Significance
Bacteria can repel invader DNA and RNA molecules by using an adaptive immunity mechanism called clustered regularly interspaced short palindromic repeats (CRISPRs)-Cas. CRISPR loci in a host genome are a repository of DNA fragments obtained from previous encounters with an invader, which can be transcribed and activated into short RNA molecules (crRNA) with sequences complementary to invader DNA or RNA. In some CRISPR-Cas systems, crRNA is assembled into a targeting complex called “Cascade” that seeks invader DNA to form an R-loop that triggers recruitment of a nuclease-helicase, Cas3, to destroy invader DNA. In this study, we show atomic resolution structures of a full-length Cas3, revealing how Cas3 coordinates binding, ATP-dependent translocation, and nuclease digestion of invader DNA.
Keywords: Cas3, CRISPR, Cascade, bacterial immunity, Cas proteins
Abstract
Mobile genetic elements in bacteria are neutralized by a system based on clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) proteins. Type I CRISPR-Cas systems use a “Cascade” ribonucleoprotein complex to guide RNA specifically to complementary sequence in invader double-stranded DNA (dsDNA), a process called “interference.” After target recognition by Cascade, formation of an R-loop triggers recruitment of a Cas3 nuclease-helicase, completing the interference process by destroying the invader dsDNA. To elucidate the molecular mechanism of CRISPR interference, we analyzed crystal structures of Cas3 from the bacterium Thermobaculum terrenum, with and without a bound ATP analog. The structures reveal a histidine-aspartate (HD)-type nuclease domain fused to superfamily-2 (SF2) helicase domains and a distinct C-terminal domain. Binding of ATP analog at the interface of the SF2 helicase RecA-like domains rearranges a motif V with implications for the enzyme mechanism. The HD-nucleolytic site contains two metal ions that are positioned at the end of a proposed nucleic acid-binding tunnel running through the SF2 helicase structure. This structural alignment suggests a mechanism for 3′ to 5′ nucleolytic processing of the displaced strand of invader DNA that is coordinated with ATP-dependent 3′ to 5′ translocation of Cas3 along DNA. In agreement with biochemical studies, the presented Cas3 structures reveal important mechanistic details on the neutralization of genetic invaders by type I CRISPR-Cas systems.
Many bacteria and archaea can eliminate invading phages and plasmids by activating a defense system that is based on processing of clustered regularly interspaced short palindromic repeats (CRISPRs) by CRISPR-associated (Cas) proteins and various additional proteins. These CRISPR-Cas systems are diverse in structure and function, but common principles of action allow their classification into three major types (I, II, and III) with some further divisions into subtypes (e.g., type IA–F) (1–6).
Common CRISPR-mediated immunity processes are usually defined into three stages: (i) acquisition of a short DNA segment (protospacer) from an invading virus or plasmid, and its insertion at the leader-proximal end of a CRISPR locus (7, 8), (ii) generation of small mature CRISPR RNAs (crRNAs) from a longer transcript of a CRISPR locus (9–11), and (iii) interference of foreign nucleic acid invaders by a crRNA-containing ribonucleoprotein effector complex (12–18).
Different interference effector complexes characterize the three major CRISPR types. Cascade (Cas complex for antiviral defense) complexes are synonymous with interference in type I CRISPRs, comprising multiple proteins in a helical structure around crRNA that targets invader DNA (13, 19, 20). Type II CRISPR systems are characterized by Cas9, a multidomain protein with a bilobed architecture harboring two nuclease sites that together catalyze cleavage of invading DNA (12, 21–24). The Csm system in CRISPR type IIIA and a related Cmr complex in type IIIB cleave DNA and RNA, respectively, forming a helical structure around crRNA that is structurally similar to Cascade (14, 17, 18, 25, 26).
Whereas Cas9 and Csm/Cmr effector complexes directly recognize and nucleolytically degrade invading genes (12, 14, 16, 17, 21), Cascade complex by itself is not a nuclease for degradation of invader DNA (13). In Escherichia coli K-12 (type IE), Cascade recognizes and binds crRNA to complimentary sequence in target DNA, generating an RNA mediated displacement loop (R-loop) of single-stranded (ss) DNA and RNA-DNA hybrid within double-stranded (ds) target DNA (13). Formation of the R-loop structure induces conformational change in Cascade subunits, triggering recruitment of Cas3 protein that is a nuclease-helicase responsible for degradation of dsDNA (27–30).
The two enzymatic units of Cas3, a histidine-aspartate (HD) nuclease and a Superfamily 2 (SF2) helicase (31), may be expressed from separate genes as Cas3′ (SF2 helicase) and Cas3′′ (HD nuclease) or may be fused as a single HD-SF2 polypeptide (32). Variation in optimal reaction conditions have also been reported, especially in cofactor requirements and substrate specificities of HD-nuclease. Streptococcus thermophilus type IE Cas3 degrades ssDNA in the presence of magnesium and transition metal ions, and uses ATP hydrolysis to translocate DNA strands with 3′ to 5′ directionality (33). Methanocaldococcus jannaschii type IA Cas3 HD nuclease cleaves ssDNA with magnesium dependence both endo-nucleolytically and 3′ to 5′ exonucleolytically, and ssRNA in the presence of Mn2+ ions (34). Thermus thermophilus HB8 type IE Cas3′′ HD nuclease degrades ssDNA endo-nucleolytically with Mn2+ or Ni2+ ions (35). Significantly for understanding CRISPR interference, E. coli type IE Cas3 protein catalyzes Mg2+ ion- and transition metal ion-dependent nicking of the displaced strand of the target dsDNA, after which ATP-dependent unwinding of the target allows for the 3′ to 5′ processive nuclease activity on the nicked ssDNA (28, 29, 33, 35, 36). Overall, the data suggests that nucleolytic activity of most Cas3 enzymes is transition metal ion-dependent.
