Abstract
The unique physico-chemical properties of gold nanoparticles (AuNPs) find manifold applications in diagnostics, medicine and catalysis. Chemical synthesis produces reactive AuNPs and generates hazardous by-products. Alternatively, plants can be utilized to produce AuNPs in an eco-friendly manner. To better control the biosynthesis of AuNPs, we need to first understand the detailed molecular response induced by AuCl−4 In this study, we carried out global transcriptome analysis in root tissue of Arabidopsis grown for 12- h in presence of gold solution (HAuCl4) using the novel unbiased Affymetrix exon array. Transcriptomics analysis revealed differential regulation of a total of 704 genes and 4900 exons. Of these, 492 and 212 genes were up- and downregulated, respectively. The validation of the expressed key genes, such as glutathione-S-transferases, auxin responsive genes, cytochrome P450 82C2, methyl transferases, transducin (G protein beta subunit), ERF transcription factor, ABC, and MATE transporters, was carried out through quantitative RT-PCR. These key genes demonstrated specific induction under AuCl4− treatment relative to other heavy metals, suggesting a unique plant-gold interaction. GO enrichment analysis reveals the upregulation of processes like oxidative stress, glutathione binding, metal binding, transport, and plant hormonal responses. Changes predicted in biochemical pathways indicated major modulation in glutathione mediated detoxification, flavones and derivatives, and plant hormone biosynthesis. Motif search analysis identified a highly significant enriched motif, ACGT, which is an abscisic acid responsive core element (ABRE), suggesting the possibility of ABA- mediated signaling. Identification of abscisic acid response element (ABRE) points to the operation of a predominant signaling mechanism in response to AuCl−4 exposure. Overall, this study presents a useful picture of plant-gold interaction with an identification of candidate genes involved in nanogold synthesis.
Keywords: Au, microarray, Arabidopsis, glutathione, plant hormone, secondary metabolism, transporters, ABA responsive element (ABRE)
Introduction
Gold, one of the least reactive metal in periodic table, possesses unique properties, namely, high free electron density, malleability and conductivity, and favors opportunities to produce stable and tuneable AuNPs for potential applications in diagnostics, biological imaging, sensors, therapeutic agent delivery, photodynamic therapy, catalysis, electronics etc. (Chen et al., 2008; Yeh et al., 2012; Spivak et al., 2013). Many chemical methods have been developed for the synthesis of AuNPs, but they produce hazardous toxic by-products, posing serious environmental health issues (Limbach et al., 2007). Alternatively, plant-based systems of nanomaterial synthesis were established recently to produce a range of AuNP shapes and sizes (Kumar and Yadav, 2009; Starnes et al., 2010; Geetha et al., 2013). Starnes et al. (2010) demonstrated in planta engineering of novel shapes in alfalfa by changing growth conditions of the plant. One of the current studies in Arabidopsis shows monodisperse spherical AuNPs formations in roots, grown hydroponically in the presence of 10 ppm of KAuCl4 (Jain et al., 2014).
Metallic gold with zero nutritive value does not cause toxicity to living organisms but higher concentrations of gold solutions (KAuCl4 or HAuCl4) may cause toxicity and impact plant growth negatively (Sharma et al., 2007; Starnes et al., 2010). Like other toxic heavy metals, for example, Cd, As, Hg, Pb, Au deactivates the proteins by binding to the sulfhydryl groups or disrupting the disulphide bonds and displacing the essential metal ion cofactors (Niemietz and Tyerman, 2002; Rodriguez et al., 2007).
A few investigations into the response of microorganisms to gold exposure have also been reported. For instance, Reith et al. (2009) characterized the mechanism of gold biomineralization in bacteria using a custom made microarray chip. Metal resistance genes, oxidative stress related genes and methyl transferases were reported to differentially express and cause reduction and detoxification of AuIII-complexes (Reith et al., 2009). Another interesting study demonstrated the involvement of a non-ribosomal small peptide—secreted as a secondary metabolite for the generation of solid gold particles from a gold resident bacterium Delftia acidovorans (Johnston et al., 2013).
Unlike the sufficient molecular information available with respect to heavy metals, little information is available in response to AuCl−4 exposure at whole transcriptome levels in plants (Taylor et al., 2014). Thus, the underlying mechanisms involved during the synthesis of AuNPs in plants remain largely elusive. To the best of our knowledge, the present study may be the first report of identifying the genes and mechanism operating in response to AuCl−4 treatment. This study will also help in predicting the changes in the associated metabolic networks.
In this study, 12 day old Arabidopsis root was exposed to chloroauric acid (HAuCl4) for 12 h, devoid of nutrient media, to record the specific response of Au. The principal finding of this study demonstrates that Arabidopsis senses the gold treatment as a strong stimulus, modulating expression of a total of 704 genes, which account for 2.5% of the whole transcriptome. The ABA- mediated signaling and glutathione binding appeared to be major possible mechanisms operating in response to AuCl−4.
Materials and methods
Plant material, growth conditions and treatments
Eighty seeds of the Arabidopsis thaliana cv. Col-0 were sterilized and germinated on mesh in each magenta box hydroponically in 0.5X MS medium for 5 days under a 16 h light (120 μmol m−2 s−1)/8 h dark photoperiod at 23°C (Jain et al., 2009). Thereafter, these seeds were transferred into 1X MS medium and grown for 7 days (Jain et al., 2009). The 12 day old seedlings were transferred to a 10 ppm AuCl−4 (HAuCl4, Sigma-Aldrich, USA) solution for 12 h at pH 4.2, in absence of nutrient medium. Another set of parallel experiments for control were carried out with distilled water at pH 4.2. Care was taken to ensure that only the roots came in contact with the 10 ppm gold solution. After 12 h of incubation in AuCl−4, the roots and shoots were separately collected for further experiment. Similarly, the Arabidopsis seedlings were treated with 10 ppm Ag+ [AgNO3; Acros, USA)] and 10 ppm Cu2+ [CuSO4.5H2O; Acros, USA] and the root and shoot tissues were collected for further experiments. Three independent biological replicates were used for each treatment and around 240 seedlings were used in each biological replicate to isolate RNA.
Total RNA isolation
Total RNA was isolated from frozen root and shoot tissues separately using RNeasy Plant Mini Kit with on column DNase digestion (Qiagen, USA), according to the manufacturer's instructions. The integrity of RNA was checked on agarose gel and concentrations were determined using Nanodrop (Wilmington, DE, USA).
Transcriptome analysis
Affymetrix GeneChip® ara ST 1.0 GeneChips (.cel files submitted to GEO, accession number GSE55436) were used to carry out the microarray experiment. Target preparation, hybridization to arrays, washing, staining, and scanning were carried out following the instructions of the manufacturer (Affymetrix, USA). Three independent biological experiments were carried out for control (no treatment) and experimental (AuCl−4 treated) Arabidopsis seedlings. The hybridization data was processed using the Affymetrix GeneChip Command Console Software (AGCC version 3.2.4). Image files were analyzed to generate the probe intensity (.cel) files, using the default settings of AGCC. Microarray experiments were performed at the Microarray Core Facility of the University of Kentucky, in Lexington, Kentucky, USA. The probe set summarization (CHP) files for gene and exons were generated from feature intensity (.cel) files using RMA algorithms provided in the Expression Console v. 1.3. software. To identify statistically significant differentially expressed genes (DEGs) or exons, a combined criterion of greater than two-fold change was adopted with the ANOVA p-value (Condition unpair) <0.05 in the analysis. The normalized CHP files were analyzed using the secondary analysis tool Transcriptome Analysis Console, software to generate the.csv files containing DEGs.
Identification of transcription factor
The identification of transcription factors were carried out using a web based tool, a transcription factor database (http://arabidopsis.med.ohio-state.edu/AtTFDB/). A separate list of transcription factors with their sub-family name and synonyms was prepared and listed in Table 2.
GO enrichment analysis of DEGs
The singular enrichment analysis (SEA, http://bioinfo.cau.edu.cn/agriGO/index.php) was carried out on up- and downregulated genes. The TAIR AGI IDs with their affiliated GO annotations were subjected to agriGO analysis using the customized annotation mode. Arabidopsis TAIR 10 database was used as a background. The hieratical graphs and GO enrichment analysis table of DEGs were prepared, using a hypergeometric test with Bonferroni correction, at P-value 0.05, with 5 minimum number of mapping entries with complete GO gene ontology. The GO abundance chart was prepared manually by selecting the biologically relevant GO terms.
