Significance
Asian rice (Oryza sativa) is among the world’s most important crops. The genus Oryza has become a model for the study of plant genome structure, function, and evolution. We have undertaken de novo, full-genome sequence analysis of five diploid AA-genome species that are closely related to O. sativa. These species are native to quite different environments, representing four continents, thus exhibiting very different adaptations. Our studies identify specific genetic changes, in both gene copy number and the degree of diversifying natural selection, that indicate specific genes responsible for these adaptations, particularly in genes related to defense against pathogens and reproductive diversification. This genome discovery and comparative analysis provide a powerful tool for future Oryza study and rice improvement.
Keywords: comparative genomics, full-genome sequencing, genomic variation, positive selection, Oryza
Abstract
Comparative genomic analyses among closely related species can greatly enhance our understanding of plant gene and genome evolution. We report de novo-assembled AA-genome sequences for Oryza nivara, Oryza glaberrima, Oryza barthii, Oryza glumaepatula, and Oryza meridionalis. Our analyses reveal massive levels of genomic structural variation, including segmental duplication and rapid gene family turnover, with particularly high instability in defense-related genes. We show, on a genomic scale, how lineage-specific expansion or contraction of gene families has led to their morphological and reproductive diversification, thus enlightening the evolutionary process of speciation and adaptation. Despite strong purifying selective pressures on most Oryza genes, we documented a large number of positively selected genes, especially those genes involved in flower development, reproduction, and resistance-related processes. These diversifying genes are expected to have played key roles in adaptations to their ecological niches in Asia, South America, Africa and Australia. Extensive variation in noncoding RNA gene numbers, function enrichment, and rates of sequence divergence might also help account for the different genetic adaptations of these rice species. Collectively, these resources provide new opportunities for evolutionary genomics, numerous insights into recent speciation, a valuable database of functional variation for crop improvement, and tools for efficient conservation of wild rice germplasm.
Drawing the landscape of genomic divergence among multiple lineages is fundamental to understanding plant gene and genome evolution (1, 2). The comprehensive comparison of closely related genomes in different chronologically ordered stages under a well-resolved phylogenetic framework could dramatically improve the inference precision and sensitivity of gene evolution studies and should allow more robust results for investigating broad-scale patterns of genomic architecture in the course of the speciation process compared with analyses of single genomes (3, 4). For instance, studies of yeast, Drosophila, and human genomes have demonstrated how comparisons of closely related genome sequences can reveal mechanisms of gene and genome evolution in fungi and animals (5–7). In plants, however, we know little about broad-scale patterns of evolutionary dynamics, differentiation, and consequences. Studies are needed of very closely related plant species that span the speciation continuum and have well-characterized biogeographic histories.
The genus Oryza, consisting of 24 species, provides a uniquely powerful system for studying comparative genomics and evolutionary biology, and can contribute to the improvement of rice, which is of pivotal significance in worldwide food production and security (8–10). Many genes involved in rice improvement are derived from wild AA-genome species, and broadening the gene pool of cultivated rice through introgression from other wild relatives of Oryza has attracted increasing attention (11). Phylogenetic analysis of the diploid AA-genome species indicated a closely spaced series of recent speciation events in this genus (12). These species span a wide range of global pantropical regions and are disjunctively distributed in Asia, Africa, Australia, and South America (13). Having diverged less than 3 Mya from a common AA-genome ancestor (12), these eight species have generated extensive adaptive and breeding traits (14, 15).
By placing multiple Oryza genome comparisons in a phylogenetic context, previous studies have recorded some of the genomic changes associated with the diversification of the rice genus (16–20). Several studies have compared orthologous genomic segments of Oryza species that represent the different genome types that have diverged over long time scales (18–20). However, none of these analyses provide the full-genome, multispecies perspective that allows comprehensive analysis of gene, genome, or trait evolution.
Here, we fully sequenced and assembled de novo the five biogeographically representative AA genomes. In comparison to the high-quality assembly of the O. sativa ssp. japonica cv. Nipponbare (SAT) genome (21), we provide a starting point for studies in the emerging field of comparative and evolutionary genomics. Despite the wealth of phenotypic diversity and adaptive differences, it seems that many attributes of these rice genomes are remarkably conserved across species. Thus, in addition to examining the relationship between sequence and phenotypic diversity, the genomes of these species provide an excellent model for studying, from specific loci to divergent genomic regions, how the evolutionary dynamics of genes and genomes can facilitate speciation processes on a genomic scale. Access to the unprecedented dataset of these Oryza genome sequences will accelerate the pace at which the untapped reservoir of agronomically important genes can be exploited for rice improvement.
Results and Discussion
Genome Assembly and Annotation.
We sequenced five Oryza nuclear genomes: Oryza nivara (NIV) from Asia, Oryza glaberrima (GLA) and Oryza barthii (BAR) from Africa, Oryza glumaepatula (GLU) from South America, and Oryza meridionalis (MER) from Australia (Fig. 1A). In each case, we performed a whole-genome shotgun sequencing (WGS) analysis with the next-generation sequencing platform from Illumina. Whole genome sequencing generated raw sequence datasets of 28.4 Gb (NIV), 21.3 Gb (GLA), 18.9 Gb (BAR), 31.9 (GLU), and 22.9 (MER), thus yielding ∼73-fold, 56-fold, 51-fold, 86-fold, and 60-fold coverage, respectively (SI Appendix, Table S2). These genomes were assembled using SOAPdenovo (22), resulting in final assembles ranging from 334.7 Mb (GLU) to 375.0 Mb (NIV). The N50 lengths of the assembled contigs varied from ∼14.6 kb (MER) to ∼25.2 kb (GLA), and scaffold N50 ranged from ∼118 kb (MER) to ∼722 kb (GLA) (Table 1 and SI Appendix, Table S4). About 85.2% (NIV), 92.5% (GLA), 87.7% (BAR), 82.5% (GLU), and 78.8% (MER) of the assemblies fall into 1,130, 881, 1,548, 2,086, and 2,148 scaffolds larger than 50 kb in length, respectively (SI Appendix, Table S5). Our assembled genomes cover less than the estimated genome sizes, with predicted unclosed gaps ranging from 16.9 Mb (BAR) to 35.3 Mb (NIV). Comparative analyses between the five assembled rice genomes with the SAT genome suggested that these gaps are primarily composed of repetitive sequences, particularly LTR retrotransposons (SI Appendix, Table S6).
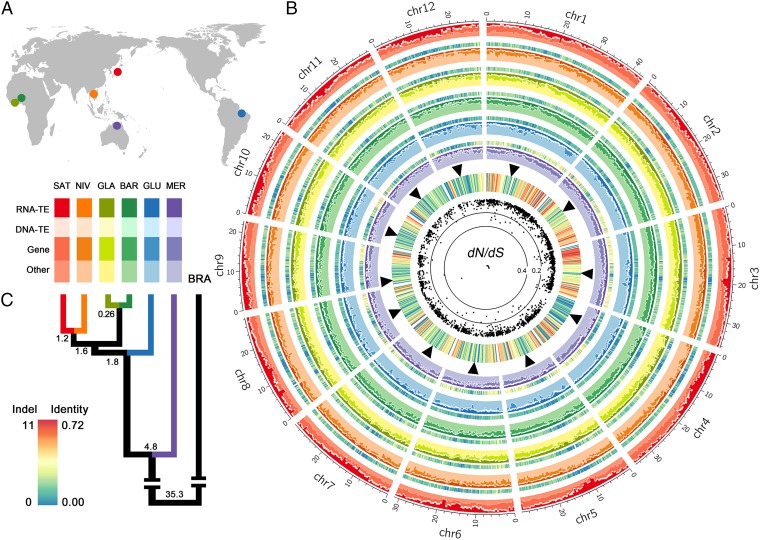
Fig. 1.