A lack of structural information on the Cas3 SF2 helicase and how it might coordinate functions with the HD nuclease domain have hindered understanding of interference in type I CRISPR immunity. Here, we present crystal structures of a full length HD-SF2 Cas3 from Thermobaculum terrenum, which has the type IE CRISPR-Cas system (Fig. S1A). Structures are solved for Cas3 both with an adenosine triphosphate analog (dATP-bound) and without dATP (dATP-free). The N-terminal HD nuclease contains an active site that is composed of two metal ions, and the dATP cofactor is bound at the interface of the two RecA-like domains (RDs). Interestingly, the HD nucleolytic active site is aligned with a predicted nucleic acid-binding tunnel running through the helicase domains. The molecular basis of DNA binding and unwinding by Cas3, and a protein module that is potentially involved in the interaction with the Cascade effector complex, can be predicted and inferred from the two structures. By reconciling these structures with previously reported biochemical features of Cas3, we propose a mechanistic model for Cas3 that includes (i) recruitment of the Cas3 to the Cascade-bound target DNA, (ii) endo-nucleolytic cleavage of the displaced DNA strand, and (iii) progressive 3′ to 5′ exonucleolytic degradation of the displaced strand and concomitant unwinding of the dsDNA target.
Results
Overall Features.
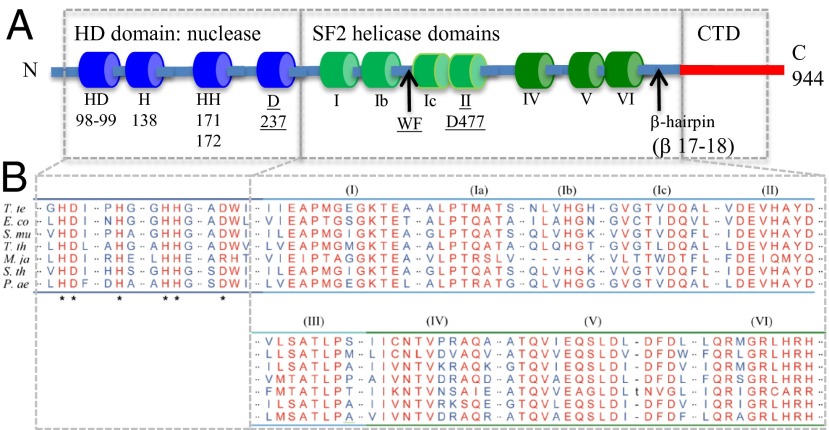
Amino acid sequence comparison of Cas3 homologs had suggested that Cas3 from T. terrenum (TteCas3) is a dual function nuclease-helicase enzyme, with an N-terminal HD nuclease domain fused to SF2 helicase domains (Fig. 1 and Fig. S1). Crystal structures of full-length TteCas3, with and without an ATP analog, were solved at resolutions of 2.8 Å and 2.5 Å, respectively (Table 1), and confirmed this general arrangement, throughout displayed with the HD nuclease domain at the upper end of the structure residing atop SF2 helicase domains and a distinct exposed C-terminal domain (CTD) (Fig. 2A).
Fig. 1.
Sequence and structural features of TteCas3. (A) Summarized organization of 944-aa TteCas3 HD nuclease, SF2 helicase, and CTD, highlighting features described in Results. Shown are five residues of the HD domain that coordinate metal ions and water for nuclease activity (Fig. 2B). Residues that were mutagenized to abolish nuclease and ATPase activities (Fig. 2 C and E) are underlined. Other features of Cas3 structure function, detailed in Results, are labeled WF motif, β-hairpin and motifs Ic, IV, and VI. (B) Sequence alignment of conserved regions. Residues that interact with catalytic metal ions and a water molecule are indicated by asterisks, and conserved helices motifs are marked (I–VI). E.co, E. coli K-12; M.ja, M. jannaschii DSM 2661; P.ae, Pseudomonas aeruginosa; S.mu, S. mutans NN2025; S.th, S. thermophilus; T.te, T. terrenum; T.th, T. thermophilus HB8.
Table 1.
Data collection and structure refinement statistics
| Data collection | dATP-bound | SeMet-derivative, dATP-free |
| Space group | C2221 | |
| Unit cell dimensions | ||
| a, b, c, Å | 105.68, 211.63, 104.27 | 104.17, 214.29, 102.10 |
| α, β, γ, ° | 90, 90, 90 | 90, 90, 90 |
| Wavelength, Å | 0.9793 | 0.9793 |
| Resolution, Å | 50–2.80 (2.85–2.80)* | 100–2.50 (2.54–2.50)* |
| Rsym | 12.3 (37.7) | 10. 9 (43.3) |
| I/σ(I) | 8.0 (1.7) | 11.2 (2.3) |
| Completeness, % | 96.4 (83.3) | 93.1 (91.8) |
| Redundancy | 3.8 (2.8) | 6.8 (4.1) |
| Figure of merit | ||
| SOLVE/RESOLVE | 0.34/0.65 | |
| Refinement | ||
| Resolution, Å | 45.7–2.80 | 93.8–2.50 |
| No. of reflections | 28267 | 37403 |
| Rwork/Rfree | 18.9 (28.5)/23.8 (35.3) | 18.5 (24.7)/22.9 (28.2) |
| No. of atoms | ||
| Protein/water/Mn/Mg/dATP | 7,207/128/2/1/1 | 7,202/283/2/0/0 |
| rmsd | ||
| Bond lengths, Å/angles, ° | 0.003/0.90 | 0.004/0.96 |
| Average B values, Å2 | ||
| Protein/water | 40.9/24.7 | 51.6/48.2 |
| Ramachandran plot, % | ||
| Favored/allowed/outliers | 96.3/3.0/0.7 | 95.9/3.5/0.6 |
| PDB ID code | 4Q2D | 4Q2C |
The numbers in parentheses are the statistics from the highest resolution shell.
