Abstract
Meiotic prophase I (MPI), is an initial stage of meiosis characterized by intricate homologous chromosome interactions, synapsis and DNA recombination. These processes depend on the complex, but poorly understood early MPI events of homologous chromosome search, alignment and pairing. Detailed molecular investigation of these early events requires isolation of individual MPI substages. Enrichment for Pachytene (P) and Diplotene (D) substages of late MPI was previously accomplished using flow cytometry. However, separation of early MPI spermatocytes, specifically, of Leptotene (L) and Zygotene (Z) substages, has been a challenge due to these cells’ similar characteristics. In this report, we describe an optimized Hoechst-33342 (Hoechst)-based flow cytometry approach for isolating individual MPI populations from adult murine testis. We get significant enrichment for individual L and Z spermatocytes, previously inseparable from each other, and optimize the isolation of other MPI substages. Our flow cytometry approach is a combination of three optimized strategies. The first is optimization of testis dissociation protocol that yields more consistent and reproducible testicular single cell suspension. The second involves optimization of flow cytometric gating protocol where a critical addition to the standard protocol for cell discrimination based on Hoechst fluorescence, involves a back-gating technique based on light scattering parameters. This step specifies selection of individual MPI substages. The third, is an addition of DNA content restriction to the gating protocol to minimize contamination from non-meiotic cells. Finally, we confirm significant enrichment of high-purity Preleptotene (PreL), L, Z, P and D MPI spermatocytes using stage-specific marker distribution. The technique will facilitate understanding of the molecular events underlying meiotic prophase I.
Keywords: Adult mouse, meiotic prophase I, Hoechst 33342, flow cytometry, cell sorting
Introduction
Spermatogenesis, a multistage process by which spermatogonial stem cells differentiate into spermatozoa, occurs in the seminiferous tubules of the adult testis. As a result of asynchronous initiation of spermatogonial stem cell differentiation, the testis contains germ cells at all phases of spermatogenesis. These include the proliferative phase, responsible for the mitotic expansion of diploid spermatogonia, the meiotic phase, which generates haploid cells from diploid progenitors by way of specialized two cell divisions preceded by one round of DNA replication, and spermiogenic phase, marked by the differentiation and maturation of haploid cells.
The cellular heterogeneity of the testis necessitates developing methods for germ cell enrichment and isolation. The study of individual stages of spermatogenesis has become more accessible with Hoechst-based flow cytometry approaches. Indeed, flow cytometry is commonly used to enrich for murine spermatogonial stem cells (1–3) and more recently, for late MPI spermatocytes (4–6). On the other hand, enrichment for high-purity early MPI populations has remained difficult, thus, these cell types have largely escaped comprehensive molecular analysis. One challenge is that testicular cell heterogeneity is not proportional with respect to the percent of each cell type present. Due to the logarithmic nature of spermatogenesis, early cell types are proportionately underrepresented. A spermatogonial stem cell can give rise to up to 1024 primary spermatocytes and four times as many haploid spermatids, and the maturation of spermatid into a mature spermatozoon is one of the lengthiest phases of spermatogenesis. As a result, post-meiotic cells occupy the majority of the total testicular volume (7). This makes it difficult to enrich for and study the less abundant and more transient early MPI populations. In fact, the purification of individual L and Z substages has not yet been accomplished to date. A major goal of this study was to achieve separation of individual L and Z spermatocytes.
Early MPI is critical for proper meiotic progression. Originally characterized in terms of changes in the chromosomal morphology (8), L and Z substages differ in a number of cellular processes. Leptonema, which follows Preleptonema where pre-meiotic S-phase occurs, involves extensive chromatin reorganization, telomere-led chromosome movement and telomere attachment to nuclear envelope. These are thought to facilitate homologous chromosome (homologue) interactions, alignment and pairing (8, 9). At this time, meiotic “programmed” double stranded breaks (DSBs) form and Axial Elements (AEs) start to assemble along the paired sister chromatids. During L to Z transition, a widely conserved telomere configuration known as meiotic bouquet can be observed, and is marked by telomere clustering at the nuclear periphery (10, 11). In Zygonema, homologues begin to synapse as AEs assemble into synaptonemal complex (SC), a proteinateous structure that will eventually juxtapose the homologs along their entire lengths in Pachynema of MPI. The events observed in early MPI, including wide-range chromosomal movements, visible changes in the distribution of heterochromatin and assembly of SC, imply extensive changes in chromatin organization. Some molecular details of these early meiotic processes have been described in C. elegans (12, 13), S. cerevisiae (14), S. pombe (15, 16) and other non-mammalian organisms. However, detailed molecular understanding of early meiotic events in mammals is still lacking. Clearly, detailed molecular and mechanistic studies, and the subsequent generation of appropriate mutants, are necessary to understand the scope of regulation and coordination of these events, necessary for proper meiosis. Thus, the isolation of homogenous populations of early meiocytes is essential for definitive molecular and biochemical studies of MPI.
Recently, refined Hoechst dye staining protocols and flow cytometric analyses, have allowed for an enrichment of several mouse MPI populations (2, 5). These efforts have led to the much needed molecular and/or genetic analyses including examination of pre-meiotic, pre-DSB homolog pairing in PreL spermatocytes (17), nucleosome profiling at recombination hotspots in spermatogonia, PreL or mixtures of L/Z and P/D cells (4) and other studies (18). However, questions that require the separation of L from Z remain unanswered, and even isolation of high-purity individual P and D populations remains a challenge, with publications resorting to analyzing mixtures of L/Z and P/D spermatocytes (4, 19). This and a number of concerns prompted us to re-examine and optimize all major steps of the published Hoechst-based flow cytometry protocols, from cell dissociation to flow cytometric analysis. One concern was the preparation of a reliable testicular single cell suspension. There are a number of published protocols available for testis dissociation and Hoechst dye staining of adult murine testicular cells (1–3, 5, 19–21). While all these protocols provide an excellent foundation for preparation of testicular single cell suspension, a challenge, still, is to obtain suspensions of consistently high quality between the experiments. In addition, since the available protocols vary from one another in a number of aspects, including the medium and the duration of testicular tissue dissociation, the duration and concentration of Hoechst staining and other parameters, the resulting Hoechst-labeled testicular suspension profiles differ between the laboratories. Another concern is cell purity. Published literature largely fails to document the purity of sorted cells, and/or note the criteria for purity determination. Consequently, practical information such as the number of cells examined, the markers used for evaluation, and the type of contaminants observed is largely unavailable. There is also an ambiguity associated with cell sorting and collection parameters, with often incomplete reporting on important parameters, including the rate of sorting and the number of populations sorted at a given time.