Comparative genomics of the six AA-genome Oryza species. (A) Geographical origins (SI Appendix, Table S1). (B) Comparative genome analysis. The outer circle represents the 12 chromosomes of SAT, along with the densities of genes, DNA transposons, RNA transposons, and other types of genome components labeled as shown in the color matrix (Left). Moving inward, the five sequenced and assembled genomes are symbolized by different colored circles. The heat map beside each circle indicates the average number of indels per kilobase in 50 kb-bins for each genome in comparison to SAT. The inner heat map illustrates the similarity among the six genomes. Blank points show the association constant dN/dS ratios of entire genes estimated by site models for 2,272 1:1 orthologous gene families. SAT centromere positions are signified by black triangles (▲). (C) Phylogeny of the six AA-genome species with BRA as an outgroup. Estimates of divergence time are given at each node, all supported with 100% bootstrap values.
Table 1.
Summary of genome assembly and annotation of the six AA-genome Oryza species
| Type | Feature | SAT | NIV | GLA | BAR | GLU | MER |
| Assembly | Depth, × | NA | 59.7 | 56.0 | 51.1 | 86.2 | 74.7 |
| Estimated genome size, Mb | 389 | 395 | 370 | 376 | 366 | 388 | |
| Assembled sequence length, Mb | 373.25 | 375.01 | 344.86 | 335.09 | 334.67 | 340.78 | |
| Scaffold N50, bp | NA | 511,541 | 722,125 | 237,573 | 129,688 | 117,674 | |
| Contig N50, bp | NA | 19,023 | 25,248 | 16,126 | 17,474 | 14,633 | |
| Predicted coverage, % | 96.0 | 94.9 | 93.2 | 89.1 | 91.4 | 87.8 | |
| Annotation | Number of predicted protein-coding genes | 55,986 | 41,490 | 41,476 | 42,283 | 41,605 | 39,106 |
| Average gene length, bp | 2,965 | 2,243 | 2,258 | 2,175 | 2,208 | 2,170 | |
| tRNAs | 726 | 551 | 491 | 588 | 621 | 598 | |
| rRNAs | 29 | 10 | 23 | 16 | 29 | 61 | |
| snoRNAs | 317 | 259 | 232 | 229 | 194 | 195 | |
| snRNAs | 112 | 116 | 121 | 120 | 125 | 117 | |
| miRNAs | 366 | 276 | 271 | 276 | 263 | 251 | |
| TEs, % | 39.36 | 27.65 | 29.35 | 29.83 | 29.91 | 29.77 |
NA, not available.
We evaluated the qualities of the WGS assemblies first through aligning the assembled genome sequences (NIV, GLA, BAR, GLU, and MER) to the SAT genome (SI Appendix, Fig. S4A). After removing repeat sequences, the mapping coverage ranged from 77.5 to 95.8% (SI Appendix, Table S7). To estimate assembly qualities accurately, we calculated the coverage of the gene-containing regions corresponding to SAT on the basis of the 2,971 orthologous genomic segments found in all six AA-genome assemblies (SAT, NIV, GLA, BAR, GLU, and MER), indicating coverage of 97.8–98.7% (SI Appendix, Table S7). To evaluate the quality of genome assemblies further, we used Nipponbare gene models to examine whether genes in our assemblies are split into multiple contigs, and found fewer than 180 cases in each species (SI Appendix, Table S7).
Alignment of the available BAC paired-end sequences from the Oryza Map Alignment Project (OMAP; www.omap.org/) to the WGS scaffolds of the four genomes (NIV, GLA, GLU, and MER) (SI Appendix, Fig. S4B), after excluding multimatched pairs probably caused by repeat sequences, revealed few potential misassemblies, ranging from 0.84% (GLU) to 1.81% (MER) (SI Appendix, Table S8). Assembly quality was finally confirmed by aligning the WGS scaffolds of GLA, GLU, and MER against their available short-arm sequences of chromosome 3 from the OMAP. After eliminating repeat sequence-masked and gap regions, pairwise alignments yielded sequence similarities of 99.7%, 99.3%, and 99.2%, respectively, and exhibited good genomic coverage at 94.4%, 83.5%, and 88.4% for GLA, GLU, and MER, respectively (SI Appendix, Fig. S5).
In combination with ab initio prediction, protein and public EST alignments, EVidenceModeler combing, and further filtering, we predict 41,490, 41,476, 42,283, 41,605, and 39,106 protein-coding genes for NIV, GLA, BAR, GLU, and MER, respectively (Table 1). After the predicted genes were functionally annotated against InterPro, Pfam, and Gene Ontology (GO) protein databases, we generated 144.01 Gb of RNA sequence (RNA-Seq) data obtained from a total of 20 libraries representing major tissue types and developmental stages to aid further annotation. RNA-Seq data revealed that alternative splicing of a substantial portion of genes was responsible for two or more isoforms, suggestive of more functional variation than represented by the gene set alone. Overall, 74.4%, 80.3%, 78.4%, 81.5%, and 81.3% of the gene models were supported by transcripts and proteins for NIV, GLA, BAR, GLU, and MER, respectively (SI Appendix, Table S12).
We also annotated noncoding RNA (ncRNA) genes, including those ncRNA genes encoding transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and microRNAs (miRNAs) (Table 1). In total, 276, 271, 276, 263, and 251 miRNA genes belonging to 203, 125, 124, 133, 125, and 123 miRNA families were identified in the NIV, GLA, BAR, GLU, and MER genomes, respectively (SI Appendix, Fig. S6 and Table S13, and Dataset S1). The annotation of repeat sequences showed that ∼27.6%, 29.4%, 29.8%, 29.9%, and 29.8% of the NIV, GLA, BAR, GLU, and MER genomes, respectively, consist of transposable elements (TEs), which is quite a bit lower than the amount (39.4%) annotated in the SAT genome with the same methods (Fig. 1B and Table 1). LTR retrotransposons were the most abundant TE type, occupying roughly 30.6%, 18.0%, 18.7%, 19.3%, 18.9%, and 19.6% of the SAT, NIV, GLA, BAR, GLU, and MER genomes, respectively. When we considered unclosed gaps, which mainly consist of repetitive sequences, the contents of TEs in these five genomes are comparable to the content in the SAT genome (SI Appendix, Tables S6 and S14), suggesting that limited TE content variation is a major contributor to their relative genome size stability (SI Appendix, Table S3). We annotated numerous simple sequence repeats that will provide valuable genetic markers to assist rice breeding programs (SI Appendix, Fig. S7 and Table S15, and Dataset S2).
Rapidly Evolving Rice AA Genomes.