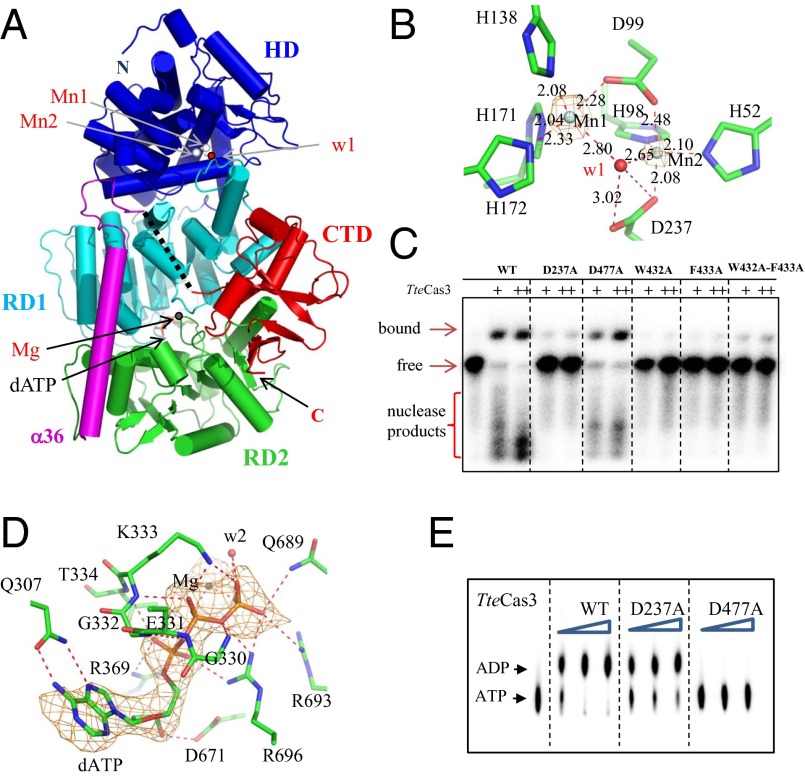
Fig. 2.
Structure of full-length TteCas3. (A) Overall structure of TteCas3. N-terminal HD-type nuclease domain (HD, blue), two RDs (RD1 and RD2, cyan and green), RD1-RD2 encompassing helix (α36, magenta), and C-terminal domain (CTD, red) are differentiated. Metal ions (Mn, Mg) and a water molecule are displayed as balls and dATP are as stick models. (B) TteCas3 HD nuclease active site. Two metal ions (Mn1 and Mn2) and a water molecule (w1) are displayed as balls in the omitted Fo-Fc electron density (orange) contoured at 20 σ. Interactions between atoms are displayed by dotted lines, and their distances are indicated. (C) Nuclease degradation of 32P end-labeled 72-mer ssDNA substrate (0.1 nM) by TteCas3 wild type (WT) compared with various mutant proteins (185 and 370 nM). Nuclease products are indicated, with uncut ssDNA and Cas3-bound ssDNA. (D) Close-up view of the ATPase site. Bound dATP at the interface of two RDs is displayed by line models in the omitted Fo-Fc difference map (orange) contoured at 7 σ, together with a Mg ion (Mg) and a water molecule (w2) of spheres. The conserved and their interaction with dATP are drawn by line models and red-dotted lines, respectively. (E) ATPase activity of wild-type (WT) TteCas3 compared with HD nuclease mutant D237A and ATPase motif II (DExD/H) mutant D477A. Formation of ADP from α[32P]ATP was compared for each protein at concentrations of 92, 185, and 370 nM in the presence of magnesium chloride (1 mM) and 72-mer ssDNA (2 nM).
The N-Terminal HD-Type Nuclease.
The N-terminal HD domain of the full-length TteCas3 (Ser20-Val282, blue in Figs. S1B and 2A) superimposes well with HD structure of the truncated Cas3 from T. thermophilus HB8, with root mean square deviation (rmsd) of 1.72 Å for the 230 aligned Cα-atoms (PDB ID code: 3SK9; Fig. S2A) (34, 35). Based on the conserved residues of the nuclease domain of TteCas3 (Fig. 1) and its divalent metal ion-dependent nucleolytic activity (Fig. S2), two of the three spherical electron densities within the HD catalytic site are assumed to correspond to Mn2+ or Ni2+ ions (Mn1 and Mn2) and the third one to a water molecule (w1) (Fig. 2B). The Mn1 ion is coordinated with the w1 water molecule and four atoms of protein ligands, corresponding to side chains of Asp99, His138, His171, and His172. The Mn2 ion interacts with amino acid side chains of His52, His98, Asp99, and Asp237, and with the w1 molecule that is hydrogen-bonded to the side-chain atom of Asp237. TteCas3 D237A mutant protein had no observable nuclease activity when incubated with ssDNA, and compared with wild-type TteCas3, it had reduced ability to bind to ssDNA (Fig. 2C and Fig. S2B), implicating a role in both ssDNA-binding and nuclease activity. The involvement of Asp237 in the nucleolytic activity is consistent with it being part of the metal-dependent active site for nucleic acid cleavage, in agreement with previous analysis of the Cas3′′ HD domain of M. jannaschii and the truncated HD domain of T. thermophilus HB8 (34, 35).
The ATPase Site.