Quantitative RT PCR validation of transcriptome
The first strand cDNA was synthesized from 4 μg of total RNA in 20 μl reaction using Superscript III Reverse Transcriptase (Invitrogen Life Technologies, USA) using oligo(dT)18 primers, according to the manufacturer's instructions. The prepared cDNA was diluted five times and 1 μl (corresponding to 40 ng total RNA) was used to perform quantitative RT-PCR (qRT-PCR) reactions using the SYBR Green (Applied Biosystem) on 7300 Real –Time PCR system (Applied Biosystems). The reaction conditions for PCR and dissociation curves were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s, 58°C–60°C for 1 min and 95°C for 15 s, 60°C for 20 s, 95°C for 15 s, 60°C for 15 s, respectively. To normalize the gene expression, beta tubulin was used as an endogenous control. Wherever possible, most of the primers were designed manually from 3′ UTR of the gene and verified using Oligo Analyzer—1.1.2. The primer sequences are listed in Supplementary Table S1. The qRT-PCR experiment was repeated two times independently and in each experiment, three technical replicates were used. Relative gene expression was calculated using the 2−ΔΔ CT values following Livak's method (Livak and Schmittgen, 2001).
Construction of heat map displaying expression profile of key genes under AuCl−4, AsV, and CdII
To differentiate the AuCl−4 specific response from toxic heavy metals, arsenate (AsV) and cadmium (CdII), 39 genes, selected on the basis of their high expression level (>5-fold) under Au exposure as well as their availability with Cd and As experiment, were analyzed using the Genevestigator perturbation tool (https://www.genevestigator.com/gv/plant.jsp). Log(2)-ratio change values of selected genes under AsV and CdII were obtained from Genevestigator program. A table was created by putting these values with the log(2)-ratio values of AuCl−4 and a heat map was constructed using an online graphing tool, plotly (https://plot.ly/plot). For the detail of the data please refer Supplementary File 4.
Analysis of biochemical pathways
The Plant MetGenMap, a web-based pathway analysis tool, was used to identify significantly changed biochemical pathways on the basis of expression profile data (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi). An input text file was prepared by putting the TAIR AGI IDs, linear fold change value and P-value as per the instruction given in Plant MetGenMap. The file was uploaded as an individual project and analyzed, as per the instructions given in the manual.
Similarly, another text file was created according to the format prescribed in Plant Metabolic Network (http://pmn.plantcyc.org/overviewsWeb/celOv.shtml) and uploaded at the Omics Data Viewer under the analysis, sub-portal of the Plant Metabolic Network.
Over-represented motif analysis in DEGs
The upstream sequences (TAIR 10 Loci Seq-1000 bp) belonging to up- and down-regulated genes were downloaded using the Bulk Data Retrieval tool in a Fasta format and saved as a tab delimited text file format (http://www.arabidopsis.org/tools/bulk/sequences/index.jsp). The search was carried out using the web based Motif finder tool: SCOPE (http://genie.dartmouth.edu/scope/). The parameters such as species, upstream sequence and strand search were selected as Arabidopsis thaliana, fixed 1000 bp and plus and minus strands both, respectively. Additionally, the TAIR Motif analysis tool (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp) was also applied on the same data to further strengthen the results.
Results and discussion
Gold exposure significantly modulates transcriptome
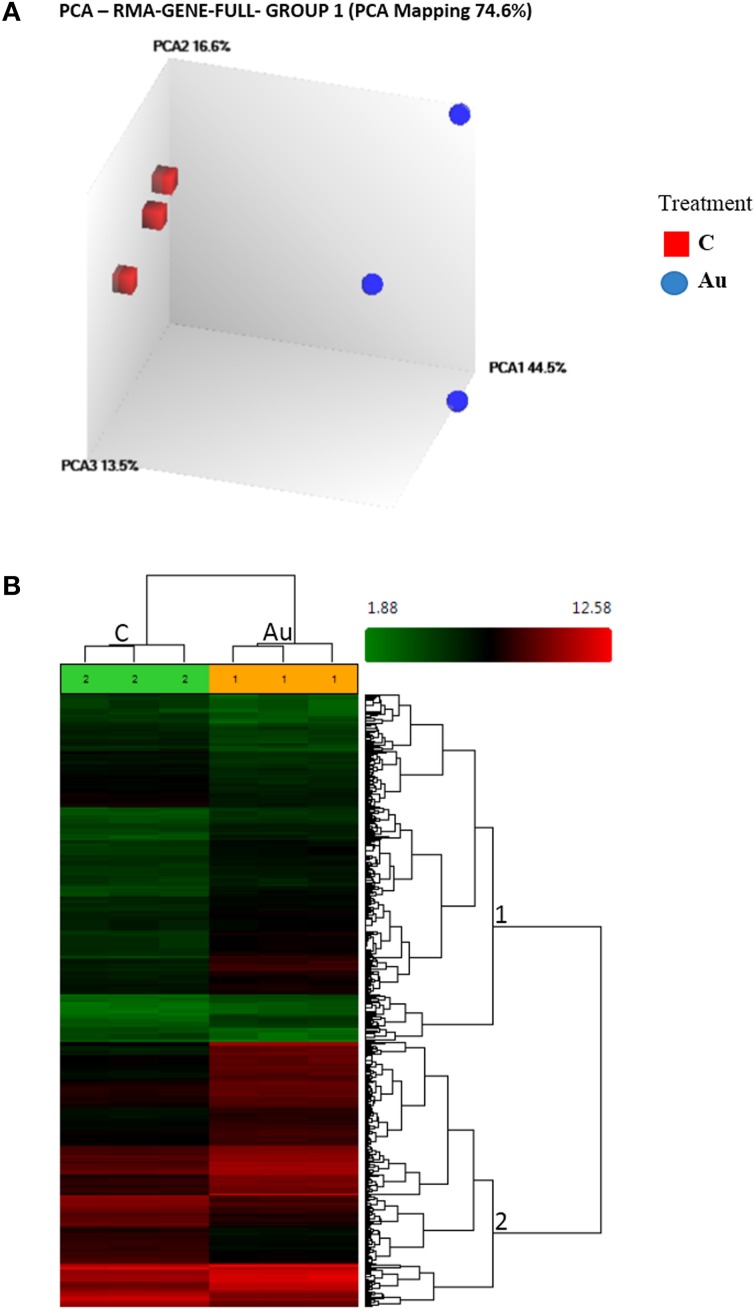
In view of the rapid reduction of Au by sucrose or a nutrient ion like Fe2+, we believe, the use of a HAuCl4 solution devoid of nutrients would give us a precise insight into the interaction of plants with gold ions. Therefore, we isolated RNA from Arabidopsis plants exposed to 10 ppm gold solution at pH 4.2 (the natural pH of a gold solution) and microarray was performed using new unbiased Ara ST 1.0 chip on Affymetrix platform. After normalizing the image files using the RMA algorithm, principal component analysis revealed a distinct position of AuCl−4 treated samples from the control, suggesting a significant change in the transcriptome (Figure 1A). The microarray data was submitted to Gene Expression Omnibus (NCBI), with accession no. GSE55436. A One-Way ANOVA model was applied to the intensity data with adopted criterion: >2-fold change turning into a statistically significant (FDR ≤ 0.16) list of 704 genes (2.5%) and 4900 exons (2.8%) (Figure 1B, Table 1, Supplementary File 1). The high percentage of differentially regulated exons indicated occurrence of a number of alternative splicing events opening the opportunity to study it further (Supplementary File 2). The number of upregulated genes (nos. 492) observed were more than double of the number of downregulated genes (nos. 212), indicating the suitability of the exposed concentration (Table 1, Supplementary File 1).
Figure 1.
Effect of gold (AuCl−4) treatment on the whole transcriptome in Arabidopsis. (A) The Principal Component Analysis (PCA) shows remarkable differences between the control and treated samples. (B) The Hierarchical Clustering of the differentially expressed Transcript Clusters (TCs) under AuCl−4 (Condition1) vs. Control (Condition2). Number on the nodes displays major gene groups based on expression pattern. Color scale shows log2 signal intensity values.
Table 1.