One advantage of sequencing genomes of multiple closely related rice species is to explore evolutionary relationships that allow a comprehensive picture of genome evolution. Using 2,305 one-to-one, single-copy orthologous genes (SI Appendix, Table S16) from the seven fully sequenced Oryza genomes, we performed phylogenomic analysis using the FF-genome species Oryza brachyantha (BRA) as an outgroup (23) (SI Appendix, Figs. S8 and S9). This robust phylogeny provides solid grounds for performing evolutionary and comparative analyses, and is consistent with the calculated phylogeny of all eight AA-genome Oryza species in an earlier study (12). With the same large gene set, we dated the speciation events leading to these major extant AA lineages to be within 4.8 million years (Myr) and 35.3 Myr between AA and FF genomes (Fig. 1C and SI Appendix, Fig. S8), agreeing both with another deep data analysis of these divergence times (24) and with recent fossil evidence suggesting the origin of the tribe Oryzeae at ∼63 Mya (25).
Genome structure is mostly well conserved across the six rice species. Genome sizes estimated by both flow cytometry and 17-mer oligonucleotide depth distribution were fairly close to previous results (10, 26, 27) and varied little across genomes (SI Appendix, Table S3), in contrast to the order of magnitude of differences among different genome types in the genus (16–18). We searched for large-scale genome rearrangements in these comparisons but did not detect any such events. To examine overall conservation of genome architecture at an intermediate scale, we constructed orthologous synteny maps and analyzed synteny relationships across species using MERCATOR and MAVID (28). In total, we obtained 2,971 orthologous genomic segments shared by all six AA-genome Oryza species, with the aligned sequences ranging from 74.7% in MER to 85.1% in SAT, of which 1,202 segments were larger than 100 kb (43.1%) (SI Appendix, Tables S17 and S18). The large number of syntenic blocks and their relatively long sizes suggested good gene colinearity across genomes. We estimated an overall extent of genome divergences between SAT and the other five AA-genome Oryza species by randomly sampling 103 segments totaling ∼15 Mb, with an average length of ∼100 kb (∼3.9% of the rice genome). Global genome divergences between genomic segments supported their phylogenetic positions in the topology but far exceeded orthologous gene sequence variation (SI Appendix, Table S19), indicating that differential TE insertion and turnover histories in the intergenic regions account for most of the sequence divergence (19, 29).
To improve the sensitivity of evolutionary inferences and study sequence evolution of the six Oryza AA-genome diploid species that diverged over a relatively short time frame, we analyzed a total of 100 orthologous genomic segments across all species on a finer scale (SI Appendix) and present two representative examples: one highly conserved, grain size 5 (GS5) (30), and one rapidly evolving, prostrate growth 1 (PROG1) (31, 32). Sequence alignment and annotation showed, to a variable extent, highly conserved gene colinearity and genomic structure across both GS5- and PROG1-orthologous regions (Fig. 2A and SI Appendix, Fig. S10), consistent with previous results (18–20). Nevertheless, there were still a number of microstructural changes, including lineage-specific differences that were concordant with the known phylogenetic relationships and random fluctuations in TE biology (SI Appendix, Tables S20 and S21). The PROG1-orthologous region represents one particularly striking example of the dynamic nature of genome microstructure in the AA genomes (SI Appendix, Tables S23 and S24). Compared with the SAT PROG1 region, many microrearrangements were identified in SAT’s wild progenitor (NIV), including a deletion spanning ∼30 kb.
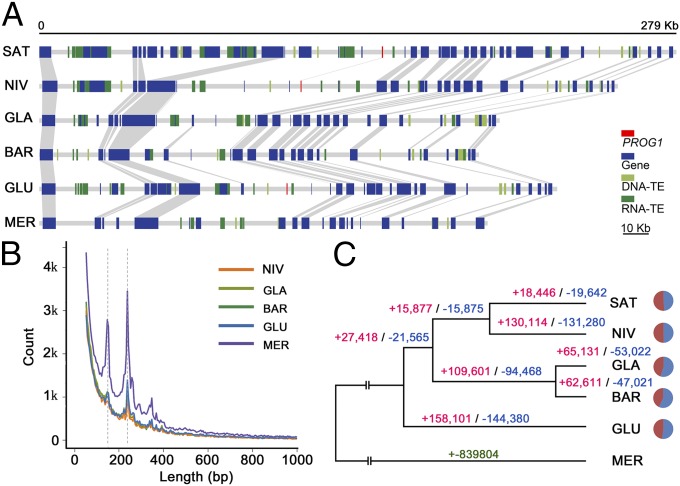
Fig. 2.
Rapid evolution of the Oryza AA genomes. (A) Gene synteny. The collinear region is located on SAT chromosome 7 (279 kb, 2,711,024–2,989,979 kb; Michigan State University 7.0; ref. 60). Orthologous genes (blue boxes), DNA transposons (green boxes), and RNA transposons (light green boxes) are connected by gray lines between genomes. (B) Distribution of genomic SVs from 50 bp to 1 kb in length for the five sequenced genomes in comparison to SAT. The peak at ∼250 bp is due to extensive TE turnover. (C) Numbers of genomic structural insertion (red) and deletion (blue) events are indicated at the terminus of each branch of the rice phylogeny while comparing the SAT genome; the pie charts at each branch terminus illustrate the insertion/deletion ratios.
The sequences of the five rice genomes, together with the published SAT genome, also allowed a comprehensive assessment of genome-wide structural variation (SV). Syntenic alignments of 2,971 orthologous genomic segments across the five rice genomes with SAT identified 232,900–514,924 putative insertions ranging from 1 to 47,650 bp. This result summed to 29.43–55.61 Mb of insertions, compared with 240,928–539,026 deletions, ranging from 1 to 40,230 bp, affecting 10.16–22.12 Mb (SI Appendix, Table S25). We performed additional in silico analyses with strict error-correction strategy to ensure high-confidence methodology of SV identification (Fig. 2B and SI Appendix, Fig. S12 and Table S25). The size distribution of SVs is consistent with previous findings (29, 33) that longer SVs were less abundant (SI Appendix, Fig. S12). Notably, several prominent peaks were observed that range in size from 100 to 300 bp (Fig. 2B), resulting from insertion of specific small DNA TEs (SI Appendix, Fig. S13). To ascertain that the lower contig N50 is not the reason for the amazingly abundant SVs in MER, we analyzed the numbers of SVs that are correlated with divergence times of the studied species (R2 = 0.987). Indeed, MER has experienced more insertion events of miniature inverted-repeat transposable elements (Tc1 and Tourist/Harbinger), resulting in higher numbers of SVs 100–300 bp in length, than other species (Fig. 2B and SI Appendix, Fig. S13). Mapping of putative indels to the SAT genome revealed an even distribution throughout all 12 chromosomes (Fig. 1B). To understand the nature of massive genomic structural variation, we characterized and classified all identified SVs by mapping them onto the phylogenetic tree of the six AA-genome species. The majority of them, ∼85.4% of insertions and ∼87.4% of deletions, could be clearly explained by their robust phylogenetic relationships, whereas the rest, about 14.6% (insertions) and 12.6% (deletions), were distributed on the phylogenetic tree in a manner subjective of incomplete lineage sorting (SI Appendix, Table S26). We dated the occurrences of SVs by locating them at the five phylogenetic nodes (Fig. 2C). Our analyses indicate that during the past 4.8 Myr, the six rice lineages, as well as their shared ancestral lineages, had 15,372–250,504 insertions and 16,368–203,931 deletions per million years, respectively. The numbers of structural variation events observed along different branches are proportional to their branch lengths and divergence times from SAT (Fig. 2C). The African branch (GLA/BAR), for example, underwent one-third fewer SVs than the Asian branch (SAT/NIV) due to the relatively recent speciation (Figs. 1C and 2C).