The ATP-binding site at the interface cleft of two RecA-like domains (RDs) was revealed from the cocrystal structure of TteCas3 and dATP (Fig. 2D). This site is similar to that of the yeast DEAD-box RNA helicase Mss116p (PDB ID code: 3I5Y; Mss Hel in Fig. S3A) (37), a closely related structural homolog with two RDs as in TteCas3 helicase (Z score 18.7) and an rmsd value of 3.12 Å for the 281 aligned Cα atoms. All three phosphate groups of bound dATP are observed in the dATP-complex structure, with extra electron density in the vicinity of the β- and γ-phosphates that was modeled with a Mg2+ ion (Mg) (Fig. 2D). Bound dATP interacts with residues of helicase motifs I, II, IV, V, and VI (Figs. S1B and 2D). Mutation of Cas3 motif II (DExD/H, alignment in Fig. 1B), to give protein D477A, resulted in abolition of ssDNA-stimulated ATPase activity, whereas ATPase activity was intact when the HD-nuclease mutation D237A was present (Figs. S3B and 2E). Comparison of dATP-bound and dATP-free structures shows no overall gross structural change in Cas3 (rmsd value of 0.5 Å for all Cα atoms), but significant focused structural changes were observed around the ATPase site involving relative positioning of helicase motifs Ia and V (Fig. S3C). In the dATP-free structure, Ser669 of motif V contacts motif Ia, but this interaction disappears when dATP is bound, as the cofactor hydrogen bonds to Asp671 of motif V (Fig. 2D). ATP-induced structural changes in motif V are important for coupling ATPase activity with nucleic acid binding and translocation in helicases as revealed by mechanistic studies on the NS3 helicase (38, 39), which is structurally similar to TteCas3, as discussed below. It is interesting to note that although the motif V Asp (Asn) residue is well conserved in Cas3 proteins, the serine residue is conserved only in the IE subtype and is replaced by glycine in Cas3 protein from other subtypes (Fig. S4).
Structural Features of the TteCas3 Helicase.
A structural homology search using both TteCas3 RD domains identified good superimposition with RD domains of archaeal Hel308 DNA helicase (rmsd 3.03 Å, PDB ID code: 2P6R) and flavivirus NS3 RNA helicase (rmsd 2.82 Å, PDB ID code: 2JLU), despite their low overall sequence identity (18 and 11% for the compared 273 and 224 residues of Hel308 and NS3 RD domains, respectively). Both Hel308 and NS3 helicases form a tunnel to accommodate single-stranded nucleic acid, binding of which induces slight rearrangement of two RDs and the CTD, but significant reordering of helicase motif Ic (40, 41) (Fig. S5). In TteCas3 helicase, three domains (RD1, RD2, and CTD) form a similar tunnel, lined by motifs Ia, Ib, IV, and VI, and an invariant tryptophan (Trp432) and a Phe433 (WF) on the RD1 and RD2 surface (gray patches in Fig. 3A). Mutations of the invariant WF residues of RD1 motif Ic resulted in decreased binding and nucleolytic activities on the 72-mer ssDNA substrate, compared with wild-type TteCas3 (Fig. 2C).
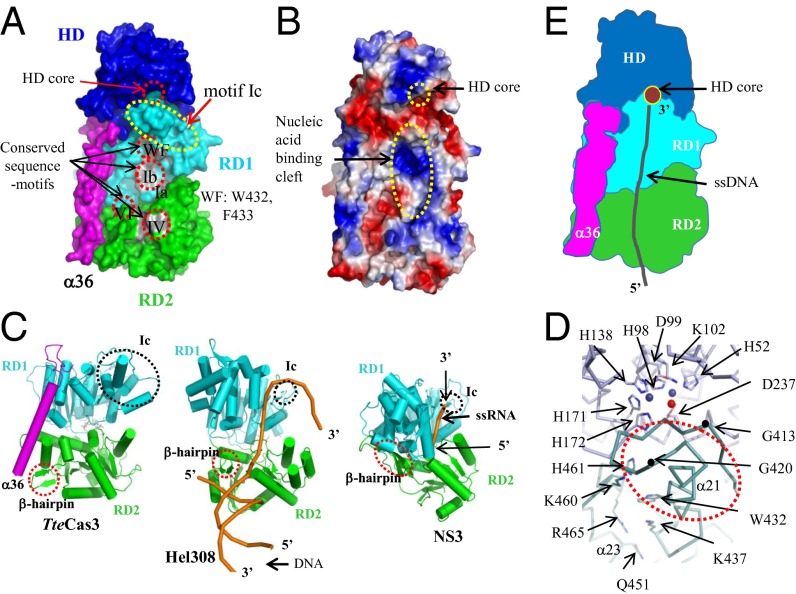
Fig. 3.
Disassembly line formed by aligned HD nuclease and helicase motifs of TteCas3. (A) Surface presentation of TteCas3. CTD has been omitted to show conserved regions on RD1 and RD2. Each segment is discerned by alternating colors. Conserved sequence motifs are labeled and highlighted by gray patches and dotted circles. (B) Surface potential map. The surface potential formed on NTD, RD1, α36, and RD2 is shown. The HD core and nucleic acid-binding cleft on both RDs were indicated. (C) Comparison of RDs from three closely related SF2 helicases. The structures of TteCas3 (Left) and Hel308 (Center) and NS3 (Right) helicases are oriented similarly. β-hairpin and motif Ic are indicated with circles. The bound nucleic acids on the Hel308 and NS3 helicases are drawn with orange ribbons. (D) The interface of HD (blue) and RD1 (cyan). A part of the TteCas3 extended Ic-motif helix is indicated. Conserved residues are displayed with stick models, and two Mn ions and a water molecule at the HD core are displayed with alternating balls. One motif Ic loop is highlighted by black dots representing residues Gly413 and Gly420. (E) Schematic diagram to show the ssDNA path to the HD core from the helicase fragment. Each domain is differentiated by colors, and the ssDNA is displayed by a black coil.
