Abstract
Background
Application of molecular diagnostic methods to the determination of etiology in suspected poxvirus-associated infections of bovines is important both for the diagnosis of the individual case and to form a more complete understanding of patterns of strain occurrence and spread. The objective of this study was to identify and characterize bovine-associated zoonotic poxviruses in Bangladesh which are relevant to animal and human health.
Findings
Investigators from the International Center Diarrhoeal Disease Research (icddr,b), the US Centers for Disease Control and Prevention (CDC), and the Bangladesh Department of Livestock Services traveled to three districts in Bangladesh—Siranjganj, Rangpur and Bhola–to collect diagnostic specimens from dairy cattle and buffalo that had symptoms consistent with poxvirus-associated infections. Bovine papular stomatitis virus (BPSV) DNA was obtained from lesion material (teat) and an oral swab collected from an adult cow and calf (respectively) from a dairy production farm in Siranjganj. Pseudocowpox virus (PCPV) DNA signatures were obtained from a scab and oral swab collected from a second dairy cow and her calf from Rangpur.
Conclusions
We report the first detection of zoonotic poxviruses from Bangladesh and show phylogenetic comparisons between the Bangladesh viruses and reference strains based on analyses of the B2L and J6R loci (vaccinia orthologs). Understanding the range and diversity of different species and strains of parapoxvirus will help to spotlight unusual patterns of occurrence that could signal events of significance to the agricultural and public health sectors.
Keywords: Bangladesh, Bovine papular stomatitis virus, Parapoxvirus, Zoonosis, Pseudocowpox virus
Findings
Introduction
Large ruminants have been implicated in the transmission of poxvirus infections to humans for centuries. In India, domestic cattle, buffalo and camels have been observed to harbor incidental infections with zoonotic poxviruses viruses from the Orthopoxvirus and/or Parapoxvirus genera [1–4]. Though many of these same types of large ruminants are found in neighboring Bangladesh, little is known about the persistence or distribution of zoonotic poxviruses in that country. A single zoonotic outbreak of suspected buffalopox was investigated—and ruled out— in southwest Bangladesh in 1976 [5]. The lack of information pertaining to the identity and burden of zoonotic poxviruses in Bangladesh suggests that both the disease burden in animals and the occupational risks posed by these viruses to animal workers remains largely undefined. The objective of this study was to identify and characterize bovine-associated poxviruses in Bangladesh which are of significance to veterinary public health and which can be transmitted to humans.
In otherwise healthy persons, zoonotic infections with bovine-associated viruses from either the Orthopoxvirus or Parapoxvirus genus can lead to painful localized pustular lesions or nodules, which generally occur on the upper extremities or face. These lesions are self-limited, resolving slowly over the course of several weeks to months [6]. Arriving at a determination of the precise etiology of a suspected poxvirus-associated infection in either humans or large ruminants necessitates the use of molecular diagnostic techniques, as serology and microscopy do not afford specificity of identification to the level of virus species and the clinical characteristics of these infections are insufficiently distinctive.
In bovines, vaccinia and buffalopox virus infections (orthopoxviruses) can engender high morbidity, with symptoms including malaise, anorexia, and pustular or ulcerated lesions or nodules on the teats and muzzles of adult and juvenile animals, respectively [1, 3, 7, 8]. Symptoms of bovine infection with bovine papular stomatitis virus (BPSV) and pseudocowpox virus (PCPV) (parapoxviruses) can be similar, involving painful erosive papules or vesicles on the muzzle, lips and teats. The exception to this is that BPSV infection in young bovines is sometimes manifest with distinctive ‘horseshoe-shaped’ papular lesions on the hard palate and oral mucosa, which can occur with or without concurrent inflammation of the gingiva [9]. Cryptic and aberrant (sometimes severe) parapoxvirus infections have also been reported in the literature further complicating clinical diagnoses [6, 10–14]. The application of molecular diagnostic methods to the determination of etiology in bovines is important not only for the diagnosis of the individual case, but also to gauge agricultural and human health risks. This by extension allows investigators to form a more complete understanding of patterns of strain occurrence and spread.