To detect recent segmental duplications in these rice genomes, we performed a genome-wide comparative analysis using whole-genome shotgun sequence detection methods (34) (SI Appendix, Table S27 and Dataset S3a). We estimated less segmental duplication variation (∼1.5%) in the rice genomes than was found in the human genome (∼5%) using equivalent methods (35). GO analysis of the genes involved in segmental duplications showed that their functional classes were significantly enriched for several specific biological functions, especially cell death and response to stress (SI Appendix, Dataset S3b). Given their very different growth environments, it is not surprising that genes involved in environmental adaptation are particularly labile over the evolutionary history of these genomes.
Evolution of Rice Gene Families.
Defining gene family emergence and extinction within closely related plants can uncover the events that underlie species adaptation and cause lineage evolution (36). With multiple rice genomes now in hand, we compared SAT, NIV, GLA, BAR, GLU, MER, and BRA proteomes, and identified 39,293 orthologous gene families comprising 211,718 genes (SI Appendix, Table S29). This comparison revealed a core set of 93,829 genes belonging to 10,576 clusters that were common to all seven rice species and represent ancestral gene families. Interestingly, a large number of clusters were found that were specific to different AA-genome species or branches, with new gene families evolving independently along each lineage, suggestive of rapid diversification from a common ancestor during the past 35.3 Myr. For instance, 1,779 gene clusters (34 GLA-specific gene clusters, 71 GLA-specific gene clusters, and 1,674 shared gene clusters between GLA and BAR; Fig. 3A) were unique to the GLA and BAR lineages, and 1,039 were unique to the SAT and NIV lineages. There were 3,184 gene clusters specific to the five wild rices (NIV, BAR, GLU, MER, and BRA), whereas 484 were specific to the two cultivated rices (GLA and SAT) (Fig. 3A). Because only one accession was analyzed for each species, we cannot be sure how many of these lineage-specific gene clusters are truly missing from or are present in accessions that we did not investigate. It is possible that lineage sorting may not be complete for this evolved difference, and further analysis of multiple haplotypes per species will be needed to resolve this issue. Analysis of GO terms for these lineage-specific families revealed differently enriched functional categories, some of which may reflect important sources of genetic traits in different AA-genome species or their different adaptations to diverse habitats (SI Appendix, Dataset S4).
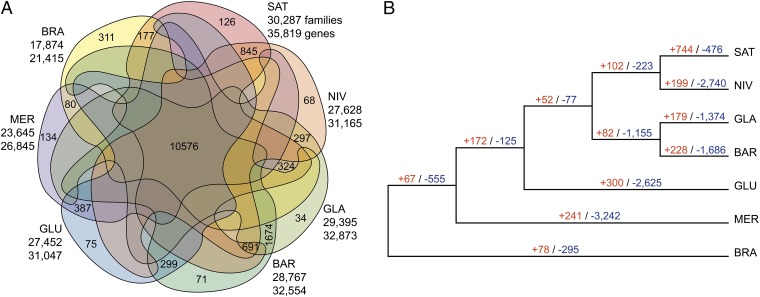
Fig. 3.
Evolutionary dynamics of rice gene families. (A) Venn diagram showing unique and shared gene families between and among the six AA-genome Oryza species plus BRA. (B) Gene family evolution among the seven Oryza species. Numerical values on each branch of the tree represent numbers of gene gain and/or loss events.
Expansion or contraction of gene families has an important role in the diversification of flowering plants (37) and all other organisms (38). To obtain in-depth insights into evolutionary rates of gene turnover among rice species, we independently used a maximum-likelihood method to estimate the rate () of change as the probability that a gene family either expands (via gene gain) or contracts (via gene loss) per gene per million years for all branches. Excluding lineage-specific families and likely annotation artifacts, 17,563 gene families, which currently contain 130,636 genes in the six AA genomes studied, were inferred to have been present in the most recent common ancestor (MRCA) of rice (SI Appendix, Dataset S5). We observed that gene families divergently evolved along different branches undergoing the most marked changes (expansion or contraction) (Fig. 3B). Notably, 552 exhibited significant expansions or contractions (P < 0.0001) (SI Appendix, Dataset S6a). Annotation of InterPro domains and GO assignments of these gene families suggested that some were involved in such key biological processes as biotic or abiotic stress responses, or the control of reproductive isolation through pollen recognition (SI Appendix, Datasets S6b and S6c). Of these rapidly evolving families, we further identified 10 with the greatest changes in copy number among the four terminal branches (SAT, NIV, BAR, and GLA) (SI Appendix, Dataset S7a). In contrast to significant contractions that occurred in their presumed wild progenitors (NIV and BAR), the two cultivated rice species, SAT and GLA, exhibited considerable expansions for all 10 families. InterPro function annotation showed that most of these 10 family genes function in recognition of pollen (GO: 0048544), transferase activity (GO: 0016758), oxidation-reduction processes (GO: 0055114), and defense response (GO: 0006952) (SI Appendix, Datasets S7b and S7c). However, determination of the significance of these differences will require more detailed analysis.
Whole-genome analysis also revealed expansion in the six AA-genome species of agronomically relevant gene families associated with disease resistance and flower development (SI Appendix, Tables S30 and S32). Using a reiterative process, we identified 631, 489, 450, 476, 392, and 416 genes encoding nucleotide-binding site (NBS) leucine-rich repeat (LRR) proteins in SAT, NIV, GLA, BAR, GLU, and MER, respectively, in contrast to 307 genes in BRA. This expansion, mainly attributable to an increase in coiled-coil–NBS and NBS domains, suggests stronger selection for enhanced disease resistance in the AA-genome species (SI Appendix, Table S30). We in silico mapped ∼98% of these orthologous R genes to specific locations across the SAT chromosomes (SI Appendix, Fig. S16). An unequal distribution of NBS-encoding genes was observed among chromosomes in the Oryza AA genomes, with chromosome 11 having by far the greatest density of R-gene candidates in every AA Oryza species (SI Appendix, Fig. S16). Analyses of genes homologous to Pid3, one of the best-characterized rice blast resistance genes, showed that pseudogenization of Pid3 occurred after the divergence of indica and japonica (SI Appendix, Fig. S17), further supporting the findings in an earlier study (39). Comparative analysis of another important rice blast R gene, Pi-ta (40), suggests that the substitution from Ile to Ser at position 6 and subsequent replacement of Ser with Ala at position 918 created the Pi-ta protein that recognizes AVR-Pita in NIV (SI Appendix, Fig. S18). We analyzed raw read sequences from 10 NIV accessions with read depths >10-fold (41), and confirmed that our observations for both Pid3 and Pi-ta are consistent across all studied accessions within NIV (SI Appendix, Dataset S8).