Several conserved positively charged patches are present in the vicinity of the tunnel and in the above mentioned sequence motifs (Figs. 3B and 1). Moreover, the average diameter of this tunnel is wide enough to accommodate single-stranded nucleic acids, which is consistent with TteCas3’s specific binding activity of ssRNA and ssDNA (Fig. S2C). Positioning of this tunnel relative to the nuclease active site is suggestive that it guides ssDNA generated at the point of helicase unwinding toward its destruction in the HD domain. A protruded β-hairpin (β17 and β18 in Fig. S1B) from Cas3 RD2 is at a structurally equivalent position to the hairpin-unwinding elements of Hel308 and NS3 (40, 41) (Fig. 3C and Fig. S5), suggesting a similar role in unwinding double-stranded nucleic acids. The β-hairpin is proximal to motif V, which is positioned for ATP binding, consistent with a coupling mechanism for ATPase-powered DNA translocation that is observed in structural studies of NS3 (39) and Hel308 (37). In the TteCas3 structure, a short α-helix (α33) is located between the hairpin and motif V and this helix may be part of a ratchet-like translocation mechanism involving conformational movement of RDs, as discussed further later.
Some unique features of TteCas3 might be relevant for its enzymatic mechanism in CRISPR interference. First, motif Ic is composed of fewer than 10 residues in NS3 and Hel308 helicases, whereas it is elongated to 80 residues in TteCas3 (Ser395-Thr471; α20-α24 and β5; Figs. S1B and S4). The significance of this elongation may arise from positioning of TteCas3 motif Ic at the interface of HD nuclease and the CTD (Fig. 3D), providing interdomain stabilization through hydrophobic and hydrophilic interactions. Additionally, motif Ic α21-helix and an adjacent Gly413-Gly420 loop partly covers the HD nuclease active site (Fig. 3D). A second unusual feature of TteCas3 is that the tunnel formed by the helicase domains that is proposed to accommodate ssDNA may be at least partially occluded by a weakly traced linker region of α36-helix and the CTD (Ser821-Glu842; a black-dotted line in Fig. 2). The length of this linker is diverse among Cas3 proteins, but its sequence is relatively well conserved (Figs. S1B and S4). Despite these differences, the overall structural features of the RDs of the TteCas3 helicase are very similar to those of Hel308 and NS3 helicases (Fig. 3C), strongly suggesting that Cas3 enzymes use an ATP-dependent translocation mechanism typical of NS3 and Hel308 SF2 helicases.
Functional Implication of the Cas3 C-Terminal Domain.
The 100-residue CTD of TteCas3 has a globular β-sandwich structure flanked by three α-helices (Fig. 2A). A structural homology search revealed highest similarity to the 60 residues of ubiquitin-like protein SMT3 (PDB ID code: 3TIX). Although the two domains share the same folding topology, their structural homology is low (Z score: 3.5) and their sequence identity is 13% for the aligned 60 residues with an rmsd value of 2.7 Å. The conserved residues are located at the hydrophobic core of the globular domain, suggesting importance in structural integrity but that the functions of Cas3 CTD and SMT3 are otherwise unrelated.
Helicase CTDs have frequently been suggested either to recruit partner proteins/complexes or to facilitate recognition of specific nucleic acid sequences (40–42). Some Cas3 proteins are predicted to exist as fusions with Cse1 protein (Cas subtype E. coli protein-1, also referred to as CasA), a subunit of Cascade interference complex. Cas3-targeting invader DNA colocalizes with Cse1 in the E. coli Cascade complex (30). Additionally, a synthetic E. coli Cas3-Cse1 fusion protein, linked between Cas3 C terminus and Cse1 N terminus, was catalytically active and formed a functional complex with the other four subunits of Cascade (28). Based on these factors, we propose that the Cas3 CTD might be the region responsible for interaction with Cse1. However, attempts to detect such an interaction with mixtures of two independently purified proteins of Cas3 and Cse1 from T. terrenum have not been successful (Fig. S6A) and similar results were obtained with mixtures of Cse1 and the truncated TteCas3 CTD (841–944; Fig. S6B). These results may suggest that stable docking of Cas3 and Cse1 requires other components of Cascade complex, perhaps to induce the conformational change that is observed in Cse1 within Cascade when DNA is bound (13, 43). It is also possible that a defined R-loop structure needs to be present to facilitate correct orientation of Cascade to allow Cas3–Cse1 interaction or other Cascade subunits are partly involved in the formation of a stable complex with Cas3. Interestingly, a synthetic Cas5-Cas3 fusion protein in type IB system degraded nucleic acids, suggesting that nuclease domain is located close to Cas5 (6), which contacts Cse1 in the crRNA–Cascade complex (43).
Discussion
In this study, we report the first crystal structures, to our knowledge, of a full-length Cas3 protein with complete HD-SF2 fusion, with and without bound ATP analog (dATP). A tunnel covered with surface residues that would allow for ssDNA binding within the helicase domains are aligned such that a target DNA strand is directed straight into the HD nucleolytic core (Fig. 3A). This domain arrangement suggests a coordinated pathway within Cas3 that passes unwound ssDNA to the HD active site through the combined activities of ATP hydrolysis, ssDNA translocation and duplex strand separation, and 3′ to 5′ exonucleolytic cleavage. By this mechanism, Cas3 would complete CRISPR interference by destroying at least one strand of invader dsDNA.
