Significance
Changing a few residues can change the function of homologous proteins. The chloride and proton affinity in the inward chloride-pumping halorhodopsin (HR) and outward proton-pumping bacteriorhodopsin (BR) are compared using classical electrostatic simulations. BR binds and releases protons from acidic residues that have been removed from HR. In the states where these acids are ionized in BR, HR binds a chloride. In the states where these acids bind a proton in BR, HR releases the chloride. Thus, BR uses static anions and mobile protons, whereas HR uses mobile ions to maintain the same charge states. The use of mobile ions makes HR more sensitive to external conditions.
Keywords: pKa, ion binding, continuum electrostatics, MCCE, buried charge
Abstract
Key mutations differentiate the functions of homologous proteins. One example compares the inward ion pump halorhodopsin (HR) and the outward proton pump bacteriorhodopsin (BR). Of the nine essential buried ionizable residues in BR, six are conserved in HR. However, HR changes three BR acids, D85 in a central cluster of ionizable residues, D96, nearer the intracellular, and E204, nearer the extracellular side of the membrane to the small, neutral amino acids T111, V122, and T230, respectively. In BR, acidic amino acids are stationary anions whose proton affinity is modulated by conformational changes, establishing a sequence of directed binding and release of protons. Multiconformation continuum electrostatics calculations of chloride affinity and residue protonation show that, in reaction intermediates where an acid is ionized in BR, a Cl– is bound to HR in a position near the deleted acid. In the HR ground state, Cl– binds tightly to the central cluster T111 site and weakly to the extracellular T230 site, recovering the charges on ionized BR-D85 and neutral E204 in BR. Imposing key conformational changes from the BR M intermediate into the HR structure results in the loss of Cl– from the central T111 site and the tight binding of Cl– to the extracellular T230 site, mirroring the changes that protonate BR-D85 and ionize E204 in BR. The use of a mobile chloride in place of D85 and E204 makes HR more susceptible to the environmental pH and salt concentrations than BR. These studies shed light on how ion transfer mechanisms are controlled through the interplay of protein and ion electrostatics.
Ions bind to proteins to carry out many essential biological reactions, including muscle function (1), signal transduction (2), protein–protein interactions (3), and oxygen transport (4). The transmembrane electrochemical gradient generated by ion and proton pumps drives key processes such as ATP synthesis and nerve transmission (5). Proton and ion transfer processes, protein–protein interactions, and enzyme catalysis often rely on charged amino acids (6). In homologous proteins, key amino acids can be replaced by bound ions that introduce enhanced sensitivity to ion concentration and pH. For example, mutation of a phosphorylation site to an anionic amino acid has been found to yield constitutive activation (7) that can trigger certain cancers (8). Amylases provide another example, where one class is dependent on the pH and Cl– concentration, with an active site Arg or Lys binding the chloride (9, 10). In contrast, amylases remove the basic amino acid and have pH- and Cl–-independent activity.
Halorhodopsin (HR), first identified in Halobacterium salinarum (11), is a light-driven transmembrane anion pump (12). It is a member of the family of archaeal rhodopsins that carry out proton or ion transport and signal transduction (13). HR and bacteriorhodopsin (BR) are from halobacteria, halophilic archaea that live in high salt (13). Like all proteins in this family, they use a Lys in the center of the transmembrane region of the protein to covalently bind a retinal. Absorption of light promotes the isomerization of all-trans retinal to its 13-cis configuration, which triggers conformational changes in the protein. In HR, retinal isomerization leads to the transport of one chloride anion inward across the cell membrane (12). In BR, it causes one proton to be pumped out of the cell (14). The protons pumped by BR are used by the F0/F1 ATPase to drive ATP synthesis, HR imports Cl– against the electrochemical gradient (15), whereas sensory rhodopsins such as SRI and SRII transmit signals to the flagellar motor (13). Channelrhodopsins, which are sensor proteins that carry out light-activated cation transport, are now exploited as optogenetic research tools (16).
BR and HR both have seven transmembrane helices with a retinal covalently attached to a buried Lys via a Schiff base (17, 18). Their sequence identity is 25–35%. The backbone coordinates have an MSD of only 1.1 Å between conserved positions in the ground state (Fig. 1). They show similar absorption spectral changes during their photocycles (17), indicating that the retinal is in a similar conformation and electrostatic environment (19). Five of six spectroscopically identified BR intermediates are found in HR (20).
Fig. 1.
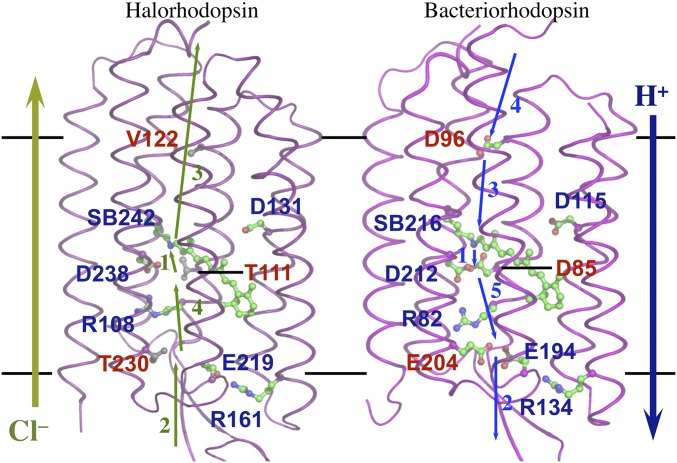
Structures of HR and BR. Buried ionizable residues in BR and the analogous conserved (blue) and not conserved (red) residues in HR. Retinal and key amino acid side chains are shown in green stick-and-ball format. SB, retinal Schiff base. The reaction cycle is well established in BR (21). The sequence of proton transfer steps is indicated by numbered arrows (1). A proton first moves from the SB to D85 within the central cluster generating the early M state: (SB0 D85H)(E194/204)−1 D96H (2). A proton is released from the extracellular cluster to generate the late M state: (SB0 D85H)(E194/204)–2 D96H (3). The proton on D96 moves to the SB yielding the N state: (SB0 D85H)(E194/204)–2 D96– (4). D96 is then reprotonated yielding the N′ state (SBH+ D85H)(E194/204)−2 D96H (5). The proton on D85 moves to the extracellular cluster acids returning the system to the ground state. (E194/204)−1 indicates a single proton is shared between the two EC glutamic acids yielding a net −1 charge. The proton may also be on a nearby water. Chloride transfer steps in HR, mirroring proton transfer in BR, are also shown. Chloride release into the cytoplasm is combined into one step as no stable chloride-binding site is found on the cytoplasmic side. This does not rule out a transient chloride binding site on the release pathway.
