Abstract
• Background and Aims If large genomes are truly saturated with unnecessary ‘junk’ DNA, it would seem natural that there would be costs associated with accumulation and replication of this excess DNA. Here we examine the available evidence to support this hypothesis, which we term the ‘large genome constraint’. We examine the large genome constraint at three scales: evolution, ecology, and the plant phenotype.
• Scope In evolution, we tested the hypothesis that plant lineages with large genomes are diversifying more slowly. We found that genera with large genomes are less likely to be highly specious – suggesting a large genome constraint on speciation. In ecology, we found that species with large genomes are under-represented in extreme environments – again suggesting a large genome constraint for the distribution and abundance of species. Ultimately, if these ecological and evolutionary constraints are real, the genome size effect must be expressed in the phenotype and confer selective disadvantages. Therefore, in phenotype, we review data on the physiological correlates of genome size, and present new analyses involving maximum photosynthetic rate and specific leaf area. Most notably, we found that species with large genomes have reduced maximum photosynthetic rates – again suggesting a large genome constraint on plant performance. Finally, we discuss whether these phenotypic correlations may help explain why species with large genomes are trimmed from the evolutionary tree and have restricted ecological distributions.
• Conclusion Our review tentatively supports the large genome constraint hypothesis.
Keywords: Evolvability, nucleotype, genome size, ecology, evolution, phenotype
INTRODUCTION
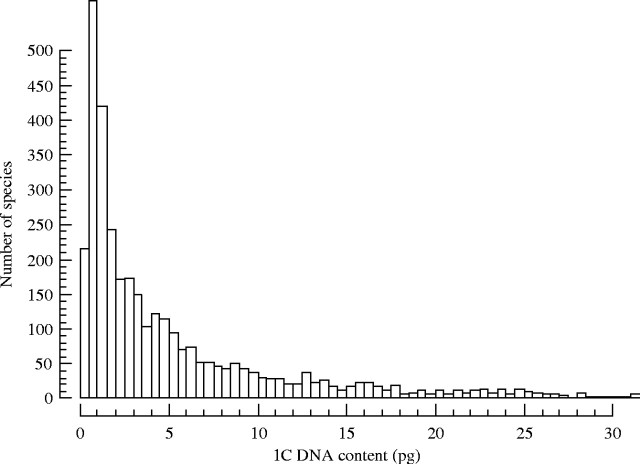
It is well known that there is significant variation in nuclear DNA content, or genome size (GS), within plants (greater than 1000-fold variation, Fig. 1), and eukaryotes in general (greater than 200 000-fold variation). Genome size varies considerably even in very closely related species. However, the evolutionary or ecological significance of this extreme variation is still largely unknown (but see Grime and Mowforth, 1982; Bennett, 1987). Much of this extreme genome size variation in plants is due to non-genic, repetitive DNA, much of which is generated by transposable elements. Given that the number of genes varies much less than GS, it appears that large genomes do not need to be large for any informational reasons. If large genomes are truly saturated with unnecessary ‘junk’ DNA, it would seem natural that there would be costs associated with accumulation and replication of this excess DNA. Here we examine the available evidence to support this hypothesis, which we term the ‘large genome constraint’.
Fig. 1.
Histogram of 3493 angiosperm genome sizes from the Plant DNA C-values database (www.rbgkew.org.uk/cval). Note that this histogram is cut off at 1C DNA content of 30 pg but the full histogram continues out to 127·4 pg.
One extreme of thinking on this issue is to reject the existence of true ‘junk’ DNA (Bennett, 1971; Cavalier-Smith, 1985, 2005). Indeed, a large number of correlations between DNA amount and cellular and physiological characters of clear functional importance is reason to believe that GS variation carries with it functional consequences (see the section on phenotype below). ‘Junk’ DNA may in fact be playing an important role, albeit non-coding in nature, but nevertheless just as important at the phenotypic level.
The other extreme is to suggest that the cost of carrying ‘junk’ DNA is so minimal, even in extreme cases, that there is no noticeable selective consequence. In this case the organism may compensate for GS effects on phenotype until the effects become deleterious. The evidence for this way of thinking comes from the ‘selfish’ nature of most of the ‘junk’ DNA (Doolittle and Sapienza, 1980; Orgel and Crick, 1980), making it more likely that its accumulation has little to do with the organism's fitness. Also, it appears that many organisms can undergo sharp increases in genome size without consequence (polyploidy formation in plants, for example). Here we present evidence to the contrary.
Within plants the distribution of genome sizes is significantly skewed, with decreasing numbers of species for every doubling of genome size (Fig. 1). One way to explain this skew is to suggest that increases in genome size are rare and that only a few lineages have experienced them. But we know that transposable elements are ubiquitous in plants and polyploidy is exceedingly common (Wendel, 2000). Both of these processes operate rapidly on evolutionary time scales. Certainly it appears that there has been enough time and ample means for all plant genomes to become large. But they are not, which in itself may provide the best argument that unchecked genomic enlargement carries maladaptive consequences.
It is also possible that genome shrinkage is a powerful and common process and can counteract the many mechanisms for genome growth (Petrov, 2002). There is evidence for genome size shrinkage and we know of mechanisms capable of causing such reduction (Petrov et al., 1996; Kirik et al., 2000; Orel et al., 2003; Bennetzen et al., 2005). But, unless one invokes selection favouring genome size reduction, it is not clear why, over time, more species would end up having small genomes. So, maybe there is selection acting against organisms with large genomes after all? However, in the case of plant species with large genomes, perhaps such selection was weak and could not stem sharp genome size increases perpetuated by fast and powerful forces of DNA addition.
In this paper we discuss the hypothesis that lineages with large genomes do pay costs. First, we may be able to detect that cost by examining the evolution of species with large genomes. Vinogradov (2003) found that species with large genomes are less likely to generate progenitor species, either through decreased speciation rates or increased rates of extinction. We re-examine this result using quantile regression analyses to show that the negative trend between the number of species in a genus and the average genome size of a genus is driven by a more significant negative relationship for genera with the largest genome sizes. We also test the relationship at higher taxonomic levels using Magallon and Sanderson's (2001) molecular clock and fossil record corrected estimates of diversification rates for several angiosperm lineages. Combined with Vinogradov's (2003) observations, these results may at least partly explain the skewed distribution of plant genome sizes.
We further explore possible ecological constraints on the distribution and abundance of species with large genomes. We find that species with large genomes are restricted to less stressful environments with longer growing seasons. Once again, careful statistical analyses are necessary to pick up trends at the edges or boundaries of these complex bi-variate distributions. These ecological constraints, combined with strong positive correlations with seed mass, may lead to species with large genomes having smaller effective population sizes, which in turn may lead to the higher probabilities of extinction in general and ‘mutational meltdown’ scenarios (Lynch et al., 1993) in particular.
Ultimately, if these ecological and evolutionary constraints are real, the genome size effect must be expressed in the phenotype and confer selective disadvantages. Therefore, we review the available data on the physiological correlates with genome size variation, present new analyses involving maximum photosynthetic rate and specific leaf area, and discuss whether these relationships help explain why species with large genomes are trimmed from the evolutionary tree and have restricted ecological distributions.