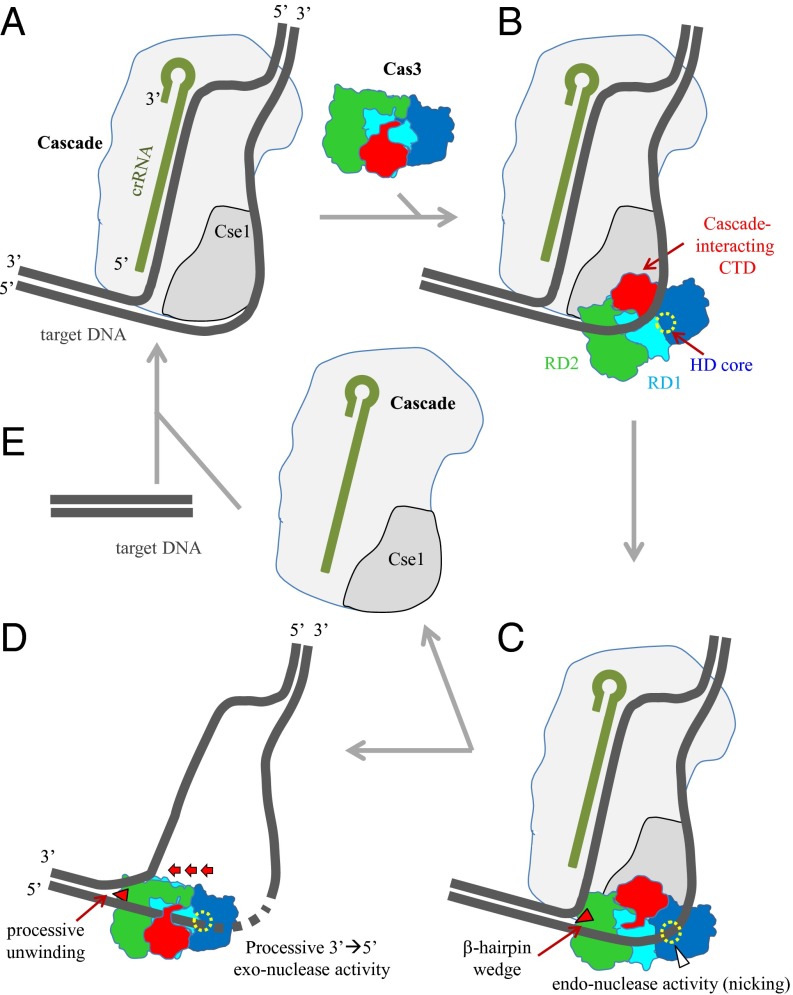
Based on our analyses, together with reported information, a detailed mechanism for ssDNA cleavage and dsDNA unwinding by Cas3 is proposed (Fig. 4). After recognition of an appropriate PAM motif by Cse1 (30), the crRNA–Cascade complex unwinds invading dsDNA in a step-wise manner (44), eventually generating an R-loop conformation with a displaced noncomplementary DNA strand and a complementary DNA strand that base pairs with the crRNA guide (28) (Fig. 4A). A conformational change of the Cascade subunit Cse1 most likely triggers recruitment of Cas3 (30, 43). The complex formation reveals a previously concealed nucleic acid-binding cleft on two RDs in the region of the α36-helix and the CTD and exposes the partly hidden HD nuclease active site by displacing the α19-helix and the associated loop of helicase motif Ic. The displaced noncomplementary ssDNA can then be located to the nucleic acid-binding cleft on the RD1 and RD2 surface (Fig. 4B). Binding of ssDNA to the HD nucleolytic core, via the Ic-motif and conserved residues Lys437, Lys460, His461, and Arg465 (Figs. S1B and S4), will initially invoke endo-nucleolytic cleavage of the noncomplementary ssDNA (Fig. 4C), possibly at residues between the 7th and 11th nucleotide into the protospacer (29, 30). Upon nicking, rearrangement of the CTD transforms the cleft into a tunnel, ensuring stable binding of the DNA strand (Fig. 4D). Binding of an ATP molecule to the RD1-RD2 interface of Cas3 induces a conformational change of motif V, resulting in relocation of motif Ia toward the ATPase site. ATP hydrolysis and ADP release will complete the cycle and rearrange these two motifs to their original spatial positions of the ATP-free state. During the cycle of ATP binding and ATP hydrolysis, the released energy is used (i) to drive the step-wise translocation of Cas3 along the nicked, displaced DNA strand into 3′ to 5′ direction, and (ii) to drive the step-wise unwinding of the dsDNA probably by using the β-hairpin (β17 and β18) as a wedge-like structural element as is the case in the related SF2 helicase Hel308. Concomitantly, the transferred 3′-end of the displaced DNA is exposed to the HD nuclease site, where it is progressively degraded in 3′ to 5′ direction by exonucleolytic activity of Cas3 (Fig. 4D). Based on the observation that Cas3 has the capacity to bind ssDNA, together with the demonstration that Cas3 can catalyze ATP-dependent unwinding of a crRNA/ssDNA hetero duplex (33, 45), it has been proposed that, although one Cas3 progressively processes the displaced DNA strand, another Cas3 molecule may release the crRNA–Cascade complex from the target DNA strand (Fig. 4E) and bind to the other strand for an endo-nucleolytic attack. Cas3-dATP binding was accompanied by structural rearrangement of motif V reminiscent of events in the translocation mechanism of the SF2 helicase NS3 (39). Translocation by NS3 includes nucleotide-dependent conformational movement of RDs orientating the helicase into “open” and “closed” positions that contribute to “ratchet-like” movement along ssDNA. Cas3-dATP lacked similar movement of RD1 and RD2 relative to nucleotide-free structures, but it is possible that DNA binding, in addition to nucleotide binding, is required to allow Cas3 RD1-RD2 movement, as observed for NS3 (39). Hel308 SF2 helicase is also thought to translocate ssDNA ratchet-like by conformational movement of RDs in response to nucleotide binding (40), and it is proposed that Cas3 conforms to this general principle of translocation by SF2 helicases.