Summary of transcriptome analysis.
| S. No. | Query type | Genes | Exons | |
|---|---|---|---|---|
| 1 | Analysis type | Gene level differential expression analysis | Exon level differential expression analysis | |
| 2 | Array Type | AraGene-1_0-st | ||
| 3 | Genome version | tair10 | ||
| 4 | Annotation file | aragene-1_0-st-v1.na33.3.tair10.transcript.csv | aragene-1_0-st-v1.na33.3.tair10.transcript.csv | |
| aragene1_0-st-v1.na33.2.tair10.probeset.csv | ||||
| 5 | Total number | 28403 | 171941 | |
| 6 | Differentially expressed | 704 | 4900 | |
| 7 | Up regulated (Exp. Vs. Con) | 492 | 2690 | |
| 8 | Down regulated (Exp. Vs. Con) | 212 | 2210 | |
| 9 | Algorithm | One-Way between-subject ANOVA (unpaired) | ||
| 10 | Default filter criteria 1 | Fold change (linear) < -2 or fold change (linear) > 2 | ||
| 11 | Default filter criteria 2 | ANOVA p-value (condition pair) < 0.05 |
General annotation of DEGs in response to AuCl−4
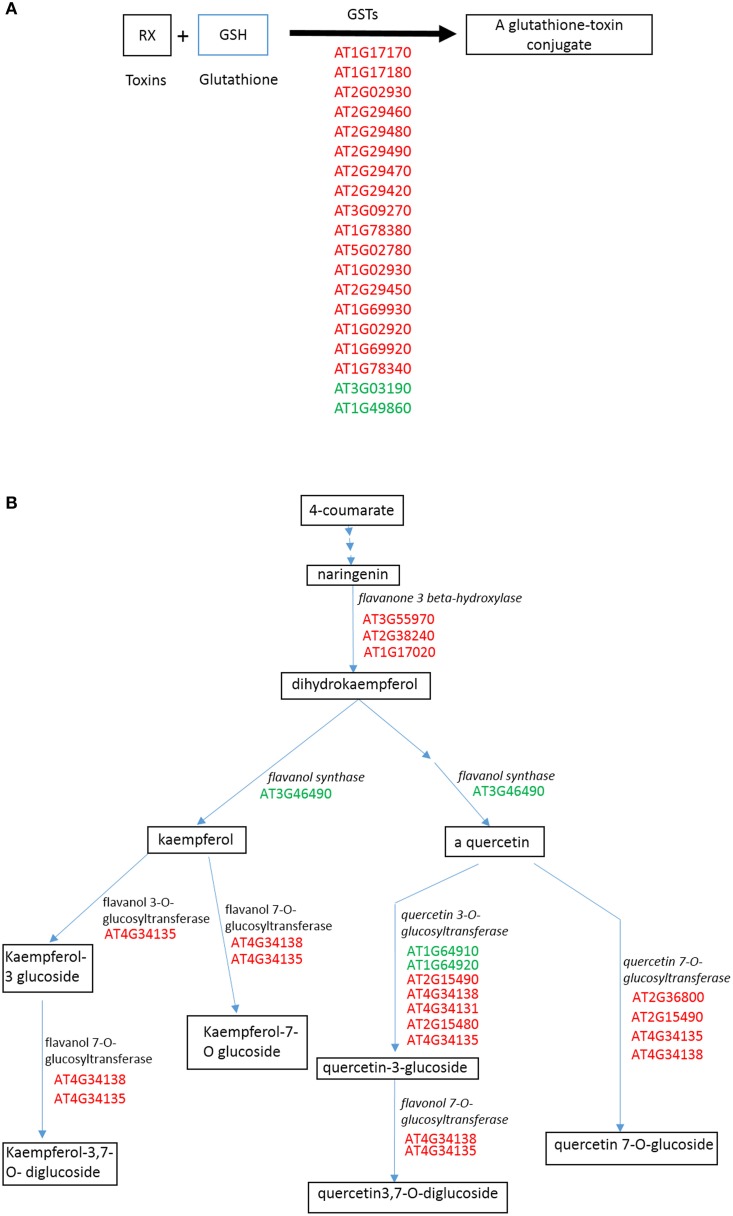
We described the functional roles of genes that shared a similar family and/or category on the basis of available literature and annotations. Glutathione-S-transferases (GSTs) were the major detoxifying enzymes that catalyze the conjugation of reduced glutathione to different cytotoxic substrates and subsequently transport into the vacuole in an ATP-dependent manner. These genes are induced by a number of metabolic processes, as well as in response to stress, heavy metals or plant hormones (auxin/jasmonic acid) (Mars, 1996; Chakrabarty et al., 2009). Additionally, some GSTs also showed glutathione peroxidase activity, strengthening their role in antioxidant metabolism (Mars, 1996). Nineteen members belonging to the GSTs family were differentially regulated (Supplementary File 1). Most of the members belong to GST-tau and GST-phi, GST-lambda class (Wagner et al., 2002). These proteins were classified under multiple GO categories, for example, metabolic process, cellular process, response to stimulus or stress or biotic stimulus, secondary metabolic process, catabolic process and lipid metabolic process (Supplementary File 3). Members of Phi class, were reported to show Co or Cu ion binding activity (Supplementary File 3). An important member of GST phi class, AT1G49860 (GSTF14), was found to be downregulated by 4.4-fold, and involved in anion transport, lateral root development, and root hair cell differentiation (Supplementary File 1). Altogether, these observations suggest a possibility of GSH-based detoxification and reduction of ionic Au species into 0 valent non-ionic, non-reactive form.
Seven members of beta-glucosidase gene family were upregulated at varying levels (Supplementary File 1). The expression patterns of these classes of genes were reported to alter according to the change in the level of sugar inside the cell (Lee et al., 2007). The beta-glucosidase 30 was expressed at the highest level (26-fold) and known for hydrolysing o-glycosyl compounds and modified cell walls (Fujiki et al., 2001). Additionally, deglucosylating activities of these enzymes may regulate the activity of a glucosylated forms of a plant hormone. Three other sugar transporters, members of the ERD6 family, were downregulated (Supplementary File 1). These observations suggest that the gold (AuCl−4) exposure could change the level of sugar inside the cell, triggering sugar associated signaling.
Cytochrome P450 is a stress responsive super gene family, comprising 267 members in Arabidopsis, and plays a variety of roles in biological processes like oxidation-reduction and detoxification of xenobiotics (Chakrabarty et al., 2009; Liu et al., 2010). These proteins share a class under metabolic, cellular process and response to stress or stimulus. Twenty members of this family were differentially regulated, in which 13 were up- and 7 were downregulated (Supplementary File 1). The highest expression was 22.7-fold for cytochrome P450, family 82, subfamily C, polypeptide2 (AT4G31970), which was known to induce under hypoxia condition and shows jasmonate-induced root growth inhibition, electron carrier, oxido-reductase and oxygen binding activity (Liu et al., 2010).
Four auxin response related genes were upregulated, signifying AuCl−4-induced auxin signaling (Supplementary File 1). AT1G60750 locus encodes an auxin-induced atb2-like protein involved in metabolic processes such as oxidation and reduction. AT5G13370, an auxin responsive GH3 family protein, was reported to induce in response to auxin stimulus (Takase et al., 2003). AT1G76520, an auxin efflux carrier protein, possesses auxin-hydrogen symporter activity and is involved in auxin polar transport and localization (Barbez et al., 2012). Recently, in one of our studies we showed the progressive reduction of primary root length and meristematic activity of primary and lateral roots under higher concentrations of AuCl−4 (Jain et al., 2014). Overall, the upregulation of these genes suggest the importance of AuCl−4 mediated-auxin response, which has the capacity to modulate the root system architecture (Jain et al., 2014).
Fourteen different methyl transferases (MTs) were differentially regulated. Six members of S-adenosyl methionine (SAM) -dependent MTs, two members of O-methyl-transferase protein and two others were upregulated (Supplementary File 1). SAM is a well-known methyl donor, suggesting the significance of the methylation event in response to gold. These results are in agreement with an earlier study in which a microorganism exposed to AuCl−4 exhibited upregulation of several MTs, suggesting the possibility of Au methylation as a conserved detoxification mechanism (Reith et al., 2009).
Three members of ABC transporter G family were upregulated (Supplementary File 1). One member, PDR12, was reported to be induced in response to auxin, ABA, ethylene, cyclopentenone, jasmonic, and salicylic acid (Lee et al., 2005; Kang et al., 2010). Additionally, it was known to take part in transportation of Pb, ABA, and terpenoids, revealing some new possibilities with respect to AuCl−4 transport (Lee et al., 2005; Kang et al., 2010).
A large gene family of at least 56 members encoding MATE-related proteins was identified from the Arabidopsis genome (Tiwari et al., 2014). MATE are known to play various roles, such as disease resistance, trafficking of secondary metabolites into the vacuole, and exporting out the small organic molecules like citric acid to sequester metal ions or iron transport and distribution in the plants (Tiwari et al., 2014). In the present study, five members of the MATE gene family were upregulated (Supplementary File 1). Of these, an important gene, ATDTX1, was earlier identified as a detoxifying efflux carrier exporting toxic compounds outside the cell (Li et al., 2002).