Methods
In April, 2007 investigators from the International Center for Diarrhoeal Disease Research (icddr,b) and the US Centers for Disease Control and Prevention (CDC), along with counterparts from the Bangladesh Department of Livestock services traveled to three locations in Bangladesh—Siranjganj and Rangpur within the Rajshahi Division and Bhola in the Khulna Division (Figure 1)—to collect diagnostic specimens from dairy cattle and buffalo that had symptoms consistent with poxvirus-associated infections. We selected the locations based on recent reports from the Department of Livestock Services of the Bangladesh Government and from a large milk producer cooperative (http://www.milkvita.org) of ‘pox-like’ illnesses in cattle and buffalo. Specimens were collected by veterinarians employed by the Bangladesh Department of Livestock services as authorized statues governing agricultural sanitation. Animal owners provided verbal consent for the collection of whole blood, oral swabs and/or lesion swab or crust specimens, which were collected from 9 symptomatic cattle from the Sirajganj and Rangpur Divisions and 4 buffalo from Bhola (Table 1). For animals that had evidence of scarring, but no active lesions, only blood was collected. Specimens were collected by veterinarians with consent from the animals’ owners. Blood specimens were assayed for the presence of orthopoxvirus antibodies by ELISA [15] and lesion, oral swab, and lesion crust specimens were examined by quantitative real-time PCR (qPCR) for the presence of genus and species level DNA signatures from zoonotic orthopoxvviruses [16] and parapoxviruses [17]. Two PCR amplicons were generated for sequences analysis. The primers and PCR conditions were described previously [18, 19].
Figure 1.
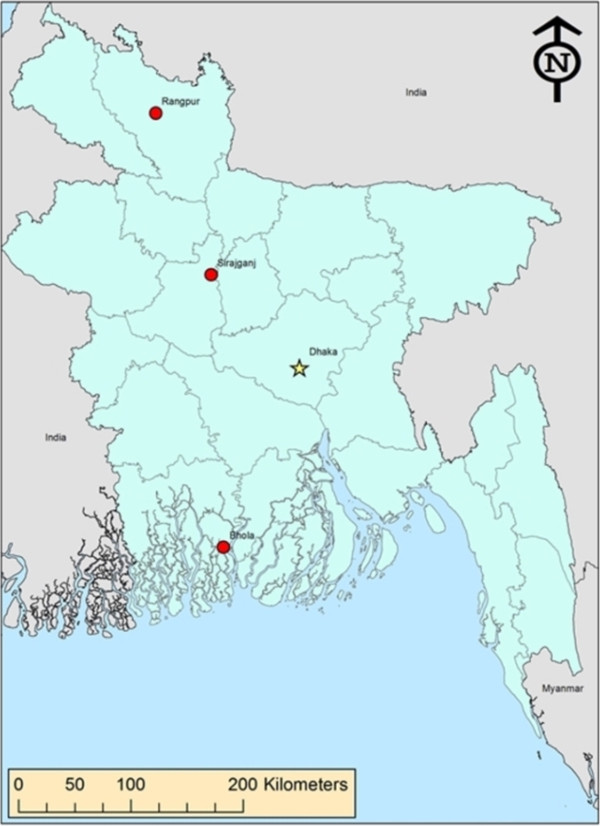
Map showing investigation sites. The locations from which specimens were collected during this investigation— Siranjganj and Rangpur within the Rajshahi Division and Bhola in the Khulna Division—are designated on the map.
Table 1.
