Significance
Upstream cell death stimuli culminate in the activation of an initiator caspase, marking the onset of apoptosis. Activation of the initiator caspase, caspase-9, is mediated by the heptameric Apaf-1 apoptosome. How Apaf-1 apoptosome facilitates the autocatalytic activation of caspase-9 has remained controversial and largely enigmatic. Two contrasting but not mutually exclusive hypotheses, proximity-induced dimerization vs. induced conformation, emphasize different aspects of initiator caspase activation. This study provides compelling evidence to support the induced conformation model for caspase-9 activation. A previously unknown interface between Apaf-1 and caspase-9 was identified to play an essential role in caspase-9 activation, and formation of a multimeric complex between Apaf-1 caspase recruitment domain (CARD) and caspase-9 was shown to be indispensable for caspase-9 activation.
Keywords: apoptosis, caspase activation, induced proximity, mechanism, CARD
Abstract
Autocatalytic activation of an initiator caspase triggers the onset of apoptosis. In dying cells, caspase-9 activation is mediated by a multimeric adaptor complex known as the Apaf-1 apoptosome. The molecular mechanism by which caspase-9 is activated by the Apaf-1 apoptosome remains largely unknown. Here we demonstrate that the previously reported 1:1 interaction between Apaf-1 caspase recruitment domain (CARD) and caspase-9 CARD is insufficient for the activation of caspase-9. Rather, formation of a multimeric CARD:CARD assembly between Apaf-1 and caspase-9, which requires three types of distinct interfaces, underlies caspase-9 activation. Importantly, an additional surface area on the multimeric CARD assembly is essential for caspase-9 activation. Together, these findings reveal mechanistic insights into the activation of caspase-9 by the Apaf-1 apoptosome and support the induced conformation model for initiator caspase activation by adaptor complexes.
Caspases are a family of highly conserved cysteine proteases that cleave their substrates after an aspartate residue (1). Apoptosis, or programmed cell death (2, 3), is triggered by a cascade of caspase activation (4). Intracellular death cue culminates in the activation of an initiator caspase such as caspase-9 in mammalian cells. Once activated, the initiator caspase cleaves and hence activates effector caspases. This process is exemplified by the activation of caspase-3 by caspase-9. The effector caspases are responsible for killing the cell by cleaving a broad range of substrate proteins (1).
Caspase-9 and caspase-3 represent the most extensively studied initiator and effector caspases, respectively. Although the activation mechanism for effector caspases is reasonably well understood (5), how an initiator caspase is activated remains enigmatic. The challenge in understanding initiator caspase activation is caused in part by the complexity of the process. Unlike an effector caspase, which is activated by a single intrachain cleavage mediated by an initiator caspase, an initiator caspase must be recruited into a multimeric adaptor protein complex, where the initiator caspase undergoes one or more autocatalytic cleavages. Such a multimeric protein complex is commonly known as an apoptosome (6). Caspase-9 is activated by the Apaf-1 apoptosome (6).
Apaf-1 contains a caspase recruitment domain (CARD) at its amino terminus, followed by a nucleotide-binding oligomerization domain (NOD) and 15 repeats of WD40 at its carboxyl terminus. In homeostatic cells, Apaf-1 exists as an inactive monomer. In apoptotic cells, cytochrome c is released from mitochondria to the cytoplasm, where it binds Apaf-1 (7, 8). Subsequent replacement of ADP by dATP/ATP in Apaf-1 triggers formation of a heptameric apoptosome (9–11). The Apaf-1 apoptosome catalyzes the autocatalytic activation of the caspase-9 zymogen. Importantly, the cleaved, mature caspase-9 remains bound to the Apaf-1 apoptosome as a holoenzyme, which displays a proteolytic activity at least two orders of magnitude higher than that of free, mature caspase-9 (12). Thus, the key for elucidating mechanism of caspase-9 activation is to understand how mature caspase-9 is allosterically activated by the Apaf-1 apoptosome. Structural analysis of the Apaf-1 apoptosome by cryo-EM revealed a wheel-shaped assembly, with CARD and NOD located at the central hub and WD40 repeats as the extended spokes (11, 13, 14).
Despite rigorous investigation and evolving hypotheses, the molecular mechanism by which caspase-9 is activated by the Apaf-1 apoptosome remains largely unknown. In the late 1990s, the induced proximity model was proposed to explain the general mechanism of initiator caspase activation (15–19). Although correct in a general sense, induced proximity applies to just about any general biological system of protein–protein interaction and fails to mention the specific underpinnings of caspase activation (20). A refined version of the induced proximity model, known as proximity-induced dimerization (21), states that multiple molecules of caspase-9 are recruited into the Apaf-1 apoptosome for increased probability of dimerization and subsequent activation. This model assumes the dimeric form of caspase-9 to be fully activated and postulates that the Apaf-1 apoptosome serves to promote caspase-9 homodimerization. At present, there is no compelling evidence to support the notion that the fully activated caspase-9 within the Apaf-1 apoptosome is homodimeric. More importantly, even if the assumption is correct, how the Apaf-1 apoptosome recruits caspase-9 and facilitates its homodimerization remains unnoted by the proximity-induced dimerization hypothesis (6, 20). An alternative model, named induced conformation, was proposed to explain caspase-9 activation; this model states that the primary function of the Apaf-1 apoptosome is to help caspase-9 attain an activated conformation through binding (20, 22).
Elucidation of the underlying mechanism of caspase-9 activation requires thorough knowledge of the interactions between caspase-9 and the Apaf-1 apoptosome. At present, the only reported interaction occurs between the CARD domains of Apaf-1 (ApCARD) and caspase-9 (C9CARD) (8, 23). The crystal structure of a 1:1 complex between ApCARD and C9CARD reveals a complementary interface that is indispensable for caspase-9 activation (23). However, the induced conformation model, but not the proximity-induced dimerization model, demands existence of additional interactions between caspase-9 and Apaf-1. Intriguingly, ApCARD alone was found to induce formation of a multimeric complex with caspase-9, in which caspase-9 exhibited markedly enhanced protease activity (24). This observation, reported 12 y ago but left unnoticed by the cell death community, has been rigorously repeated and unequivocally demonstrates the existence of additional specific interactions between ApCARD and caspase-9. However, the nature of such interactions, or the relation of these interactions with caspase-9 activation, has remained elusive.
In this paper, we provide conclusive evidence that caspase-9 activation by the Apaf-1 apoptosome requires at least two additional specific interfaces between caspase-9 and Apaf-1. Thus, the previously reported 1:1 complex between ApCARD and C9CARD (23), although indispensable, fails to recapitulate the complete set of interactions between Apaf-1 and caspase-9 in the apoptosome holoenzyme. Our experimental evidence demonstrates that ApCARD and C9CARD assemble into a higher-order oligomer, which may further recruit the protease domain of caspase-9 for activation. These findings constitute strong evidence in support of the induced conformation model for initiator caspase activation (20, 22).