Previous studies often used juvenile testes as the means of reducing cellular heterogeneity of the starting material and/or maximizing the yield of early MPI spermatocytes. Our method is optimized for the adult murine testis, which provides an enriched source of all MPI substages. An additional important reason for using adult versus juvenile testis in our analysis, is that the first round of spermatogenesis is thought to substantially differ from all subsequent rounds, and has been linked to differences in undifferentiated spermatogonia (Spg) (22) as well as associated with increased apoptosis (23, 24). As the differences in the first wave of spermatogenesis may suggest differences in the first meiotic wave, this raises a question of whether first meiocytes are distinct from the subsequent ones. Furthermore, our approach should allow sorting from adult testes of mutant mouse lines deficient in various aspects of male germ cell differentiation.
In this report we present an optimized flow cytometric analysis of Hoechst testicular single-cell suspension that greatly improves the purity and reproducibility of the sorted spermatogenic cell populations from adult murine testis.
Materials and Methods
Animals
Adult C57BL/6 male mice (Jackson Laboratory) were raised in our animal facilities. 2–5 month old mice were used as a source of adult testes. All experimental procedures were performed in compliance with ethical regulations and approved by the IACUC of Carnegie Institution for Science.
Materials, reagents and solutions for testicular cell suspension preparation
Materials: 15 ml conical tubes (BD Falcon, #352097); Shaking Water Bath (VWT, #89032); Disposable transfer pipet (VWR); 100 μm nylon cell strainer (BD Falcon, #352360); 40 μm nylon cell strainer (BD Falcon, #352340); 12×75mm tube with 35-μm cell strainer cap (BD Falcon, #352235). Reagents: Collagenase Type I (Worthington Biochemical, #LS004196) DNAse I (Invitrogen, #18068-015); Gey’s Balanced Salt Solution (GBSS) (Sigma-Aldrich); 2.5% Trypsin (10X) (Gibco, #15090-046); Hoechst 33342 (10 mg/ml solution in water) (Life Technologies, #H3570); Newborn Calf Serum (NCS) (Life Technologies); Propidium Iodide (PI) (1 mg/ml) (Sigma-Aldrich, #P4864). Stock solutions: DNAse I (1mg/ml solution in 50% glycerol was made from 552 Kunitz units/mg powder and stored at −20°C); Freshly prepared solutions: “Collagenase I/Dnase I” (Collagenase type I [200 U/ml] and DNAse I [5 μg/ml] in GBSS); “Collagenase I/Dnase I/Trypsin” (Collagenase type I [200 U/ml], DNAse I [5 μg/ml], and Trypsin (0.025%) in GBSS).
Testicular single-cell suspensions and Hoechst-33342 (Hoechst) staining
Testis dissociation was based on a recently described method (5) with modifications. Our final protocol was conducted based on six consecutive steps described below.
Testis digestion: After the removal of tunica albuginea, each testis was placed in a 15 ml conical tube on ice with 6 ml “Collagenase I/Dnase I” solution. The tube was sealed with parafilm, shaken in horizontal position at 150 rpm for 10 min at 35°C. The temperature and agitation speed were the same for all subsequent incubation steps. Halfway into the 10 min incubation, the testis were gently pipetted, up and down twice to help tubule dispersion. This and all other pipetting steps were done using disposable transfer pipets. By the end of this step, tubules appeared thin and dispersed.
Somatic cell removal: Tubules were allowed to settle for 2 min at room temperature (RT) by standing the tube vertically. The supernatant, enriched in interstitial testicular cells was removed, leaving just enough liquid to cover the settled tubules.
Seminiferous tubule digestion: 6 ml of pre-heated “Collagenase I/Dnase I/Trypsin” solution was added to the tube and the tubules were gently pipetted up and down 10 times. Halfway into the 25 min digestion period, 60 μl of 2.5% Trypsin was added, and the tubules were pipetted again 10 times. At the end of the incubation time, pipetting was repeated 10 times. The tubules appeared fragmented and solution dense with cells. The resulting suspension was passed through a 100 μm nylon cell strainer. At this point, a 100 μl aliquot was removed to be processed for cell counting and viability estimation (see Materials and Methods below) while the rest of filtered cell suspension was pre-stained with Hoechst dye (during step 4 below).
Pre-staining with 100 μg Hoechst dye: To the resulting filtered cell suspension, 10 μl of DNAse I (1 mg/ml) and 10 μl of Hoechst dye (10 mg/ml) were added. The suspension was pipetted up and down 10 times and incubated for 20 min. Halfway into the 20 min period, the suspension was pipetted again. At the end of incubation, 600 μl of NCS was added to inactivate trypsin and the suspension was pipetted up and down 5 times.
Staining with Hoechst: After determining the cell number, the suspension was spiked with 10 μl of DNAse I (1 mg/ml), and stained with Hoechst dye for the final 6 μg Hoechst/million cells. The suspension was pipetted up and down 10 times and incubated for 25 min. Halfway into the incubation period, the solution was pipetted again, and then once more at the end of the incubation. Finally, the suspension was passed through a 40 μm nylon cell strainer. The suspension was kept on ice until sorting, which usually proceeded within 30 min to one hour after the completion of step 5.
Staining with PI: Immediately prior to sorting, 2 ml cell suspension was removed into a 5 ml polypropylene culture tube, the cells were stained with 10 μl of PI at room temperature and filtered into a tube with 35 μm cell strainer cap.
Cell Counting and Viability Estimation
Prior to Hoechst staining, viability and total yield of testicular single cells were determined. For trypan blue exclusion test of cell viability, 100 μl of the cell suspension was combined with 200 μl GBSS and 300 μl 0.4% trypan blue and analyzed using hemocytometer. Cell viability was found to be more than 95% in all cases. The total number of alive cells per testis (50 to 90 million cells depending on age) was calculated based on the total viable cells per ml. Note, that these counts underestimate the total number of cells per testis, since the counts were performed after filtration through 100 μl nylon cell strainer, a procedure that eliminates an uncertain number of elongated spermatozoa. Additionally, small size and the mobility of sperm cells introduce some difficulty to accurate sperm quantification, potentially further underestimating cell counts.
Prior to cell sorting, cell viability of the ready-to-sort samples was examined based on staining of cells with PI described above. The total cell viability, based on the number of PI-positive cells, ranged between 86 and 94 percent (Supplementary Figure 1).
Flow Cytometric Analysis and Fluorescence Activated Cell Sorting
Data analysis was done using BD FACSDiva software. Hoechst was excited using 375 nm laser, and the dye’s wide emission spectrum detected in two distinct channels: the “Ho Blue” (450/40 nm band-pass filter) and the “Ho Red” (670 nm long pass filter). The latter was also used to detect PI. A dichroic mirror (610 nm long pass filter) was used to split these emission wavelengths. Forward Scatter (FSC-A) and Side Scatter (SSC-A) were detected using 488 nm laser. Two-way sorting was performed using a seventy-micron nozzle size. Sorting Flow rate was adjusted to 2000–3500 events/second. A minimum of 500,000 events, were pre-recorded before setting of the gates. Cells were sorted into 5 ml polypropylene round-bottom tubes coated with (by pipetting) and containing 1 ml of 5% NCS in GBSS.