Using 77 previously identified rice MADS-box genes as queries (42), we found more such genes in AA-genome species (70 in NIV, 73 in GLA, 72 in BAR, 72 in GLU, and 70 in MER) than in BRA (60 in BRA) (SI Appendix, Table S32). MADS-box genes are thought to be involved in the regulation of flower development, and specific expansion of MADS-box genes in rice and other AA-genome relatives may have contributed to the observed flowering-time phenotypic variation that is crucial for reproductive success and contributes a key component of adaptation in the remarkably diverse range of habitats that these Oryza species inhabit.
Gene Loss.
Gene gain and loss have been proposed to be two of the primary contributors to functional changes that differentiate lineages (43). Summing across all of the inferred family sizes at the root of the tree, we estimated that the MRCA of the seven Oryza species contained at least 20,873 genes (SI Appendix, Dataset S5). This number may be an underestimation because many genes have been present in gene families that no longer exist in extant lineages. Overall in the seven genomes, a greater amount of gene contraction than expansion was inferred for the majority of branches (Fig. 3B), suggesting that loss of function has an especially important role in functional evolution. Of 6,987 families inferred to have been present in the MRCA that have no copy in at least one extant genome (SI Appendix, Dataset S9), 798 families are present in the MRCA but appear entirely lost in the SAT genome, of which 431 are still present in NIV. We measured potential gene extinction events by estimating the number of gene families that were entirely lost in SAT while being retained in other species, and found that ∼547 genes (488 families × 1.12 genes per family) were lost in SAT after the divergence from a common ancestor with NIV. To identify gene loss events and/or novel gene creations in SAT, we performed de novo assemblies of unmapped reads from the other five AA-genome species using a previously reported methodology, with some modifications (41). We found 490 genes that were lost in the SAT genome, in agreement with the estimated number based on the loss of gene families across the AA-genome Oryza species (SI Appendix, Dataset S10a). Functional annotation of lost genes from the five non-SAT species indicated that they were significantly enriched (P < 0.001) for disease-related genes, such as those genes encoding nucleotide-binding adaptor shared by APAF-1, R proteins, and CED-4 (IPR002182), tyrosine-protein kinase (IPR020635), and LRR (IPR013210) proteins (SI Appendix, Dataset S10a). These findings support the hypothesis that numerous disease resistance genes have been lost during rice domestication (44), making other AA-genome relatives particularly attractive as germplasm sources for future rice improvement.
To obtain first insights into patterns of gene gain and loss in the six rice species, we screened the 31 agronomically important genes that have been functionally well characterized in rice (SI Appendix, Table S34). To reflect a full evolutionary history of these genes within all eight AA-genome species of Oryza, we added O. sativa ssp. indica (IND), O. sativa ssp. tropical japonica (TRJ), O. rufipogon (RUF) and O. longistaminata (LON) for comparisons. Using whole-genome reads mapping, we computationally identified four genes, namely GS5, PROG1, S5, and SaF, that exhibited gene gain and loss along different lineages, supported by RNA-Seq data (SI Appendix, Tables S35 and S36). Besides, we further verified the evolutionary dynamics of five speciation genes (S5, SaM, SaF, mtRPL27, and HWH1) that underlie reproductive barriers (RIs) (45). Comparative sequence analyses revealed the loss of S5 (ORF5) in African cultivated rice (GLA) and its wild progenitor (BAR), also supported by RNA-Seq data (SI Appendix, Figs. S19–S23 and Table S36). We further examined the other two ORFs (ORF3 and ORF4) at this locus that work with ORF5 to form a killer–protector system (46). In the high-quality assemblies of S5 orthologous genomic regions for all five newly sequenced genomes, our results showed that ORF3 was fragmented and ORF4 was not detected in both GLA and BAR genomes. Thus, loss of multiple components of S5 causes its nonfunctional status in African cultivated rice (GLA) and its wild progenitor (BAR). The recent loss of S5, which sometimes causes female sterility in O. sativa subsp. indica-japonica hybrids (47), suggests that this locus has played an important role in the formation of RIs among these Oryza species at intersubspecific or interspecific levels.
We next analyzed PROG1, a gene that controls several agronomic traits, including prostrate growth in Asian cultivated rice (31, 32). We performed detailed computational and experimental confirmation, as well as synteny alignments of orthologous genomic regions (SI Appendix, Fig. S25). All obtained results together demonstrated loss of the PROG1 genes in BAR, GLA, and MER (SI Appendix, Table S35). The loss of PROG1 may account for the erect growth habit that is specific to BAR, GLA, and MER plants among the AA-genome species. These findings provide insights into the evolutionary consequences of frequent gene gain and loss that are of significance in the adaptation, divergence, and speciation of Oryza species.
Positive Darwinian Selection on Rice Genes.
The six closely related rice genomes provide a phylogenetic framework to examine protein-coding genes that show an accelerated molecular evolution in rice species and detect signals of positive selection on genes that are involved in adaptive divergence. We identified 2,272 high-confidence 1:1 orthologous gene families to find genes that show accelerated evolution in each of these six rice lineages (SI Appendix). Average synonymous (dS) and nonsynonymous (dN) gene divergence values varied but are comparable to the branch lengths that account for lineage divergence (Fig. 1B and SI Appendix, Table S38). Overall, the observed branch-specific ω values [nonsyonymous/synonymous rate ratio (dN/dS)] were 0.47986, 0.45181, 0.40689, and 0.28136 for SAT, NIV, GLU, and MER, respectively, suggesting relaxed selective constraints compared with MER. Inconsistent with the above-mentioned species that experienced strong purifying selection, we detected evidence for positive selection on GLA (ω = 1.995) and BAR (ω = 1.62731), with distinctly lowered levels of both dN and dS (SI Appendix, Figs. S26 and S27A). Previous studies suggested that the rate and tempo of molecular evolution might be promoted by speciation (48). Our results indicate that in the short period after speciation, where GLA and BAR diverged from a common African ancestor, numerous protein-coding genes show accelerated rates of evolution.
To test the hypothesis that these rapidly evolving genes showing increased dN/dS ratios have been under positive selection, we looked for such footprints using likelihood ratio tests for the same orthologous gene set in the six AA genomes. All tests identified a total of 537 nonredundant positively selected genes (PSGs; false discovery rate of <0.05) (SI Appendix, Table S39 and Datasets S11 and S12). In addition to 268 PSGs in the site model tests for all branches (SI Appendix, Fig. S27A), branch-specific PSGs widely varied from 20 (the ancestral Asian rice branch) to 234 (MER) (SI Appendix, Fig. S27 B–I). Comparing previous genome-wide scans for positive selection (49), we detected strikingly large proportions of PSGs in the overall phylogeny of rice species (23.6%, n = 537) and lineage-specific PSGs for each species (55.7% on average) that might be associated with the process of speciation and subsequent adaptation to particularly unstable environments.