The paper is divided into three sections discussing genome size effects on (1) evolution, (2) the distribution and abundance of species (ecology), and (3) phenotype. While we present some new analyses, this paper is intended primarily to define concepts and to review the large body of research in these areas. Fitting with this purpose we combine our methods, results and discussion into one section for each of the three topics.
EVOLUTION
Recently Vinogradov (2003) reported a negative correlation between the number of species in a genus and the average GS of that genus, suggesting a genome size constraint on evolvability. Vinogradov (2003) also consulted a conservation database and found that species listed as rare or endangered tended to have larger genomes (rarity status was determined both locally and world-wide by the United Nations Environment Program World Conservation and Monitoring Center, UNEP-WCMC). Both of these lines of evidence suggest that large genomes are maladaptive at the species level, and reduce the abundance of species with large genomes.
To further probe this interesting relationship, we re-tabulated the data on the genus-level diversity and the genus-average genome size. We used the Plant DNA C-values database compiled at the Royal Botanical Gardens at Kew by Bennett et al. (2001; www.rbgkew.org.uk/cval) to acquire the average GS for 761 genera. We related these values to the number of species in each genus using the database compiled by Mabberley (1999). While genus is a somewhat arbitrary grouping, it is currently the best system available for this type of anaylsis. Some genera have been studied more than others, perhaps leading to specious genera as a result of investigative effort. In addition, taxonomic ‘splitters’ and ‘lumpers’ abound. However, by studying 761 genera we expect that errors introduced by these effects are random.
Similar to Vinogradov (2003), we found a very weak negative Spearman's rank correlation (r = −0·065, one-tailed P = 0·036). However, for our data, the relationship is only significant with a one-tailed test. A randomization test also showed the presence of a weak negative correlation of similarly marginal statistical significance (3·2 % of randomized samples generated as strong, or stronger negative correlation as the data). It is noteworthy that the correlation coefficient in Vinogradov (2003), although also negative and weak (r = −0·11; P < 0·001), was nevertheless stronger than ours. Perhaps the source for this discrepancy involves our use of Mabberly (1999) to tabulate the number of species in a genus while Vinogradov used the International Plant Names Index. These differences notwithstanding, either analysis shows at best a weak relationship between genus-level diversity and GS (Fig. 2A).
Fig. 2.
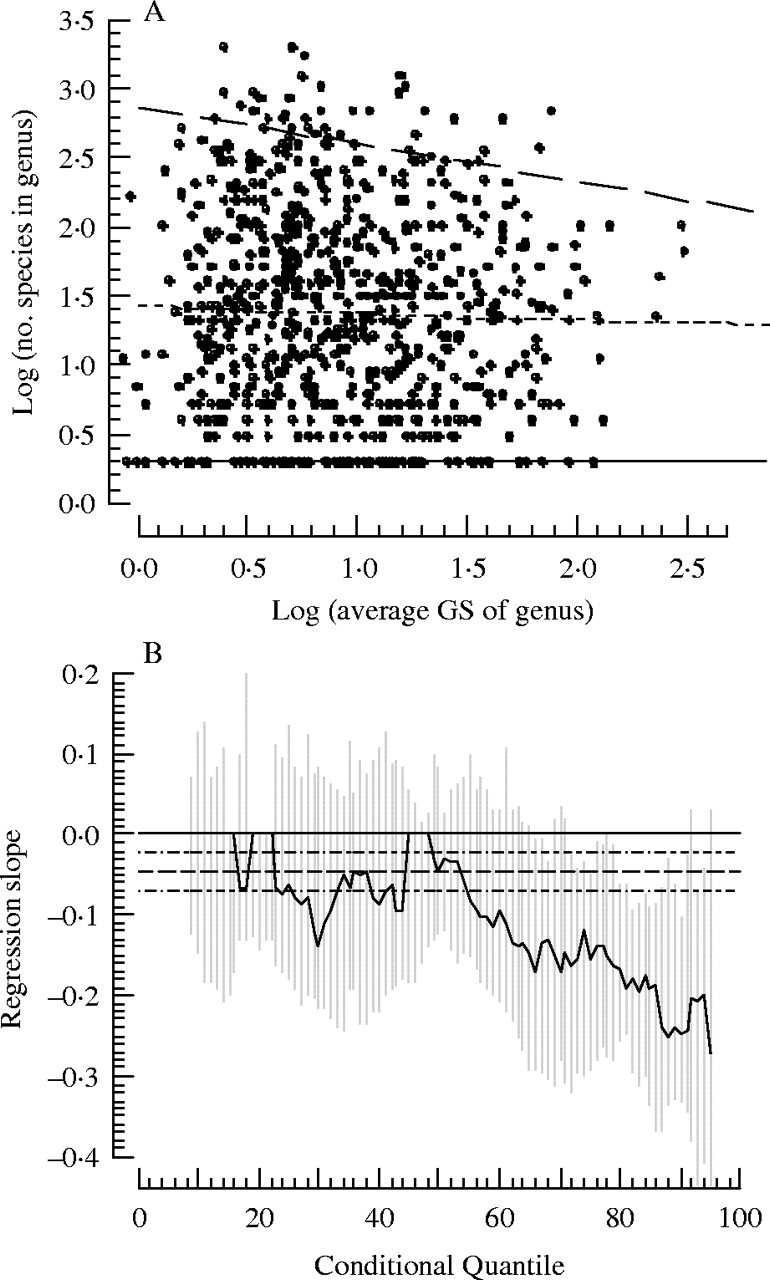
(A) Scatter plot of the log of the average genome size (1C gigabase pairs) of a genus versus the log of the number of species in that genus. (B) Quantile regression analysis of (A) showing a decreasing regression slope with increasing quantiles. The lines in (A) correspond to the 5th quantile (thin solid line), the 50th quantile (thin dashed line) and the 95th quantile (thick dashed line). The lines in (B) correspond to the least squares estimate for the linear relationship between x and y in (A) (dashed line) and the confidence interval of that estimate (double-dashed line). The grey bar surrounding the quantile dependent regression slope estimate is the standard error of the estimate.
We wanted to further explore these data to address the model of a large genome constraint on evolvability more specifically. The correlation statistics employed above test for the existence of a relationship through the means (or centre) of the data distribution. We have a more specific model. Our hypothesis suggests that there should be little signal for species with small GS. In those cases, many other determinants of diversification rates may come into play. More specifically, we would like to test whether species with large or very large genomes are less likely to attain high diversification rates and whether they are more likely to exhibit higher extinction rates.
To test this more specific model, we employed quantile regression analyses. Each quantile regression estimate involves every point in a bi-variate data distribution, but the points above the regression line are weighted by the quantile (for instance 0·65 for the 65th quantile) while the points falling below the regression line are weighted by one minus the quantile (corresponding to 0·35 for the same 65th quantile). The 65th quantile regression also implies that 65 % of the observations fall below the regression line while 35 % of the observations fall above the line. The 50th quantile regression estimate is the same as the traditional least-squares regression estimate with the condition that equal numbers of points fall above and below the line (here both groups are equally weighted by 0·5). Because of the quantile-dependent partitioning of data points above and below the regression line, quantile regression is a non-parametric technique. Results for log-transformed or untransformed data give identical results. Koenker and Bassett (1978), Cade and Richards (1996), Cade et al. (1999), Cade and Guo (2000), Koenker and Hallock (2001), Knight and Ackerly (2002) and Cade and Noon (2003) provide detailed discussion of quantile regression methods.