Meiotic spreads and Immunostaining
To estimate percent purity of individual sorted populations, an aliquot of each was processed for immunofluorescence staining. Nuclear spreads were prepared as described (25), with minor modifications. Briefly, a 50 μl aliquot of sorted cells was mixed with 50 μl hypotonic buffer (30 mM Tris-Cl, 50 mM sucrose, 17 mM sodium citrate dehydrate, 5 mM EDTA in water) and incubated at RT for 30 min. Cells were pelleted at 5000 rpm for four minutes, 80 μl supernatant was removed and cells re-suspended with 65 μl of 100 mM sucrose, pH 8.2. Next, 30 μl of the suspension was applied to a glass slide (Superfrost Plus, VWR) dipped just before in a solution of 1% paraformaldehyde, pH9.2, supplemented with 0.1% Triton X-100. The pH of all solutions was set using 50 mM sodium borate. Nuclei were dried overnight (O/N) in a slightly opened humid box over water. Finally, the slides were washed for one minute in 0.2% Photoflo (Kodak), dried at RT, and stored at minus 20°C. Frozen slides were thawed at 42°C, followed by serial washes, five minutes each, of 0.5% Triton X-100 in 1X PBS, 0.05%Triton X-100 in 1X PBS and 1X PBS. Slides were treated with blocking buffer (2% BSA, 0.05% Triton X-100) for 30 minutes and incubated O/N at RT with primary antibodies (1:750 anti-SCP3, Abcam, #ab15093 and 1:1000 anti-phospho-Histone γH2A.X, Millipore, #05-636) in blocking buffer supplemented with 10% normal donkey serum). After washes as above, cells were incubated with secondary antibodies (1:500 donkey anti-rabbit 594, Life Technologies, #A21207 and 1:1000 donkey anti-mouse 488, Life Technologies, #A21202). Slides were subsequently washed as described above and counterstained for 5 min in 0.1 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in PBS. After a rinse in PBS, the slides were mounted in Vectashield (Vector Laboratories) and sealed with nail polish. To quantify percent purity, between 100 and 250 spread cells from each sorted cell population, were scored and classified after each sort.
Fluorescence Microscopy Imaging
Confocal images were acquired with Leica DM6000 microscope and analyzed with ImageJ. All images for quantification of cell purity were acquired with an upright fluorescence microsope (Olympus BX61) and analyzed with ImageJ. Images of spread preparations represent single optical sections.
Results and Discussion
Preparation of adult murine testicular cell suspension, Hoechst dye staining and flow cytometric analysis
A crucial first step towards successful flow sorting is preparation of a reliable testicular single cell suspension. Guided by the available published protocols, and predominantly by the one developed by Getun and colleagues (5), here, we optimize and present an improved testis dissociation and Hoechst dye staining protocol of adult murine testicular cells (detailed in the Materials and Methods section). The protocol has been optimized to produce a well-defined and consistent profile of MPI populations. We have incorporated a number of points in the testis dissociation and Hoechst dye staining procedures that facilitate the reproducibility of subsequent sorting results. For example, to minimize variability in cell dissociation efficiency, we performed all steps in a larger reaction volume and we have optimized the concentration of Collagenase I, DNAse I and Trypsin used. To avoid cell and DNA clumping, we periodically mix cells throughout the cell dissociation step and spike the testicular suspension with Trypsin and DNAse I. Perhaps the most critical optimized aspect of our protocol involves adjusting Hoechst concentration per total cell number counted per testis. We use 6 µg Hoechst dye per million cells, and depending on total cells counted, a different amount of Hoechst dye will be added to a particular testicular cell suspension. This is in contrast to, for example, adding a fixed amount of Hoechst dye per testis. Indeed, variability in Hoechst staining protocol easily leads to discrepancies in testicular Hoechst profile between the experiments. Although this protocol, like any other that involves Hoechst dye staining, exhibits sensitivity to variations that may influences Hoechst equilibration and subsequent Hoechst-stained cellular profile (26), our protocol represents an improvement towards consistency and reproducibility between experiments. This point is illustrated by similar Hoechst profiles of different testicular digests (Supplementary Fig. 2.)
Subsequently, Hoechst- and PI- dye stained testicular cell suspension is processed for flow cytometric analysis. It is essential to minimize noise from the unwanted sources such as debris. This is done on the basis of the cells’ light scattering parameters, namely, the Forward Scatter (FSC) and the Side Scatter (SSC), proportional to the cell size and cell granularity, respectively. Since most debris particles are typically of small size, the debris is excluded by setting a gate that excludes signals with low FSC intensity (Fig. 1A). Due to the very small size of elongated spermatozoa, exclusion based on low FSC also eliminates much of their contribution from the analysis and sorting.
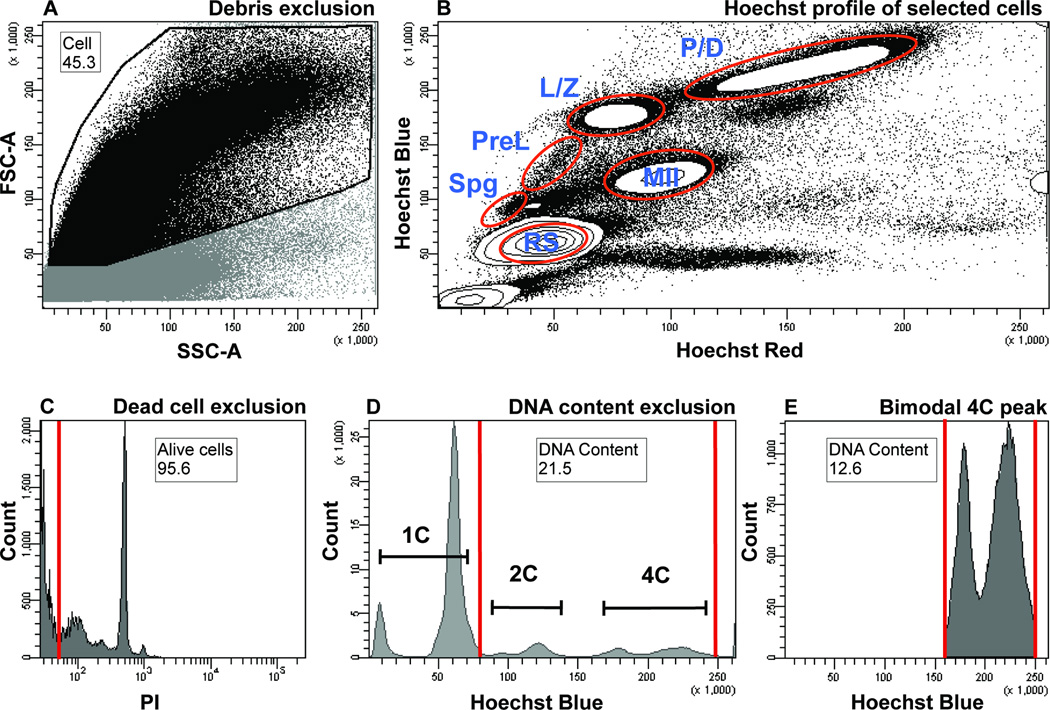
Figure 1.