Laboratory findings and characteristics of animals from which specimens were collected, Bangladesh, 2007
| Animal number | Type/breed | Age | Sex | Condition | Specimen | qPCR* | Serology OPXV ‡ | ||
|---|---|---|---|---|---|---|---|---|---|
| Parapoxvirus (Genus assay) | BPSV † | PCPV † | |||||||
| 1 | Cow/Jersey | 2 mo. | M | Lesion, oral cavity | Oral swab | pos | pos | -- | -- |
| Serum | -- | -- | -- | neg | |||||
| 2 | Cow/Frisian | 7 yr. | F | Lesion, teat | Teat scabs | pos | pos | -- | -- |
| Serum | -- | -- | -- | neg | |||||
| 3 | Cow/Frisian | 4 yr. | F | Lesion, teat | Teat scabs | neg | -- | -- | -- |
| Serum | -- | -- | -- | neg | |||||
| 4 | Cow/Jersey | 1 yr. | F | No lesions evident | Serum | -- | -- | -- | neg |
| 5 | Cow/Frisian | 8 yr. | F | Lesion, teat | Swabs/Scabs | neg | -- | -- | -- |
| Serum | -- | -- | -- | -- | |||||
| 6 | Cow/indigenous | 4 yr. | F | No lesions evident | Serum | -- | -- | -- | -- |
| 7 | Cow/indigenous | 5 yr. | F | Lesion, udder | Scabs | neg | -- | -- | -- |
| 8 | Cow/indigenous | 4 yr. | F | Lesion, udder | Scabs | pos | -- | pos | -- |
| Serum | -- | -- | -- | neg | |||||
| 9 | Cow/indigenous | 1 mo. | M | Lesion, oral cavity, muzzle | Swab | pos | -- | pos | -- |
| 10 | Buffalo/Monipuri | 8 yr. | F | Lesion, udder | Serum | -- | -- | -- | neg |
| 11 | Buffalo/Monipuri | 8 yr. | F | Lesion, udder | Serum | -- | -- | -- | neg |
| 12 | Buffalo/Monipuri | 13 yr. | F | Scar, udder | Serum | -- | -- | -- | neg |
| 13 | Buffalo/Monipuri | 16 yr. | F | No lesions evident | Serum | -- | -- | -- | neg |
*All negative for vaccinia-specific CrmB and Opx-generic E9L (negative >45 CT).
†BPXV, bovine papular stomatitis virus; PCPV, pseudocowpox virus.
‡Sera were examined for the presence of anti-OPX immunolglobulin by ELISA12 at 1:100 and 1:400 dilutions.
-- Testing not performed.
Phylogenetic analyses were performed using the Bayesian analysis software package (v1.75, http://beast.bio.ed.ac.uk), BEAST, BEAUti, and Tracer [20]. The analyses run MCMC chain length of 8,000,000 with an HKY nucleotide rate substitution model, strict molecular settings and sampling of every 1,000 states. A strict clock was used and the position of the root was estimated in the tree. The DNA sequences were aligned using BioEdit (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html) and Clustal alignment programs [21]. Data supporting the results of this article are available in GeneBank; accession numbers are included below. The sequences used in the phylogenetic analysis of the B2L amplicon were selected from available parapoxviruses or other high G + C content poxviruses in genbank, include isolates: PCPV_F05_990C [GenBank: JF773694], PCPV_VR634 [GQ329670], PCPV_IT_1303 [JN171852], PCPV_GE3_07 [KF478804], PCPV_JP_IW2010H [AB921003], PCPV_BR_SV721 [KC896641], ORFV_IND 67_04 [DQ263305], ORFV_FIN_F07_3748S [JN773702], ORFV_CHN_Gansu388 [KC485343], ORFV_NZ2 [U06671], ORFVIA82 [AY386263], ORFV_SA00 [AY386264], ORFV_D1701 [HM133903], BPSV_AR02 [AY386265], BPSV_BR_SV819 [JN629089], BPSV_BR_SV716 [KC896639], BPSV_GE_V660 [KF478805], BPSV_JP_IW2010E [AB921002], BPSV_JP_IW2010F [AB921001], SEAV_V842 [AY952943], BPSV_IT_9108 [JN162119], MOCV_T1 [U60315], MOCV_T2_369 [HE977615], SQRV_UK [HE601899]. From the CDC repository, The B2L amplicons were generated from PCPV isolates Bangladesh PCPV_BSH07012, PCPV_BSH07013 (from this study, marked with asterisk), Virginia PCPV_VA0904, and orf isolates ORFV_VA2010910054, BPSV isolates BPSV_VA0982, BPSV_VA09186, BPSV_BSH07005 (GeneBank accession numbers are KF830854, KF830855, KF830856, KF830857, KF830859, KF830860, KF830858, respectively; isolates from this study, marked with asterisk). The sequences used in the phylogenetic analysis of the J6R amplicon include isolates: MOCV_UK [Genebank: JQ269324], MOCV_2008_031 [GQ902057], CROV_Nile [DQ356948], PCPV isolates Bangladesh PCPV_BSH07012 [GQ902051], PCPV_BSH07013 [GQ902052], PCPV_MD06025 [GQ902049], BPSV_BSH07005 [GQ902054], BPSV_WA07058 [GQ902053], PCPV_FIN00_120R [GQ329669], SQRV_UK [HE601899]. From the CDC repository: Virginia PCPV_VA0904J, PCPV_GA08024J, orf isolates ORFV_VA2010910054J, BPSV isolates BPSV_VA0982, BPSV_VA09186J, (GeneBank accession numbers are KF830862, KF830863, KF830861, KF830864, KF830865, respectively).