Buried ionizable residues are needed for BR proton transfers and HR chloride transfers (Fig. 1). In BR, there are nine ionizable residues. The Schiff base (SB), BR-D85, and D212 form the central cluster and E194 and E204 comprise the extracellular cluster, whereas BR-D96 on the inner side of the protein transfers a proton from the cytoplasm to the SB (14, 21). Each of these groups change charge as BR pumps protons from the high-pH cytoplasm to the low-pH extracellular space (Table 1) (6, 21). In HR, six of the buried BR ionizable residues are conserved. However, BR-D85, E204, and D96 are replaced by HR-T111, T230, and V122, respectively (Fig. 2 A and B and Fig. S1). These changes remove one anion from each of the cytoplasmic, central and extracellular clusters and replace it with a small, polar or nonpolar side chain.
Table 1.
MCCE-calculated residue and chloride binding-site charges
| Cluster | HR | BR | ||||||||||
| Residue | Ground state | N state | Residue | Ground state | Late M state | N′ state | ||||||
| Ideal | 1E12 | Ideal | Chimera | Ideal | 1C8R | Ideal | Chimera | 1C8S | Ideal | |||
| CC | Schiff base | 1 | 0.94 | 1 | 0.78 | Schiff base | 1 | 0.98 | 0 | 0.35 | 0.73 | 1 |
| Cl– | −1 | −0.94 | 0 | 0.00 | BR-D85 | −1 | −0.98 | 0 | −0.25 | −0.13 | 0 | |
| HR-D238 | −1 | −1.00 | −1 | −1.00 | BR-D212 | −1 | −1.00 | −1 | −1.00 | −0.92 | −1 | |
| Net charge | −1 | –1.00 | 0 | −0.22 | Net charge | −1 | –1.00 | −1 | −0.90 | −0.32 | 0 | |
| EC | HR-E219 | −1 | −0.99 | −1 | −1.00 | BR-E194 | 1/0 | −0.53 | −1 | −0.81 | −0.99 | −1 |
| Cl– | 0 | −0.15 | −1 | −1.00 | BR-E204 | 1/0 | −0.47 | −1 | −0.54 | −0.74 | −1 | |
| Net charge | −1 | –1.14 | −2 | −2.00 | Net charge | −1 | –1.00 | −2 | −1.35 | −1.73 | −2 | |
MCCE-calculated residue ionization and chloride binding-site charge (−1 × Cl– occupancy) in HR and BR at pH 7. See Fig. 1 for a description of the ideal ionization states. BR data are from ref. 38. See Fig. 4 and Fig. S2 for the effect of pH on proton and Cl– affinity. The chimera structures replace the ground-state trans-retinal Schiff base and BR-R82 (HR-R108) conformer with the cis-retinal SB and BR-R82 from the late M BR structure 1C8S. The resulting structures were minimized with GROMACS and the Cl– and proton distributions calculated.
Fig. 2.
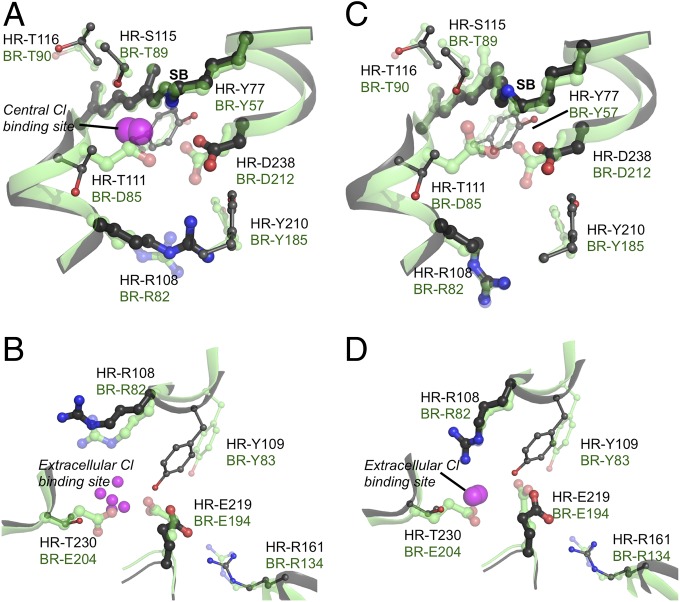
Best alignment of the key residues in the (A) central and (B) extracellular cluster of HR (black) and BR (green) in the crystal structures HR-1E12 and BR-1C8R. The backbone Cα RMSD for conserved residues is 1.1 Å. Alignment in (C) central and (D) extracellular cluster chimera structures. Here, the cis-retinal and rotated Arg BR-R82 are imposed into the ground-state crystal structures for HR and BR and subjected to GROMACS minimization.
Canonical Pumping Cycle in BR and HR
Spectroscopic, mutagenesis, and computational studies have led to a consensus reaction mechanism for proton pumping with defined roles for key residues in BR (Fig. 1) (14, 22, 23). In the ground state, the BR-SB, D85, D212 are all ionized, D96 is neutral, and BR-E194/E204 share a proton or are both ionized with an associated hydronium (22, 24). Following isomerization of the retinal from all-trans to 13-cis, the first proton transfer shifts the proton from the SB to D85 (early M state), and then the proton is lost to the periplasm from E194/E204 (late M state). The proton is replaced on the SB from the cytoplasm through transient deprotonation of D96 (N state). Reprotonation of D96 forms the N′ state. Following reisomerization of the retinal (O state), proton transfer from D85 to the extracellular cluster recovers the ground state. In summary, the first proton transfer step moves a proton within the central cluster. A proton is then released into the low-pH, extracellular space. Then, a proton is bound from the high-pH cytoplasm, into the central cluster via D96. Finally, a proton is transferred from the central cluster to the extracellular cluster.