GO classification showed that the detected PSGs spanned a wide range of functional categories (SI Appendix, Table S40). Notably, the inclusion of multiple rice genomes for all nonredundant PSGs yields a statistically significant enrichment for GO categories in response to developmental processes. Of these PSGs, 94 genes involved in “response to stimulus” and 47 in “reproduction” categories showed evidence for positive selection, which is significantly higher than the background levels of positive selection across the genomes (P < 0.05) (SI Appendix, Fig. S29). Flower development-related traits, often leading to reproductive isolation, and other adaptations to specific environments are expected to be key drivers of the divergence of the AA-genome species residing on different continents. Hence, it is interesting that genes involved in flower development, reproduction, and resistance-related processes have been influenced by positive selection in these species. With this fact in mind, we further examined functional enrichment for branch- or species-specific datasets of PSGs, which varied greatly in both number and type across different lineages (SI Appendix, Fig. S29 and Table S40). Consistent with previously reported genome-wide positive selection scans (49), many candidate PSGs were significantly overrepresented in categories related to flower development, embryogenesis, reproduction, pathogen defense, disease resistance, and stress resistance processes, and varied between different rice clades (SI Appendix, Fig. S30 and Table S42). We found that 14 genes known to play an important role in flower development pathways were PSGs (CRY2, LHY, PFT1, VIL1, ASHH2, PEP, HEN1, XTH, PG, MOS3/SAR3, PDIL, TAF6, TMS1, and SEC5). Of these 14 genes, five key genes (PG, PDIL, TAF6, TMS1, and SEC5) that show signs of positive selection are associated with pollination (Fig. 4).
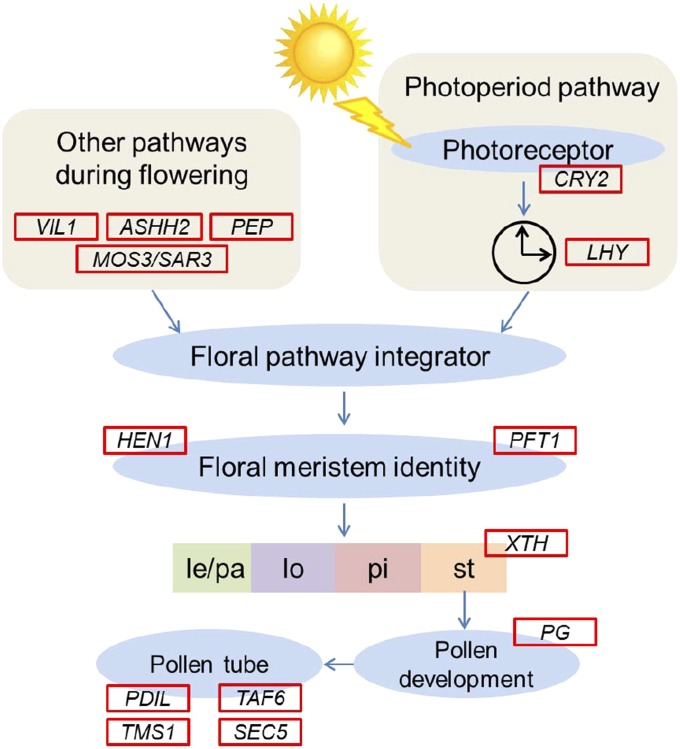
Fig. 4.
PSGs involved in the predicted integrated pathways of rice flower development. Within the genes assigned to categories of “flower development” (GO: 0009908) and “pollination” (GO: 0009856), the 14 genes that are homologs to reproduction-related genes or associated with the flowering process are shown with evidence of positive selection (in red boxes). ASHH2, ABSENT, SMALL, OR HOMEOTIC DISCS 1 HOMOLOG 2; CRY2, CRYPTOCHROME 2; HEN1, HUA ENHANCER 1; le, lemma; lo, lodicule; LHY, LATE ELONGATED HYPOCOTYL; MOS3/SAR3, MODIFIER OF SNC1,3/SUPPRESSOR OF AUXIN RESISTANCE 3; pa, palea; PDIL, PROTEIN DISULFIDE ISOMERASE-LIKE; PEP, PEPPER; PFT1, PHYTOCHROME AND FLOWERING TIME 1; PG, POLYGALACTURONASE; pi, pistil; SEC5, EXOCYST COMPLEX COMPONENT; st, stamen; TAF6, TATA BOX BINDING PROTEIN-ASSOCIATED FACTOR 6; TMS1, THERMOSENSITIVE MALE STERILE 1; VIL1, VERNALIZATION INSENSITIVE 3-LIKE 1; XTH, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE.
Noncoding RNA Gene Family Evolution.
Using the obtained ncRNA gene annotations (SI Appendix, Table S13), comparisons of ncRNA gene families showed that they varied in number across the phylogeny of the six rice species, although these changes were not generally dramatic (SI Appendix, Fig. S6). GO analysis of predicted miRNA target genes (SI Appendix, Table S43 and Dataset S1), most of which were verified by RNA-Seq data (SI Appendix, Tables S9 and S10), indicates that the majority of miRNA target genes were conserved in a wide range of important functional categories across all species (SI Appendix, Fig. S31). Several biological processes or molecular functions were significantly enriched in some target genes of all six rice species (P < 0.05), such as death (GO: 0016265), cellular process (GO: 0009987), and response to stimulus (GO: 0050896). However, some GO terms, for example, cellular component organization (GO: 0016043), macromolecular complex (GO: 0032991), organelle (GO: 0043226), and organelle part (GO: 0044422), were only enriched in MER (SI Appendix, Fig. S32). Comparative sequence analysis of all classes of ncRNA genes across the six rice species indicated that snRNA genes exhibit relatively high nucleotide substitution rates (2.61%) (SI Appendix, Fig. S33 and Table S44). Individual species-based analyses confirmed the overall conserved pattern of sequence evolution for every class of ncRNA genes (SI Appendix, Fig. S34), but we did observe that, except for some housekeeping ncRNA genes (tRNAs, snoRNAs, and snRNAs), divergence rates of pre-miRNAs, mature miRNAs, and miRNA targets in the two cultivated rice species, consistent with genomic background components, were comparatively lower than in their counterpart wild progenitors, probably resulting from bottleneck effects during the domestication of cultivated rice. Among the six AA-genome species, MER exhibited the highest rates of sequence evolution for all ncRNA genes and genomic background components.
MiRNA genes have emerged as master regulators of plant growth, particularly in plant development processes and a variety of stress responses (50). Comparisons of nucleotide substitutions in miRNA genes and target sites across the six rice species showed that most were conserved without mutations, whereas a small proportion (98 of 227 in miRNA and 560 of 1,598 in targets) of them were variable for one or more nucleotide substitutions (SI Appendix, Fig. S35). A systematic survey of the 21 miRNA gene families related to flower development indicated a wide range of variation in copy number among the six Oryza species (SI Appendix, Table S45). In addition to the expansion of the MADS-box gene family (SI Appendix, Table S32), copy number variation in several miRNA gene families may partially account for the characteristic differences of flower development and reproduction, providing new resources for studying regulation by miRNA genes. Random sampling of four miRNA genes (osa-miR159a, osa-miR159e, osa-miR159f, and osa-miR1847), belonging to two miRNA gene families (miRNA159, miRNA1847), uncovered differential amplification in one or more of the other five non-SAT species. In addition to functional enrichment of protein phosphorylation (GO: 0006468) of the lineage-specific gene families in SAT and NIV (SI Appendix, Dataset S4), the detected nucleotide mutations in certain lineages may be responsible for phenotypic variation and different ecological adaptations, such as tolerance of drought conditions and phosphate starvation (SI Appendix, Figs. S37 and S38 and Table S43).