Our quantile regression results (Fig. 2) show that the weak negative trend for standard correlation coefficients must be due to the increasingly negative trend in the larger quantiles. Quantile regression estimates between the 5th and 70th quantile are not significantly different from zero. However, quantile regression estimates between the 70th and 95th quantile are negative and significantly different from zero. The negative relationships in these quantiles (corresponding to the species with larger GS) must drive the statistical significance of Spearman rank correlations. These results provide support for the model of large genome constraint. Not only does it show that much of the signal is in the low diversity of large GS genera, but also that large genomes can reduce diversity quite substantially.
Despite the apparent success in the quantile regression statistics for pin-pointing the cause of the negative relationship, there are several potential sources of error. What we are really after is the genome size of the ancestral species that either did or did not diversify. Whether or not the average genome size of extant species captures this ancestral value is not clear. The average genome size of a genus often does not take into consideration every species in the genus and some genera are under-represented compared to other genera, both for sampling reasons (i.e. in some cases, such as Atriplex, only 10 out of 300 species have been measured) or for taxonomic reasons. For example, three of the four Milium species have been estimated, but with a maximum possible sample size of four there is little confidence in the ancestral genome state. But removing these lineages from the analyses would bias results towards high-diversity clades. In addition, different genera may have been diversifying for very different amounts of time. Ideally one could perform an independent contrast test with a complete molecular-clock-corrected genus-level phylogeny. This type of phylogeny is one of the last gaps in the Angiosperm Phylogeny Working Group's (APG) endeavour to produce a comprehensive phylogeny for angiosperms (The Angiosperm Phylogeny Group, 2003). Historically, individual investigators have focused on molecular phylogenies within genera. Recently, family-level phylogenies have received considerable interest (The Angiosperm Phylogeny Group, 2003). The placement of genera is still uncertain. In the future, independent contrast tests for the relationship between genome size and the number of species in a genus may ascertain whether the relationship is truly robust.
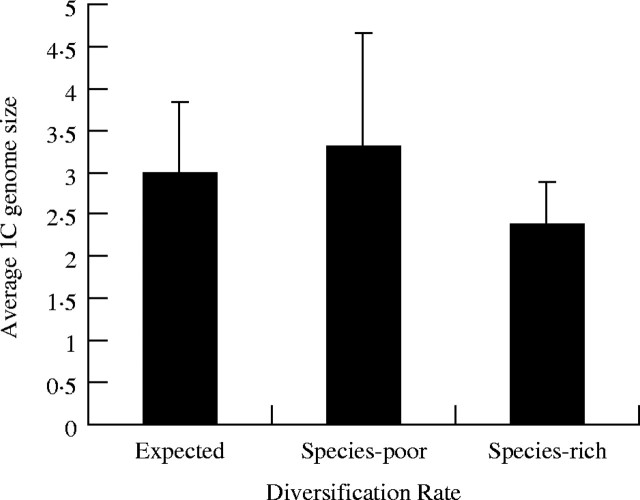
We can, however, use a family-level phylogeny where the diversification rates have been estimated using a molecular clock and the fossil record (Magallon and Sanderson, 2001). The Magallon and Sanderson (2001) study provided estimates of diversification rates for several major angiosperm groups. They identified ten lineages that were diversifying significantly faster, 13 lineages that were diversifying significantly slower, and 17 lineages classified as having the expected diversity based on their age. We tabulated the GS for these groups and found no significant GS effect for diversification rate (Spearman's rank correlation, r = 0·10, P > 0·05, Fig. 3). The lineages that were identified as species-rich did have lower GS compared to both the species-poor clades and the clades diversifying at the expected or average rate. However, these results were also not statistically significant. It is possible that the GS effect is too weak to be picked up in such a small dataset (40 lineages). In addition, family-level means may saturate any signal for potential causative effect on diversification rates. Molecular-clock and fossil-record-corrected phylogenies at the genus level should help disentangle some of these ambiguities.
Fig. 3.
Average 1C genome size (gigabase pairs) for angiosperm clades identified by Magallon and Sanderson (2001) as being species-rich or species-poor compared with the expected diversity based on molecular clock and fossil record corrected estimates of the age of clades (compared to the rest of the angiosperms).
Self-incompatibility may be one of the factors favouring persistence and diversification of lineages because of fitness advantages to out-crossing (Richards, 1997). In most cases polyploidy breaks down self-incompatibility due to the genetic interaction of diploid pollen grains with the haploid egg (Richards, 1997). Polyploids can buffer the effects of inbreeding better than diploid species because of their increased heterozygosity (Husband and Schemske, 1997; Lande and Schemske, 1985). However, in changing environments self-compatible species may be at a disadvantage. It is estimated that between 47 and 70 % of flowering plants are the descendants of polyploid ancestors (Masterson, 1994). Therefore, if genome size increases are brought about by polyploidy (with associated re-diploidization), perhaps one of the large genome constraints involves the breakdown of self-incompatibility.
Given the analyses presented above, at best we can suggest that there is a tentative GS effect for the generation of plant species diversity that is worthy of further investigation. Lineages with small or average GS may diversify at both fast and slow rates. However, when the analysis is restricted to lineages with the largest GS (the quantile regression analysis) the constraint on diversification rate becomes more pronounced.
ECOLOGY
In an attempt to provide an ecological significance to the pronounced variation in plant genome size (GS), early investigators used altitude and latitude as proxies for abiotic selection pressures putatively acting on GS. A summary of GS/altitude studies reveals that nine found positive correlations, eight found negative correlations, and six were inconclusive or not statistically significant (see Table 1 for references). On first impression it would seem that there is no general relationship between altitude of origin and GS. However, it may be that the trend is not linear, and mean regression statistics across the whole range of environments may not fully capture the relationship. Rayburn (1990) found both positive and negative correlations for 23 populations of Zea mays when comparing their altitude of origin in Mexico. Rayburn's observations suggest that species with large genomes occur at intermediate elevations and species with small genomes tend to occur at both low (sea-level) and high (2440 m) elevations. Perhaps results for altitude have been inconclusive because the real trend is not linear but rather hump-shaped or unimodal.
Table 1.