The HR photocycle has been less well studied and is generally analyzed by analogy to BR (17, 18). Many studies monitor the spectral signatures of the retinal molecule, which respond to isomerization and to the shifting electrostatic influence of nearby charges (25). There is no evidence for a state analogous to the BR M state with a deprotonated SB in the HR reaction cycle (17). Rather, a Cl– stays associated with the protonated SB NH+ (18). This Cl– is later released in the N state (17), which is analogous to the BR N′ state (26). The SB proton and nearby bound Cl– are accessible to the extracellular side of the protein in the trans state and to the intracellular side after light absorption has generated cis-retinal. However, in HR, there is little known about the mechanism of Cl– uptake from the extracellular side and release to the cytoplasm, which take place far from the retinal.
Despite many similarities, HR and BR might be viewed as carrying out opposing functions—one moving an anionic chloride into the cell, the other moving a cationic proton out of the cell. These two ion pumps could use different transport mechanisms or BR could act as an inward anion hydroxyl pump in analogy to HR (27). Under certain conditions, however, the functions of these two proteins can be rather easily interchanged. At low pH, wild-type BR is an inward chloride pump (28). A single BR-Asp85 to Ser (29), Thr (29), or Asn (30) substitution converts BR into a chloride pump. On the other hand, it has proved harder to mutate HR to a proton pump (31). An HR with six BR amino acids added along the transfer pathway does not pump protons. The authors suggested that, even with these mutations, HRs do not have a necessary strongly hydrogen-bonded water. However, wild-type HR will pump protons in the presence of the mobile anionic azide ion (32).
BR (22, 23) and HR (33) have been the subject of many computational studies using a wide variety of methods. Programs that use a continuum electrostatics (CE) analysis of protein interactions with Monte Carlo sampling have shed light on the Boltzmann distribution of protonation states at equilibrium in a given protein structure (6, 34). CE calculations have followed proton transfers in BR trapped in the ground and later intermediate structures (22, 23) and in other proteins (35, 36). These calculations show how the BR structural changes in trapped intermediates change the proton affinity of key sites through the reaction cycle (37). Multiconformation continuum electrostatics (MCCE) calculations of BR have determined the minimum conformational changes needed to generate the pKa shifts that yield proton release and proton uptake leading to pumping (23, 38). It was shown that conformational changes of BR-R82 and BR-E194 that are triggered by retinal isomerization are necessary and largely sufficient to stabilize the later ionization states. Methods that use CE analysis and Monte Carlo sampling are also particularly well suited to investigate the coupling of proton and ion binding, as each comes to equilibrium at a given pH and ionic strength in the same calculation. Such CE studies have identified the Cl– binding site in Na+/Cl–-dependent transporters (39) and modeled the Cl– affinity as a function of pH in membrane-bound and soluble proteins (10) and in different redox states of photosystem II (40).
In the work presented here, MCCE-calculated ground-state residue ionization states and chloride binding in HR are compared with previous BR calculations (23, 38). MCCE allows the HR protonation states to remain in equilibrium with the bound Cl–. This is possible with the Monte Carlo sampling strategy used here, but is very difficult to accomplish in standard molecular-dynamics (MD) simulations of ion binding to proteins. It is found that Cl– is bound near two small polar groups in HR that replace ionized buried acidic residues in BR. Thus, the electrostatic factors that stabilize the ionized BR-D85 and E204 are retained to stabilize a bound Cl– in HR. Two conformational changes are then made to the HR structure to mimic those found in the trapped BR M-state structure: the Schiff base is isomerized and an Arg buried between the central and extracellular clusters is rotated toward the outside. The calculated changes in Cl– and proton binding in HR are then compared with the changes in protonation states found in similarly modified BR ground-state and true BR M-state structures. This work sheds light on how the interplay between charged amino acids and bound external ions can dictate protein function. These findings can be applied to others proteins where electrostatic interactions play important roles.
Results and Discussion
Cl– Binding Sites in HR.
MCCE calculations of HR start with 179 Cl– placed in cavities in the structure (Fig. S3). Cl– occupancy, residue protonation, and conformation are subjected to Monte Carlo sampling at the same time (10). At pH 7 and below chloride concentrations of ∼1 M, two chloride binding sites are found within the protein (Fig. 2 A and B). One is on the extracellular side of the central cluster, near HR-T111. This chloride is found in the crystal structure (Cl–501 in 1E12 and 2JAG) (41, 42), and in earlier MD simulations (33). It is near the position of the carboxylate side chain of D85 in BR. A new Cl– binding site is found near HR-T230 and R108, which replaces BR-E204 in the extracellular cluster (Fig. 2B). This Cl– is ∼7 Å away from a more surface accessible Cl– in the HR X-ray crystal structure (Cl–502 in 2JAG) (42). At pH 4 with ∼4 M chloride, ∼20% of the protein molecules have a third Cl– bound in one of two sites, each near an acid that binds a proton along with the Cl–. One binding site is between HR-R108 and D238. The charge on the Cl– replaces that of D238. The other lies between HR-Y109 and E219, again, replacing a charge on a now protonated E219. At low Cl– concentrations, both D238 and E219 have low pKa values and are ionized. No Cl– ions are found near V111, which is in the position analogous to BR-D96. Asp-96 is neutral in the BR ground state and only transiently deprotonated during the reaction cycle (43). As this region does not stabilize a buried Asp– in BR, a bound Cl– is not expected in an equilibrated structure.
Calculated Protonation and Cl– Binding in the BR and HR Ground States.