Fig. 4.
Proposed DNA interference mechanism by Cas3 in association with Cascade effector complex. (A) R-loop formation by Cascade–crRNA complex. crRNA in Cascade forms an RNA-DNA hybrid duplex by base-pairing to the complementary DNA strand of invading dsDNA, resulting in displacement of the noncomplementary DNA strand. (B) Cas3 recruitment. The Cas3 CTD is proposed to interact with the Cse1 subunit, which opens the nucleic acid-binding cleft on RD1 and RD2 and exposes the HD core. The displaced DNA strand is bound by the nucleic acid-binding cleft on the RD1 and RD2 surface. (C) Initial endo-nucleolytic cleavage of the displaced DNA strand. The probable dsDNA-unwinding β-hairpin wedge is indicated with a filled triangle. Tight binding of ssDNA by residues at the interface of RD1 and in the vicinity of HD core invokes an initial nicking. (D) Processive exonucleolytic cleavage of the ssDNA and dsDNA unwinding. CTD rearranges, resulting in formation of a tunnel structure to trap the cleaved fragment of the ssDNA. ATPase-driven conformational changes drive the translocation of the Cas3 protein along the nicked DNA strand into the 3′ to 5′ direction, whereas dsDNA is further unwound. The ssDNA is degraded in a step-wise manner by 3′ to 5′ exonuclease activity. (E) Cascade targets invading DNA. The cycle is completed when the Cascade–crRNA complex is released, which detects and interacts with another molecule of invading DNA.
From the analysis of the reported cleavage patterns, it was suggested that Cas3 has endo- nucleolytic and exonucleolytic activities (33, 34). Several products of different size from the dsDNA substrate of partial duplex that has a 5′-end–labeled ssDNA region indicates that the endo-nucleolytic activity of Cas3 appears not to be processive. Simultaneously, their cleavage patterns in the absence of ATP were provided as evidence for Cas3’s exonucleolytic activity. However, these exonucleolytic products may not result from processive degradation because one would not expect the translocation activity of Cas3 without ATP, suggesting that its exonucleolytic activity is also, at least to some extent, distributive. To resolve this issue, future studies are required on Cas3 in complex with Cascade and/or R loop to understand precise mechanisms of DNA interference by type I Cascade/Cas3 complexes.
While this paper was in revision, the crystal structure of an iron-dependent Cas3 from Thermobifida fusca (TfusCas3) was published in complex with ssDNA derived from the expression host (46). In TfusCas3, ssDNA binding causes differences in relative orientation of RD2 and CTD compared with the structure we observe in TteCas3 (Fig. S7A), and significantly, the α21-helix WF motif is rotated almost 90° from the HD domain, exposing the HD core to the bound ssDNA (Fig. S7B). This observation is in full agreement with the aforementioned prediction for TteCas3. Our mutational studies on TteCas3 (D237A and the WF motif) showed decreased ssDNA binding and cleavage activities, which has not been reported in TfusCas3. TteCas3 WF motif is present as WL in TfusCas3, and, although WL does not contact DNA, it contributes to both formation of the stable nucleic acid-binding pocket near the HD core and correct positioning of conserved positively charged residues that do contact DNA. Therefore, the complementary studies on two full-length bacterial Cas3 proteins provide a major gain of insight in the molecular mechanism of type I CRISPR-Cas interference.
Materials and Methods
Cloning and Protein Production.
The gene encoding full-length TteCas3 was amplified by PCR from a template of chromosomal DNA from T. terrenum, using primers designed for ligation-independent cloning (47). The PCR product was treated with T4 DNA polymerase (New England Biolabs) and inserted into a vector derived from the pET21a plasmid (Novagen). This vector was designed to express the cloned gene fused to a (His)6 tag, and the Tobacco etch virus cleavage sequence at the N terminus. The E. coli BL21 Star (DE3) Solu strain transformed with the expression construct was grown in the Luria Bertani medium. After induction by adding 0.5 mM isopropyl β-d-1-thiogalactoside, cells were cultivated for 15 h at 37 °C. The harvested cells were resuspended and disrupted by ultrasonication in a buffer solution containing 20 mM Tris⋅HCl (pH 7.5) and 500 mM NaCl (buffer A). The supernatant was loaded onto HisTrap chelating column (GE Healthcare). The column was extensively washed with buffer A, and the bound proteins were eluted with a linear gradient from 0 to 500 mM imidazole in buffer A. The eluted sample was dialyzed against a buffer solution containing 20 mM Tris⋅HCl (pH 7.5) and 100 mM NaCl (buffer B). TteCas3 was further isolated by using a HiTrapQ anion exchange column (GE Healthcare) operated with a linear gradient from 0 to 1 M NaCl in buffer B. Seleno-l-methionine (SeMet)-substituted TteCas3 was prepared similarly as the native protein. The purified protein was concentrated up to 6 mg/mL after a buffer change to 20 mM Tris⋅HCl (pH 8.5) and 200 mM NaCl.
Crystallization, Data Collection, and Structure Determination.
