(26):9761–9766. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J Biol Chem. 2003;278(23):20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 33.Ma L, Janetopoulos C, Yang L, Devreotes PN, Iglesias PA. Two complementary, local excitation, global inhibition mechanisms acting in parallel can explain the chemoattractant-induced regulation of PI(3,4,5)P3 response in Dictyostelium cells. Biophys J. 2004;87(6):3764–3774. doi: 10.1529/biophysj.104.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karunarathne WKA, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci USA. 2013;110(17):E1575–E1583. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iglesias PA, Devreotes PN. Navigating through models of chemotaxis. Curr Opin Cell Biol. 2008;20(1):35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Huang C-H, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15(11):1307–1316. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satulovsky J, Lui R, Wang YL. Exploring the control circuit of cell migration by mathematical modeling. Biophys J. 2008;94(9):3671–3683. doi: 10.1529/biophysj.107.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neilson MP, et al. Chemotaxis: A feedback-based computational model robustly predicts multiple aspects of real cell behaviour. PLoS Biol. 2011;9(5):e1000618. doi: 10.1371/journal.pbio.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houk AR, et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148(1-2):175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pouthas F, et al. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci. 2008;121(14):2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- 41.Guo W, Wang Y-L. Micropatterning cell-substrate adhesions using linear polyacrylamide as the blocking agent. Cold Spring Harb Protoc. 2011;2011(3):prot5582. doi: 10.1101/pdb.prot5582. [DOI] [PubMed] [Google Scholar]
- 42.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116(22):4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 43.Lele TP, et al. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207(1):187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 44.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171(2):209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luxton GWG, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23(5):579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zieve GW, Turnbull D, Mullins JM, McIntosh JR. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp Cell Res. 1980;126(2):397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 47.Mikhailov A, Gundersen GG. Relationship between microtubule dynamics and lamellipodium formation revealed by direct imaging of microtubules in cells treated with nocodazole or Taxol. Cell Motil Cytoskeleton. 1998;41(4):325–340. doi: 10.1002/(SICI)1097-0169(1998)41:4<325::AID-CM5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Chrzanowska-Wodnicka M, Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J Cell Biol. 1996;133(6):1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rape AD, Guo W-H, Wang Y-L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials. 2011;32(8):2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stricker J, Aratyn-Schaus Y, Oakes PW, Gardel ML. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys J. 2011;100(12):2883–2893. doi: 10.1016/j.bpj.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279(5350):519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 52.Firestone AJ, et al. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484(7392):125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittmann T, Waterman-Storer CM. Cell motility: Can Rho GTPases and microtubules point the way? J Cell Sci. 2001;114(21):3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- 54.Kortholt A, Keizer-Gunnink I, Kataria R, Van Haastert PJM. Ras activation and symmetry breaking during Dictyostelium chemotaxis. J Cell Sci. 2013;126(19):4502–4513. doi: 10.1242/jcs.132340. [DOI] [PubMed] [Google Scholar]
- 55.Lee S, Shen Z, Robinson DN, Briggs S, Firtel RA. Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis. Mol Biol Cell. 2010;21(11):1810–1824. doi: 10.1091/mbc.E10-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gail MH, Boone CW. Effect of colcemid on fibroblast motility. Exp Cell Res. 1971;65(1):221–227. doi: 10.1016/s0014-4827(71)80070-8. [DOI] [PubMed] [Google Scholar]
- 57.Ganguly A, Yang H, Sharma R, Patel KD, Cabral F. The role of microtubules and their dynamics in cell migration. J Biol Chem. 2012;287(52):43359–43369. doi: 10.1074/jbc.M112.423905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Balzer EM, et al. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26(10):4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charras G, Paluch E. Blebs lead the way: How to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9(9):730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 60.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 61.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109(5):611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 62.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290(5490):333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 63.Itoh RE, et al. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22(18):6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dujardin DL, et al. A role for cytoplasmic dynein and LIS1 in directed cell movement. J Cell Biol. 2003;163(6):1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camley BA, Zhao Y, Li B, Levine H, Rappel WJ. Periodic migration in a physical model of cells on micropatterns. Phys Rev Lett. 2013;111(15):158102. doi: 10.1103/PhysRevLett.111.158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184(4):481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.