Results
Oligomerized ApCARD Potently Activates Caspase-9.
ApCARD and caspase-9 assemble into a multimeric complex, within which caspase-9 exhibits drastically increased protease activity (24). This ApCARD–caspase-9 complex, named a miniapoptosome, appeared to be unstable and slowly dissociated over gel filtration chromatography (24). The concentration-dependent formation of the miniapoptosome provides a plausible explanation to the observation that the protease activity of caspase-9 in the miniapoptosome is lower than that in the intact Apaf-1 apoptosome. Because the ApCARD–caspase-9 complex contains no artificial oligomerization motif or exogenous fusion protein, it may faithfully recapitulate the core elements of the caspase-9–Apaf-1 apoptosome holoenzyme. Under this scenario, we hypothesize that the main function of the Apaf-1 apoptosome is to provide a stable scaffold to prevent dissociation of the ApCARD–caspase-9 complex.
This hypothesis predicts that any ApCARD-linked stable scaffold, ideally of sevenfold symmetry, may suffice to activate caspase-9 to a similar extent as the Apaf-1 apoptosome. To examine this scenario, we fused ApCARD (residues 1–105) to the amino terminus of the chaperonin GroES (residues 1–97). GroES is known to form a stable heptamer (25); the carboxyl-terminal 10 residues of ApCARD are flexible enough to allow ApCARD sufficient freedom in binding caspase-9. We purified the fusion protein ApCARD–GroES to homogeneity (Fig. S1), along with caspase-9 and a truncated Apaf-1 (residues 1–591, referred to as Apaf1-591 hereafter) that retains the ability to form a functional apoptosome (26).
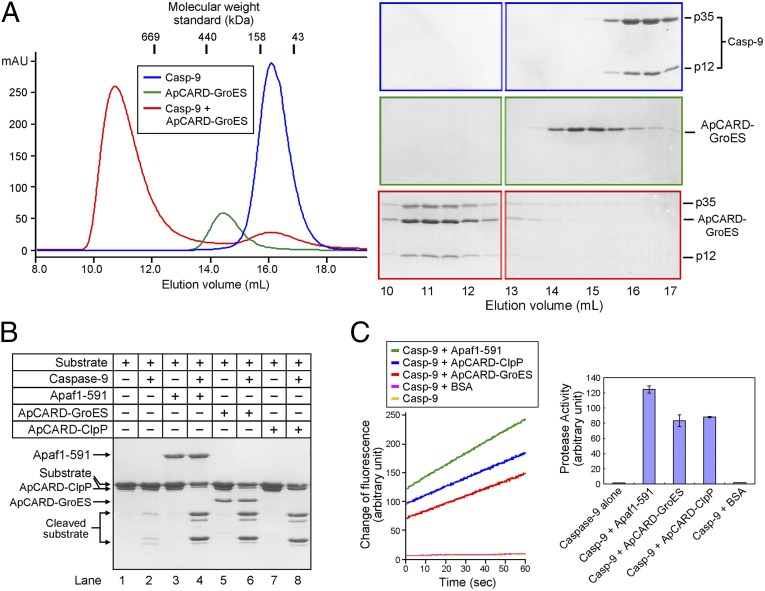
The predicted molecular mass for a heptameric ApCARD–GroES complex is ∼160 kDa. The 22-kDa fusion protein ApCARD–GroES was eluted from gel filtration with a molecular mass of approximately 250 kDa (Fig. 1A). The larger-than-predicted molecular mass of the heptameric ApCARD–GroES complex is likely caused by the increased radius of hydration caused by the flexible sequences between the heptameric GroES base and the seven ApCARD modules. Incubation of ApCARD–GroES with an excess amount of caspase-9 resulted in the formation of an even larger complex, which exhibited a molecular mass greater than 700 kDa. Visualization and subsequent quantification of this complex by SDS/PAGE indicate that the molar ratio of caspase-9 over ApCARD–GroES is less than 1 (Fig. 1A), suggesting that fewer than seven molecules of caspase-9 were bound to each heptameric complex of ApCARD–GroES.
Fig. 1.
Oligomerized Apaf-1 CARD (i.e., ApCARD) potently activates caspase-9 to a similar level as the Apaf-1 apoptosome. (A) Caspase-9 and the heptameric ApCARD–GroES complex form a multimeric assembly. Peak fractions from the color-coded chromatograms of gel filtration (Left) were visualized on SDS/PAGE gels by Coomassie blue staining (Right). The PAGE gels are identified by the same colors as those for the gel filtration chromatograms. Blue, caspase-9 alone; green, heptameric ApCARD–GroES complex alone; red, caspase-9 plus the ApCARD–GroES complex. (B) The heptameric ApCARD–GroES or ApCARD–ClpP complex stimulates the protease activity of caspase-9 to a similar level as the preassembled Apaf1-591 apoptosome. The Apaf1-591 apoptosome has the same ability to activate caspase-9 as the full-length Apaf-1 apoptosome (26). Shown here is a representative SDS/PAGE gel. The single-chain caspase-3 (C163A) was used as the substrate. (C) The heptameric ApCARD–GroES or ApCARD–ClpP complex potently activates the caspase-9 to a similar level as the preassembled Apaf1-591 apoptosome. The fluorogenic peptide Ac-LEHD-AFC was used as the substrate. Shown here are results of the caspase-9 protease activities from the original experiments (Left) and their quantification (Right). Each experiment described in this study was independently repeated at least three times. The error bar represents the SD of the observed values.
Next, we examined whether caspase-9 assembled onto the ApCARD–GroES scaffold is catalytically activated by using an in vitro protease activity assay whereby caspase-3 (residues 1–277; C163A) was used as the substrate (Fig. 1B). Free caspase-9 exhibited a basal level of protease activity (Fig. 1B, lane 2), which was drastically increased by the presence of the preassembled Apaf1-591 apoptosome (Fig. 1B, lane 4). Confirming our prediction, incubation of caspase-9 with the ApCARD–GroES heptamer also led to marked stimulation of the protease activity (Fig. 1B, lane 6).
To better quantify the results, we used the fluorogenic peptide substrate Ac-LEHD-AFC in the protease assay (Fig. 1C). Corroborating the aforementioned conclusion, caspase-9 displayed a similar level of protease activity in the presence of the Apaf1-591 apoptosome or the heptameric ApCARD–GroES complex. In either case, the protease activity is drastically higher than that of free caspase-9 (Fig. 1C). Importantly, caspase-9 activity remained essentially unchanged in the presence of BSA, ruling out protein crowding effect as a contributing factor by the heptameric ApCARD–GroES complex.