Results
Evidence of BPSV DNA was obtained from lesion material (teat) and an oral swab collected from a single adult cow and calf (respectively) from a dairy production farm in Siranjganj (Table 1, Figure 2). PCPV DNA signatures were obtained from a scab and oral swab collected from a second dairy cow and her calf from Rangpur. No evidence of prior (antibody) or current (virologic) orthopoxvirus infection was detected in any of the 13 animals examined (Table 1).
Figure 2.
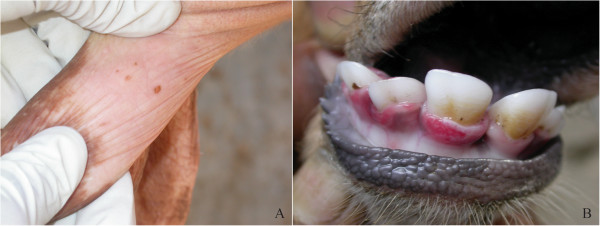
Photograph of BPSV infection in dairy cattle from Siranjganj. Panel (A) shows parapoxvirus lesion on the teat of animals number 2 described in Table 1. Panel (B) shows erosions on the gingiva of animal number 1 from Table 1.
DNA sequence fragments corresponding to the orthologs of vaccinia B2L and J6R (594 and 630 bp, respectively) were amplified from three of the parapoxvirus-positive specimens described in this study. Primer sets and amplification conditions described elsewhere [18, 19]. The B2L PCR assay has been used frequently for the diagnosis and typing of parapoxviruses, including sealpoxviruses [22]. The pan_pox highGC (J6R) PCR assay has also been frequently employed for the diagnosis and typing of parapoxvirus from clinical samples; one advantage of J6R assay is that it amplify other high G + C content chordopoxvirus, including molluscum contagiousum virus (MOCV) and crocodilepox virus (CROV), and other novel high G + C content poxviruses The J6R assay has been used to amplify an molluscum like poxvirus from an donkey (Fox et al. [23]) and two novel poxviruses in central US (Osadebe et al., Emerging Infectious Diseases accepted). The J6R assay is likely to amplify the UK red squirrel poxviruses based on the primer sequences alignment (unpublished results).
Phylogenetic trees using both B2L and J6R amplicon DNA sequences were generated to compare the BPSV and PCPV viruses from this study (BPSV_BSH07005, PCPV_BSH07012, PCPV_BSH07013) and other poxviruses to those of other reference viruses and to various viruses from around the world (Figure 3). Panel (A) depicts the phylogenetic tree based on analysis of a 594 bp amplified fragment of the B2L locus [18]; panel (B) the phylogenetic tree based on 630 bp fragment of the RNA polymerase subunit gene J6R [19]. DNA amplicons prepared from viruses in the CDC repository and from sequences derived from Genbank were used for analysis and comparison. Those from the CDC repository include: BPSV_VA9186, _VA0982, ORFV_VA2010910054 [24]; BPSV_WA07058, PCPV_GA08024, PCPV_MD06025 [6]; PCPV_VA0904 [25]. Sequences derived from Genbank are associated with the following strains: PCPV_F05_990C [26]; PCPV_VR634 [27]; BSPB_AR02, ORFVSA00, ORFV_IA82 [28]; BSPB_ITA9108; ORFV_NZ2 [29]; ORFV_D1701 [30].
Figure 3.