Previous studies on the relationship between genome size with altitude (Alt.) and latitude (Lat.)
| Variable |
Correlation |
Level |
Description |
Authors |
|---|---|---|---|---|
| Alt. | + | Pop. | 15 Dasypyrum villosum in Italy | Caceres et al., 1998 |
| Alt. | + | Sp. | 4 Crambe and 3 Sonchus in Macronesia | Suda et al., 2003 |
| Alt. | + | Pop. | 12 Zea mays southwestern USA | Rayburn and Auger, 1990 |
| Alt. | + | Sp. | Crepis praemorsa | Godelle et al., 1993 |
| Alt. | + | Sp. | Hypocheris | Cerbah et al., 1999 |
| Alt. | + | Sp. | Secale species | Bennett, 1976a; Smith et al., 1976 |
| Alt. | + | Pop. | Teosinte | Laurie and Bennett, 1985 |
| Alt. | + | Sp. | 11 Salix (mountain species with greatest DNA content) | Thibault, 1998 |
| Alt. | − | Sp. | 8 Argyranthemum in Macronesia | Suda et al., 2003 |
| Alt. | − | Pop. | 20 Arachis duranensis in Argentina/Bolivia | Temsch and Greilhuber, 2001 |
| Alt. | − | Pop. | 8 Dactylis glomerata in Spain | Creber et al., 1994 |
| Alt. | − | Pop. | 17 Dactylis glomerata in France and Italy | Reeves et al., 1998 |
| Alt. | − | Pop. | 12 Zea mays in New Mexico | Rayburn, 1990 |
| Alt. | − | Pop. | Corn (knob number) | Mangelsdorf and Cameron, 1942; Longley and Kato, 1965; Wellhausen et al., 1952; Bennett, 1976b |
| Alt. | − | Pop. | 24 Berberis in Patagonia | Bottini et al., 2000 |
| Alt. | − | Pop. | 15 Zea mays | Poggio et al., 1998 |
| Alt. | − | Pop. | 11 Arachis duranensis | Singh et al., 1996 |
| Alt. | ∧ | Pop. | 23 Zea mays Southwestern USA. | Rayburn, 1990 |
| Alt. | ns | Sp | 4 Silene and 5 Micromeria in Macronesia | Suda et al., 2003 |
| Alt. | ns | Pop. | 10 Sesleria albicans European | Lysak et al., 2000 |
| Alt. | ns | Sp. | 21 Artemisia | Torrel and Valles, 2001 |
| Alt. | ns | Sp. | 51 neotropical Lonchocarpus trees | Palomino and Sousa, 2000 |
| Alt. | ns | Pop. | 5 Dactylis glomerata in Slovenian Alps | Vilhar et al., 2002 |
| Alt. | ns | Sp. | 7 Echeandia | Palomino, 1993 |
| Lat. | + | Pop. | Several Picea sitchensis | Miksche, 1967, 1971 |
| Lat. | + | Sp. | Tropical vs. temperate grasses | Avdulov, 1931 |
| Lat. | + | Sp. | 329 tropical vs. 527 temperate plants | Levin and Funderburg, 1979 |
| Lat. | + | Sp. | 17 Poaceae and 15 Fabaceae crops | Bennett, 1976a |
| Lat. | + | Pop. | 24 Berberis in Patagonia | Bottini et al., 2000 |
| Lat. | − | Sp. | 20 Arachis duranensis in Argentina/Bolivia | Temsch and Greilhuber, 2001 |
| Lat. | − | Pop. | Several Festuca arundinacea | Ceccarelli et al., 1992 |
| Lat. | − | Pop. | North American cultivars of Zea mays | Rayburn et al., 1985 |
| Lat. | − | Sp. | 162 British plants | Grime and Mowforth, 1982 |
| Lat. | − | Sp. | 23 Arctic plants | Bennett et al., 1982 |
| Lat. | − | Pop. | 22 North American Zea mays | Rayburn et al., 1985 |
| Lat. | − | Pop. | 11 North American Zea mays | Laurie and Bennett, 1985 |
| Lat. | ns | Sp. | 18 pines | Joyner et al., 2001 |
| Lat. | ns | Pop. | 6 Allium cepa cultivars | Bennett et al., 2000 |
| Lat. | ns | Pop. | Several Picea glauca | Teoh and Rees, 1976 |
| Lat. | ns | Pop. | 10 Dactylis glomerata | Creber et al., 1994 |
| Lat. | ns | Sp. | 11 Tropical vs. temperate Pines | Hall et al., 2000 |
| Lat. | ns | Sp. | 19 Helianthus | Sims and Price, 1985 |
Correlations are either +, −, not significant (ns), or non-linear (∧). Studies were classified into different levels: those dealing with different populations of the same species (Pop.) or multiple species (Sp.).
Results for GS and latitude of origin mirror those for altitude. Five studies found positive, seven found negative, and five found non-significant correlations (see Table 1 for references). Levin and Funderburg (1979) did a discrete test between tropical and temperate species and found that temperate species had nearly double the GS of tropical plants, while Bennett et al. (1982) and Bennett (1987) suggest that angiosperm species with large GS are progressively excluded from northern latitudes—again suggesting a unimodal or hump-shaped distribution for GS across latitudinal gradients, similar to Rayburn's observations for altitude.
Confusion about the relationship between GS and altitude or latitude may arise because factors such as temperature and precipitation, which may more accurately represent selection pressures acting on GS, do not vary linearly with altitude or latitude. In addition, most studies did not consider a full range of elevations or latitudes—from sea-level to mountain tops and from the tropics to the arctic. Finally, even using the correct underlying variables over the whole range of values, we may still find that the relationship is truly non-linear. Results from Rayburn (1990), Levin and Funderburg (1979) and Bennett et al. (1982; Bennett, 1987) suggest that the true relationship between GS and altitude or latitude may more accurately be represented by a unimodal distribution where species with low DNA content may exist at any elevation or latitude but species with the largest DNA contents may be excluded from the extremes.
Knight and Ackerly (2002) confirmed this prediction using GS trends across environmental gradients of temperature and precipitation in the California flora. They used a geographic information system to calculate mean July maximum temperature and annual precipitation inside the geographic range of 401 species in the California flora and compared these values to tabulated measurements of GS for these species (taken from the Plant DNA C-values database). Their findings show that species with large genomes tend to be excluded from extreme environments with shorter growing seasons (high or low July maximum temperatures, or reduced annual precipitation). Similar to the results presented in the evolution section, these results were not obvious with mean regression statistics. The trends only became apparent when quantile regression analyses were applied. In this case, a quadratic quantile regression was used to model the predicted unimodal trend. With increasing quantiles, the quadratic coefficient became more negative, implying increased concavity of the inverted parabolic function. We re-analysed the relationships presented by Knight and Ackerly (2002) with 20 additional species (421 in total). These analyses were not significantly different from the original interpretation (Fig. 4), and again support the hypothesis that species with large genomes are progressively excluded from habitats with extreme July maximum temperatures and decreased annual precipitation. Qualitatively, results were similar when performed with basic genome size (2C values divided by ploidy level) and 1C DNA contents.
Fig. 4.