The calculated protonation states and chloride occupancies in the ground-state HR and BR structures are compared with each other and with the canonical charge distributions in the two proteins (Table 1) (Fig. S2) (21, 23, 38). As has been shown previously, calculations carried out with the BR ground-state structures reproduce the expected protonation states, stabilizing the ionized SB, BR-D85 and D212, a proton shared by BR-E194 and E204 and a neutral D96 (Table 1) (22, 23). In the BR ground state, the lowest energy central cluster proton distribution is BR-SB+D212–D85– (23). HR calculations yield the analogous HR-SB+D238–Cl–, where a bound Cl– substitutes for BR-D85– (Figs. 2A and 3). Monte Carlo sampling yields these states 98% of the time in BR and 94% of the time in HR (Fig. 4A and Fig. S2). The −1.0 net charge in the equilibrated HR or BR central clusters is stabilized by interactions conserved between both proteins (23). Contributors include the backbone dipoles, positively charged HR-R108 (BR-R82) and R161 (BR-R134) and polar HR-Y77 (BR-Y57), Y210 (BR-Y185), S115 (BR-T89), and T116 (BR-T90). HR-D141 remains neutral as does the homologous residue BR-D115. As there are three groups in the central cluster, there are several microstates with a −1 charge. In both proteins, the protonated SBH+ is stabilized by the nearby HR-D238−, T111, and S115 (BR-D212−, D85−, and T89) (Figs. 2 A and B, and 3). In both BR and HR, there is a small fraction of deprotonated SB at pH 7 in the ground-state structure. However, the net −1 central cluster charge is retained. In BR, the proton is transferred within the central cluster to either D85 or D212. In HR, when the SB is deprotonated, the Cl– is lost (Fig. 4A and Fig. S2).
Fig. 3.
Electrostatic potential of the interior surface of (A and C) BR and (B and D) HR in the (A and B) ground state, and (C) late M state or (D) chimera state mimicking the structural changes captured in the BR late M state. The electrostatic potential is calculated with BR-D85 and E204 set to be neutral, and no Cl– in HR. The rest of the protein has MCCE-calculated ionization states and protein conformation at pH 7. Color scale of the surface potential is in kT/e, where k is the Boltzmann constant, T is 297.15 Kelvin, and e is the unit electron charge. Key residues are shown as sticks, and bound chlorides are shown in spheres. The arrows highlight the change in electrostatic potential at each Cl– or proton binding site due to the conformational differences between the ground and intermediate states, which changes the proton and Cl– affinity. A blue region stabilizes a deprotonated D85 or E204 in BR or a bound Cl– in HR.
Fig. 4.
Calculated pH dependence of the fractional charge on residues and chloride-binding sites for (A and C) central cluster (CC) and (B and D) extracellular cluster (EC) in the (A and B) ground state and (C and D) chimera state. (A and C) Central cluster, Schiff base (brown circles), HR-D238 (red squares), and chloride (green triangles); (B and D) extracellular cluster, HR-E219 (red squares) and chloride (green triangles). The solid line is the cluster net charge. Calculations use a chloride concentration of 50 mM, which leads to a calculated Schiff base pKa of 8.5 (45).
BR uses two Glu residues, BR-E194 and E204, along with a water network to form a pathway for proton release on the extracellular side (Fig. 2B) (21–23). In the BR ground state, the cluster has a net charge of −1 with a proton bound between the two acids or to a water cluster associated with the acids (22). This proton is released in the subsequent M state (Fig. S2). In the ground state, the pKa for proton release is above 10 in calculations (23, 38) and experiments (21).
The HR extracellular cluster retains one Glu, HR-E219 (BR-E194), whereas HR-T230 replaces BR-E204 (Fig. 2B). Although there is no ordered Cl– in this site in the HR X-ray crystal structure, Monte Carlo sampling at pH 7 shows several overlapping locations in the cavity near HR-T230, R108 (BR-R82) and E219 (BR-E194) that can bind Cl– (Table 1). In the ground state, Cl– binds ∼75 times more weakly to the extracellular site than to the central cluster. A negative charge in the extracellular cluster provided by either an HR-Cl– or a BR-Glu, is stabilized by the backbone, the charges on HR-R108 and R161 (BR-R82 and R134), and hydrogen bonds to HR-S104 (BR-I78), Q105 (BR-Y79), Y109 (BR-Y83), and T230 (BR-E204), and waters. The second sphere of stabilizing residues is spread out, forming a hydrogen bond network. In HR, this allows for a more delocalized Cl– binding site.
In calculations at pH 7 with the default Cl– concentration of ∼50 mM, ∼15% of the external cavity binding sites have a Cl– bound even when HR-E219 is deprotonated (Table 1 and Fig. 4C). HR is better able to stabilize two proximal negative charges (E219 and Cl–) than is BR. In the ground state, BR-E194 and E204 are within hydrogen-bonding distance of each other. In HR, the homologous residues, HR-E219 and HR-T230, point away from each other. T230 participates in a salt bridge with R161 (BR-R134) (Fig. 2B), separating HR-E219 and the Cl– cavity by 6.5 Å. A large water cavity in HR screens the electrostatic repulsion between Cl– and HR-E219. In addition, the mobile Cl– can shift toward the positively charged HR-R108 with flexibility that BR-E194 does not have. As a result, the average HR-Glu–-Cl– repulsion is calculated to be ∼8 kcal/mol. In contrast, the BR-Glu–-Glu– repulsion is close to 20 kcal/mol, raising the pKa for proton release to 10. Additionally, HR has a unique distortion of the HR-L110 ∼ A113 helix (18, 41) that allows for a hydrogen bond between HR-Y109 and E219 in the ground state, to stabilize the deprotonated HR-E219. In BR, the analogous hydrogen bond can only be made in structures with cis-retinal and is therefore only formed in the M state where it will help stabilize proton loss from the extracellular cluster.
Moving HR Beyond the Ground State.