To further examine the robustness of the conclusion, we engineered another fusion protein ApCARD–ClpP, in which ApCARD was fused to the amino terminus of the heptameric protease ClpP (residues 1–194), and purified ApCARD–ClpP to homogeneity (Fig. S1). The fusion protein ApCARD–ClpP formed a stable heptameric complex on gel filtration (Fig. S2). Incubation of caspase-9 with ApCARD–ClpP resulted in the formation of a large complex in which caspase-9 was present in a substoichiometric quantity (Fig. S2). The protease activity of caspase-9 was markedly stimulated by the presence of the ApCARD–ClpP complex, and the degree of caspase-9 activation is similar to that by the Apaf1-591 apoptosome (Fig. 1 B and C). Taken together, these experimental observations demonstrate that caspase-9 can be activated by a heptameric scaffold, in which the amino terminus of each protomer is fused to ApCARD for caspase-9 recruitment.
ApCARD and C9CARD Assemble into Multimeric Complexes.
ApCARD and C9CARD were thought to form a 1:1 heterodimer (23). The conclusion was reached by using recombinant, amino-terminally tagged C9CARD (23). During a recent discussion about possible mechanisms of caspase-9 activation by the Apaf-1 apoptosome, we serendipitously came to the realization that, in ApCARD and C9CARD, the amino-terminal methionine residue is buried, with its amino group inaccessible for peptide bond linkage (23). Thus, any peptide tag at the amino terminus of ApCARD or C9CARD is likely to cause a local structural perturbation. This analysis suggests that any specific interaction(s) mediated by the amino-terminal region of ApCARD or C9CARD would likely be overlooked if the amino-terminally tagged proteins were used for such studies.
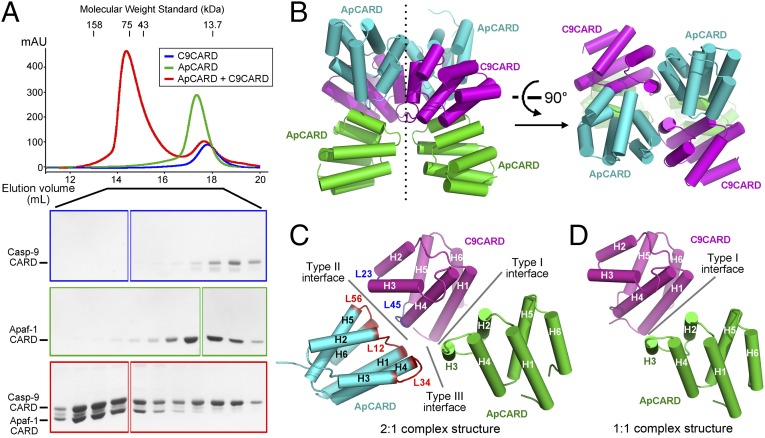
Motivated by this rationale, we reexamined complex formation between ApCARD and C9CARD by using untagged ApCARD (residues 1–97) and carboxyl-terminally His6-tagged C9CARD (residues 1–100). The result was striking: rather than forming a 1:1 heterodimer as previously reported (23), ApCARD and C9CARD assembled into a multimeric complex with an apparent molecular mass in excess of 75 kDa (Fig. 2A). Given the calculated molecular weight of 11.1 kDa for ApCARD and 12.7 kDa for C9CARD, the observed ApCARD–C9CARD complex likely contains seven or eight molecules of CARD. Formation of this complex appeared to be unstable and concentration-dependent, as suggested by the asymmetric appearance of its peak on gel filtration (Fig. 2A). Consistent with this analysis, decreased concentrations of ApCARD and C9CARD led to the assembly of a complex with a smaller molecular mass of ∼50 kDa (Fig. S3).
Fig. 2.
ApCARD and C9CARD form a multimeric complex through three types of interfaces. (A) ApCARD and C9CARD form a multimeric complex on gel filtration. The apparent molecular mass of the ApCARD–C9CARD complex exceeds 75 kDa, likely containing seven or eight copies of the CARD modules. The chromatograms (Upper) and the PAGE gels (Lower) are color-coded. (B) Structure of a multimeric complex between ApCARD and C9CARD. There are six molecules of CARD in each asymmetric unit, constituting two basic repeats. Each of the two repeats contains two molecules of ApCARD and one molecule of C9CARD. Two perpendicular views are shown here. C9CARD is colored magenta, whereas ApCARD is displayed in cyan and green. (C) Each basic unit of the ApCARD–C9CARD complex uses three types of interface. The type I and type II interfaces occur between ApCARD and C9CARD, whereas the type III interface involves two ApCARD modules. The interhelical loop L12 refers to the intervening sequences between α-helices H1 and H2. Loops L23, L45, and L56 are similarly defined. (D) The type I interface was previously observed in the 1:1 complex between ApCARD and C9CARD (23). All structural figures were prepared with PyMOL (www.pymol.org).
These experimental observations demonstrate that, in addition to the observed interface between ApCARD and C9CARD (23), each domain must use at least one more specific surface epitope for interaction with the other domain. We attempted to determine the molar ratio between ApCARD and C9CARD in the complexes of varying molecular masses. Unfortunately, we failed to detect a consistent molar ratio in response to varying concentrations of ApCARD and C9CARD. This observation suggests that the heptameric Apaf-1 apoptosome may serve as a stable scaffold to govern caspase-9 recruitment and hence its defined stoichiometry.
Structure of a Multimeric Complex Between ApCARD and C9CARD.
To gain insights into the additional interfaces between ApCARD and C9CARD, we sought to crystallize a multimeric complex. After numerous efforts, we were able to generate crystals in the space group P21. The structure was determined at 2.1-Å resolution (Fig. 2B and Table S1). Each asymmetric unit contains four molecules of ApCARD and two molecules of C9CARD, which assemble into a butterfly-shaped structure (Fig. 2B). These six CARD molecules are arranged into two basic units, each containing two molecules of ApCARD and one molecule of C9CARD (Fig. 2C). These two units are related to each other by a twofold pseudosymmetry (Fig. 2B).
CARD belongs to a superfamily of death domain fold proteins, which are characterized by six α-helices arranged in a typical Greek key topology (27). These homotypic interaction motifs are thought to interact with each other through three types of interfaces (28). Remarkably, the two molecules of ApCARD and one molecule of C9CARD in each unit associate with each other by using all three types of interfaces (Fig. 2C). First, C9CARD uses two α-helices H1/H4 to closely stack against helices H2/H3 from an ApCARD, constituting a type I interface (23, 28). This interface is nearly identical to that previously reported between C9CARD and ApCARD (23) (Fig. 2D). Second, the same C9CARD employs helix H4 and the interhelical loops L23/L45 to contact helix H6 and the interhelical loops L12/L56 from the other ApCARD, representing a type II interface (28) (Fig. 2C). Finally, the two ApCARD molecules within the same unit interact with each other through a type III interface, involving helix H3 from ApCARD and L34/H4 from the other ApCARD (28).