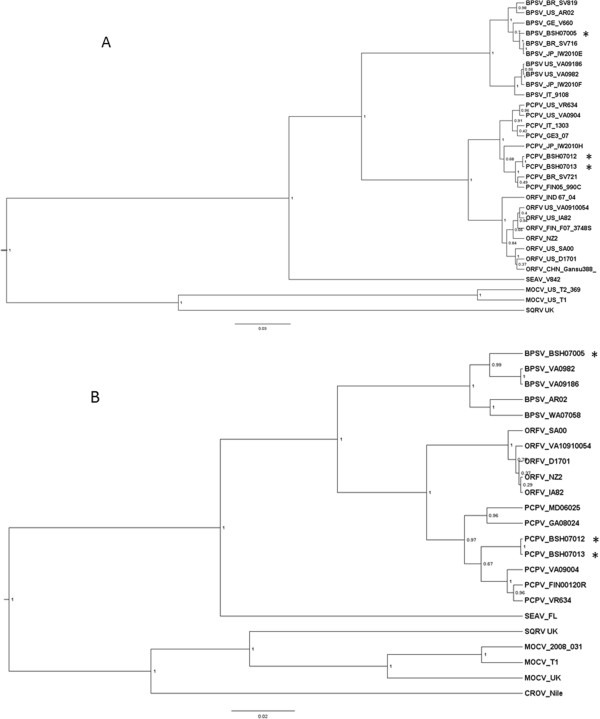
Phylogenetic trees rendering the relationships between the viruses identified in this study. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to construct the phylogenetic tree. The posterior probabilities were labeled at the each branch with probability values between 0 and 1. (A) The phylogenetic trees were constructed from 594 nucleotide of B2L PCR amplicon. (B) The phylogenetic tree constructed from 630 nucleotide sequences of J6R PCR amplicons. Sequences from Bangladesh are marked with asterisks.
The B2L tree and J6R tree yield similar topologies at the virus species level for ORFV, PCPV and BPSV: BPSV sequences form a cluster divergent from the PCPV and ORFV clades, while Sealpox virus and MOCV are yet further diverged from those traditional “barn yard” parapoxviruses. Interestingly, Figure 3A shows that PCPV sequence from Bangladesh forms a cluster with PCPV isolates of Brazil (PCPV_BR_SV721) and Finland (PCPV_FIN05_990C), and BPSV sequence from Bangladesh forms a cluster with BPSV isolates from Germany (BPSV_GE_V660), Brazil (BPSV_BR_SV716) and Japan (BPSV_JP_IW2010E), there are no clear geographic linkages for the isolates of both PCPV and BPSV. (This contrasts to the situation for orthopoxviruses, which generally group based on their geographic origins [31].) Individual variations between the two trees may reflect the different levels of sequence conservation between the two loci under consideration. Between closely related sequences PCPV_BSH07012 and PCPV_BSH07013, there is 1 single-nucleotide-polymorphism (SNP) between the B2L amplicons sequences and no SNPs for the J6R amplicon sequences.
Discussion
In summary, two species of zoonotic parapoxviruses were identified in specimens obtained from symptomatic cattle in Bangladesh. This marks the first occasion that parapoxviruses have been identified in Bangladesh. Unlike capripoxviruses (sheep pox and goat pox), which are endemic to Bangladesh, parapoxviruses can be transmitted to humans [32]. This underscores the importance of determining the infectious etiology of pox-like dermal lesions on domestic bovines to avoid confusion with infections caused by more serious zoonotic pathogens (e.g., buffalopox virus, Bacillus anthracis), or with agents that cause severe, notifiable animal diseases (e.g., FMD or Bluetongue) and to ensure that appropriate medical and veterinary interventions are employed. The feasibility of such an approach would be increased in low-resource countries such as Bangladesh by the development of an inexpensive rapid test, such as has been developed for detection of Orthopoxvirus antigen in clinical specimens [33].
People at risk for acquisition of parapoxvirus infections are those whose occupations or food preferences involve direct interaction with infected animals. In most instances human infections are the result of direct contact with infected animals, via milking, feeding, or handling of pelts and carcasses [34, 35]. Humans who have occupational exposure to bovines in Bangladesh –through dairy or meat production or other husbandry activities—should take precautions when handling animals with lesions or gingival symptoms. Particular care should be given to attending to wounds on the forearms, hands and fingers as these are the most common sites of parapoxvirus infections in humans [34].
Conclusions
Aberrant and often serious clinical manifestations of parapoxvirus infections in bovines have been increasingly recognized across the globe [10–13, 36, 37], even as new parapoxviruses are being identified in new large ruminant hosts [25, 38, 39]. Understanding the range and diversity of different species and strains of parapoxvirus will help to spotlight any unusual patterns of occurrence that could signal events of significance to either the agricultural or public health sectors. Insights relevant to the phylogeny and genetic diversity of these viruses may as well impact the design of future animal vaccines. And finally, the findings from this study enable us to hypothesize that zoonotic poxviruses are not uncommon in Bangladesh, however, further studies are needed to determine the prevalence of infection and to identify infection risks for both animals and humans.