Time-resolved optical and Fourier transform infrared (FTIR) spectroscopic data of BR and HR have suggested that the conformational changes of the retinal and its surrounding environment in the reaction cycles are similar in the two proteins (17, 18). Analysis of BR is greatly aided by a number of good-quality X-ray crystal structures of the protein trapped in different reaction intermediates (14, 26). A previous study created a series of hybrid, chimera structures of BR by imposing the SB isomerization and BR-R82 side-chain rotation observed in the late M structure [Protein Data Bank (PDB) 1C8S] onto the ground-state structure (PDB 1C8R) (38). The aim was to identify the minimum set of conformational changes required to advance BR through the reaction cycle. These two imposed structural changes modify the site proton affinities, resulting in proton transfer from the BR-SB to D85, proton release from the extracellular cluster forming the late M state, and proton uptake to the SB to move into the N′ state (Table 1). In the BR ground state/late M chimera, the equilibrium ionization pattern shifts to 65% early and late M. In contrast, equilibrium ionization calculations with the X-ray crystal structure of the M-state intermediate (PDB 1C8S) indicate ∼70% N′ and ∼30% M-state protonation, suggesting that relaxation to later intermediates could occur in the trapped crystal structure (38) (Table 1 and Fig. 1A). The same changes in the retinal isomer and BR-R82 (HR-R108) are imposed on the HR ground-state structure to generate an HR chimera.
Proton and Cl– Binding to the Central Cluster in the HR Ground/M-State Chimera.
MCCE calculations of the chimera HR structure show a reordering of the relative energy of the ionization states analogous to what is found in the BR chimera or true BR M-state structures (Table 1, Figs. 3 and 4 C and D, and Fig. S2). The Cl– affinity in the HR chimera central cluster is >104 weaker than in the ground-state structure. Retinal isomerization moves the SB away from the Cl– binding cavity, reducing their interaction. The SB can retain its proton even as the Cl– counterion is lost as found in the BR N′ state (Table 1). There are several factors that stabilize the central cluster state HR-SBH+ D238− state with no Cl–. One is that shifting HR-R108 away from the central cluster destabilizes the −1 charge state found when Cl– is bound. In addition, the extracellular cluster has a −2 charge when HR-E219 is ionized and Cl– bound. This lowers the Cl– affinity of the central cluster by long-range electrostatic interactions.
The loss of the Cl– does lower the SB pKa so the SB is deprotonated 22% of the time at pH 7, which is not seen experimentally (44). Spectroscopic measurements indicate that the Cl– should follow the SB NH+ from the extracellular side to be lost to the intracellular compartment (18). MD simulations also showed that this Cl– stays bound to the SB after the isomerization (33). In the fixed backbone chimera structure, there is no cavity near the isomerized SB on the cytoplasmic side of the protein, near the position in the MD simulations (33). Additional structural relaxation of the HR backbone is needed to open up a binding site. Cavities are found ∼6.5 Å away from the isomerized SB, on the intracellular side of the retinal (Fig. S3). These binding sites are too far from the SB to strongly stabilize the Cl–SBH+ state and there is little Cl– bound (<0.01 occupancy).
Proton and Cl– Binding to the Extracellular Cluster in the HR Chimera.
BR releases a proton from the extracellular cluster in the M state. In BR, the resultant −2 change on the two deprotonated Glu is stabilized by BR-R82 moving closer to the two acids and by the acids moving apart, breaking the hydrogen bond between them. In the HR chimera with the cis-retinal and extracellular-pointing HR-R108, the affinity of the extracellular Cl– binding site increases by 50-fold. As in BR, the conformational change of HR-R108 stabilizes the E219–Cl– state by ∼13 kcal/mol, compensating for the ∼8 kcal/mol repulsion between the bound Cl– and E219−. This −2 charge state is stable between pH 5 and 10 (Fig. 4D). Thus, the retinal isomerization and the HR-R108 conformational change favors chloride uptake on the extracellular side of the Arg, and chloride release from the central cluster, analogous to the BR-N′ state (Fig. 3). The chimera moves 78% of HR into a state with a −2 charge in the extracellular cluster, protonated SB and deprotonated HR-D238 (BR-D212). The extracellular Cl– is tightly bound over a wide pH range (Fig. 4D).
The pH Dependence of Cl– and Proton Binding.
Cl– binding in the HR ground state is dependent on both the pH and Cl– concentration, as found in other ion-binding systems (9, 10). At low Cl– concentrations, no Cl– is bound and the HR absorption spectrum is blue-shifted, indicating that the trans retinal SB is deprotonated (45). As Cl– is added, the protonated spectrum is recovered. The FTIR C=NH+ stretching frequency indicates a strong interaction between the ionized Schiff base and Cl– in both ground and L states and changes with different halides (46). The SB is deprotonated in the simulations at high pH (Fig. 4A) and at low Cl– concentration. Proton and Cl– binding is also calculated to be coupled together in the extracellular cluster. Here, Cl– is bound at low pH when E219 is protonated and lost at higher pH when E219 is deprotonated to maintain a cluster charge near −1 (Fig. 4B).
The pKa of the HR Schiff base is measured to be 8.5 at 0.2 M chloride (45). In MCCE, the Cl– concentration is included in the energy terms for Monte Carlo sampling (Eq. S1) (10). Using the energy for the Cl– solution chemical potential, optimized for a similar buried binding site in α-amylase, the SB pKa is calculated to be 8.5 at 0.050 M Cl– (Fig. 4A) and 9.0 at 0.2 M Cl–. Thus, the Cl– affinity for the central cluster is 0.8 kcal/mol tighter than that determined experimentally. This is in remarkably good agreement with experiment, particularly given that no parameters were changed from previous calculations of proton and Cl– binding (10). The ground-state BR SB pKa is measured (47) and calculated to be >12 (23). Because the counterion for the SB is the covalently bound D85, SB deprotonation at high pH forms SB0D85–D212–, which has an unstable charge of −2. In contrast, in HR, a Cl– and a proton are lost together from the central cluster, forming the more accessible SB0D238− state at pH 9.5, retaining the favored −1 charge.