Sulfate transporters are shown to be induced under sulfur starvation, as well as against the exposure of heavy metals (Chakrabarty et al., 2009). Sulfur is the vital part of S-rich non-protein thiols like GSH and phytochelatins, known to exhibit heavy metal tolerance and accumulation (Shukla et al., 2012, 2013a,b). Interestingly, the expression of two members of the phytochelatin synthase (PCS) gene family was modulated <2-fold in the present study, indicating the role of PCs in the detoxification of Au (data not shown). In this case, sulfate transporter 3.1 encoding a chloroplast localized transmembrane protein was also upregulated by 6.5-fold (Supplementary File 1), similar to that reported by Cao et al. (2013). Another member, sulfate transporter 3.5 was upregulated by 4.8-fold and known to be involved in root to shoot transport of sulfates. Induction of these transporters may suggest the enhanced requirement of S-containing non-protein thiols against the oxidative stress caused by AuCl−4 exposure.
A phosphate transporter 3.2, which encodes a mitochondrial phosphate transporter 2 (MPT2), reported to be induced in response to ethylene stimulus, was upregulated (Supplementary File 1). Nodulin, a group of MTN21-like transporter family proteins involved in the transport of multiple amino acids, were upregulated (Supplementary File 1). Similarly, a transmembrane amino acid transporter, (AT1G08230), which codes for an H+ driven, high affinity gamma-aminobutyric acid (GABA) transporter localized at plasma membrane, was upregulated (Supplementary File 1). Some putative oligo-peptide transporters (OPTs), known for the transport of peptides/nitrates having the probability of metal binding, were upregulated (Supplementary File 1). Interestingly, two heavy metal associated domain containing transporters, were also up- and downregulated. It appears that the treatment of gold (Au) also upregulated some transporters, which are relatively new or less characterized.
Identification of transcription factors from DEGs
In order to shed light on the regulatory genes modulated by the AuCl−4 treatment, we separated the list of transcription factors from DEGs using an Arabidopsis transcription factor identification web-based tool (http://arabidopsis.med.ohio-state.edu/AtTFDB/). We found a total of 57 transcription factors that were differentially expressed in the present experiment (Table 2). The high number of expressed regulatory proteins in this experiment indicates their important role in transcriptome modulation. Of these, the maximum number (10) was represented by the members of AP2 domain containing ethylene-responsive element binding proteins (AP2-EREBP) and NAC domain containing proteins. Generally, the AP2EREBP gene family was known to be involved in plant responses to drought, cold, salt, and ABA signaling (Mishra et al., 2013, 2014). Interestingly, a member of the AP2-EREBP family (At5g64750), was reported to induce by ABA treatment but functions as a transcriptional repressor of ABA response (Pandey et al., 2005). Members of the NAC gene family were reported to associate with plant senescence (Shibuya et al., 2014). Recently, Christiansen and Gregersen (2014) showed that the NAC TF binds with the CGT motif. It is worth stating that CGT motif shares a part with the abscisic acid responsive core element (ABRE), ACGT. Six members of the R2R3-MYBs gene family were differentially expressed in Arabidopsis; these are known to be involved in abiotic stress response. Five members of the WRKY gene family were differentially regulated as well, of which WRKY70 and WRKY63 were earlier reported to be involved negatively in leaf senescence and ABA response, respectively (Ren et al., 2010; Besseau et al., 2012). WRKY8, and WRKY31 were reported to be involved in jasmonate signaling (Schluttenhofer et al., 2014). WRKY28 is responsive to drought, salt, and oxidative stress (Tripathi et al., 2014). Some other members of different regulatory gene families were also expressed, such as C2H2 zinc-finger, Homeobox HDZIP, bHLH, C3H, Trihelix, C2C2-CO-like and GRAS etc. Overall, a common signaling event associated with these regulatory proteins is senescence. Hence, it may be possible that abscisic acid (ABA) is playing a key role in mediating plant responses against the AuCl−4 challenge.
Table 2.
List of transcription factors regulated during AuCl−4 treatment.
| TF family name | TF locus Id | Protein name | Sub family | Gene name, synonym |
|---|---|---|---|---|
| AP2-EREBP | At1g71520 | NA | ||
| AP2-EREBP | At5g64750 | NA | ABR1 | |
| AP2-EREBP | At2g38340 | NA | ||
| AP2-EREBP | At2g33710 | NA | ||
| AP2-EREBP | At2g40340 | NA | ||
| AP2-EREBP | At1g22810 | NA | ||
| AP2-EREBP | At1g22985 | NA | ||
| AP2-EREBP | At3g23230 | NA | ||
| AP2-EREBP | At3g11020 | NA | DREB2, DREB2B | |
| AP2-EREBP | At4g13620 | NA | ||
| NAC | At1g02230 | NA | ANAC004 | |
| NAC | At1g52890 | NA | ANAC019 | |
| NAC | At5g39610 | NA | ANAC092, ATNAC2, ATNAC6 | |
| NAC | At2g43000 | NA | anac042 | |
| NAC | At3g15500 | NA | ANAC055, ATNAC3 | |
| NAC | At1g77450 | NA | anac032 | |
| NAC | At5g63790 | NA | ANAC102 | |
| NAC | At1g28470 | NA | ANAC010, SND3 | |
| NAC | At4g27410 | NA | ANAC072, RD26 | |
| NAC | At5g39820 | NA | anac094 | |
| MYB | At1g68320 | AtMYB62 | R2R3-MYBs | AtMYB62, BW62B, BW62C, MYB62 |
| MYB | At1g48000 | AtMYB112 | R2R3-MYBs | AtMYB112, MYB112 |
| MYB | At2g47190 | AtMYB2 | R2R3-MYBs | ATMYB2, MYB2 |
| MYB | At5g14750 | AtMYB66 | R2R3-MYBs | ATMYB66, WER, WER1 |
| MYB | At4g01680 | AtMYB55 | R2R3-MYBs | AtMYB55, MYB55 |
| MYB | At4g25560 | AtMYB18 | R2R3-MYBs | AtMYB18, LAF1 |
| WRKY | At4g22070 | AtWRKY31 | NA | AtWRKY31, WRKY31 |
| WRKY | At1g66600 | AtWRKY63 | NA | AtWRKY63, WRKY63 |
| WRKY | At4g18170 | AtWRKY28 | NA | AtWRKY28, WRKY28 |
| WRKY | At5g46350 | AtWRKY8 | NA | AtWRKY8, WRKY8 |
| WRKY | At3g56400 | AtWRKY70 | NA | ATWRKY70, WRKY70 |
| bHLH | At4g25400 | AtbHLH118 | NA | |
| bHLH | At1g62975 | AtbHLH125 | NA | |
| bHLH | At4g20970 | AtbHLH162 | NA | |
| C2C2-CO-like | At4g27310 | NA | ||
| C2C2-CO-like | At5g57660 | NA | COL5 | |
| C2C2-CO-like | At5g54470 | NA | ||
| C2C2-Dof | At1g69570 | NA | ||
| C2H2 | At5g60470 | NA | ||
| C2H2 | At3g28210 | NA | PMZ | |
| C2H2 | At2g21400 | NA | SRS3 | |
| C2H2 | At3g49930 | NA | ||
| C2H2 | At3g05155 | NA | ||
| C2H2 | At3g05160 | NA | ||
| C3H | At5g05530 | NA | ||
| C3H | At5g42200 | NA | ||
| C3H | At1g14200 | NA | ||
| C3H | At5g55970 | NA | ||
| Homeobox | At1g70920 | HD-Zip II | ATHB18 | |