The three interfaces, particularly types I and II, involve a large number of specific interactions. As previously reported (23), the type I interface observed here contains a number of important interdomain hydrogen bonds (i.e., H-bonds), exemplified by those between Asp40 of ApCARD and Arg13 of C9CARD and between Asp27 of ApCARD and Arg52 of C9CARD (Fig. S4A). At the type II interface, Glu78 of ApCARD accepts a pair of charge-stabilized H-bonds from Arg36 of C9CARD; Arg65 of C9CARD interacts with Tyr80 of ApCARD through cation-π interactions while donating a H-bond to the carbonyl oxygen of Glu78 (Fig. S4B). At the type III interface, Glu41 of ApCARD accepts three charge-stabilized H-bonds, two from Arg52 of the other ApCARD and one from the main chain amide of Gln49 (Fig. S4C).
In contrast to the dense interactions at the three types of interfaces, association between the two basic units appears to be relatively weak (Fig. S5). Except for four direct H-bonds, all other H-bonds are mediated by three sulfate ions that are present in the crystallization buffer. Intriguingly, two sulfate ions are located next to Ser31, raising the possibility that phosphorylation of the solvent-exposed residue Ser31 might bear a similar consequence as the presence of sulfates. We acknowledge that this analysis is highly speculative and lacks relevant physiological relevance.
An Essential Role for Caspase-9 Activation by the Type II Interface.
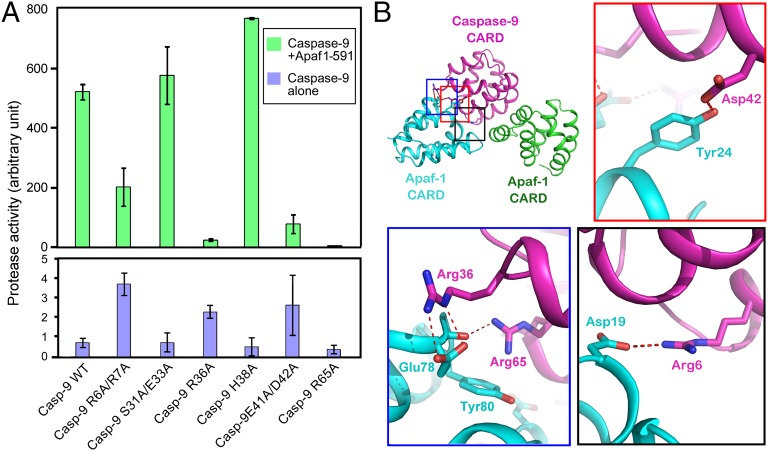
Four molecules of ApCARD and two molecules of C9CARD assembled into a heteromeric complex in the crystals. There is no experimental evidence to suggest the molar ratio or the size of the ApCARD–C9CARD complex in the crystals to be physiologically relevant. Nonetheless, the observed three types of interfaces in the crystals may recapitulate true structural features in the caspase-9–bound Apaf-1 apoptosome holoenzyme. To examine this scenario, we generated six missense variants of caspase-9, each containing one or two point mutations that target the type II interface. We individually purified these variant proteins to homogeneity (Fig. S6A) and evaluated the impact of the mutations on the ability of caspase-9 to be activated by the Apaf1-591 apoptosome by using an in vitro caspase-9 protease activity assay (Fig. 3A).
Fig. 3.
The type II interface between ApCARD and C9CARD plays an essential role in caspase-9 activation by the Apaf-1 apoptosome. (A) Characterization of six caspase-9 variants each containing one or two missense mutations at the type II interface. The six caspase-9 variants examined here are classified into three categories in terms of phenotype: no effect (H38A and S31A/E33A), mild effect (R6A/R7A and E41A/D42A), and crippling effect (R36A and R65A). Two mutations in caspase-9 R36A and R65A, which target key residues at the type II interface, nearly abrogated the ability for these caspase-9 variants to be activated by the Apaf1-591 apoptosome. The Apaf1-591 apoptosome has the same ability to activate caspase-9 as the full-length Apaf-1 apoptosome (26). (B) Three close-up views of the interdomain H-bonds at the type II interface. Arg36 and Arg65 appear to anchor the interface by each mediating more than one H-bond. H-bonds are represented by red dashed lines.
By using the fluorogenic peptide Ac-LEHD-AFC as the substrate, the protease activity of WT caspase-9 was stimulated nearly 700-fold by the presence of the Apaf1-591 apoptosome (Fig. 3A). The six caspase-9 variants exhibited three categories of phenotypes that are faithfully corroborated by structural observations. First, two variants, H38A and S31A/E33A, behaved similarly as the WT caspase-9, displaying greatly stimulated protease activities in the presence of the Apaf1-591 apoptosome (Fig. 3A). Consistent with the WT-like phenotype for the two variants, His38 makes no direct interactions at the type II interface, whereas Ser31/Glu33 also contributes little to the interface (Fig. S4B).
Second, in sharp contrast to WT caspase-9, two variants, each containing a missense mutation R36A or R65A, exhibited a severely crippled ability to be activated by the Apaf1-591 apoptosome (Fig. 3A). In the crystal structure, Arg36 of C9CARD donates a pair of charge-stabilized H-bonds to Glu78 of ApCARD, whereas Arg65 of C9CARD mediates strong cation-π interactions with Tyr80 of ApCARD while donating an H-bond to the main chain carbonyl oxygen of Glu78 (Fig. 3B). Thus, Arg36 and Arg65 of C9CARD appear to play an anchoring role at the type II interface, explaining why their mutations led to a crippled phenotype. Notably, although these two caspase-9 variants retained the ability to form a multimeric complex with the Apaf1-591 apoptosome, the sizes of such complexes are noticeably smaller than that of the WT caspase-9–Apaf1-591 apoptosome holoenzyme (Fig. S7).
Third, each of the remaining two caspase-9 variants, R6A/R7A and E41A/D42A, displayed a mildly stimulated level of protease activity in the presence of the Apaf1-591 apoptosome (Fig. 3A). The affected residues help stabilize the type II interface (Fig. 3B), although the interactions are not as extensive as those mediated by Arg36 or Arg65. Correspondingly, the phenotypes for these two variants are not as severe as that for R36A or R65A. The sizes of the complexes between these two caspase-9 variants and the Apaf1-591 apoptosome are slightly smaller than that of the WT caspase-9–Apaf1-591 apoptosome holoenzyme, but larger than that for caspase-9 R36A or R65A (Fig. S6).