Interchanging BR and HR Functions.
Understanding the conservation of the electrostatic patterns in BR and HR that change the Cl– and proton affinities through their reaction cycles help to explain how only a few mutations can determine whether HR and BR is a proton or chloride pump (28). For example, at low pH, D85 with a pKa ∼3 is protonated (21, 23), and a proton is not released to the extracellular side until later in the reaction cycle (Fig. S2) (23, 38). With D85 and E194/E204 protonated, BR resembles HR where these residues have been replaced by polar side chains. BR still favors a negative charge at these sites even at low pH so chloride ions are bound and transferred during the reaction, turning BR into an inward Cl– pump (28).
Light-activated inward Cl– pumping is also found with in BR variants with BR-D85T, D85S (48), or S85N (30) mutations. These mutations mimic HR-T111 in the central cluster. Conversely, in HR, replacing the Cl– binding site residues with acidic groups such as azide, which has a solution pKa of 4.8 (49), results in proton pumping (50). The expectation is that azide binds near HR-T111 and HR-T230 replacing Cl–. However, because azide has a pKa similar to Asp or Glu, it can carry out a protonation/deprotonation reaction to transfer protons.
There have been efforts to convert HR to a proton pump using mutagenesis. This has proved to be more difficult than turning BR to a chloride pump (31). Both proton and Cl– pumping require changes in affinity, which has been explored here. They also require control of the accessibility to intracellular and extracellular compartments through the reaction cycle, which has not been considered in the work reported here. In addition, proton pumping requires long-range hydrogen bonded networks to transfer the H+, which are not needed for ion transfer (31). Based on the finding reported here, we would suggest a double-mutant BR-D85T/E204T may be a more efficient Cl– pump than the single-mutant D85T, as it should bind chloride more effectively. In HR, if T230 were mutated to an Asp or Glu, proton loss from the extracellular cluster would be expected to be easier, which could impair the efficiency of Cl pumping.
Conclusion
Small residues in HR replace three key acidic residues of BR: BR-D85, D96, and E204. In the ground state, BR-D85 is ionized, BR-E204 shares a negative charge with E194, and BR-D96 is neutral (21, 23). As seen here, at pH 7, two Cl– binding sites in HR replace the two acids that would be ionized in BR. The Cl– in the central cluster, observed in the crystal structure, binds strongly in the ground state, but is lost in a chimera ground/M-state structure with a cis-retinal and HR-R108 rotation, which models later intermediates. The Cl– in the extracellular cluster binds weakly in the ground state but tightly in the chimera structure. Thus, HR preserves the same charge distribution as BR in the ground and later states in the key central and extracellular clusters by exchanging a Cl– for a charged acidic residue.
In BR, the active-site Asp and Glu are static anions with mobile protons acting to change the cluster site charges during the reaction cycle. In contrast, in HR the anions move throughout the cycle. The same charge shifts result in a chloride moving inward in HR and a proton moving outward in BR. This charge similarity is exemplified by how easy it is to convert BR into an anion pump by neutralizing Asp-85 by mutation to Ser or Thr (48) or lowering the pH (28), and HR can be turned into a proton pump by adding azide (50). Comparison between the pH dependences of BR and HR shows that HR is more sensitive to the external condition of pH and ionic concentration than is BR. However, at the pH and ionic concentration range in which ion translocation is carried out, the electrostatic interactions that stabilize different charge states in each cluster in each stage of the cycle are conserved in these two related proteins.
Methods
The structure of HR (PDB 1E12) (41) was obtained from PDB. MCCE (51) was used to sample residue ionization states, rotamer positions, and chloride occupancies as a function of pH (23, 38) and chloride concentration (10) as described previously. BR calculations were conducted with 1C8R (52) and 1C8S (52). See SI Methods for more details of the MCCE analysis.
The HR Chimera Structure.
There are no X-ray crystal structures of later intermediates in the HR pumping cycle. Cl– binding in a putative M state was investigated by making an HR ground/M-state chimera structure by pasting the late M-state retinal Schiff base and the side-chain conformation of BR-R82 from the BR structure 1C8S into the HR ground-state structure 1E12 (38). The resulting structure was minimized with GROMACS (Fig. 2 C and D). The late M-state structure of BR 1C8S (52) was aligned with the HR ground-state structure 1E12 with PyMol. The Cα RMSD is 0.98 Å for the 153 structurally aligned residues. The HR retinal, Lys-242, and Arg-108 are then replaced with the 13-cis retinal, the SB Lys, and Arg-82 from 1C8S. The retinal is kinked with C13 moving toward the cytoplasm and the two ends moving toward the extracellular side. The Arg is rotated around the bond CG-CD by 80°, moving the guanidinium from the central cluster toward the extracellular cluster.
The HR chimera state structure is then minimized with GROMACS (53) without water, Cl–, or lipids. Asp-141 is protonated, all His are neutral in the ND tautomer except for His-95, which has NE protonated, and all other Asp, Glu, Arg, and Lys ionized. The ionization states used here are from the ground state of HR and BR so that the chloride binding in the chimera state is not biased by the structure minimization step. Energy minimization uses steepest decent. The initial minimization step is 0.1 Å, and converged within 190 steps with a force threshold of 0.024 kcal⋅mol−1⋅Å−1. IPECE (23) is then used to add 179 Cl– to the optimized structure (52). See SI Methods and Fig. S3 for more details. Only small changes in the structure were observed. The backbone Cα RMSD between the HR ground-state and BR M-state crystal structure is 1.17 Å. After minimization of the HR structure with the BR late M retinal, Lys and Arg coordinates, the Cα RMSD between the chimera HR structure and the BR M-state structure is reduced to 0.79 Å, as the retinal C20 is pushed toward the extracellular side by W207. Similar compression of the central cluster geometry around BR cis-retinal pasted into the trans-retinal ground-state backbone was seen in previous calculations (38). The close association of the SB and counterion BR-D85 in the BR chimera is one reason that the ground-state protonation pattern is maintained in 25% of the proteins. Overall, minimization of the HR chimera changes the cis Schiff base by 0.61 Å (all heavy-atom RMSD), R108, moves from the BR M-state configuration, by 0.56 Å and the rest of the protein by 0.10 Å. The movement of R108 yields a larger central cluster cavity and a smaller extracellular cavity.