| Homeobox | At3g61890 | AtHB12 | HD-Zip I | ATHB-12, ATHB12 |
| Trihelix | At5g01380 | NA | GT-3a | |
| Trihelix | At1g12040 | NA | LRX1 | |
| MADS | At1g17310 | TypeI | AGL100 | |
| NLP | At2g43500 | AtNLP8 | NA | |
| RAV | At3g25730 | NA | ||
| G2-like | At5g45580 | NA | ||
| GRAS | At3g13840 | AtGRAS16 | NA |
Singular enrichment analysis (SEA) of DEGs
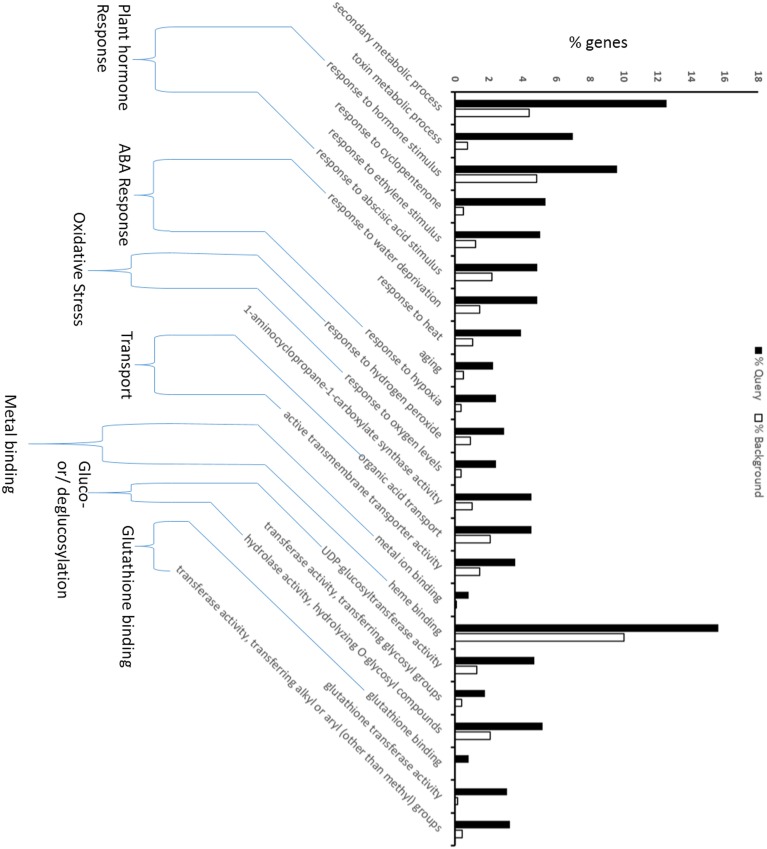
To discern the relevant biological meaning, SEA was performed on the DEGs using an AgriGO analysis tool (Supplementary File 3). Seventy-nine significantly (FDR < 0.04) enriched GO terms were grouped under three functional categories: biological process (BP); cellular component (CC); and molecular function (MF) (Supplementary Figure 1). Of these, 23 GO terms, which were biologically relevant with the present experiment and objective, were displayed in Figure 2. The response of plant hormones, such as cyclopentenone, an intermediate of jasmonic acid, ethylene, and abscisic acid, appears to be upregulated. The ABA associated response, oxidative stress, metal ion binding, transport and glutathione binding were also found to be upregulated. This data suggest that AuCl−4 treatment may trigger ABA specific response, which could potentially modulate the expression of large set of genes. Water deprivation, heat, hypoxia and aging are associated with the ABA-mediated signaling, strengthening the ABA specific response. Moreover, ethylene and ABA do cross talk with each other at several points and together may generate these responses. Toxicity of gold solution may produce oxidative stress similar to other toxic heavy metals, and upregulation of H2O2 response is a sign of oxidative stress. The plant produces non-enzymatic antioxidant molecules such as GSH, ascorbic acid and secondary metabolites to combat the oxidative stress. In this study, glutathione appeared to be a key player for reducing the oxidative stress. A significant upregulation of GST activity opens the possibility of conjugation of GSH with toxic metabolites like ROS, or possibly AuCl−4, to sequester them into the vacuole for detoxification (Figure 2).
Figure 2.
GO enrichment analysis for differentially regulated genes during gold treatment (AuCl−4). The analysis shows the ABA and Glutathione related processes were affected. The Singular Enrichment Analysis (SEA) of up- and downregulated genes with their GO IDs was performed using the agriGO analysis tool under customized mode, P < 0.05 and minimum 5 mapping entries. The black bar shows the fraction of query genes for particular GO term out of total no. of query genes, and the white bar provides the similar information with respect to genes present in background (whole genome). Please see details in Materials and Methods.
Comparative expression profile of key genes under AuCl−4 and toxic heavy metal
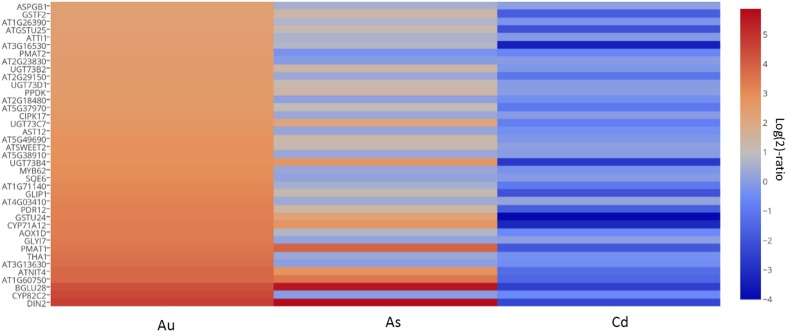
The effect of AuCl−4 on plant gene expression is largely elusive, however, the research on plant-based synthesis of Au nanoparticles has started to show results. In order to differentiate the AuCl−4 specific genetic response from toxic heavy metal(loid)s like arsenic (As) and cadmium (Cd), we constructed a heat map by taking 39 genes on the basis of high expression cut off (>5-fold) under AuCl−4 exposure as well as their presence under Cd and As exposure (Supplementary File 4). In general, all these genes were highly upregulated in the presence of AuCl−4. Contrastingly, they were found to be downregulated in presence of AsV or CdII (Figure 3). These genes showed a significantly distinct expression profile in the presence of AuCl−4, compared to AsV and CdII, suggesting that the interaction of AuCl−4 with plants is different. Since Au ions or nanoparticles possess distinct—physicochemical properties, it may generate a unique signature on the plant transcriptome.
Figure 3.
Expression profile of higly expressed genes demonstrates a distinct Au response in comparison to AsV and CdII. Thirty nine genes, selected on the basis of their expression cut off (>5-fold) under the conditions of Au and heavy metal exposures, demonstrate significant differences in expression patterns under the three conditions. The log(2)-ratio expression values for AsV and CdII downloaded from the known database using the Genevestigator were combined with the values from the present data and a heat map was prepared using an online plot tool. Please refer to the Materials and Methods for more information.
Validation of array data using qRT-PCR under AuCl−4, as well as for Ag and Cu exposure
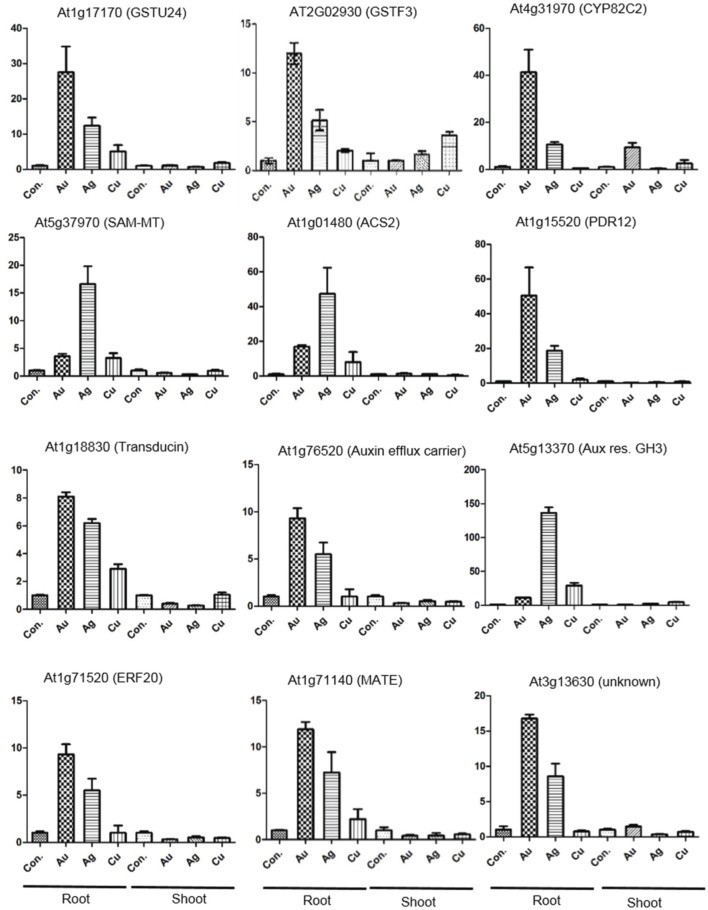
Twelve key genes, were selected on the basis of their possible role in Au- reduction, detoxification, plant hormone responses, signaling, transport, transcriptional regulation and unknown function for the validation of microarray data through quantitative RT-PCR (Figure 4). We performed the qRT-PCR separately for the root and shoot tissues to resolve tissue-specificity. Including Au, we chose metals sharing similar group in the periodic table (i.e., Ag, Cu) to test the metal specificity. The quantitative gene expression results correlated with the trend of regulation observed in the microarray experiment. In general, the expression level of genes was higher in root as compared to shoot (Figure 4). The transporters (PDR-12, MATE), and GSTs were more responsive to AuCl−4, compared to Ag and Cu, indicating probably their role in Au reduction or detoxification (Figure 4). Cytochrome P450 (AT4g31970, CYP82C2) was upregulated significantly in response to AuCl−4, indicating their involvement in detoxification (Figure 4). This enzyme possesses properties like oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (Supplementary File 3). Expression of an auxin efflux carrier (AT1G76520) suggests the triggering of auxin mediated signaling. Expression of ethylene biosynthesis and signaling related genes: ACS2 (AT1G01480) and ERF20 (AT1G71520) points toward the Au-mediated induction of ethylene and ABA signal transduction (Figure 4). Surprisingly, a beta subunit of G protein, named here as transducin (AT1G18830), was upregulated in response to AuCl−4, Ag and Cu in a decreasing order (Figure 4). G protein coupled receptors are known to be involved in ABA reception (Nitta et al., in press). It can be hypothesized that the plant uses an ABA receptor, associated with G protein, and transduces signaling, modulating the expression of a large set of genes for adaptation purpose under AuCl−4 exposure (Yadav et al., 2013, 2014).