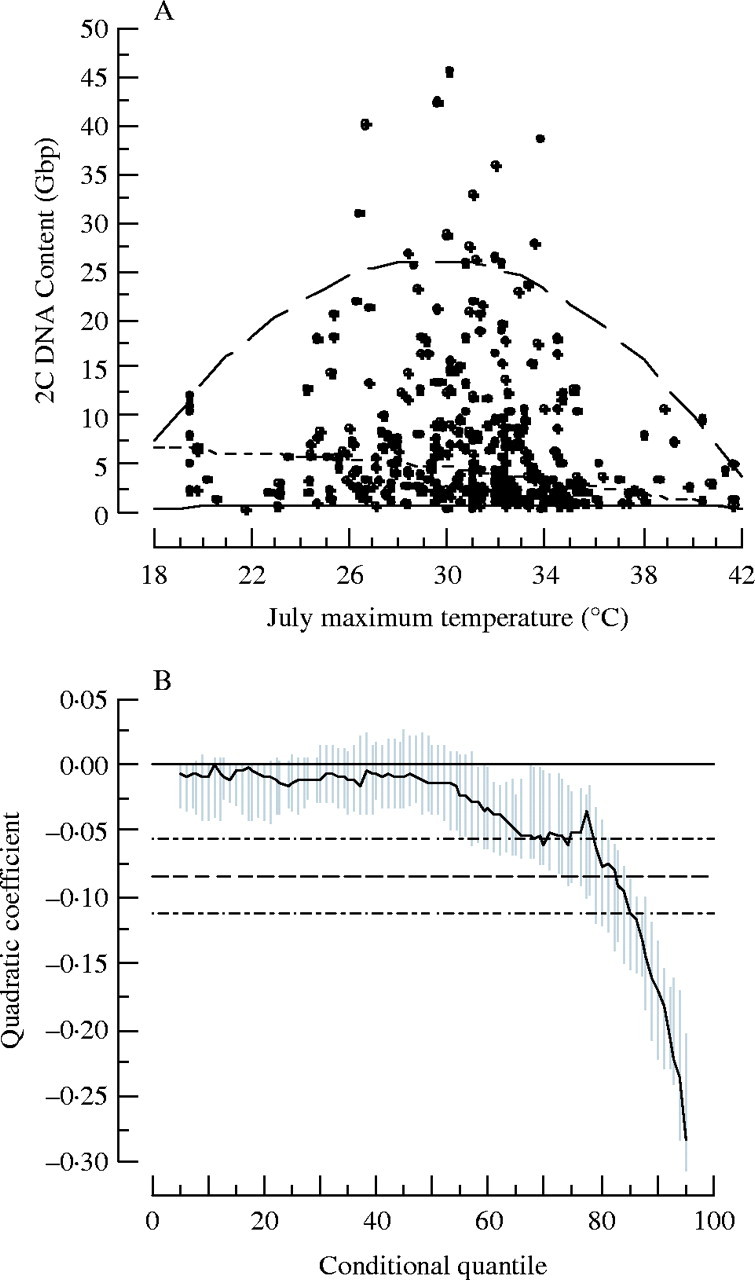
(A) Scatter plot of the mean July maximum temperature inside the range of 421 species in the California flora versus the mean 2C DNA content of those species in gigabase pairs (Gbp). (B) Quantile regression analysis of (A), showing a decreasing quadratic coefficient for increasing quantiles. The lines in A correspond to the 5th quantile (thin solid line), the 50th quantile (thin dashed line) and the 95th quantile (thick dashed line). The lines in (B) correspond to the least-squares estimate for the normal mean quadratic function for the relationship depicted in (A) (dashed line) and the confidence interval of that estimate (double-dashed line). The grey area depicts the quantile dependent confidence interval for the quadratic coefficient.
Other investigators have used mean temperatures inside species' geographic ranges as a correlate with GS (see Table 2 for complete references). Suda et al. (2003) found both positive and negative relationships in the Macronesian flora with mean annual temperature. Turpeinen et al. (1999) found a positive correlation between GS and mean January temperatures in populations of wild barley in Israel, and Wakamiya et al. (1993) found a negative correlation in pines for the highest spring mean monthly air temperature (Table 2). Combining geographic information system (GIS) analyses with species-specific plant functional traits, such as genome size, should continue to be a fruitful endeavour for the analysis of putative abiotic selection pressures operating on GS.
Table 2.
Previous studies on the relationship between genome size and temperature
| Correlation |
Level |
Description |
Authors |
|---|---|---|---|
| +a | Sp. | 8 Argyranthemum, 4 Silene and 5 Micromeria in Micronesia | Suda et al., 2003 |
| +b | Pop. | 10 Hordeum spontaneum in Israel | Turpeinen et al., 1999 |
| +a | Pop. | Several Festuca arundinacea | Ceccarelli et al., 1992 |
| +d | Pop. | 45 Dactylis glomerata | Bretagnolle and Thompson, 1996 |
| −c | Pop. | Herbaceous plants in UK | MacGillivray and Grime, 1995 |
| −a | Sp. | 4 Crambe and 3 Sonchus in Micronesia | Suda et al., 2003 |
| −e | Sp. | 18 North American pines | Wakamiya et al., 1993 |
| −f | Pop. | 7 white clover | Campbell et al., 1999 |
| −j | Pop. | 8 maize populations | McMurphy and Rayburn, 1991 |
| −g | Sp. | 131 British angiosperms | Thompson, 1990 |
| −f | Sp. | 30 Grasses and forbs in UK | Grime et al., 1985 |
| −f | Sp. | 24 UK geophytes and grasses | Grime and Mowforth, 1982 |
| −g | Sp. | 43 UK angiosperms | Grime et al., 1997 |
| ∧h | Sp. | 401 California Angiosperms | Knight and Ackerly, 2002 |
| nsi | Pop. | 6 Hordeum spontaneum in Israel | Kalendar et al., 2000 |
Correlations are either +, −, not significant (ns), or non-linear (∧). Studies were classified into different levels: those dealing with different populations of the same species (Pop.) or multiple species (Sp.). Footnotes a–j describe the temperature variable used.
Mean annual temperatures.
mean January temperature
lowest temperature survived (frost tolerance) – higher values equals less frost tolerance
ability for winter growth
highest mean monthly spring air temperature
timing of spring growth
minimum germination temperature
July maximum temperature
north-to-south facing transects (individuals on south-facing slope had slightly larger genome)
cold tolerance and frost tolerance.
Other temperature variables have been used as correlates with GS, including (1) the timing of spring growth, (2) germination temperatures, and (3) frost tolerance. Grime and Mowforth (1982) and Grime et al. (1985) measured the timing of spring growth for several species in the UK flora. Species that delayed growth until the warmer spring and summer months tended to have smaller GS. Those that grew early in the spring had larger GS (here recorded as a negative correlation with temperature, Table 2.). Campbell et al. (1999) found the same correlation for populations of white clover. However, Bretagnolle and Thompson (1996) found the opposite relationship for sympatric Dactylis glomerata populations. MacGillivray and Grime (1995) measured frost tolerance and found that species that survived colder temperatures tended to have larger GS (also recorded as a negative correlation with temperature in Table 2). Thompson (1990) and Grime et al. (1997) measured optimal germination temperatures for several species in the UK flora and found that species that germinated at lower temperatures tended to have larger GS (also recorded as a negative correlation with temperature in Table 2).
Grime and Mowforth (1982) suggested that early spring growth is facilitated by large GS and the associated larger cell sizes (see section on phenotype). Their model suggests that growth at low temperatures is facilitated by cell expansion driven by turgor pressure and not cell division. The theory is supported by observations that cell size is positively correlated with GS (Rees et al., 1966; Bennett et al., 1983; Anderson et al., 1985) and the fact that cell division is inhibited at lower temperatures (Haber and Luippold, 1960).