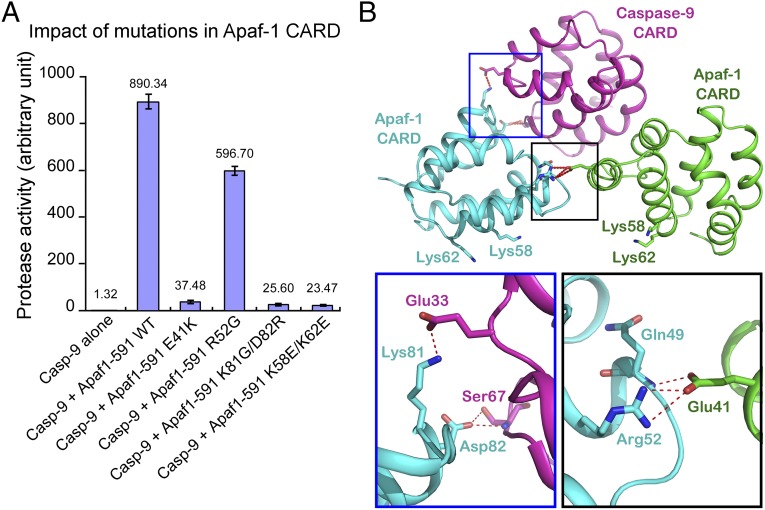
We also created and purified a variant of Apaf1-591 that contains two missense mutations K81G/D82R targeting the type II interface (Fig. S6B). Compared with WT Apaf1-591, this variant exhibited a greatly diminished ability to stimulate the protease activity of WT caspase-9 (Fig. 4A). At the type II interface, Lys81 of ApCARD donates a H-bond to Glu33 of C9CARD whereas Asp82 of ApCARD makes a pair of H-bonds to the main chain and side chain of Ser67 in C9CARD (Fig. 4B). This Apaf1-591 variant retained the ability to form a multimeric complex with WT caspase-9, but the size of this complex is smaller than that of the WT caspase-9–Apaf1-591 apoptosome holoenzyme (Fig. S8). Taken together, our experimental observations demonstrate that the type II interface between ApCARD and C9CARD plays an essential role in the activation of caspase-9 by the Apaf-1 apoptosome.
Fig. 4.
Functional characterization of the interfaces between ApCARD and C9CARD. (A) Characterization of four Apaf1-591 variants each containing one or two missense mutations. Of the four variants, K81G/D82R targets the type II interface whereas E41K and R52G affect the type III interface. The Apaf1-591 variant K58E/K62E contains two mutations that affect residues not located at the three types of interfaces. (B) Two close-up views of the interdomain H-bonds at the type II interface (Lower Left) and type III interface (Lower Right). Unlike the other two types of interfaces, the type II interface involves two ApCARD modules.
Characterization of the Type III Interface for Caspase-9 Activation.
Unlike the type I or type II interface, the observed type III interface in our crystal structure involve only interactions between two ApCARD modules. In addition, the type III interface contains considerably fewer H-bonds compared with the type I or type II interface (Fig. S4). This analysis suggests that, compared with the other two types of interfaces, the type III interface may play a less important role in activation of caspase-9 by the Apaf-1 apoptosome. To examine this scenario, we generated two variants of Apaf1-591, E41K and R52G, each containing a single missense mutation. Both variants were purified to homogeneity (Fig. S6B) and were evaluated for their ability to activate caspase-9 by using an in vitro caspase-9 protease activity assay (Fig. 4).
Although Arg52 directly interacts with Glu41 at the type III interface (Fig. 4B), the Apaf1-591 variant E41K, but not R52G, exhibited a greatly reduced ability to mediate caspase-9 activation compared with WT Apaf1-591 (Fig. 4A). The different phenotypes may be explained by two factors. First, compared with Arg52, Glu41 mediates one additional H-bond between its side chain and the main chain amide of Gln49 (Fig. 4B and Fig. S4C). Second, the mutation E41K places a positively charged residue Lys next to positively charged Arg52, which may repel each other and thus actively sabotage this interface. By contrast, the mutation R52G introduces a Gly, which would not be in contact with Glu41. The Apaf1-591 variant E41K or R52G retained the ability to form a multimeric complex with WT caspase-9, which exhibited a smaller size compared with that of the WT caspase-9–Apaf1-591 apoptosome holoenzyme (Fig. S8).
Identification of a Key Surface Epitope on ApCARD.
Our experimental evidence demonstrates that caspase-9 activation requires the assembly of an oligomeric CARD complex between Apaf-1 and caspase-9. We speculate that the primary function of the CARD complex is to provide a docking site for the protease domain of caspase-9 to bind and such binding results in formation of an activated active site conformation. To examine this scenario, we performed systematic mutagenesis studies on ApCARD and C9CARD, with the goal of identifying a mutation that has no impact on caspase-9 recruitment but abolishes caspase-9 activation. This effort led to the identification of two missense mutations K58E/K62E in Apaf1-591. The variant Apaf1-591 K58E/K62E forms a normal apoptosome holoenzyme with caspase-9, with an identical elution volume as that for the holoenzyme between WT Apaf1-591 and caspase-9 (Fig. S8). Strikingly, however, the Apaf1-591 K58E/K62E apoptosome exhibited a severely crippled ability to stimulate caspase-9 activity (Fig. 4A). We speculate that the surface epitope defined by Lys58 and Lys62 may be intimately involved in caspase-9 activation, presumably through direct interactions with the catalytic domains of caspase-9.
Stoichiometry Between Caspase-9 and Apaf-1.
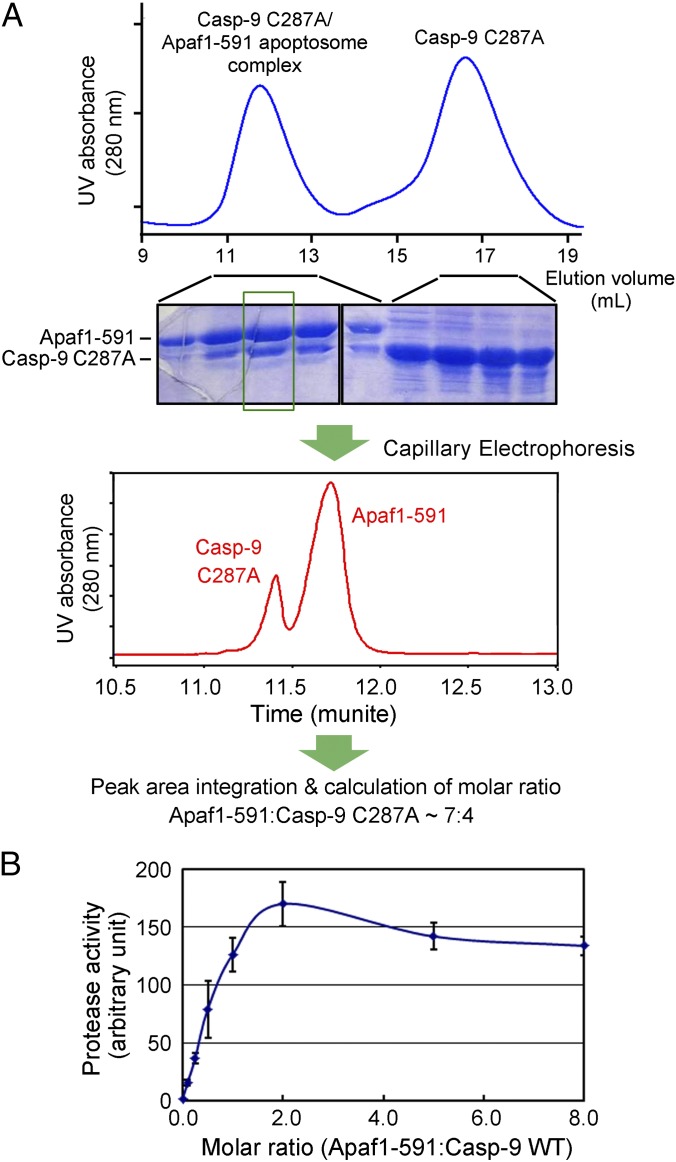
Our experimental evidence indicates that ApCARD and C9CARD form a multimeric complex. To determine the precise stoichiometry between Apaf-1 and caspase-9 in the apoptosome holoenzyme, we isolated a stable complex between the single-chain caspase-9 (C287A) and the Apaf1-591 apoptosome by gel filtration (Fig. 5A). Then, an aliquot of the stable complex was subjected to capillary electrophoresis under denaturing condition (Fig. 5A). On the basis of UV absorbance at 280 nm, the peak areas that correspond to caspase-9 and Apaf-1 were deconvoluted, integrated, and calculated for their relative molar ratio. This experiment was independently repeated three times, with the results nicely converging with each other. Quantification of the results identified the molar ratio of Apaf-1 over caspase-9 to be ∼1.66 ± 0.11, which corresponds to 4.2 ± 0.3 molecules of caspase-9 for each Apaf1-591 heptamer (Fig. 5A). This analysis suggests a 7:4 molar ratio between Apaf-1 and caspase-9 in the apoptosome holoenzyme.