Flow cytometric analysis of adult murine testicular cells based on Hoechst and PI fluorescence and light scattering parameters. Numbers on plots represent percent of parent population, the latter identified within a figure legend below. Gate name is found above the number. Both, the number and the gate name are encased by black box.
A) Debris exclusion based on low light scattering parameters. Cells are distinguished from debris based on the FSC and SSC, proportional to the cell size and cell granularity, respectively. A dot plot shows debris exclusion gate (“Cell” gate, black outline) that includes the cells (black dots) and excludes the debris (grey dots), which exhibit low FSC intensity. The excluded region also contains some elongated spermatozoa, whose small size is a major contributor to the low FSC signal. The parent of the “Cell” gate includes all the cells.
B) Hoechst profile of testicular cells. Cells selected in Figure 2A are visualized in a “Hoechst Blue”/“Hoechst Red” contour plot, in which the density of the cells is displayed as contour lines that form circular contours upon high cell density. The main subpopulations visualized are contained within the white densities outlined in red. Spg, spermatogonia; PreL, preleptotene spermatocytes; L/Z, leptotene/zygotene spermatocytes; P/D, pachytene/diplotene spermatocytes; MII, meiosis II spermatocytes; RS, round spermatids.
C) Dead cell exclusion based on PI fluorescence. Alive, PI-negative cells are found within an “Alive cells” gate to the left of the red line and include over 94% of all cells (most are pushed off the x-axis). Cells positive for PI (to the right of the red gate) are excluded from the analysis. The parent of the “Alive cells” gate is the “Cell” gate from Figures 1A and 1B.
D) DNA content exclusion based on “Hoechst Blue” fluorescence. Populations that fall within the red gate called “DNA Content” are included in the analysis (2C and 4C DNA contents are labeled). Haploid cells with 1C DNA content are outside of the gate and are excluded from the analysis. The parent of the “DNA Content” gate is the “Alive cells” gate from figure 1C. In this example, the “DNA Content” gate represents 43.3 percent of all cells.
E) Bimodal distribution of cells with 4C DNA content shows L/Z and P/D populations. The left and right peaks encompassed by the red gate (a restricted “DNA Content” gate) correspond to L/Z and P/D populations, respectively. The parent of the “DNA Content” gate is the “Alive cells” gate from figure 1C. In this example, the restricted “DNA Content” gate represents 5.4 percent of all cells.
Subsequent visualization of the selected cells simultaneously in Hoechst blue and red channels reveals a complex fluorescence profile (Fig. 1B) that is consistent with published literature (1, 2, 17, 19). Major populations that can be identified include those expected to be enriched in spermatogonia (Spg), Pre-leptotene spermatocytes (PreL), Leptotene/Zygotene (L/Z) spermatocytes, Pachytene/Diplotene (P/D) spermatocytes, meiosis II spermatocytes (MII) and post-meiotic haploid round spermatids (RS) (Fig. 1B, outlined in red). Importantly, our experimental and cytometric analysis setups yield “tight” demarcation of these spermatogenic populations. This is evident from the formation of distinct L/Z, P/D, MII and RS populations, when viewed using contour plot of one million cells (Fig. 1B, red circles).
To the debris-excluded cells, dead cell exclusion is applied based on PI fluorescence (Fig. 1C). Next, we can additionally exclude unwanted cells based on DNA-content (C-value) by setting boundaries on the Hoechst Blue gate (Fig. 1D). Routinely, we exclude haploid cells with 1C DNA content from our analysis (Figure 1D). Notably, the prominent delineation between L/Z and P/D in our analysis is marked by the presence of the bimodal 4C DNA content peak on the Hoechst Blue histogram (Fig. 1E), where left peak corresponds to the L/Z and right peak to the P/D population. Recently, a similar bimodal peak has been documented in guinea pig spermatocytes, whose testis exhibit a higher proportion of L/Z cells, as compared to mouse and rat (27). Our analysis suggests that despite the underrepresentation of early meiocytes in adult murine testis, consistent dissociation and Hoechst staining conditions, followed by careful discrimination of debris, and noise like elongated spermatozoa, can significantly improve the resolution of the testicular single cell Hoechst profile.
Gating strategy and back-gating strategy for isolating high-purity MPI populations
Previously published sorting approaches of murine MPI populations were based on gates specified on Hoechst dye fluorescence, similar to the gating approach in Figure 1A–C. This selection alone, however, does not discriminate sufficiently to obtain high-purity L, Z, P or D cells, and, in our experience, these cell populations were found contaminated with each other and with other spermiogenic cell types like haploid spermatozoa. To specify each MPI population more accurately, we wanted to better define the flow cytometric selection gates to minimize contamination. To this end, we have developed an analysis workflow of Hoechst dye-labeled testicular cell suspension that utilizes a back-gating approach to enrich for high-purity MPI substages. The back-gating approach involves setting a gate around the cells with certain fluorescence characteristics, and then asking what are the physical characteristics of these selected cells, based on their FSC and SSC criteria (28). Ultimately, this approach allows to define a particular cell population of interest not only in terms of two Hoechst fluorescence parameters (Hoechst Blue and Hoechst Red), but also, in terms of the light scattering parameters (FSC and SSC), allowing a more precise definition of that population.