Acknowledgements
We would like to thank Benjamin Monroe and Whitni Davidson (both from CDC) for their assistance with creation of a map and specimen management, respectively.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EL, SUK, SL, and MR conceived of the study. EL and SUK performed the field investigation and collected the data. HZ, ZB, KK, and ID generated and interpreted laboratory findings. JG, YL generated and analyzed phylogenetic data. MR, EL, SUK and SL wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Edith Lederman, Email: erlederman@yahoo.com.
Salah Uddin Khan, Email: m.khan@ufl.edu.
Stephen Luby, Email: sluby@stanford.edu.
Hui Zhao, Email: cgz9@cdc.gov.
Zachary Braden, Email: zab5@cdc.gov.
JinXin Gao, Email: xbd5@cdc.gov.
Kevin Karem, Email: kdk6@cdc.gov.
Inger Damon, Email: iad7@cdc.gov.
Mary Reynolds, Email: nzr6@cdc.gov.
Yu Li, Email: lay4@cdc.gov.
References
- 1.Kolhapure RM, Deolankar RP, Tupe CD, Raut CG, Basu A, Dama BM, Pawar SD, Joshi MV, Padbidri VS, Goverdhan MK, Banerjee K. Investigation of buffalopox outbreaks in Maharashtra State during 1992–1996. Indian J Med Res. 1997;106:441–446. [PubMed] [Google Scholar]
- 2.Nagarajan G, Ghorui SK, Kumar S, Pathak KM. Complete nucleotide sequence of the envelope gene of pseudocowpox virus isolates from Indian dromedaries (Camelus dromedarius) Arch Virol. 2010;155:1725–1728. doi: 10.1007/s00705-010-0784-z. [DOI] [PubMed] [Google Scholar]
- 3.Singh RK, Hosamani M, Balamurugan V, Satheesh CC, Shingal KR, Tatwarti SB, Bambal RG, Ramteke V, Yadav MP. An outbreak of buffalopox in buffalo (Bubalus bubalis) dairy herds in Aurangabad, India. Rev Sci Tech. 2006;25:981–987. [PubMed] [Google Scholar]
- 4.Venkatesan G, Balamurugan V, Prabhu M, Yogisharadhya R, Bora DP, Gandhale PN, Sankar MS, Kulkarni AM, Singh RK, Bhanuprakash V. Emerging and re-emerging zoonotic buffalopox infection: a severe outbreak in Kolhapur (Maharashtra), India. Vet Ital. 2010;46:439–448. [PubMed] [Google Scholar]
- 5.Tarantola D, Huq F, Hoque K, Selivanov Y, Wilson C. A Suspected Outbreak of Buffalopox in Bangladesh. 1978. WHO/SE/78.103, 1–7. [Google Scholar]
- 6.MacNeil A, Lederman E, Reynolds MG, Ragade NJ, Talken R, Friedman D, Hall W, Shwe T, Li Y, Zhao H, Smith S, Davidson W, Hughes C, Damon IK. Diagnosis of bovine-associated parapoxvirus infections in humans: molecular and epidemiological evidence. Zoonoses Public Health. 2010;57:e161–e164. doi: 10.1111/j.1863-2378.2009.01317.x. [DOI] [PubMed] [Google Scholar]
- 7.de Sant'Ana FJ, Leal FA, Rabelo RE, Vulcani VA, Moreira CA, Jr, Cargnelutti JF, Flores EF. Coinfection by Vaccinia virus and an Orf virus-like parapoxvirus in an outbreak of vesicular disease in dairy cows in midwestern Brazil. J Vet Diagn Invest. 2013;25:267–272. doi: 10.1177/1040638713475799. [DOI] [PubMed] [Google Scholar]
- 8.Rivetti AV, Jr, Guedes MI, Rehfeld IS, Oliveira TM, Matos AC, Abrahão JS, Kroon EG, Lobato ZI. Bovine vaccinia, a systemic infection: evidence of fecal shedding, viremia and detection in lymphoid organs. Vet Microbiol. 2013;162:103–111. doi: 10.1016/j.vetmic.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Dal Pozzo F, Martinelle L, Gallina L, Mast J, Sarradin P, Thiry E, Scagliarini A, Büttner M, Saegerman C. Original findings associated with two cases of bovine papular stomatitis. J Clin Microbiol. 2011;49:4397–4400. doi: 10.1128/JCM.05281-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cargnelutti JF, Flores MM, Teixeira FR, Weiblen R, Flores EF. An outbreak of pseudocowpox in fattening calves in southern Brazil. J Vet Diagn Invest. 2012;24:437–441. doi: 10.1177/1040638711435408. [DOI] [PubMed] [Google Scholar]