Fig. 5.
Determination of the molar ratio between Apaf1-591 and caspase-9. (A) Determination of the molar ratio between Apaf1-591 and caspase-9 by capillary electrophoresis. A stable complex between the single-chain caspase-9 (C287A) and the Apaf1-591 apoptosome was isolated from gel filtration (Top) and visualized on SDS/PAGE by Coomassie blue staining (Middle). The peak fraction that corresponds to the caspase-9–Apaf1-591 apoptosome holoenzyme was subjected to capillary electrophoresis under denaturing condition. The peak areas were deconvoluted, integrated, and converted to molar ratio between Apaf1-591 and caspase-9. The molar ratio between Apaf1-591 and caspase-9 was determined to be 1.66 ± 0.11 or 7:4.2 ± 0.3. (B) Enzymatic characterization of caspase-9 supports the molar ratio of 7:4 between Apaf1-591 and caspase-9. Upon incubation with increasing concentrations of preassembled Apaf1-591 apoptosome, the protease activities of caspase-9 were measured and plotted against the molar ratios between Apaf1-591 and caspase-9. The highest protease activity was recorded at a molar ratio of 2:1 between Apaf1-591 and caspase-9.
The suggested 7:4 stoichiometry between Apaf-1 and caspase-9 agrees well with enzymatic characterization of caspase-9 activation by the Apaf-1 apoptosome (Fig. 5B). A fixed amount of WT caspase-9 was incubated with seven different concentrations of preassembled Apaf1-591 apoptosome to allow assembly of the caspase-9 holoenzyme. Then, the protease activity of caspase-9 was measured and plotted against the molar ratios of Apaf1-591 over caspase-9 (Fig. 5B). Caspase-9 activity increased rapidly with increasing molar ratios of 0.1, 0.2, 0.5, and 1.0. Importantly, caspase-9 exhibited the highest level of protease activity at the molar ratio of 2.0, which is ∼37% higher than that at the Apaf-1:caspase-9 molar ratio of 1.0 (Fig. 5B). This analysis suggests that, at the molar ratio of 1.0, extra caspase-9 may remain unbound by the Apaf1-591 apoptosome and thus yield a lower protease activity. Notably, caspase-9 activity was slightly reduced at molar ratios higher than 2.0 (Fig. 5B), which can be explained by the notion that the competition for caspase-9 recruitment by excess amount of the Apaf1-591 apoptosome resulted in substoichiometric binding of caspase-9 to each apoptosome.
Discussion
Activation of caspase-9 consists of two steps. First, autocatalytic cleavage of the caspase-9 zymogen is greatly facilitated by the Apaf-1 apoptosome. Second, the mature caspase-9 remains bound to the Apaf-1 apoptosome as a holoenzyme, within which caspase-9 exhibits a stimulated level of protease activity that is at least two orders of magnitude higher than that of free caspase-9. The first step of caspase-9 was once thought to be unimportant, because mature caspase-9 and the single-chain uncleaved caspase-9 seemed to exhibit a similar level of protease activity in the presence of the Apaf-1 apoptosome (29–31). In these studies (29–31), caspase-3–specific substrate was used for the measurement of caspase-3 activity, which served as an indirect readout of caspase-9 activity. Unfortunately, the conclusion derived from such studies may be incorrect under the condition that saturating levels of caspase-3 activity can be caused by markedly different levels of caspase-9 activity. This conclusion was overturned by a recent investigation, which, using caspase-9–specific substrate as a direct readout of caspase-9 activity, showed the proteolytic processing of the caspase-9 zymogen to be essential for caspase-9 activation by the Apaf-1 apoptosome (32). The essence of the second step is allosteric activation of mature caspase-9 by the Apaf-1 apoptosome. Obviously, both steps of caspase-9 activation strictly depend on the physical interactions between caspase-9 and the Apaf-1 apoptosome. The present study focuses on uncovering such previously unknown interactions.
In this paper, we demonstrate that the previously observed 1:1 complex between ApCARD and C9CARD (23), involving the type I interface, is insufficient for the activation of caspase-9 by the Apaf-1 apoptosome. Missense mutations targeting key H-bonds at the type II interface, exemplified by R36A or R65A in caspase-9, abrogated caspase-9 activation by the Apaf-1 apoptosome. By contrast, mutations affecting noninteracting residues at the type II interface, such as H38A in caspase-9, had no impact on caspase-9 activation. Thus, the severity of mutations in terms of affecting interactions in the type II interface directly correlates with the extent of caspase-9 activation by the Apaf-1 apoptosome. These experimental findings unequivocally show that a specific type II interface between caspase-9 and Apaf-1 is indispensable for the activation of caspase-9 by the Apaf-1 apoptosome. The observed type III interface may also play an important role in caspase-9 activation, as the mutation E41K in Apaf-1 resulted in a severely reduced ability for the Apaf1-591 apoptosome to stimulate caspase-9 activity.