Figure 4.
Validation of the array data through qRT-PCR. The expression analysis of the selected genes shows the positive correlation with the microarray data. qRT-PCR was carried out with root and shoot tissues of Arabidopsis exposed to AuCl−4, Ag+, and Cu2+, under similar experimental conditions used for the microarray analysis.
Prediction of plant metabolic shift in response to AuCl−4
To visualize the effect of AuCl−4 in the context of metabolic pathways, we analyzed the DEGs using web-based analysis program, Plant MetGenMap (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi) and Plant Metabolic Network (http://pmn.plantcyc.org). Glutathione mediated detoxification, flavones and derivative biosynthetic, ethylene biosynthetic, cytokinins-O-glucoside biosynthetic, jasmonic acid biosynthetic, and terpenoids pathways were predicted to be significantly changed (Figure 5, Supplementary Figures 2, 3, Supplementary File 5). It appears that the glutathione conjugation was used as a primary tool to cope up with toxicity. Flavones and derivative biosynthetic pathways were predicted to be upregulated. Up-regulation of glucosyltransferases indicates the glucosylation of compounds, which could change the biological activity significantly. Flavones like quercetin and Kaempferol possess antioxidative properties, thus they might reduce the stress effects (Winkel-Shirley, 2002). We predicted glucosylation of cytokinins leading to the inhibition of cytokinin activity (Supplementary Figure 2), as in an earlier study (Hou et al., 2004). Similarly ABA was predicted to be conjugated with glucose, turning it into an inactive form (Supplementary Figure 2). These modifications are often required to maintain a fine tuned equilibrium between active and inactive forms of the plant hormones. Interestingly, lutein (terpenoids) biosynthesis was also predicted to be upregulated. It is reported to have unique medicinal properties and used as an antioxidant (Tian et al., 2004). Overall, these biochemical predictions indicate a fine-tuned regulation of secondary metabolites to address the challenge posed by the AuCl−4 treatment.
Figure 5.
Prediction of changes in plant biochemical pathways on the basis of changed gene expression profile. A web based tool, Plant MetGenMap (http://bioinfo.bti.cornell.edu/cgi-bin/MetGenMAP/home.cgi) was used to predict the changes. (A) Glutathione mediated detoxification of xenobiotics. (B) Flavones and derivative biosynthetic pathway. Red color shows the upregulation of involved genes while green shows down. Please see Materials and Methods for more detail.
Identification of over-represented motifs in upstream sequences of DEGs in response to AuCl−4
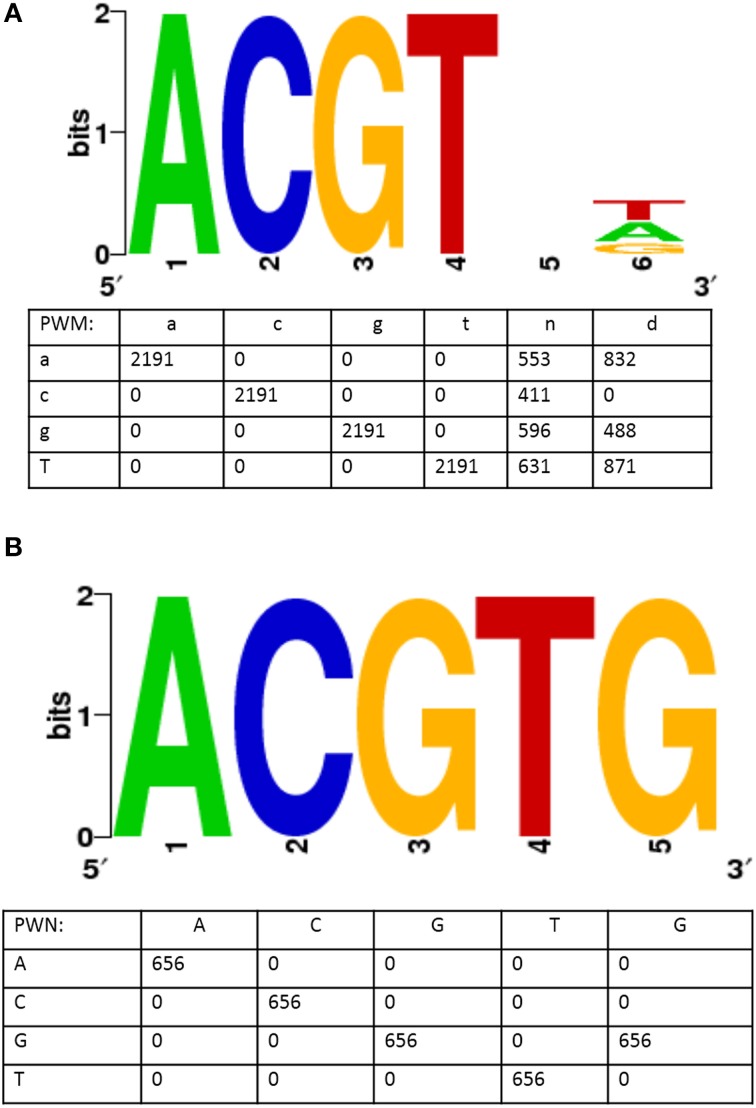
The question of identification of over-represented cis-regulatory elements is important as it sheds light on the regulatory mechanism active in the particular environmental condition. To address this, we pulled out the 1000 bp upstream sequences of the up- or downregulated genes from the Tair10 database and submitted them to the SCOPE motif finder tool (http://genie.dartmouth.edu/scope/) to identify the enriched motifs. We identified an abscisic acid response core element, (ABRE): ACGT, in the 1000 bp upstream sequences of 419 upregulated genes (Table 3, Figure 6). The same motif was also identified by the TAIR motif finder tool (http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp, data not shown). Interestingly, the top two statistically significant motifs represented ABRE cis-regulatory elements (Figure 6). Table 3 describes the general information about the motif. For example, count indicates the number of occurrence of the motif in the input data. Coverage indicates the fraction of genes containing the motif, and the algorithm used to identify the motif.
Table 3.
Summary features of 5 most significant motifs enriched by the SCOPE tool.
| Consensus sequence | Count | Sig value | Coverage (%) | Algorithm |
|---|---|---|---|---|
| Acgtnd | 2191 | 200.2 | 94.3 | PRISM |
| Acgtg | 656 | 73.6 | 69.3 | PRISM |
| Rktcwahv | 840 | 55.0 | 88.9 | PRISM |
| Aatat | 3378 | 51.3 | 98.8 | PRISM |
| Ataaa | 3764 | 48.2 | 99.8 | PRISM |
Figure 6.
ABA Responsive cis-regulatory element (ABRE) identified in upstream region of genes upregulated during AuCl−4 treatment. Figure shows two statistically highly significant consensus sequence generated by a motif finder tool, SCOPE (http://genie.dartmouth.edu/scope/). (A) It shows a motif similar to ABA responsive core element. (B) It shows a motif representing a complete ABA responsive element. The numeric values in the position weight metrix (PWMs) indicates the number of times a particular base present in the respective site.