- 11.de Sant’Ana FJ, Rabelo RE, Vulcani VA, Cargnelutti JF, Flores EF. Bovine papular stomatitis affecting dairy cows and milkers in midwestern Brazil. J Vet Diagn Invest. 2012;24:442–445. doi: 10.1177/1040638711434799. [DOI] [PubMed] [Google Scholar]
- 12.Inoshima Y, Nakane T, Sentsui H. Severe dermatitis on cattle teats caused by bovine papular stomatitis virus. Vet Rec. 2009;164:311–312. doi: 10.1136/vr.164.10.311-b. [DOI] [PubMed] [Google Scholar]
- 13.Leonard D, Otter A, Everest D, Wood A, McInnes C, Schock A. Unusual bovine papular stomatitis virus infection in a British dairy cow. Vet Rec. 2009;164:65. doi: 10.1136/vr.164.2.65. [DOI] [PubMed] [Google Scholar]
- 14.Hessman BE, Sjeklocha DB, Fulton RW, Ridpath JF, Johnson BJ, McElroy DR. Acute bovine viral diarrhea associated with extensive mucosal lesions, high morbidity, and mortality in a commercial feedlot. J Vet Diagn Invest. 2012;24:397–404. doi: 10.1177/1040638711436244. [DOI] [PubMed] [Google Scholar]
- 15.Hutson CL, Lee KN, Abel J, Carroll DS, Montgomery JM, Olson VA, Li Y, Davidson W, Hughes C, Dillon M, Spurlock P, Kazmierczak JJ, Austin C, Miser L, Sorhage FE, Howell J, Davis JP, Reynolds MG, Braden Z, Karem KL, Damon IK, Regnery RL. Monkeypox zoonotic associations: insights from laboratory evaluation of animals associated with the multi-state US outbreak. Am J Trop Med Hyg. 2007;76:757–768. [PubMed] [Google Scholar]
- 16.Li Y, Olson VA, Laue T, Laker MT, Damon IK. Detection of monkeypox virus with real-time PCR assays. J Clin Virol. 2006;36:194–203. doi: 10.1016/j.jcv.2006.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao H, Wilkins K, Damon IK, Li Y. Specific qPCR assays for the detection of orf virus, pseudocowpox virus and bovine papular stomatitis virus. J Virol Methods. 2013;194:229–234. doi: 10.1016/j.jviromet.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 18.Inoshima Y, Morooka A, Sentsui H. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods. 2000;84:201–208. doi: 10.1016/S0166-0934(99)00144-5. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Meyer H, Zhao H, Damon IK. GC content-based pan-pox universal PCR assays for poxvirus detection. J Clin Microbiol. 2010;48:268–276. doi: 10.1128/JCM.01697-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tryland M, Klein J, Nordoy ES, Blix AS. Isolation and partial characterization of a parapoxvirus isolated from a skin lesion of a Weddell seal. Virus Res. 2005;108:83–87. doi: 10.1016/j.virusres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Fox R, Thiemann A, Everest D, Steinbach F, Dastjerdi A, Finnegan C. Molluscum contagiosum in two donkeys. Vet Rec. 2012;170:649. doi: 10.1136/vr.100721. [DOI] [PubMed] [Google Scholar]
- 24.Roess AA, McCollum AM, Gruszynski K, Zhao H, Davidson W, Lafon N, Engelmeyer T, Moyer B, Godfrey C, Kilpatrick H, Labonte A, Murphy J, Carroll DS, Li Y, Damon IK. Surveillance of parapoxvirus among ruminants in Virginia and Connecticut. Zoonoses Public Health. 2013;60(8):543–548. doi: 10.1111/zph.12036. [DOI] [PubMed] [Google Scholar]