How do the additional interfaces contribute to caspase-9 activation? A conclusive answer to this question ultimately constitutes the long sought-after mechanism of caspase-9 activation by the Apaf-1 apoptosome. Although this paper does not provide a straightforward answer to this question, we provide tantalizing clues. First, the additional interfaces are used to form a multimeric complex between ApCARD and caspase-9, within which caspase-9 activity is greatly stimulated (24). In fact, the fusion protein ApCARD–GroES or ApCARD–ClpP forms a stable heptamer, which potently stimulates the protease activity of caspase-9 to a similar level as the Apaf-1 apoptosome. This finding argues that the primary function of the Apaf-1 apoptosome is to stabilize the multimeric complex between ApCARD and caspase-9.
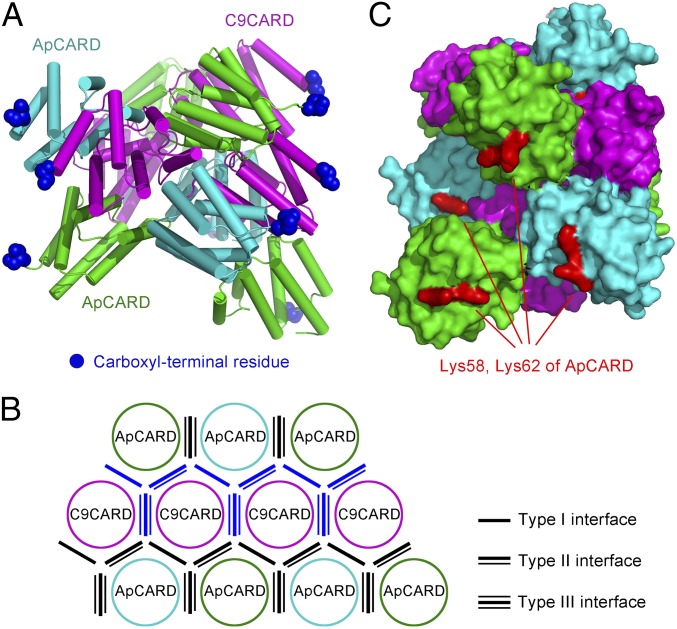
Second, within the caspase-9–bound Apaf-1 apoptosome, the molar ratio of Apaf-1 over caspase-9 is likely to be 7:4. This conclusion is supported by two lines of experimental evidence: direct quantification by capillary electrophoresis and enzymatic characterization (Fig. 5). The substoichiometric molar ratio for caspase-9 in the Apaf-1 apoptosome holoenzyme was previously reported in another study (29), although the precise ratio was not determined. Notably, in the multimeric complex with ApCARD–GroES or ApCARD–ClpP, the molar ratio of caspase-9 was also less than 1 (Fig. 1A and Fig. S2). How can ApCARD and caspase-9 assemble into a 7:4 complex? We attempted to build an atomic model by using the observed structural information. Much to our surprise, simple propagation of the three types of specific interfaces in the ApCARD–C9CARD complex gave rise to a 11-mer complex, with seven molecules of ApCARD and four molecules of C9CARD (Fig. 6A). Within this 11-mer complex, there are a total of 14 previously observed, specific interfaces, including four type I interfaces, four type II interfaces, and six type III interfaces (Fig. 6B). In addition, there are nine interfaces of the reversed polarity: three for each type of the three interfaces (Fig. 6B). For example, in the type I interface of reversed polarity, ApCARD uses its H1/H4 to stack against H2/H3 from C9CARD; whereas, in the normal type I interface, H1/H4 of C9CARD contact H2/H3 of ApCARD. Remarkably, all carboxyl termini of the 11 CARD domains are located on the outer surface of the assembly, available for covalent linkage to the ensuing sequences. This model is consistent with the observed CARD disk above the Apaf-1 apoptosome as revealed by cryo-EM analysis (14), and shares common structural features reported in the helical assemblies for other death domain modules (33–35).
Fig. 6.
A hypothetical model on the assembly of the multimeric complex between ApCARD and C9CARD. (A) An overall view of the 11-mer complex between ApCARD and C9CARD. This complex contains seven molecules of ApCARD (cyan and green) and four molecules of C9CARD (magenta). The three types of specific interfaces observed in our crystal structure (Fig. 2C) were used to generate this model. Notably, all carboxyl termini (blue spheres) of the 11 CARD domains are located on the outside surface of the helical assembly. (B) A schematic planar view of the 11-mer complex between ApCARD and C9CARD. The helical assembly is unwounded to show the interfaces among the 11 CARD molecules. There are a total of 23 interfaces, of which 14 belong to the normal three types of interface (black lines) and the other nine exhibit reversed polarity (blue lines). (C) Lys58 and Lys62 (red spheres) from four ApCARD molecules are located on the outside surface of the modeled ApCARD–C9CARD assembly. These residues appear to define a binding epitope, perhaps for caspase-9.
Third, how can assembly of the CARD complex facilitate caspase-9 activation? We have identified two key residues on ApCARD, Lys58 and Lys62, neither of which is involved in the three observed types of interfaces. Despite the ability of Apaf1-591 K58E/K62E to form a normal apoptosome holoenzyme with caspase-9 (Fig. S8), the resulting apoptosome has nearly abrogated the ability to stimulate caspase-9 activity (Fig. 4A). Notably, Lys58 and Lys62 from four ApCARD molecules are located close to each other and appear to define a binding epitope on the outer surface of the CARD assembly (Fig. 6C). We speculate that this surface might be responsible for docking the catalytic domains of caspase-9.
Our experimental observations provide a strong support for the induced conformation model of initiator caspase activation. The confirmed additional interfaces between caspase-9 and Apaf-1, the observed multimeric complex between ApCARD and C9CARD, the calculated molar ratio between Aapf-1 and caspase-9, and the putative docking site on Lys58/Lys62 of Apaf-1 are all consistent with each other and with the induced conformation model. These lines of experimental evidence go beyond what has been described in published literature and reveal significant mechanistic insights into caspase-9 activation.
Materials and Methods
Protein Preparation.
Apaf-1 CARD (i.e., ApCARD; residues 1–97) was cloned into the vector pET21b (Novagen) without any tag, overexpressed in Escherichia coli Bl21(DE3), and purified to homogeneity by anion-exchange chromatography. Caspase-9 CARD (i.e., C9CARD; residues 1–100) was cloned into the vector pBB75 (36) with a C-terminal hexahistidine (6× His) tag, over-expressed in E. coli Bl21(DE3), and purified by Ni2+-NTA affinity column (Qiagen) and Source-15Q column (GE Healthcare). The ApCARD–C9CARD complex for crystallization was obtained by coexpression in E. coli Bl21(DE3). The other proteins, including caspase-3 (C163A), ApCARD–GroES, ApCARD–ClpP, and WT and mutant Apaf-1 or caspase-9, were cloned into the vector pET-29b (Novagen) with a carboxyl-terminal 6× His tag, overexpressed, and purified as C9CARD. All proteins were further purified by gel filtration (Superdex-200, 10/30; GE Healthcare). The fluorogenic substrate Ac-LEHD-AFC was purchased from Enzo Life Sciences, and dATP was from Sigma.