In our standard workflow, we begin by setting the gates on Hoechst dye fluorescence profile (Fig. 2A). Information on gate statistics for Figure 2 can be found in Supplementary Figure 3. Within a large MPI gate, we assign PreL, L, Z, P and D gates based on previously published and empirical observations. We also gate on non-meiotic populations, including pre-meiotic diploid spermatogonia (gate Spg), diploid spermatocytes II (gate MII), and haploid, post-meiotic spermatids (gate RS) (Fig. 2A). Sorting based on the debris and dead cell exclusion, and on Hoechst dye fluorescence, led to an extensive compromise in purity. Extensive cross-contamination from P was observed in most fractions examined, with Z and D fractions often containing more than 30% of P spermatocytes. Additionally, Spg, PreL and L fractions were found heavily cross-contaminated. Furthermore, most fractions contained some elongated spermatozoa. The overall contamination is largely explained by the proximity and overlap of the contaminating populations based on FSC and SSC parameters, or the overlap in cell size and granularity. To best illustrate this point, we can examine L and Z populations (Fig. 2B). When L cells from Hoechst dye profile (Fig. 2A, gate L, green) are viewed on the light scatter plot (Fig. 2B, panel i, green), and the same is done with the Z population (Fig. 2A and 2B, panel ii, pink), together, the profiles exhibit a large region of overlap (Fig. 2B, panel iii). Additional gates on the FSC versus SSC profile can be set to more conservatively describe L or Z populations, namely, gates “L-A” and “Z-A”, respectively (Fig. 2B, panel iv). These back-gates minimize cross-contamination based on light scattering parameters, and also, incorporate discrimination based on Hoechst dye parameters (Fig. 2B, panel iv). Thus, in this back-gating approach, a gate set on “Hoechst Blue”/“Hoechst Red” fluorescence plot is used to set a gate based on the “FSC”/“SSC” plot. This approach effectively discriminates cells based on both Hoechst dye fluorescence and light scatter parameters.
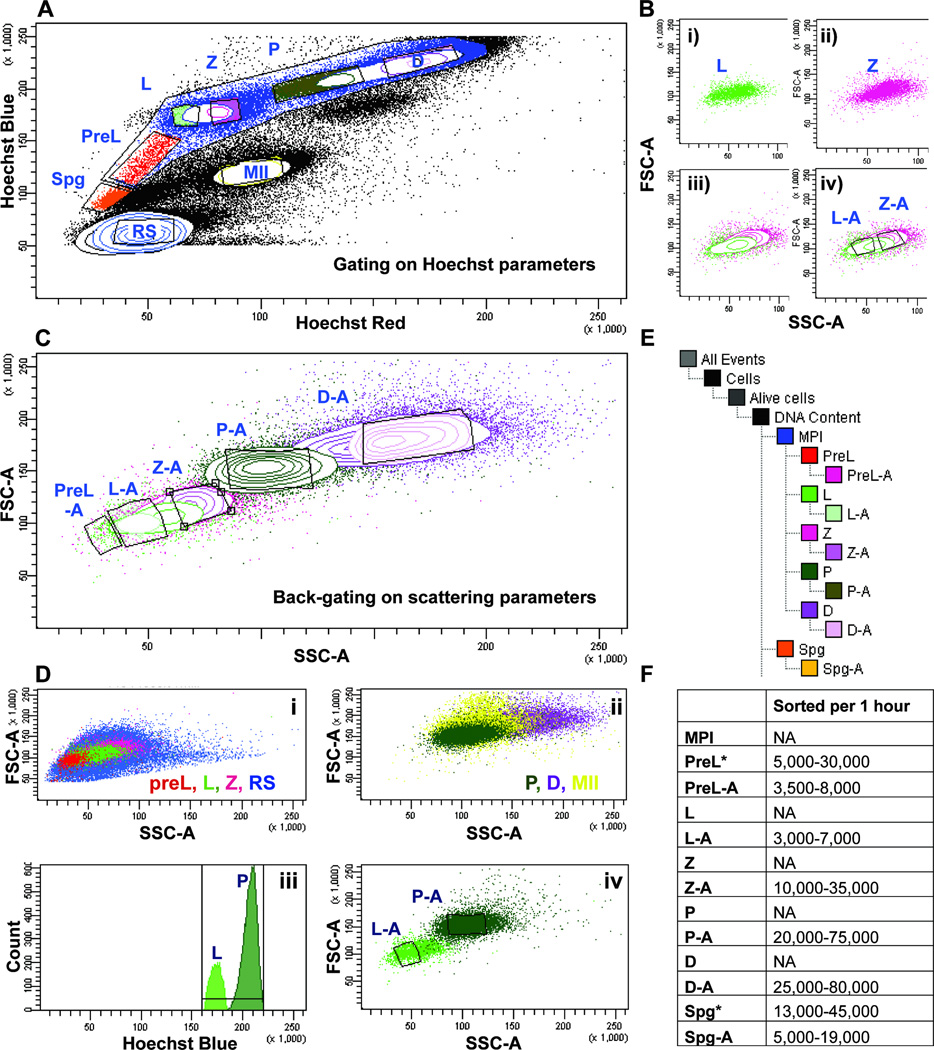
Figure 2.
Gating and back-gating strategies for isolating individual MPI populations.
A) Gating on individual spermatogenic populations based on Hoechst fluorescence. A large meiotic gate encompasses smaller gates containing cells of individual MPI substages, including preleptotene- (PreL, red), leptotene- (L, green), zygotene- (Z, pink), pachytene- (P, dark green) and diplotene- (D, magenta) spermatocytes. Gates enriched in pre-meiotic spermatogonia (Spg, orange), round spermatids (rSP, blue) and meiosis II spermatocytes (MII, yellow) are also outlined.
B) Back-gating approach. A particular gate on the Hoechst fluorescence plot (Figure 3A) can be viewed on the FSC vs. SSC plot. Here, L spermatocytes (panel i) and Z spermatocytes (panel ii) defined by a fluorescence gate in Figure 3A, display particular characteristics on the “FSC”/“SSC” plot (green and pink dots, respectively). iii) When viewed on the same plot, L and Z share similar light scattering parameters and partly overlap. iv) Based on regions of minimal overlap on the “FSC”/“SSC” plot, “L-A” gate is created to restrict contamination from the Z gate, and “Z-A” gate is made to restrict contamination from the L gate.
C) Back-gating approach applied to all MPI substages. Individual spermatogenic populations defined by gates on Hoechst fluorescence plot (“Hoechst Blue”/“Hoechst Red”, Figure 3A) are used to set gates on the light scattering plot (“FSC”/“SSC”). A gate set on “FSC”/“SSC” plot and appended with “-A” (e.g., P-A) marks a “back-gate” of a gate (e.g. “P”) set on the “Hoechst Blue”/“Hoechst Red” plot.
D) DNA content-restricting gate helps eliminate contamination from non-MPI cell types. The light scattering profile of MPI spermatocytes overlaps with that of other spermatogenic cells in the testis. (i) Abundant and morphologically diverse haploid rSP cells (blue) overlap with PreL, L and Z cells. (ii) MII spermatocytes (yellow) overlap with P and D cells. (iii) A gate specifying DNA content on “Hoechst Blue” histogram can be restricted to include only the 4C content where L and P cells are found, and to exclude haploid and diploid cells that fall outside of this gate. Thus, for example, a two-way sort for L and P involves exclusion based on “Hoechst Blue” parameters and on (iv) specification of back-gates “L-A” (light green, outlined subset) and “P-A” (dark green, outlined subset).