Supporting Information Available.
A complete sequence alignment comparing BR and HR with calculated ionization states in BR and a figure of chlorides filling cavities in HR are provided with SI Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge helpful discussions with Janos K. Lanyi. We gratefully acknowledge the financial support of National Science Foundation Grant MCB 0517589 and Research Centers in Minority Institutions–NIH Grant RR03060 for infrastructure support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411119111/-/DCSupplemental.
References
- 1.Berchtold MW, Brinkmeier H, Müntener M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol Rev. 2000;80(3):1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 2.Feske S, Skolnik EY, Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat Rev Immunol. 2012;12(7):532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumetz AC, Snellinger-O’Brien AM, Kaler EW, Lenhoff AM. Patterns of protein protein interactions in salt solutions and implications for protein crystallization. Protein Sci. 2007;16(9):1867–1877. doi: 10.1110/ps.072957907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prange HD, Shoemaker JL, Jr, Westen EA, Horstkotte DG, Pinshow B. Physiological consequences of oxygen-dependent chloride binding to hemoglobin. J Appl Physiol (1985) 2001;91(1):33–38. doi: 10.1152/jappl.2001.91.1.33. [DOI] [PubMed] [Google Scholar]
- 5.Gerencser GA, Zhang J. Existence and nature of the chloride pump. Biochim Biophys Acta. 2003;1618(2):133–139. doi: 10.1016/j.bbamem.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 6.Gunner MR, Amin M, Zhu X, Lu J. Molecular mechanisms for generating transmembrane proton gradients. Biochim Biophys Acta. 2013;1827(8-9):892–913. doi: 10.1016/j.bbabio.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W, Erikson RL. Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc Natl Acad Sci USA. 1994;91(19):8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta. 2003;1653(1):25–40. doi: 10.1016/s0304-419x(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 9.Feller G, Bussy O, Houssier C, Gerday C. Structural and functional aspects of chloride binding to Alteromonas haloplanctis α-amylase. J Biol Chem. 1996;271(39):23836–23841. doi: 10.1074/jbc.271.39.23836. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Gunner MR. Using multiconformation continuum electrostatics to compare chloride binding motifs in α-amylase, human serum albumin, and Omp32. J Mol Biol. 2009;387(4):840–856. doi: 10.1016/j.jmb.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Matsuno-Yagi A, Mukohata Y. ATP synthesis linked to light-dependent proton uptake in a rad mutant strain of Halobacterium lacking bacteriorhodopsin. Arch Biochem Biophys. 1980;199(1):297–303. doi: 10.1016/0003-9861(80)90284-2. [DOI] [PubMed] [Google Scholar]
- 12.Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257(17):10306–10313. [PubMed] [Google Scholar]
- 13.Oesterhelt D. The structure and mechanism of the family of retinal proteins from halophilic archaea. Curr Opin Struct Biol. 1998;8(4):489–500. doi: 10.1016/s0959-440x(98)80128-0. [DOI] [PubMed] [Google Scholar]
- 14.Lanyi JK. Bacteriorhodopsin. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 15.Müller V, Oren A. Metabolism of chloride in halophilic prokaryotes. Extremophiles. 2003;7(4):261–266. doi: 10.1007/s00792-003-0332-9. [DOI] [PubMed] [Google Scholar]
- 16.Nagel G, et al. Channelrhodopsin-1: A light-gated proton channel in green algae. Science. 2002;296(5577):2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 17.Váró G. Analogies between halorhodopsin and bacteriorhodopsin. Biochim Biophys Acta. 2000;1460(1):220–229. doi: 10.1016/s0005-2728(00)00141-9. [DOI] [PubMed] [Google Scholar]
- 18.Essen LO. Halorhodopsin: Light-driven ion pumping made simple? Curr Opin Struct Biol. 2002;12(4):516–522. doi: 10.1016/s0959-440x(02)00356-1. [DOI] [PubMed] [Google Scholar]
- 19.Kloppmann E, Becker T, Ullmann GM. Electrostatic potential at the retinal of three archaeal rhodopsins: Implications for their different absorption spectra. Proteins. 2005;61(4):953–965. doi: 10.1002/prot.20744. [DOI] [PubMed] [Google Scholar]
- 20.Váró G, et al. Photocycle of halorhodopsin from Halobacterium salinarium. Biophys J. 1995;68(5):2062–2072. doi: 10.1016/S0006-3495(95)80385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balashov SP. Protonation reactions and their coupling in bacteriorhodopsin. Biochim Biophys Acta. 2000;1460(1):75–94. doi: 10.1016/s0005-2728(00)00131-6. [DOI] [PubMed] [Google Scholar]
- 22.Spassov VZ, Luecke H, Gerwert K, Bashford D. pKa calculations suggest storage of an excess proton in a hydrogen-bonded water network in bacteriorhodopsin. J Mol Biol. 2001;312(1):203–219. doi: 10.1006/jmbi.2001.4902. [DOI] [PubMed] [Google Scholar]
- 23.Song Y, Mao J, Gunner MR. Calculation of proton transfers in Bacteriorhodopsin bR and M intermediates. Biochemistry. 2003;42(33):9875–9888. doi: 10.1021/bi034482d. [DOI] [PubMed] [Google Scholar]
- 24.Rammelsberg R, Huhn G, Lübben M, Gerwert K. Bacteriorhodopsin’s intramolecular proton-release pathway consists of a hydrogen-bonded network. Biochemistry. 1998;37(14):5001–5009. doi: 10.1021/bi971701k. [DOI] [PubMed] [Google Scholar]
- 25.Pal R, Sekharan S, Batista VS. Spectral tuning in halorhodopsin: The chloride pump photoreceptor. J Am Chem Soc. 2013;135(26):9624–9627. doi: 10.1021/ja404600z. [DOI] [PubMed] [Google Scholar]