Caspase-9 Assay.
The Apaf1-591 apoptosome was assembled by incubating Apaf1-591 with 1 mM dATP at 4 °C overnight, and purified by gel filtration before use. The buffer used in the assays contained 100 mM KCl, 20 mM Hepes, pH 7.5, and 5 mM DTT. For the caspase-9 protease activity assay using Ac-LEHD-AFC as the substrate, WT caspase-9 or the variants, each at 0.2 μM concentration, was mixed with the Apaf1-591 apoptosome (containing 0.4 μM Apaf1-591 protomer) and incubated at 22 °C for 10 min. Then, the substrate Ac-LEHD-AFC was added to a final concentration of 200 μM, and the fluorescence intensity was monitored in a fluorescence spectrophotometer (F-4600; Hitachi). The excitation and emission wavelengths were 400 nm and 505 nm, respectively. The activity of caspase-9 was also measured using caspase-3 (C163A) as the substrate. Caspase-3 (C163A) at 40 μM was incubated with the Apaf1-591 apoptosome (containing 2 μM Apaf1-591 protomer) and caspase-9 (1 μM) for 1 h at 4 °C, and additional 30 min at 22 °C. Reaction was stopped by adding an equal volume of 2× SDS loading buffer and immediately boiling at 96 °C for 5 min. The samples were loaded into a 16% (wt/vol) SDS/PAGE gel, which was stained by Coomassie blue.
Gel Filtration Analysis.
Superdex-200 (10/30; GE Healthcare) or Superose-6 (10/30; GE Healthcare) was used in this study. The column was preequilibrated with 100 mM KCl, 20 mM Hepes, pH 7.5, 5 mM DTT, and calibrated with molecular weight standards (GE Healthcare). After injection, the samples were eluted with a flow rate of 0.4 mL/min.
Capillary Electrophoresis.
The Apaf1-591 apoptosome/caspase-9 C287A complex was assembled in the presence of 1 mM AMPPNP and purified by gel filtration. The peak fraction was collected and denatured by SDS and then separated by capillary electrophoresis. The detection wavelength was 280 nm.
Crystallization.
Crystals of ApCARD (residues 1–97)–C9CARD (residues 1–100) complex were grown at 18 °C using the hanging-drop vapor-diffusion method by mixing 1 μL of the complex protein with 1 μL of the reservoir solution containing 13% PEG 3000, 0.1 M MES, pH 5.5, and 0.2 M ammonium sulfate. The crystals were directly flash-frozen in a cold nitrogen stream at 100 K.
Data Collection and Structural Determination.
The data were collected at the Rigaku CCD Saturn 944. All data sets were integrated and scaled by using the HKL2000 package (37). Further processing was carried out by using programs from the CCP4 suite (38). Data collection statistics are summarized in Table S1. The crystal structure of the 1:1 complex of Apaf-1 CARD and caspase-9 CARD [Protein Data Bank ID code 3YGS (23)] was used as the initial model, and our structure was solved by molecular replacement by using PHASER (39) and manually refined with COOT (40) and PHENIX (41).
Supplementary Material
Acknowledgments
This work was funded by National Natural Science Foundation of China Projects 30888001 (to Y.S.), 31021002 (to Y.S.), 31130002 (to Y.S.), and 31200549 (to D.W.) and a postdoctoral fellowship from the Tsinghua-Peking Joint Center for Life Sciences (to D.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID code 4RHW).
See Profile on page 16234.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418000111/-/DCSupplemental.
References
- 1.Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281(5381):1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 2.Horvitz HR. Worms, life, and death (Nobel lecture) ChemBioChem. 2003;4(8):697–711. doi: 10.1002/cbic.200300614. [DOI] [PubMed] [Google Scholar]
- 3.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 4.Yan N, Shi Y. Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol. 2005;21:35–56. doi: 10.1146/annurev.cellbio.21.012704.131040. [DOI] [PubMed] [Google Scholar]
- 5.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5(11):897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 6.Chai J, Shi Y. Apoptosome and inflammasome: Conserved machineries for caspase activation. National Science Review. 2014;1(1):101–118. [Google Scholar]
- 7.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell. 1996;86(1):147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 8.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91(4):479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 9.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102(49):17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao Q, Lu W, Rabinowitz JD, Shi Y. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol Cell. 2007;25(2):181–192. doi: 10.1016/j.molcel.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Acehan D, et al. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9(2):423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez J, Lazebnik Y. Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 1999;13(24):3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu X, et al. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure. 2005;13(11):1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Yuan S, et al. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18(5):571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCorkle RA, Freeman KW, Spencer DM. Synthetic activation of caspases: artificial death switches. Proc Natl Acad Sci USA. 1998;95(7):3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1(7):949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 17.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J Biol Chem. 1998;273(5):2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 18.Yang X, Chang HY, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell. 1998;1(2):319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 19.Salvesen GS, Dixit VM. Caspase activation: The induced-proximity model. Proc Natl Acad Sci USA. 1999;96(20):10964–10967. doi: 10.1073/pnas.96.20.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi Y. Caspase activation: Revisiting the induced proximity model. Cell. 2004;117(7):855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11(2):529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 22.Chao Y, et al. Engineering a dimeric caspase-9: A re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 2005;3(6):e183. doi: 10.1371/journal.pbio.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin H, et al. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399(6736):549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 24.Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci USA. 2002;99(7):4197–4202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt JF, Weaver AJ, Landry SJ, Gierasch L, Deisenhofer J. The crystal structure of the GroES co-chaperonin at 2.8 A resolution. Nature. 1996;379(6560):37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 26.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434(7035):926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn DE, Rodriguez J, Lazebnik Y, Joshua-Tor L. Crystal structure of Apaf-1 caspase recruitment domain: An alpha-helical Greek key fold for apoptotic signaling. J Mol Biol. 1999;293(3):439–447. doi: 10.1006/jmbi.1999.3177. [DOI] [PubMed] [Google Scholar]
- 28.Weber CH, Vincenz C. A docking model of key components of the DISC complex: Death domain superfamily interactions redefined. FEBS Lett. 2001;492(3):171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- 29.Malladi S, Challa-Malladi M, Fearnhead HO, Bratton SB. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009;28(13):1916–1925. doi: 10.1038/emboj.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivasula SM, et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;2001;410(6824):112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 31.Stennicke HR, et al. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274(13):8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 32.Hu Q, Wu D, Chen W, Yan Z, Shi Y. Proteolytic processing of the caspase-9 zymogen is required for apoptosome-mediated activation of caspase-9. J Biol Chem. 2013;288(21):15142–15147. doi: 10.1074/jbc.M112.441568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrao R, Wu H. Helical assembly in the death domain (DD) superfamily. Curr Opin Struct Biol. 2012;22(2):241–247. doi: 10.1016/j.sbi.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park HH, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128(3):533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiozaki EN, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11(2):519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.