Considering the proposed importance of cell size, turgor pressure and cell expansion for predicting responses of large GS species to low temperatures, it is not surprising that several studies have also found positive correlations between GS and estimates of water availability or mean annual precipitation within species ranges (see Table 3). Knight and Ackerly (2002) reported positive correlations between GS and annual precipitation for 401 species in the California flora. The relationship was steeper and more significant as GS increased (see quantile regression analyses presented above). Price et al. (1981), and Castro-Jimenez et al. (1989) also found positive relationships. Bottini et al. (2000) report a negative correlation, but water availability differed between sites independently of the annual average rainfall. Populations that occurred at sites with high water availability tended to have larger GS (no statistics were presented to support this claim). Other investigators have found negative or inconclusive results. For example, Wakamiya et al. (1993) found a negative correlation between annual precipitation and GS for 18 North American pines. Suda et al. (2003) also found a negative correlation for several species in the Micronesian flora. Sims and Price (1985) found no significant relationship between GS and estimates of mean annual precipitation within the ranges of 16 Helianthus species. Taken together, one might conclude that there is no interaction between annual precipitation or water availability with genome size. However, the sample sizes were small for many of these analyses. In addition, the combined effects of water stress and high temperature stress may amplify putative environmental effects on nuclear DNA content (annual precipitation and July maximum temperature are strongly correlated at the high temperature extreme).
Table 3.
Previous studies on the relationship between genome size and precipitation
| Correlation |
Level |
Description |
Authors |
|---|---|---|---|
| + | Sp. | 5 Microseris bigelovii and 5 M. laciniata | Castro-Jimenez et al., 1989 |
| + | Sp. | 4 Crambe and 3 Sonchus in Micronesia | Suda et al., 2003 |
| +a | Pop. | 24 Microseris douglasii | Price et al., 1981 |
| + | Sp. | 401 California angiosperms | Knight and Ackerly, 2002 |
| − | Sp. | 8 Argyranthemum, 4 Silene and 5 Micromeria in Micronesia | Suda et al., 2003 |
| −b | Sp. | 18 North American Pinus | Wakamiya et al., 1993 |
| −c | Pop. | 24 Berberis in Patagonia | Bottini et al., 2000 |
| ns | Sp. | 19 Helianthus | Sims and Price, 1985 |
Correlations are either +, −, not significant (ns). Studies were classified into different levels: those dealing with different populations of the same species (Pop.) or multiple species (Sp.).
Temporal variation within a single population
lowest mean monthly precipitation
annual rainfall, but there was a positive correlation with water availability (no statistical analyses shown).
Studying six pine species, Wakamiya et al. (1996) found that species with higher turgor-loss points (i.e. greater water stress sensitivity) had larger GS. These species would be less able to survive drought and therefore excluded from areas with decreased water availability. Further studies on turgor-loss point, cell size and DNA content, with measures of water availability, should be performed to further characterize the effects of DNA content on the distribution and abundance of species with large genomes across gradients of precipitation.
PHENOTYPE
Although the causal nature of the large genome constraint outlined in the previous sections is uncertain, it may arise from correlated effects beginning at the cellular level. Here we discuss several well-established cellular correlations with GS: the strong positive correlation with seed size, GS correlations with life history parameters (annual vs. perennial growth habit, relative growth rate and generation time), and new analyses dealing with GS effects on maximum photosynthetic rate and specific leaf area.
Cellular correlations
Early investigators found several fascinating cellular and developmental correlations with GS (see Bennett, 1973, 1987, and Cavalier-Smith, 1985, for reviews). First, nuclear DNA content is positively and quite directly correlated with chromosome size, nuclear size and cell size (Rees et al., 1966; Baetecke, 1967; Edwards and Endrizzi, 1975; Bennett et al., 1983; Lawrence, 1985). Second, nuclear DNA content is positively and strongly correlated with the duration of cell division (Bennett, 1971, 1977; Evans et al., 1972; Van't Hof, 1975). The causal nature of these correlations may arise from simple first principles. More DNA may require a larger container and require more time for replication. Gregory (2002) suggests that these phenomena may be manifest by GS interaction with cell cycle regulation via an unknown mechanism. It seems plausible that a species with a small genome might have a larger cell size or longer cell cycle, but that is rarely the case. These simple predictions may manifest at higher phenotypic levels, first and perhaps most directly, with seed mass.
Seed mass
Seed mass (also called seed size, the oven dry mass of the average seed) is thought to be a significant factor affecting seedling survival (Willson, 1983; Westoby et al., 1992). The positive correlation between seed size and GS has been shown repeatedly, both between populations of the same species (Caceres et al., 1998; Chung et al., 1998) and across large numbers of species (see Table 4, and Fig. 5). The consistency of these results is remarkable in comparison to the results for altitude, latitude and temperature (see Tables 1 and 2). Mowforth (1985) showed that the relationship between seed size and GS was a filled triangle rather than simply a linear positive trend. As seed mass increases there is a larger range of observed GS, but species with the largest GS always have large seeds.
Table 4.
Previous studies on the relationship between genome size and seed mass
| Correlation |
Level |
Description |
Authors |
|---|---|---|---|
| + | Pop. | 15 Dasypyrum villosum | Caceres et al., 1998 |
| + | Pop. | 12 Soybean strains | Chung et al., 1998 |
| + | Sp. | 131 British angiosperms | Thompson, 1990 |
| + | Sp. | 43 British plants | Grime et al., 1997 |
| + | Sp. | 22 Crepis | Jones and Brown, 1976 |
| + | Sp. | 12 Allium and 6 Vicia | Bennett, 1972 |
| + | Sp. | 18 North American pines | Wakamiya et al., 1993 |
| + | Sp. | 19 Mediterranean annuals | Maranon and Grubb, 1993 |
| + | Sp. | 148 species in California flora | Knight and Ackerly, 2002 |
| + | Sp. | Several Poaceae and Fabaceae | Mowforth, 1985 |
| Ns | Sp. | 16 grassland species, UK | Leishman, 1999 |
Correlations are either positive (+) or not significant (ns). Studies were classified into different levels: those dealing with different populations of the same species (Pop.) or multiple species (Sp.).
Fig. 5.