ABA, known as an abiotic stress hormone of plants, mediates several signaling reactions in response to altered environmental conditions. The motif ACGT was earlier identified in the regulatory regions of the genes induced by environmental related factors. Moreover, ABA response is associated with Ca2+ ion mediated signaling and this motif was reported to be involved in the Ca2+ mediated signaling. Recently, it has been demonstrated that the promoter sequences of Ca2+ ATPases harbors ABA related cis-acting elements (ABRE), indicating ABA mediated Ca2+ ion response (Huda et al., 2013). Taken together, these findings suggest that the AuCl−4 response is mediated through ABA signaling, modulating the large set of genes for the adaptation purposes.
We also identified the enriched represented motifs in upstream sequences of downregulated genes, as shown in Supplementary Figure 4. However, these motifs were unknown and not assigned to any important known regulatory element or part of the signal transduction mechanisms.
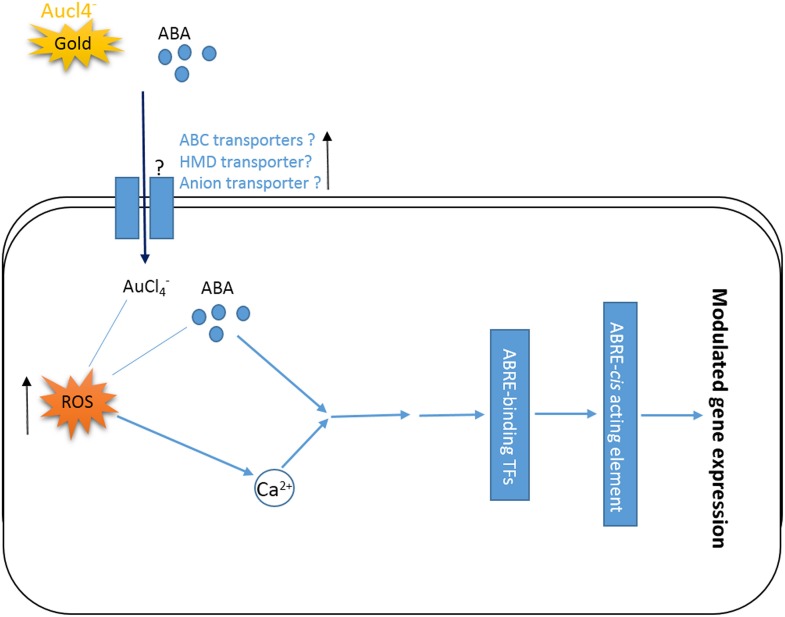
A hypothetical model displaying AuCl−4 signaling
On the basis of the results obtained in the present work and earlier reported information, a hypothetical model was proposed (Figure 7). This model shows how AuCl−4 treatment triggers signals modulating the global gene expression in a short period of time. AuCl−4, like the other heavy metal, may be taken inside the cells through ABC transporters, heavy metal domain containing transporters and/or anion transporters (Chakrabarty et al., 2009; Socha and Guerinot, 2014). ABC transporters are also known for the ABA influx to the plant cells (Fujii, 2014). Both heavy metal (AuCl−4) and ABA, potentially could increase the level of ROS (Pottosin et al., 2014) and alter the Ca2+ level inside the cells, inducing ABRE binding transcription factor (Kaplan et al., 2006; Finkler et al., 2007) and eventually regulating the global gene expression by binding on ABRE element (Galon et al., 2010; Huda et al., 2013).
Figure 7.
A hypothetical model displaying ABA mediated signaling during gold (AuCl−4) exposure. AuCl−4 could trigger ABA mediated signaling. AuCl−4 and or ABA might be taken inside the cells through various transporters and could elevate the level of ROS triggering the changes in Ca2+ level. Eventually ABA and/or Ca2+ may induce ABRE-binding transcription factors through some intermediates and regulate the global gene expression. ↑Shows the upregulation of related genes.
Authors' contributions
Devesh Shukla carried out bioinformatics, analyzed the data and drafted the manuscript. Sneha Krishnamurthy performed the experiments and contributed in data discussion. Shivendra V. Sahi contributed intellectually with all the aspect of this research work and helped in finalizing the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ms. Donna Wall, Staff Analyst, for her help in carrying out microarray experiment at the University of Kentucky. This research was supported by a grant from The National Science Foundation (Award No. MCB-1158507) to Shivendra V. Sahi. Partial financial support from a WKU-RCAP Category I grant to Shivendra V. Sahi is also acknowledged. We acknowledge Dr Nilesh Sharma for critically reviewing the manuscript, and the WKU Biotechnology Centre for providing the core facilities to carry out this project.
Glossary
Abbreviations
- DEGs
Differentially expressed genes
- AsV
Arsenate
- Cd
Cadmium
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00652/abstract
References
- Barbez E., Kubeš M., Rolčík J., Béziat C., Pěnčík A., Wang B., et al. (2012). A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. 10.1093/jxb/err450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M. J., Wang Z., Wirtz M., Hell R., Oliver D. J., Xiang C. B. (2013). SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 73, 607–616. 10.1111/tpj.12059 [DOI] [PubMed] [Google Scholar]
- Chakrabarty D., Trivedi P. K., Misra P., Tiwari M., Shri M., Shukla D., et al. (2009). Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 74, 688–702. 10.1016/j.chemosphere.2008.09.082 [DOI] [PubMed] [Google Scholar]
- Chen P. C., Mwakwari S. C., Oyelere A. K. (2008). Gold nanoparticles: from nanomedicine to nanosensing. Nanotechnol. Sci. Appl. 1, 45–66. 10.2147/NSA.S3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen M. W., Gregersen P. L. (2014). Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J. Exp. Bot. 65, 4009–4022. 10.1093/jxb/eru046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkler A., Ashery-Padan R., Fromm H. (2007). CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. 10.1016/j.febslet.2007.07.051 [DOI] [PubMed] [Google Scholar]
- Fujii H. (2014). Abscisic acid implication in plant growth and stress responses, in Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications, eds Tran L. S. P., Pal S. (New York, NY: Springer; ), 37–54. [Google Scholar]
- Fujiki Y., Yoshikawa Y., Sato T., Inada N., Ito M., Nishida I., et al. (2001). Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by sugars. Physiol. Plant 111, 345–352. 10.1034/j.1399-3054.2001.1110312.x [DOI] [PubMed] [Google Scholar]
- Galon Y., Finkler A., Fromm H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3, 653–669. 10.1093/mp/ssq019 [DOI] [PubMed] [Google Scholar]
- Geetha R., Ashokkumar T., Tamilselvan S., Govindaraju K., Mohamed Sadiq A., Singaravelu G. (2013). Green synthesis of gold nanoparticles and their anticancer activity. Cancer Nanotech. 4, 91–98 10.1007/s12645-013-0040-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B., Lim E. K., Higgins G. S., Bowles D. J. (2004). N–glucosylation of Cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279, 47822–47832. 10.1074/jbc.M409569200 [DOI] [PubMed] [Google Scholar]
- Huda K. M. K., Banu M. S. A., Pathi K. M., Tuteja N. (2013). Reproductive organ and vascular specific promoter of the rice plasma membrane Ca2+ ATPase mediates environmental stress responses in plants. PLoS ONE 8:e57803. 10.1371/journal.pone.0057803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Poling M. D., Smith A. P., Nagarajan V. K., Lahner B., Meagher R. B., et al. (2009). Variations in the composition of gelling agents affect morphophysiological and molecular responses to deficiencies of phosphate and other nutrients. Plant Physiol. 150, 1033–1049. 10.1104/pp.109.136184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Sinilal B., Starnes D. L., Sanagala R., Krishnamurthy S., Sahi S. V. (2014). Role of Fe-responsive genes in bioreduction and transport of ionic gold to roots of Arabidopsis thaliana during synthesis of gold nanoparticles. Plant Physiol. Biochem. 84, 189–196. 10.1016/j.plaphy.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Johnston C. W., Wyatt M. A., Li X., Ibrahim A., Shuster J., Southam G., et al. (2013). Gold biomineralization by a metallophore from a gold-associated microbe. Nat. Chem. Biol. 9, 241–243. 10.1038/nchembio.1179 [DOI] [PubMed] [Google Scholar]
- Kang J., Hwang J. U., Lee M., Kim Y. Y., Assmann S. M., Martinoia E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M. R., Fluhr R., et al. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18, 2733–2748. 10.1105/tpc.106.042713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Yadav S. K. (2009). Plant-mediated synthesis of silver and gold nanoparticles and their applications. J. Chem. Tech. Biotech. 84, 151–157 10.1002/jctb.2023 [DOI] [Google Scholar]
- Lee E. J., Matsumura Y., Soga K., Hoson T., Koizumi N. (2007). Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 48, 405–413. 10.1093/pcp/pcm009 [DOI] [PubMed] [Google Scholar]