E) Gating tree. The tree indicates the sequential gating and back-gating procedure applied to Hoechst-labeled testicular single cell suspension before sorting. Gates that have “-A” appended to them were the final sorting gates, such that sorting of L spermatocytes involved collecting cells from the “L-A” gate.
F) The range of numbers of cells collected from ten different sorts. The wide range largely reflects the difference in the ages of mice used (2–5 months old) and the adjustment, from experiment to experiment, in the size of gates and back-gates set. Populations that can be sorted without back-gating are indicated by “*”.
As MPI substages represent successive stages of germ cell development, there is an extensive overlap on FSC versus SSC plot, between all adjacent MPI populations, not only L and Z. Consequently, we applied the back-gating protocol to all MPI populations (Fig. 2C). In addition, on the “FSC”/“SSC” plot, RS population (Fig. 2D, panel i, blue), which is abundant and morphologically diverse, overlaps with PreL, L and Z populations (Fig. 2D, panel i). Similarly, MII cells (Fig. 2D, panel ii, yellow), overlap with P and D populations (Fig. 2D, panel ii). To accommodate for the overlaps from RS and MII cell types, we additionally discriminate at the level of DNA content, on the “Hoechst Blue” histogram (Fig. 2D, panel iii). Thus, our typical two-way sort, e.g. of L and P populations, will involve a) Debris and dead cell exclusion, b) Exclusion based on DNA content, where the gate is limited to 4C on “Hoechst Blue” histogram (Fig. 2D, panel iii) and c) setting of the back-gates “L-A” and “P-A” on the “FSC”/“SSC” dot plot to specify the final L and P populations before sorting (Fig. 2D, panel iv). Our final sorting tree is depicted in Fig. 3E. The range of cell numbers collected in an hour, can be found in Fig. 2F, and contain the numbers from up to ten successful sorts, using mice of various ages. Typically, we couple the sorting of PreL or L with P, and Z with D. Indeed, some populations like P and D can be collected much faster, without much sacrifice to purity, if they are sorted without the rate-limiting L or Z cells. Also note, that Spg and PreL populations can be sorted without back-gating (Fig. 2F, as indicated by “*”), if sorted alone and with strictly limited DNA content gate, in which case, the numbers collected would also rise.
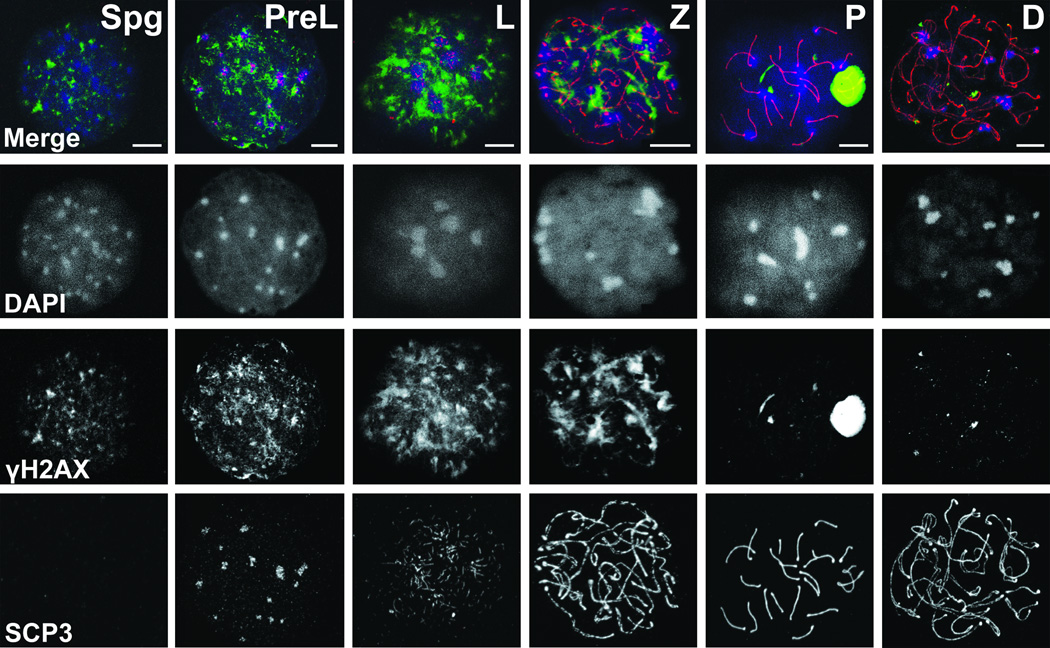
Figure 3.
Immunofluorescence analysis of MPI progression. Spread nuclei were double labeled with γH2AX (green) and SYCP3 (red) and counterstained with DAPI (blue). Staging was deduced from changing, stage-specific labeling patterns of these markers. Fluorescence images generated by confocal microscopy show representative MPI substages including the preleptotene- (PreL), leptotene- (L), zygotene- (Z), pachytene- (P), diplotene- (D) spermatocytes, and an example of pre-meiotic Spermatogonia (Spg). Bar - 10 µm.
Detailed immunofluorescence (IF)-based characterization and purity assessment of sorted MPI cells
To assess the identity and the extent of homogeneity of sorted MPI cell populations, we performed co-immunostaining of sorted cells with well-characterized nuclear markers of MPI. Specifically, we examined the distribution of phosphorylated histone H2AX (γH2AX) and synaptonemal complex protein 3 (SYCP3), markers of double stranded-breaks (DSBs) and meiosis-specific synaptonemal complex (SC), respectively. We also used DNA stain DAPI to visualize these cells’ dynamic MPI chromatin distribution. Representative MPI substages are shown in Figure 3, as imaged by the confocal fluorescence microscopy. Profound changes in chromatin dynamics are marked by the changing shape and number of DAPI-labeled chromocenters (Fig. 3). The assembly of axial elements to which sister chromatids are attached, and the SC, which connects homologous chromosomes, are marked by progressive elongation and thickening of SYCP3. Indeed, SYCP3 aggregates are observed in PreL spermatocytes and long thick SYCP3 fibers in P spermatocytes, the latter indicative of fully synapsed autosomes with assembled SCs (Fig. 3). Increasing amounts and intensity of γH2AX signal in early meiocytes, namely, the PL, L and Z, mark DSB initiation and accumulation, while disappearance of γH2AX in late MPI reflects DSBs resolution, with an exception of an intense, well defined γH2AX signal in P spermatocytes corresponding to partially synapsed sex chromosomes within the sex body (Fig. 3).