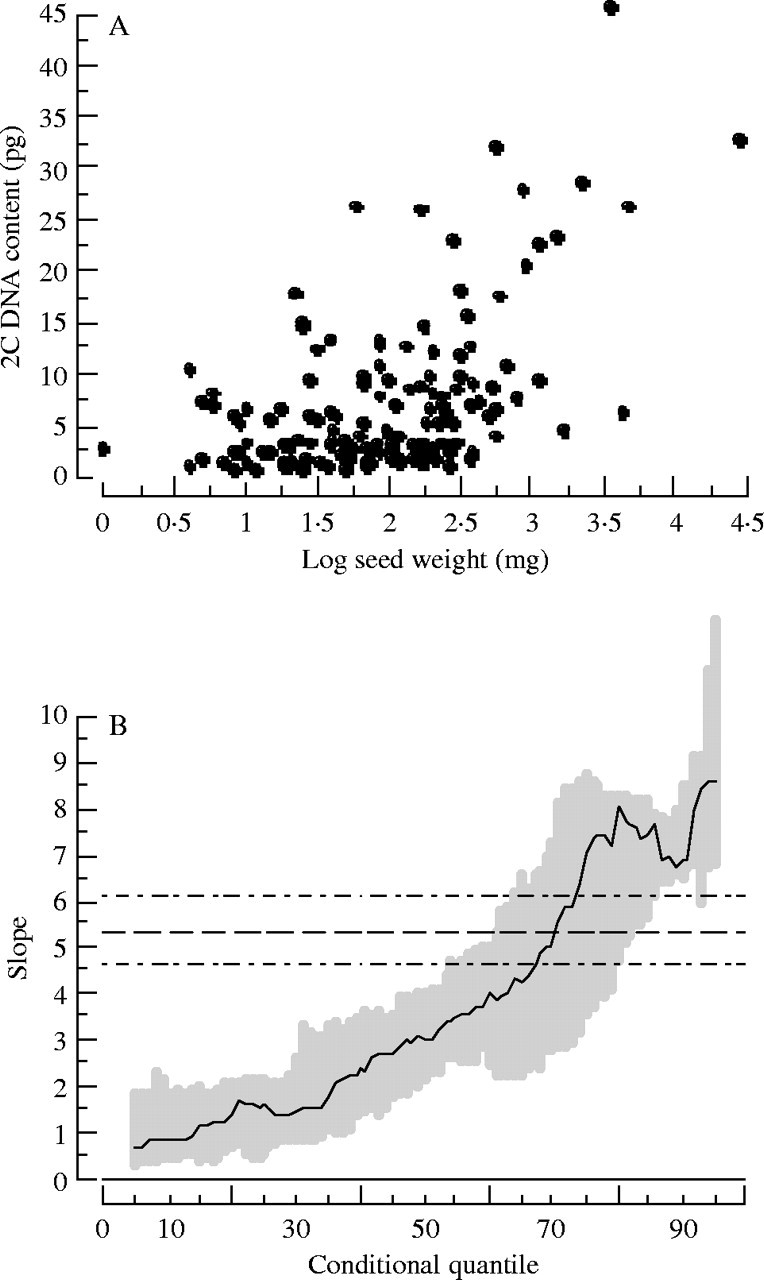
(A) 2C DNA content variation with seed size for 148 angiosperms in the California flora. Notice that there is an absence of species with large 2C DNA contents and small seeds. Species with small genome sizes can have a range of seed sizes while species with larger genome sizes are restricted to having larger seed sizes. (B) A quantile regression plot of (A) showing how the linear slope between DNA content and the log of seed weight changes with increasing quantiles. The dashed line is the least-squares regression with confidence intervals (double-dashed lines).
We reanalysed Knight and Ackerly's (2002) seed mass/GS data using a semi-log plot. Displayed this way, their data take on the triangular appearance shown by Mowforth (1985, Fig. 5). We also performed a quantile regression analysis of these data. This demonstrated a threshold or limit for the relationship between GS and seed size. With increasing quantiles the slope of the linear regression became steeper. The quantile regression analysis supports the idea that there is a GS-dependent constraint on attainable seed masses. There was a shallow positive slope for the lowest quantiles, with significantly steeper slope estimates for the largest quantiles. As GS increases, cell sizes increase, which may naturally force seed sizes to become larger. There may be some mechanisms for counteracting this tendency, but these mechanisms may evolve more slowly than the accumulation of DNA.
Species that produce small seeds can increase reproductive output because small seeds are produced in greater numbers (Cornelissen et al., 2003), which may also lead to greater dispersal ability. However, the positive fitness consequences of having large seeds have also been examined. It is thought that large-seeded species tend to produce larger seedlings (Leishman et al., 2000) with greater reserves for growth, enabling seedlings to survive under shaded canopies (Grime and Jeffery, 1965; Leishman and Westoby, 1994a; Saverimuttu and Westoby, 1996), in dry soil (Leishman and Westoby, 1994b) and in low-nutrient environments (Milberg et al., 1998). Large-seeded species may also compete better for resources (Black, 1958; Gross and Werner, 1982; Reader, 1993) and better withstand herbivory and pathogen attack (Armstrong and Westoby, 1993; Harms and Dalling, 1997). Large-seeded species, and thus large genome species, may increase their probability of regeneration in their current environment at the expense of dispersal into new environments.
If the small-genome plants can evolve larger seeds but rarely do, it would suggest that having a larger number of smaller seeds is the superior strategy. The large-genome plants may often be unable to counteract the increase of seed size due to the increase in GS and have to evolve a suite of traits that ameliorate such negative consequences. In any case, the greater dispersal ability brought about by the increased seed number may contribute to the reduced rates of extinction of small-genome species (see the evolution section) and may also contribute to the increased dispersal into extreme habitats and thus increase the probability of allopatric speciation events.
Leaf anatomical traits
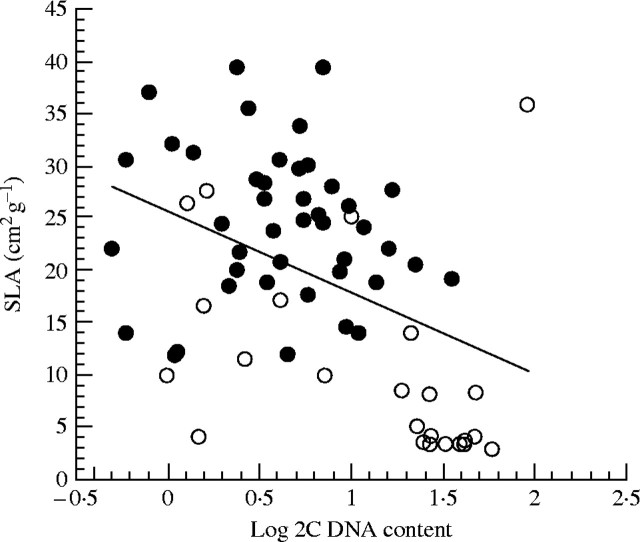
The rate of cell division and cell size could have significant effects on leaf morphology. Other investigators have found both positive and negative correlations between GS and leaf area, length or width (see Table 5). The relationship of leaf area to leaf mass (specific leaf area, SLA) is a trait at the nexus of a suite of co-varying traits related to the efficiency of carbon gain and leaf longevity (Reich et al., 1997, 1998). Given the cellular correlations presented above and the potential functional associations to be gleaned from a relationship between GS and SLA, we examined this relationship using two functional trait databases (that of Grime et al., 1997 and Reich et al., 1998). We obtained SLA estimates for 67 species with known 2C DNA content. We found a significant negative correlation between GS and SLA (r = −0·42, P < 0·0001; Fig. 6). Species with low SLA (typically smaller, thicker leaves) tended to have larger GS.
Table 5.
Previous studies on the relationship between genome size and leaf anatomical traits
| Correlation |
Level |
Description |
Authors |
|---|---|---|---|
| + | Sp. | 5 Microseris bigelovii and 5 M. laciniata (leaf area) | Castro-Jimenez et al., 1989 |
| + | Sp. | 12 Soybean strains (leaf length and width) | Chung et al., 1998 |
| + | Sp. | 18 North American Pinus (needle fresh and dry mass) | Wakamiya et al., 1993 |
| − | Pop. | 15 Dasypyrum villosum in Italy (leaf length) | Caceres et al., 1998 |
| − | Sp. | 43 British plants (leaf width) | Grime et al., 1997 |
| − | Pop. | 30 Festuca arundinacea (leaf length) | Ceccarelli et al., 1993 |
| Ns | Sp. | 43 British plants (leaf area) | Grime et al., 1997 |
Correlations are either +, −, or not significant (ns). Studies were classified into different levels: those dealing with different populations of the same species (Pop.) or multiple species (Sp.).
Fig. 6.