- 25.Roess AA, Galan A, Kitces E, Li Y, Zhao H, Paddock CD, Adem P, Goldsmith CS, Miller D, Reynolds MG, Zaki SR, Damon IK. Novel deer-associated parapoxvirus infection in deer hunters. N Engl J Med. 2010;363:2621–2627. doi: 10.1056/NEJMoa1007407. [DOI] [PubMed] [Google Scholar]
- 26.Hautaniemi M, Vaccari F, Scagliarini A, Laaksonen S, Huovilainen A, McInnes CJ. Analysis of deletion within the reindeer pseudocowpoxvirus genome. Virus Res. 2011;160:326–332. doi: 10.1016/j.virusres.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 27.FRIEDMAN-KIEN AE, ROWE WP, BANFIELD WG. Milker’s nodules: isolation of a poxvirus from a human case. Science. 1963;140:1335–1336. doi: 10.1126/science.140.3573.1335. [DOI] [PubMed] [Google Scholar]
- 28.Delhon G, Tulman ER, Afonso CL, Lu Z, de la Concha-Bermejillo A, Lehmkuhl HD, Piccone ME, Kutish GF, Rock DL. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J Virol. 2004;78:168–177. doi: 10.1128/JVI.78.1.168-177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson AJ, Ellis G, Balassu T. The genome of orf virus: restriction endonuclease analysis of viral DNA isolated from lesions of orf in sheep. Arch Virol. 1982;71:43–55. doi: 10.1007/BF01315174. [DOI] [PubMed] [Google Scholar]
- 30.Mayr A, Herlyn M, Mahnel H, Danco A, Zach A, Bostedt H. [Control of ecthyma contagiosum (pustular dermatitis) of sheep with a new parenteral cell culture live vaccine] Zentralbl Veterinarmed B. 1981;28:535–552. doi: 10.1111/j.1439-0450.1981.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci U S A. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitching RP, McGrane JJ, Hammond JM, Miah AH, Mustafa AH, Majumder JR. Capripox in Bangladesh. Trop Anim Health Prod. 1987;19:203–208. doi: 10.1007/BF02242117. [DOI] [PubMed] [Google Scholar]
- 33.Townsend MB, MacNeil A, Reynolds MG, Hughes CM, Olson VA, Damon IK, Karem KL. Evaluation of the Tetracore Orthopox BioThreat(R) antigen detection assay using laboratory grown orthopoxviruses and rash illness clinical specimens. J Virol Methods. 2013;187:37–42. doi: 10.1016/j.jviromet.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson AJ, Petersen GV. Orf virus infection of workers in the meat industry. N Z Med J. 1983;96:81–85. [PubMed] [Google Scholar]
- 35.Uzel M, Sasmaz S, Bakaris S, Cetinus E, Bilgic E, Karaoguz A, Ozkul A, Arican O. A viral infection of the hand commonly seen after the feast of sacrifice: human orf (orf of the hand) Epidemiol Infect. 2005;133:653–657. doi: 10.1017/S0950268805003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao K, Song D, He W, Lu H, Zhang B, Li C, Chen K, Gao F. Identification and phylogenetic analysis of an Orf virus isolated from an outbreak in sheep in the Jilin province of China. Vet Microbiol. 2010;142:408–415. doi: 10.1016/j.vetmic.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 37.Oem JK, Lee EY, Lee KK, Kim SH, Lee MH, Hyun BH. J Vet Med Sci. 2013. Bovine Papular Stomatitis Virus (BPSV) infections in Korean native cattle. [DOI] [PubMed] [Google Scholar]
- 38.Hautaniemi M, Ueda N, Tuimala J, Mercer AA, Lahdenpera J, McInnes CJ. The genome of pseudocowpoxvirus: comparison of a reindeer isolate and a reference strain. J Gen Virol. 2010;91:1560–1576. doi: 10.1099/vir.0.018374-0. [DOI] [PubMed] [Google Scholar]
- 39.Scagliarini A, Vaccari F, Turrini F, Bianchi A, Cordioli P, Lavazza A. Parapoxvirus infections of red deer, Italy. Emerg Infect Dis. 2011;17:684–687. doi: 10.3201/eid1704.101454. [DOI] [PMC free article] [PubMed] [Google Scholar]