Classification of sorted cells, based on the intensity and distribution of DAPI, γH2AX and SYCP3 signals, can be straightforward when the observed cells exhibit known distribution of these markers. Indeed, many of these marker patterns have been previously observed (17, 29–31). However, examination of hundreds of cells after sorting reveals a number of “non-typical” cells, commonly observed in our sorts, whose MPI substage classification can be challenging. These cells, which have not been well documented in literature, include cells that transition from one MPI substage to another and those at the very beginning or the end of a substage. Thus, the “non-typical” category may include early and late PreL cells, early and late L cells and late L cells transitioning to Z.
Routine and extensive post-flow cytometry IF analysis of cells based on DAPI staining, SYCP3 and γH2AX, allowed us to better document the diversity of MPI cells. For such routine examination, we used upright fluorescent microsope, since, confocal microscopy, while producing images of very high quality and resolution, is an involved technique that precludes quick cell-by-cell analysis of tens or hundreds of sorted cells, as is required after each sort. Based on IF staining, we defined approximate early-, mid- and late- boundaries of early MPI substages, as they appeared in our partly squashed cell preparations of meiotic nuclei (Fig. 4). Indeed, early through late PreL, L and Z spermatocytes exhibit unique patterns of DAPI, γH2AX and SYCP3 staining (Fig. 4 and Figure Legend) and a combination of these patterns can be used as a staple of a particular substage. As there is no ambiguity related to the identification of P and D spermatocytes, we do not present their characterization here.
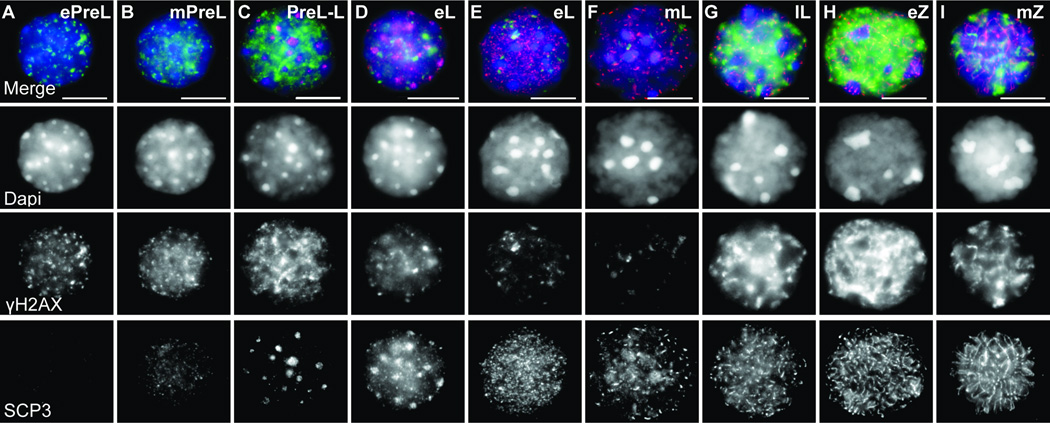
Figure 4.
Detailed immunofluorescence characterization of early MPI sorted cells.
Images show examples of sorted PreL- (A–C, preleptotene), L- (D–G, leptotene) and Z- (H–I, zygotene) spermatocytes. Staging was deduced from DAPI, SYCP3 and γH2AX patterns. Early to mid PreL cells exhibit numerous peripheral DAPI chromocenters corresponding to satellite DNA (A–C). (A) Early PreL (ePreL) nucleus shows punctate γH2AX foci and absence of SYCP3, (B) mid PreL (mPreL) nucleus showing foci and patches of γH2AX and weak and diffuse SCP3 staining and (C) a nucleus with late PreL and L characteristics (PreL-L) exhibits numerous SYCP3 aggregates and a decrease in DAPI foci along the rim of the nuclear periphery. PreL-L cells often exhibit intense γH2AX signal. (D–G) From early L (eL) to late L (lL) nuclei exhibit a progression from short to longer stretches of SYCP3 and from sparse foci to large intense and partly homogenous γH2AX. At least two types of eL cells can be observed, one PreL-like (D) but with lower γH2AX signal, and a more typical one (E). (H,I) In early Z (eZ), long, interrupted SYCP3 fibers are observed throughout the cell. Polarized concentration of thickening SYCP3 fiber ends marks telomere bouquet base. Mid Z (mZ) exhibits long and thin SYCP3 stretches. By late Z, chromosome axes are fully formed and appear as long thing fibers, and levels of γH2AX decrease. Bar - 10 µm.
The categorization based on IF is both, consistent with, and elaborates on previous descriptions of MPI substage specifications, with only a few examples of the latter shown in Figure 4. For instance, we observe that late PreL (lPreL) cells often exhibit diffuse and irregular SYCP3 patterns. We label these as late PreL cells because they most often appear in our PreL-specific sorted fraction. However, these lPreL cells can also be considered as transitional PreL to L cells, since they exhibit few to numerous SYCP3 aggregates (Fig. 4C). Consistently, similar cells, with SYCP3 aggregates have been previously placed in either PreL (9, 32) or L (31, 33) category. Some ambiguities associated with substage identification still remain. One is the difficulty to distinguish with certainty SCP3-negative and γH2AX-negative early PreL cells from differentiated B Spg, which give rise to the PreL cells.
Extensive categorization based on the described markers was critical for subsequent quantification of percent purity of cells after sorting (Table 1). Significantly, most spermatogenic populations sorted by our approach are of very high-purity, with a range of 75 to 95 percent (for individual data and statistical analysis see Supplementary Figure 4). In all the sorted fractions, we were able to greatly reduce contamination from non-meiotic and post-meiotic cell types. Importantly, we were able to enrich for individual L and Z spermatocytes, enabling separation of now all individual MPI spermatocytes in high-purity form.
Table 1.
Percent purity quantification based on immunofluorescence analysis after cell sorting. MPI substage purity was calculated as (cell type observed/total cells counted by IF)*100. MPI substage purity was assessed based on at least eight different sorts, with a range of 100-250 cells counted per sorted population. The smaller counts are representative of very pure populations, where minimal cell counts were required to establish purity.
| COLLECTED | Spg | PreL | L | Z | P | D |
|---|---|---|---|---|---|---|
| OBSERVED | ||||||
| Spermatogonia (Spg) | 80–91% | 5–10% | 3% | 0 | 0 | 0 |
| Preleptotene (PreL) | 5–10% | 75–92% | 5–10% | 0 | 0 | 0 |
| Leptotene (L) | 0 | 5–10% | 60–80% | 5–10% | 0 | 0 |
| Zygotene (Z) | 0 | 0 | 10–15% | 75–90% | 3% | 2% |
| Pachytene (P) | 0 | 0 | 3% | 10–15% | 81–95% | 4–10% |
| Diplotene (D) | 0 | 0 | 0 | 0 | 5–7% | 82–95% |
| Other | 4–5% | 1–2% | 1% | 1% | 1–2% | 2–3% |
Conclusion
In summary, we provide an optimized Hoechst-based flow cytometry method for isolating individual MPI populations from Hoechst dye labeled testicular cell suspension. We offer improvements in sample preparation and show that back-gating technique and DNA content restriction can significantly improve the resolution and separation of early MPI meiocytes. It should be noted, however, that a user of the protocol will probably have to do several flow cytometry sorting experiments to best adjust the gates and back-gates. To evaluate sorted MPI cell purity, we urge users of flow cytometry methodology to employ immunofluorescence staining routinely, and resolve ambiguities with the markers mentioned above, and other meiotic and spermatogonial markers that are available.