Concluding Remarks
We present high-quality reference genome sequences for O. nivara, O. glaberrima, O. barthii, O. glumapatula, and O. meridionalis. Over the past century, the introduction of genes from AA-genome relatives into Asian cultivated rice has helped expand rice genetic diversity important to the generation of environmentally resilient and high-yielding varieties. This approach led to the discovery of the “wild-abortive rice” genes from O. rufipogon, which allowed the facile production of hybrid rice, a landmark in rice improvement (51). Similarly, we expect that the genome sequences described in this article will become valuable resources for more extensive exploitation of wild Oryza germplasms to enhance rice breeding programs. With the high-quality sequence of the Nipponbare genome of O. sativa as a guide, our multispecies comparisons of the five AA rice genomes, aided by the recently reported FF genome of O. brachyantha as an outgroup (23), afford novel opportunities for addressing a wealth of questions about gene and genome evolution in rice. The use of multispecies orthology and careful annotations of functional features provide convincing evidence in strong support of gene models, including both protein-coding and ncRNA genes.
Overall genome size, number of genes, contents of TEs, and genomic architecture and gene colinearity are all fairly conserved across these rice genomes. However, MER is an outlier by several criteria, including evidence for greater divergence at levels of protein-coding genes, ncRNA genes, and structural variation, plus accelerated molecular evolution and specific functional enrichment of miRNA target genes. This greater divergence in MER is at least partly caused by the fact that it is more than 2.5-fold more distantly related to the other AA genomes studied than they are to each other, but it may also be an outcome of unique environmental demands for MER adaptation in its Australian home.
Abundant microstructural rearrangements and lineage-specific gene gains and/or losses are expected to be largely responsible for the functional dynamics of adaptation over evolutionary time in these closely related rice species. This expectation is particularly well-exemplified in the PROG1-orthologous regions, where numerous cases of large-scale SVs were observed and, for example, the PROG1-encoding gene was deleted in several species, thereby leading to their current erect growth habit. Segmental duplications that are abundant across these Oryza genomes have played an important role in driving rice genome evolution. The genes within these duplications were significantly enriched for important biological functions, thus providing essential sources of the gene dosage variation that can permit subfunctionalization or neofunctionalization.
The rapid evolution of gene families, particularly a sizeable fraction showing fast and/or lineage-specific expansions and contractions, exhibiting different functional enrichment, is also expected to contribute to the adaptive evolution of Oryza species to globally diverse habitats. We detected a considerable portion of lineage-specific protein-coding genes under positive selection that were involved in flower development, reproduction, and biotic- and abiotic defenses. These genes are particularly good candidates to investigate for adaptive evolution to changing environments. Future studies should use advanced genetic tools (e.g., gene replacement and other forms of reverse genetics) to determine which of these genes are involved in which adaptations. Significant GO enrichment of defense-related genes was detected in the five non-SAT species, indicating that AA-genome relatives of SAT serve as reservoirs for novel disease resistance alleles that may enhance rice breeding programs.
Because these comparative genomic analyses were empowered by being embedded in the context of a robust phylogeny, we were able to discover that the African branch (GLA/BAR) differs in several evolutionary behaviors from the Asian branch (SAT/NIV). The African branch exhibits one-third fewer genomic SVs; remarkable evidence for positive selection with distinctly lowered levels of both dN and dS values; more gene loss (PROG1 and S5); and larger numbers of PSGs significantly overrepresented in categories mainly related to flower development, reproduction, and defense processes than the Asian branch. This finding indicates accelerated molecular evolution after their very recent speciation to adapt quickly to specific environments in Africa. Comparative analysis also found some instances where both SAT and GLA diverged in similar ways from their counterpart wild progenitors NIV and BAR, as shown by significantly more expansions for all top 10 gene families of reproductive and ecological significance and lowered sequence evolutionary rates of ncRNA genes in the domesticated lineages. Considering the quick extinction and endangered status of natural populations due to severe deforestation in worldwide tropical and subtropical regions (52), it is our hope that the genome assemblies and genomic variation data presented here will provide valuable information to aid the global conservation of these precious wild rice species.
Methods
Genome Sequencing, Assembly, and Validation.
We sequenced the five AA-genome Oryza species by using a WGS strategy with next-generation sequencing technologies from paired-end, small-insert libraries (∼300 bp and ∼500 bp) and mate-pair, large-insert libraries (2 kb, 4 kb, 6 kb, and 8 kb). The whole-genome de novo assemblies were built using SOAPdenovo (22). To assess the quality of the assembled genomes, we used three evaluation approaches. First, we separately mapped our assembled genome sequences (NIV, GLA, BAR, GLU, and MER) to the SAT genome to estimate the coverage of the assemblies using MUMmer (53). We further compared orthologous genomic segments based on the aligned orthology map of the six AA-genome Oryza species (SAT, NIV, GLA, BAR, GLU, and MER) and calculated the coverage of the gene-containing genomic regions corresponding to the SAT genome for each species. We next evaluated the assembly quality by using Nipponbare gene models to examine whether they are split into multiple contigs into the assemblies. Second, we took the four species (NIV, GLA, GLU, and MER) with BAC end sequencing (BES) data from OMAP and aligned these BES data with our de novo assembled genomes. Of these species, BES datasets for NIV, GLA, and BAR were separately mapped to our de novo assembled scaffolds before the de novo assemblies were improved by BES integration. Finally, by running both MUMmer (53) and Blastz (54), the assembly quality was further evaluated by aligning our assembled genomes against sequences from the short arms of chromosome 3 released by the OMAP.
Genome Annotation.
Putative protein-coding loci were identified based on ab initio annotation, homology-based gene prediction, and EST-aided annotation, and quality validation of the gene model was performed by using transcriptome, EST, and homologous peptide evidence. De novo searches for and annotation of TEs were completed by integrating RepeatMasker (www.repeatmasker.org), LTR_STRUCT (55), RECON (56), LTR_Finder (57), and RepeatScout (58). The five different types of ncRNA genes, including tRNA, rRNA, snoRNA, snRNA, and miRNA genes, were predicted using de novo and homology search methods. We annotated miRNAs using two steps. First, we downloaded the existing rice miRNA entries from miRBase, release 18.0 (59). Then, the conserved miRNAs were identified by mapping all miRBase-recorded SAT miRNA precursor sequences against the assembled NIV, GLA, BAR, GLU, and MER genomes using nucleotide–nucleotide BLAST (blastn) with cutoffs at an E value of <1e−5, identity of >80%, and query coverage of >80%. Second, additional miRNA genes were identified by aligning all miRBase-recorded grass miRNA precursor sequences against our assembled genomes using blastn with cutoffs at an E value of <1e−5, identity of >60%, and query coverage of >60%, because deep-sequencing genome projects have found numerous new miRNAs in grasses. When a miRNA was mapped to a target Oryza genome, flanking sequences were next checked for hairpin structures. Those loci that fulfilled miRNA precursor secondary structures were annotated as additional miRNA genes. We excluded miRNA genes that identified multiple hits, most likely repeat sequences, in the six assembled rice genomes.