- 26.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757(8):1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Herzfeld J, Lansing JC. Magnetic resonance studies of the bacteriorhodopsin pump cycle. Annu Rev Biophys Biomol Struct. 2002;31:73–95. doi: 10.1146/annurev.biophys.31.082901.134233. [DOI] [PubMed] [Google Scholar]
- 28.Dér A, et al. Alternative translocation of protons and halide ions by bacteriorhodopsin. Proc Natl Acad Sci USA. 1991;88(11):4751–4755. doi: 10.1073/pnas.88.11.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sasaki J, et al. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269(5220):73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 30.Ganea C, Tittor J, Bamberg E, Oesterhelt D. Chloride- and pH-dependent proton transport by BR mutant D85N. Biochim Biophys Acta. 1998;1368(1):84–96. doi: 10.1016/s0005-2736(97)00173-9. [DOI] [PubMed] [Google Scholar]
- 31.Muroda K, Nakashima K, Shibata M, Demura M, Kandori H. Protein-bound water as the determinant of asymmetric functional conversion between light-driven proton and chloride pumps. Biochemistry. 2012;51(23):4677–4684. doi: 10.1021/bi300485r. [DOI] [PubMed] [Google Scholar]
- 32.Bamberg E, Tittor J, Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci USA. 1993;90(2):639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruia AD, Bondar AN, Smith JC, Fischer S. Mechanism of a molecular valve in the halorhodopsin chloride pump. Structure. 2005;13(4):617–627. doi: 10.1016/j.str.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Gunner MR, Mao J, Song Y, Kim J. Factors influencing the energetics of electron and proton transfers in proteins. What can be learned from calculations. Biochim Biophys Acta. 2006;1757(8):942–968. doi: 10.1016/j.bbabio.2006.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y, Michonova-Alexova E, Gunner MR. Calculated proton uptake on anaerobic reduction of cytochrome C oxidase: Is the reaction electroneutral? Biochemistry. 2006;45(26):7959–7975. doi: 10.1021/bi052183d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Z, Gunner MR. Energetics of quinone-dependent electron and proton transfers in Rhodobacter sphaeroides photosynthetic reaction centers. Biochemistry. 2005;44(1):82–96. doi: 10.1021/bi048348k. [DOI] [PubMed] [Google Scholar]
- 37.Onufriev A, Smondyrev A, Bashford D. Proton affinity changes driving unidirectional proton transport in the bacteriorhodopsin photocycle. J Mol Biol. 2003;332(5):1183–1193. doi: 10.1016/s0022-2836(03)00903-3. [DOI] [PubMed] [Google Scholar]
- 38.Song Y, Gunner MR. Identifying the key conformational changes coupled to proton pumping in bacteriorhodopsin. J Phys Chem B. in press. [Google Scholar]
- 39.Forrest LR, Tavoulari S, Zhang YW, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl−-dependent transporters. Proc Natl Acad Sci USA. 2007;104(31):12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivalta I, et al. Structural-functional role of chloride in photosystem II. Biochemistry. 2011;50(29):6312–6315. doi: 10.1021/bi200685w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 A resolution. Science. 2000;288(5470):1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 42.Gmelin W, et al. The crystal structure of the L1 intermediate of halorhodopsin at 1.9 angstroms resolution. Photochem Photobiol. 2007;83(2):369–377. doi: 10.1562/2006-06-23-RA-947. [DOI] [PubMed] [Google Scholar]
- 43.Miller A, Oesterhelt D. Kinetic optimization of bacteriorhodopsin by aspartic acid-96 as an internal proton donor. Biochim Biophys Acta. 1990;1020(1):57–64. [Google Scholar]
- 44.Blanck A, Oesterhelt D. The halo-opsin gene. II. Sequence, primary structure of halorhodopsin and comparison with bacteriorhodopsin. EMBO J. 1987;6(1):265–273. doi: 10.1002/j.1460-2075.1987.tb04749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schobert B, Lanyi JK, Oesterhelt D. Effects of anion binding on the deprotonation reactions of halorhodopsin. J Biol Chem. 1986;261(6):2690–2696. [PubMed] [Google Scholar]
- 46.Walter TJ, Braiman MS. Anion-protein interactions during halorhodopsin pumping: Halide binding at the protonated Schiff base. Biochemistry. 1994;33(7):1724–1733. doi: 10.1021/bi00173a015. [DOI] [PubMed] [Google Scholar]
- 47.Druckmann S, Ottolenghi M, Pande A, Pande J, Callender RH. Acid-base equilibrium of the Schiff base in bacteriorhodopsin. Biochemistry. 1982;21(20):4953–4959. doi: 10.1021/bi00263a019. [DOI] [PubMed] [Google Scholar]
- 48.Kalaidzidis IV, Kaulen AD. Cl−-dependent photovoltage responses of bacteriorhodopsin: Comparison of the D85T and D85S mutants and wild-type acid purple form. FEBS Lett. 1997;418(3):239–242. doi: 10.1016/s0014-5793(97)01390-2. [DOI] [PubMed] [Google Scholar]
- 49.Brown LS, Lanyi JK. Determination of the transiently lowered pKa of the retinal Schiff base during the photocycle of bacteriorhodopsin. Proc Natl Acad Sci USA. 1996;93(4):1731–1734. doi: 10.1073/pnas.93.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Váró G, Brown LS, Needleman R, Lanyi JK. Proton transport by halorhodopsin. Biochemistry. 1996;35(21):6604–6611. doi: 10.1021/bi9601159. [DOI] [PubMed] [Google Scholar]
- 51.Song Y, Mao J, Gunner MR. MCCE2: Improving protein pKa calculations with extensive side chain rotamer sampling. J Comput Chem. 2009;30(14):2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999;286(5438):255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 53.Lindahl E, Hess B, Van Der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7(8):306–317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.