Supplementary Material
Acknowledgements
This work was performed with the support from Carnegie Institution for Science. We thank Dr. Hao Zhang (at the Johns Hopkins Bloomberg School of Public Health) for the help in early stages of the project and Dr. Phillipe Bois for personal communication and advise on flow cytometry.
Footnotes
The authors declare no conflict of interest.
Literature Cited
- 1.Lassalle B, Bastos H, Louis JP, Riou L, Testart J, Dutrillaux B, Fouchet P, Allemand I. 'Side Population' cells in adult mouse testis express Bcrp1 gene and are enriched in spermatogonia and germinal stem cells. Development. 2004;131:479–487. doi: 10.1242/dev.00918. [DOI] [PubMed] [Google Scholar]
- 2.Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. Cytometry A. 2005;65:40–49. doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- 3.Barroca V, Lassalle B, Coureuil M, Louis JP, Le Page F, Testart J, Allemand I, Riou L, Fouchet P. Mouse differentiating spermatogonia can generate germinal stem cells in vivo. Nat Cell Biol. 2009;11:190–196. doi: 10.1038/ncb1826. [DOI] [PubMed] [Google Scholar]
- 4.Getun IV, Wu ZK, Khalil AM, Bois PR. Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep. 2010;11:555–560. doi: 10.1038/embor.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Getun IV, Torres B, Bois PR. Flow cytometry purification of mouse meiotic cells. J Vis Exp. 2011 doi: 10.3791/2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fallahi M, Getun IV, Wu ZK, Bois PR. A Global Expression Switch Marks Pachytene Initiation during Mouse Male Meiosis. Genes. 2010;1:469–483. doi: 10.3390/genes1030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russell LD. Histological and histopathological evaluation of the testis. Clearwater, Fl: Cache River Press; 1990. pp. xiv–286. [Google Scholar]
- 8.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 9.Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scherthan H. A bouquet makes ends meet. Nat Rev Mol Cell Biol. 2001;2:621–627. doi: 10.1038/35085086. [DOI] [PubMed] [Google Scholar]
- 11.Bass HW. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell Mol Life Sci. 2003;60:2319–2324. doi: 10.1007/s00018-003-3312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nabeshima K. Collaborative homologous pairing during C. elegans meiosis. Worm. 2012;1:116–120. doi: 10.4161/worm.19528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rog O, Dernburg AF. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr Opin Cell Biol. 2013;25:349–356. doi: 10.1016/j.ceb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess SM. Homologous chromosome associations and nuclear order in meiotic and mitotically dividing cells of budding yeast. Adv Genet. 2002;46:49–90. doi: 10.1016/s0065-2660(02)46004-x. [DOI] [PubMed] [Google Scholar]
- 15.Ding DQ, Haraguchi T, Hiraoka Y. From meiosis to postmeiotic events: alignment and recognition of homologous chromosomes in meiosis. FEBS J. 2010;277:565–570. doi: 10.1111/j.1742-4658.2009.07501.x. [DOI] [PubMed] [Google Scholar]
- 16.Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, Yamamoto M, Hiraoka Y. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336:732–736. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- 17.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev Cell. 2013;24:196–205. doi: 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009;5:e1000648. doi: 10.1371/journal.pgen.1000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 20.Falciatori I, Borsellino G, Haliassos N, Boitani C, Corallini S, Battistini L, Bernardi G, Stefanini M, Vicini E. Identification and enrichment of spermatogonial stem cells displaying side-population phenotype in immature mouse testis. FASEB J. 2004;18:376–378. doi: 10.1096/fj.03-0744fje. [DOI] [PubMed] [Google Scholar]
- 21.Shimizu Y, Motohashi N, Iseki H, Kunita S, Sugiyama F, Yagami K. A novel subpopulation lacking Oct4 expression in the testicular side population. Int J Mol Med. 2006;17:21–28. [PubMed] [Google Scholar]
- 22.Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- 23.Kluin PM, Kramer MF, de Rooij DG. Spermatogenesis in the immature mouse proceeds faster than in the adult. Int J Androl. 1982;5:282–294. doi: 10.1111/j.1365-2605.1982.tb00257.x. [DOI] [PubMed] [Google Scholar]
- 24.Mori C, Nakamura N, Dix DJ, Fujioka M, Nakagawa S, Shiota K, Eddy EM. Morphological analysis of germ cell apoptosis during postnatal testis development in normal and Hsp 70-2 knockout mice. Dev Dyn. 1997;208:125–136. doi: 10.1002/(SICI)1097-0177(199701)208:1<125::AID-AJA12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 25.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 26.Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8:136–147. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Casuriaga R, Geisinger A, Santinaque FF, Lopez-Carro B, Folle GA. High-purity flow sorting of early meiocytes based on DNA analysis of guinea pig spermatogenic cells. Cytometry A. 2011;79:625–634. doi: 10.1002/cyto.a.21067. [DOI] [PubMed] [Google Scholar]
- 28.Givan AL. Flow cytometry : first principles. New York: Wiley-Liss; 2001. pp. xviii–273. [Google Scholar]
- 29.Liebe B, Petukhova G, Barchi M, Bellani M, Braselmann H, Nakano T, Pandita TK, Jasin M, Fornace A, Meistrich ML, et al. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp Cell Res. 2006;312:3768–3781. doi: 10.1016/j.yexcr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 30.Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Ishiguro K, Kim J, Fujiyama-Nakamura S, Kato S, Watanabe Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011;12:267–275. doi: 10.1038/embor.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellegrini M, Di Siena S, Claps G, Di Cesare S, Dolci S, Rossi P, Geremia R, Grimaldi P. Microgravity promotes differentiation and meiotic entry of postnatal mouse male germ cells. PLoS One. 2010;5:e9064. doi: 10.1371/journal.pone.0009064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki A, Schoenmakers S, Baarends WM. DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics. 2010;5:255–266. doi: 10.4161/epi.5.4.11518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.