More methods and details of data collection are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the International Rice Research Institute (Manila, Philippines) for providing rice germplasm. This work was supported by the Top Talents Program of Yunnan Province (Grant 20080A009), the Hundreds Oversea Talents Program of Yunnan Province, the project of the Yunnan Innovation Team, the Key Project of the Natural Science Foundation of Yunnan Province (Grant 2010CC011), the Hundreds Talents Program of the Chinese Academy of Sciences (CAS), a grant from the CAS (Grant KSCX2-YW-N-029), a grant from the Chinese Department of Science and Technology (973 Program Grant 2007CB815703), and a startup grant from the Kunming Institute of Botany, CAS (to L.-Z.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information database, www.ncbi.nlm.nih.gov (BioProject nos. PRJNA264484, PRJNA264480, PRJNA264485, PRJNA264496, and PRJNA264483 for O. nivara, O. glaberrima, O. barthii, O. glumaepatula, and O. meridionalis, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418307111/-/DCSupplemental.
References
- 1.Bennetzen JL. Patterns in grass genome evolution. Curr Opin Plant Biol. 2007;10(2):176–181. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- 3.Presgraves DC. The molecular evolutionary basis of species formation. Nat Rev Genet. 2010;11(3):175–180. doi: 10.1038/nrg2718. [DOI] [PubMed] [Google Scholar]
- 4.Strasburg JL, et al. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):364–373. doi: 10.1098/rstb.2011.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark AG, et al. Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450(7167):203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 6.Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440(7082):341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- 7.Scally A, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483(7388):169–175. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang TT. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica. 1976;25(1):425–441. [Google Scholar]
- 9.Lu BR, Sharma SD, Shastry SVS. Taxonomy of the genus Oryza (Poaceae): Historical perspective and current status. Int Rice Res Notes. 1999;24:4–8. [Google Scholar]
- 10.Ammiraju JSS, et al. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16(1):140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brar DS, Singh K. In: Oryza. Wild Crop Relatives: Genomic and Breeding Resources, Cereals. Kole C, editor. Berlin: Springer; 2011. pp. 321–365. [Google Scholar]
- 12.Zhu T, et al. Phylogenetic relationships and genome divergence among the AA-genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol Phylogenet Evol. 2014;70:348–361. doi: 10.1016/j.ympev.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan DA. The Wild Relatives of Rice: A Genetic Resources Handbook. International Rice Research Institute; Manila, Philippines: 1994. [Google Scholar]
- 14.Oka HI. Origin of Cultivated Rice. Scientific Societies Press/Academic Press; Tokyo, Japan: 1988. [Google Scholar]
- 15.Morishima H, Sano Y, Oka H. Evolutionary studies in cultivated rice and its wild relatives. Oxf Surv Evol Biol. 1992;8:135–184. [Google Scholar]
- 16.Zuccolo A, et al. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol. 2007;7:152–167. doi: 10.1186/1471-2148-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ammiraju JSS, et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007;52(2):342–351. doi: 10.1111/j.1365-313X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- 18.Ammiraju JSS, et al. Dynamic evolution of oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20(12):3191–3209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F, et al. Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci USA. 2009;106(6):2071–2076. doi: 10.1073/pnas.0812798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanyal A, et al. Orthologous comparisons of the Hd1 region across genera reveal Hd1 gene lability within diploid Oryza species and disruptions to microsynteny in Sorghum. Mol Biol Evol. 2010;27(11):2487–2506. doi: 10.1093/molbev/msq133. [DOI] [PubMed] [Google Scholar]
- 21.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 22.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Che JF, et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun. 2013;4:1595. doi: 10.1038/ncomms2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou XH, Yang Z, Doyle JJ, Ge S. Multilocus estimation of divergence times and ancestral effective population sizes of Oryza species and implications for the rapid diversification of the genus. New Phytol. 2013;198(4):1155–1164. doi: 10.1111/nph.12230. [DOI] [PubMed] [Google Scholar]
- 25.Prasad V, et al. Late Cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nat Commu. 2011;2:480. doi: 10.1038/ncomms1482. [DOI] [PubMed] [Google Scholar]
- 26.Uozu S, et al. Repetitive sequences: Cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol. 1997;35(6):791–799. doi: 10.1023/a:1005823124989. [DOI] [PubMed] [Google Scholar]
- 27.Myiabayashi T, et al. Genome size of twenty wild species of Oryza determined by flow cytometric and chromosome analyses. Breed Sci. 2007;57:73–78. [Google Scholar]
- 28.Bray N, Pachter L. MAVID: Constrained ancestral alignment of multiple sequences. Genome Res. 2004;14(4):693–699. doi: 10.1101/gr.1960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA. 2004;101(34):12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011;43(12):1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 31.Jin J, et al. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40(11):1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 32.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40(11):1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 33.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43(5):476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297(5583):1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 35.Marques-Bonet T, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457(7231):877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitreva M, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43(3):228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purugganan MD, Rounsley SD, Schmidt RJ, Yanofsky MF. Molecular evolution of flower development: Diversification of the plant MADS-box regulatory gene family. Genetics. 1995;140(1):345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohno S. Evolution by Gene Duplication. Springer; New York: 1970. [Google Scholar]
- 39.Shang J, et al. Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site—Leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics. 2009;182(4):1303–1311. doi: 10.1534/genetics.109.102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Jia Y, Shu QY, Wu D. Haplotype diversity at the Pi-ta locus in cultivated rice and its wild relatives. Phytopathology. 2008;98(12):1305–1311. doi: 10.1094/PHYTO-98-12-1305. [DOI] [PubMed] [Google Scholar]
- 41.Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat Biotechnol. 2012;30(1):105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 42.Arora R, et al. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242–263. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13(10):2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai H, Itoh T. Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genomics. 2010;11:121–144. doi: 10.1186/1471-2164-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rieseberg LH, Blackman BK. Speciation genes in plants. Ann Bot (Lond) 2010;106(3):439–455. doi: 10.1093/aob/mcq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science. 2012;337(6100):1336–1340. doi: 10.1126/science.1223702. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA. 2008;105(32):11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venditti C, Pagel M. Speciation as an active force in promoting genetic evolution. Trends Ecol Evol. 2010;25(1):14–20. doi: 10.1016/j.tree.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Pentony MM, et al. The plant proteome folding project: Structure and positive selection in plant protein families. Genome Biol Evol. 2012;4(3):360–371. doi: 10.1093/gbe/evs015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 51.Tanksley SD, McCouch SR. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science. 1997;277(5329):1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 52.Gao L. Population structure and conservation genetics of wild rice Oryza rufipogon (Poaceae): A region-wide perspective from microsatellite variation. Mol Ecol. 2004;13(5):1009–1024. doi: 10.1111/j.1365-294X.2004.02108.x. [DOI] [PubMed] [Google Scholar]
- 53.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz S, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13(1):103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarthy EM, McDonald JF. LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics. 2003;19(3):362–367. doi: 10.1093/bioinformatics/btf878. [DOI] [PubMed] [Google Scholar]
- 56.Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12(8):1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Z, Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–i358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 59.Kozomara A, Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawahara Y, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6(1):4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.