- Lee M., Lee K., Lee J., Noh E. W., Lee Y. (2005). AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 138, 827–836. 10.1104/pp.104.058107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., He Z., Pandey G. K., Tsuchiya T., Luan S. (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. 10.1074/jbc.M108777200 [DOI] [PubMed] [Google Scholar]
- Limbach L. K., Wick P., Manser P., Grass R. N., Bruinink A., Stark W. J. (2007). Exposure of engineered nanoparticles to human lung epithelial cells: influence of chemical composition and catalytic activity on oxidative stress. Environ. Sci. Tech. 41, 4158–4163. 10.1021/es062629t [DOI] [PubMed] [Google Scholar]
- Liu F., Jiang H., Ye S., Chen W. P., Liang W., Xu Y., et al. (2010). The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Cell Res. 20, 539–552. 10.1038/cr.2010.36 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) methods. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mars K. A. (1996). The functions and regulation of glutathione-S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–158. 10.1146/annurev.arplant.47.1.127 [DOI] [PubMed] [Google Scholar]
- Mishra M., Kanwar P., Singh A., Pandey A., Kapoor S., Pandey G. K. (2013). Plant omics: genome-wide analysis of ABA Repressor1 (ABR1) related genes in rice during abiotic stress and development. Omics J. Int. Biol. 17, 439–450. 10.1089/omi.2012.0074 [DOI] [PubMed] [Google Scholar]
- Mishra S., Shukla A., Upadhyay S., Sharma P., Singh S., Phukan U. J., et al. (2014). Identification, occurrence, and validation of DRE and ABRE cis-regulatory motifs in the promoter regions of genes of Arabidopsis thaliana. J. Int. Plant Biol. 56, 388–399. 10.1111/jipb.12149 [DOI] [PubMed] [Google Scholar]
- Niemietz C. M., Tyerman S. D. (2002). New potent inhibitors of aquaporins: silver and gold compounds inhibit aquaporins of plant and human origin. FEBS. Let. 531, 443–447. 10.1016/S0014-5793(02)03581-0 [DOI] [PubMed] [Google Scholar]
- Nitta Y., Ding P., Zhang Y. (in press). Heterotrimeric G proteins in plant defense against pathogens and aba signaling. Environ. Exp. Bot. 10.1016/j.envexpbot.2014.06.01119054360 [DOI] [Google Scholar]
- Pandey G. K., Grant J. J., Cheong Y. H., Kim B. G., Li L., Luan S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. 10.1104/pp.105.066324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottosin I., Velarde-Buendía A. M., Bose J., Zepeda-Jazo I., Shabala S., Dobrovinskaya O. (2014). Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. J. Exp. Bot. 65, 1271–1283. 10.1093/jxb/ert423 [DOI] [PubMed] [Google Scholar]
- Reith F., Etschmann B., Grosse C., Moors H., Benotmane M. A., Monsieurs P., et al. (2009). Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U.S.A. 106, 17757–17762. 10.1073/pnas.0904583106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E., Parsons J. G., Peralta-Videa J. R., Cruz-Jimenez G., Romero-Gonzalez J., Sanchez-Salcido B. E., et al. (2007). Potential of Chilopsis linearis for gold phytomining: using XAS to determine gold reduction and nanoparticle formation within plant tissues. Int. J. Phyto. 9, 133–147. 10.1080/15226510701232807 [DOI] [PubMed] [Google Scholar]
- Schluttenhofer C., Pattanaik S., Patra B., Yuan L. (2014). Analyses of Catharanthus roseus and Arabidopsis thaliana WRKY transcription factors reveal involvement in jasmonate signaling. BMC Genomics 15:502. 10.1186/1471-2164-15-502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N. C., Sahi S. V., Nath S., Parsons J. G., Gardea-Torresdey J. L., Pal T. (2007). Synthesis of plant-mediated gold nanoparticles and catalytic role of biomatrix-embedded nanomaterials. Environ. Sci. Tech. 41, 5137–5142. 10.1021/es062929a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K., Shimizu K., Niki T., Ichimura K. (2014). Identification of NAC transcription factor, EPHEMERAL1, that controls petal senescence in Japanese morning glory. Plant J. Cell Mol. Biol. 79, 1044–1051. 10.1111/tpj.12605 [DOI] [PubMed] [Google Scholar]
- Shukla D., Kesari R., Mishra S., Dwivedi S., Tripathi R. D., Nath P., et al. (2012). Expression of phytochelatin synthase from aquatic macrophyte Ceratophyllum demersum L. enhances cadmium and arsenic accumulation in tobacco. Plant Cell Rep. 31, 1687–1699. 10.1007/s00299-012-1283-3 [DOI] [PubMed] [Google Scholar]
- Shukla D., Kesari R., Tiwari M., Dwivedi S., Tripathi R. D., Nath P., et al. (2013b). Expression of Ceratophyllum demersum phytochelatin synthase, CdPCS1, in Escherichia coli and Arabidopsis enhances heavy metal(loid)s accumulation. Protoplasma 250, 1263–1272. 10.1007/s00709-013-0508-9 [DOI] [PubMed] [Google Scholar]
- Shukla D., Tiwari M., Tripathi R. D., Nath P., Trivedi P. K. (2013a). Synthetic phytochelatins complement a phytochelatin-deficient Arabidopsis mutant and enhance the accumulation of heavy metal(loid)s. Biochem. Biophy. Res. Comm. 434, 664–669. 10.1016/j.bbrc.2013.03.138 [DOI] [PubMed] [Google Scholar]
- Socha A. L., Guerinot M. L. (2014). Mn-euvering manganese: the role of transporter gene family members in manganese uptake and mobilization in plants. Front. Plant Sci. 5:106. 10.3389/fpls.2014.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak M. Y., Bubnov R. V., Yemets I. M., Lazarenko L. M., Tymoshok N. O., Ulberg Z. R. (2013). Development and testing of gold nanoparticles for drug delivery and treatment of heart failure: a theranostic potential for PPP cardiology. EPMA. J. 4:20. 10.1186/1878-5085-4-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starnes D. L., Jain A., Sahi S. V. (2010). In planta engineering of gold nanoparticles of desirable geometries by modulating growth conditions: an environment-friendly approach. Environ. Sci. Tech. 44, 7110–7115. 10.1021/es101136q [DOI] [PubMed] [Google Scholar]
- Takase T., Nakazawa M., Ishikawa A., Manabe K., Matsui M. (2003). DLF 2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 44, 1071–1080. 10.1093/pcp/pcg130 [DOI] [PubMed] [Google Scholar]
- Taylor A. F., Rylott E. L., Anderson C. W., Bruce N. C. (2014). Investigating the toxicity, uptake, Nanoparticle formation and genetic response of plants to gold. PLoS ONE 9:e93793. 10.1371/journal.pone.0093793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D. (2004). the Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc. Natl. Acad. Sci. U.S.A. 101, 402–407. 10.1073/pnas.2237237100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M., Sharma D., Singh M., Tripathi R. D., Trivedi P. K. (2014). Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 4:3964. 10.1038/srep03964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi P., Rabara R. C., Rushton P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. 10.1007/s00425-013-1985-y [DOI] [PubMed] [Google Scholar]
- Wagner U., Edwards R., Dixon D. P., Mauch F. (2002). Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 49, 515–532. 10.1023/A:1015557300450 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. 10.1016/S1369-5266(02)00256-X [DOI] [PubMed] [Google Scholar]
- Yadav D. K., Shukla D., Tuteja N. (2013). Rice heterotrimeric G-protein alpha subunit (RGA1): in silico analysis of the gene and promoter and its upregulation under abiotic stress. Plant Physiol. Biochem. 63, 262–271. 10.1016/j.plaphy.2012.11.031 [DOI] [PubMed] [Google Scholar]
- Yadav D. K., Shukla D., Tuteja N. (2014). Isolation, in silico characterization, localization and expression analysis of abiotic stress-responsive rice G-protein beta subunit (RGB1). Plant Sig. Behav. 9:e28890. 10.4161/psb.28890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. C., Creran B., Rotello V. M. (2012). Gold nanoparticles: preparation, properties, and applications in bionanotechnology. Nanoscale 4, 1871–1880. 10.1039/c1nr11188d [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.