Crystallization attempts of TteCas3 were performed at 22 °C by the sitting-drop vapor-diffusion method. The initial crystals were obtained from the precipitant solution containing 2 M NaCl, 0.1 M NaH2PO4, 0.1 M KH2PO4, and 0.1 M Mes (pH 6.5). This condition was optimized by a grid search by using 24-well Linbro plates and the hanging-drop vapor-diffusion method at 22 °C where 1 μL of protein sample and 1 μL of precipitant were mixed together and equilibrated with 0.2 mL of precipitant. Suitable crystals for diffraction experiments were obtained from the precipitant containing 1.5 M NaCl, 0.07 M NaH2PO4, 0.07 M KH2PO4, and 0.1 M Bis-Tris (pH 5.5). For cocrystals of TteCas3 and dATP, the protein was mixed with dATP at the molar ratio of 1–1.5. SeMet-substituted protein and dATP-complex crystals were obtained under the same crystallization conditions. For diffraction experiments, crystals were briefly immersed into the precipitant solution containing additional 50% (vol/vol) glycerol as the cryoprotectant and immediately placed in the 100 K nitrogen-gas stream. Using a SeMet-labeled TteCas3 crystal, single-wavelength anomalous dispersion data were collected at the wavelength of 0.9793 Å on the beamline 7A SB1 at Pohang Accelerator Laboratory (Korea) with the per-frame oscillation of 1° and per-frame exposure of 2 s. A dataset for ATP complex was collected on the same beamline with the per-frame oscillation of 1° and per-frame exposure of 3 s. In both cases, a total of 180 images were collected on the ADSC-Q270 CCD detector. The indexing, integration, and scaling of the reflections were conducted by using HKL2000 suite (48). Twenty-one of the expected 21 Se sites in the asymmetric unit were identified at a resolution of 2.60 Å by using SOLVE (49). Electron density modification was performed by using PHENIX (50) combined with RESOLVE (51), resulting in automatic modeling of ∼60% of the residues. Further model building was performed manually by using WinCoot (52), and subsequent refinement was performed with PHENIX (50). The data and refinement statistics are summarized in Table 1.
Electrophoretic Mobility Shift Assay and Nuclease Activity Assay.
The sequences of different nucleic acids used in this study are tabulated (Table S1). The synthesized RNA and DNA strands were purchased from ST Pharm (Korea). The 5′-ends of RNAs and DNAs were labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Roche). To remove unincorporated [γ-32P]ATP, the mixture was desalted with an RNase free Sephadex G-25 column. The labeled probes (0.1 nM) were incubated with TteCas3 proteins for 30 min at 25 or 50 °C with or without divalent metal ions in a buffer solution containing 20 mM Tris⋅HCl (pH 8.0), 100 mM KCl, and 100 ng/μL BSA. The mixtures were loaded onto a 15% (wt/vol) nondenaturing polyacrylamide gel (40:1), and electrophoresis was conducted at 70 V for 60 min at 25 °C in a Tris/Borate/EDTA buffer (89 mM Tris, 89 mM boric acid, and 2 mM EDTA). The results were visualized by using a Fuji PhosphorImager.
ATPase Assay.
The [α-32P]ATP (5 Ci/mmol, 0.1 nM) (IZOTOP) was mixed with TteCas3 proteins for 10 min at 45 °C in the same buffer solution that was used for nuclease activity assay. Of a total reaction volume of 10 μL, 5 μL was spotted onto a polyethyleneimine-cellulose thin layer plate (Sigma) and separated by chromatography in a 325 mM KH2PO4 (pH 3.5), which followed by phosphorimager visualization.
Production of the TteCas3 Mutants.
TteCas3 mutants were generated by using a protocol based on the QuikChange II site-directed mutagenesis kit (Agilent technologies), and the mutations were confirmed by DNA sequencing. The primer sequences used for site-directed mutagenesis are in Table S2. The mutant proteins were purified, similarly as the native protein.
Supplementary Material
Acknowledgments
The X-ray diffraction experiments used Beamline 7A at the Pohang Accelerator in Korea and Beamline BL1A at Photon Factory. This work was supported by Basic Science Research National Research Foundation Grant 2014R1A2A2A01004915 funded by the Ministry of Education, Science, and Technology of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4Q2C and 4Q2D).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410806111/-/DCSupplemental.
References
- 1.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14(3):321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 5.Reeks J, Naismith JH, White MF. CRISPR interference: A structural perspective. Biochem J. 2013;453(2):155–166. doi: 10.1042/BJ20130316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van der Oost J, Westra ER, Jackson R, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12(7):479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 8.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 9.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22(24):3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 14.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nam KH, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20(9):1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, et al. Mycobacterium tuberculosis complex CRISPR genotyping: Improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J Med Microbiol. 2010;59(Pt 3):285–294. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 17.Staals RH, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52(1):135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spilman M, et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52(1):146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lintner NG, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286(24):21643–21656. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Duijn E, et al. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics. 2012;11(11):1430–1441. doi: 10.1074/mcp.M112.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell. 2013;52(1):124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45(3):303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinkunas T, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32(3):385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288(31):22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hochstrasser ML, et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA. 2014;111(18):6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the Helicase Family Tree. Curr Opin Struct Biol. 2014;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinkunas T, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30(7):1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beloglazova N, et al. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30(22):4616–4627. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286(36):31896–31903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westra ER, et al. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012;9(9):1134–1138. doi: 10.4161/rna.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35(5):598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annual Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 39.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107(2):521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 41.Luo D, et al. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82(1):173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karow AR, Klostermeier D. A structural model for the DEAD box helicase YxiN in solution: Localization of the RNA binding domain. J Mol Biol. 2010;402(4):629–637. doi: 10.1016/j.jmb.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 43.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46(5):606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howard JA, Delmas S, Ivančić-Baće I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439(1):85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 46.Huo Y, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol. 2014;21(9):771–777. doi: 10.1038/nsmb.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terwilliger TC. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.