The relationship between specific leaf area (SLA) versus log of the 2C DNA content (gigabase pairs) The solid line depicts the normal least squares regression line. Data from Grime et al. (1997, solid circles) and Reich et al. (1998, open circles).
The association between GS and SLA suggests that several other plant traits may also be associated with GS given their strong interdependence with SLA. These include maximum photosynthetic rate, dark respiration rate, leaf nitrogen content and leaf lifespan (Reich et al., 1997, 1998). Below we test for relationships with maximum photosynthetic rate. The endeavour of joining results from plant functional trait databases with estimates of GS has considerable promise to shed new insight into the phenotypic and physiological consequences of GS variation in plants.
Photosynthetic rate
Previous investigators have examined the effect of within-species ploidy variation on photosynthetic rate. Both positive (Randall et al., 1977; Joseph et al., 1981) and negative (Garrett, 1978; Setter et al., 1978; Austin et al., 1982; Wullschleger et al., 1996) correlations have been reported. To our knowledge, no cross-species comparisons have been made. Here we test for GS-dependent variation in maximum photosynthetic rate using data from a published plant functional trait database (Reich et al., 1998). We compiled estimates of mass-based maximum photosynthetic rate for 24 species with known GS. These analyses revealed a significant negative correlation. 6 Species with large genomes tend to have lower maximum photosynthetic rates.
These results parallel observations for mammals and birds where metabolic rate is negatively correlated with GS (Vinogradov, 1995, 1997; Gregory, 2002). These complementary results suggest similar scaling mechanisms for metabolic efficiency that are somehow associated with GS. While the causative nature of this relationship is still uncertain in both cases, the implications are far-reaching. The result may help to explain the relationship between GS and minimum generation time. Species with small seeds also tend to have greater mass-based photosynthetic rates because they must acquire resources rapidly on emergence rather than relying on seed stores. Species with small genomes are thus able to complete their life cycle faster. The negative correlation between SLA and GS may be a consequence of overlapping adaptive strategies selecting for a well-described suite of physiological traits fine-tuned to achieve rapid growth.
Growth rate and generation time
The relative growth rate and growth interval before reproduction are both significant factors predicting the life history and regeneration niche of a species. Species that grow fast and reproduce in short intervals are more likely to be weedy or invasive and are opportunists that occupy disturbed sites. Several studies have found negative correlations for GS and relative growth rate (RGR) and positive correlations between GS and generation time (days to flowering, seed-bearing age, flowering date; Table 6). These correlations are equivalent since species that grow faster also tend to reproduce earlier. However, some studies have found the opposite relationships for RGR and generation time (Table 6), making generalizations difficult. Bennett et al. (1998) found that invasive species tend to have smaller GS. Annual species also typically have smaller GS (see Table 6 for references). Nonetheless, decreased growth rate and increased generation time may, in some cases, form the physiological link for the observed constraints on the distribution of large genome species across environmental gradients, as well as the evolutionary constraints outlined above.
Table 6.
Previous studies on the relationship between genome size (GS) and generation time (Gen.) or relative growth rate (RGR)
| Variable |
Correlation |
Description |
Authors |
|---|---|---|---|
| Gen. | + | GS annuals <GS perennialsin 45 spieces of Vicia | Chooi, 1971 |
| Gen. | + | GS annuals <GS obligate perennials in 271 angiosperms | Bennett, 1972 |
| Gen. | + | GS annuals <GS perennials in 22 species of Crepis | Jones and Brown, 1976 |
| Gen. | + | GS annuals <GS perennials in 18 species of Helianthus | Sims and Price, 1985 |
| Gen. | + | GS annuals <GS perennials in 10 Arachis sp | Singh et al., 1996 |
| Gen. | + | GS annuals <GS perennials in 565 angiosperms (genus level) | Vinogradov, 2001 |
| Gen. | + | GS annuals <GS perennials in 24 species of Lathyrus | Nandini and Murray, 1997 |
| Gen. | ns | Annuals vs. perennials in 401 CA angiosperms | Knight and Ackerly, 2002 |
| Gen. | + | Length of vegetative period in maize strains | Tito et al., 1991 |
| Gen. | + | Helianthus annuus | Natali et al., 1993 |
| Gen. | + | Days to flowering in 30 populations of Festuca arundinacea | Ceccarelli et al., 1993 |
| Gen. | + | Minimum seed-bearing age in 18 North American pines | Wakamiya et al., 1993 |
| Gen. | + | Maize | Rayburn, 1994 |
| Gen. | − | Initial month flowering in 42 Allium species | Labani and Elkington, 1987 |
| Gen. | − | Tetraploids flowered earlier than diploids, populations of D. glomerata | Bretagnolle and Thompson, 1996 |
| Gen. | − | First month flowering in 28 Allium species | Baranyi and Greilhuber, 1999 |
| Gen. | ns | 162 British annuals and perennials | Grime and Mowforth, 1982 |
| Gen. | ns | Pisum sativum | Cavallini et al., 1993 |
| Gen. | ns | Seed germination to anthesis, 15 populations of Dasypyrum villosum | Caceres et al., 1998 |
| RGR | + | Rate of leaf extension at low temperatures for 24 UK species | Grime et al., 1985 |
| RGR | + | Pisum sativum | Cavallini et al., 1993 |
| RGR | + | Epicotyl length 2–3 d after germination in 10 populations of Vicia faba | Minelli et al., 1996 |
| RGR | + | Growth at low temperatures for 7 population of Trifolium repens | Campbell et al., 1999 |
| RGR | + | 16 grassland species in UK | Leishman, 1999 |
| RGR | − | 6 weeks growth for 44 tillers of Poa annua | Mowforth and Grime, 1989 |
| RGR | − | 10 d after germ. in 30 populations of Festuca arundinaceae | Ceccarelli et al., 1993 |
| RGR | − | 12 Southwestern Zea mays | Biradar et al., 1994 |
| RGR | − | Relative growth rate for 30 pines | Grotkopp et al., 1998 |
| RGR | ns | Helianthus annuus | Natali et al., 1993 |
| RGR | ns | Growth 7–21 d following germination for 43 species in the UK | Grime et al., 1997 |
Correlations are either +, −, or not significant (ns).
CONCLUSION AND FUTURE DIRECTIONS
Having reviewed the current data on the macroevolutionary, conservation, ecological and functional significance of GS variation, what can we say about the viability of the large genome constraint hypothesis? We believe that we can endorse it, but rather cautiously at present. A recurrent theme is that strict correlation analyses often do not tell the whole story. Sophisticated statistical techniques that highlight larger genome sizes are needed to understand more clearly constraints on evolution, species distribution and phenotype. In this paper we have demonstrated the utility of quantile regression methods for addressing these complexities.
Macroevolutionary and conservation data show that lineages with the largest genomes have slower than average rates of diversification and disproportionately higher extinction risk (Vinogradov, 2003). However, these relationships are non-linear. For example, lineages with small genomes are not uniformly specious and are often as depurate as lineages with very large average genome sizes. However, lineages with large genomes are rarely highly specious. It appears that diversity is constrained in some way in these lineages.
A very similar relationship is evident when examining trends across environmental gradients. Species with small genomes tend to be found in widely varying habitats. However, lineages with very large genomes appear to be excluded from the most extreme habitats. Again, it does not appear that genome size is generating a consistent causative effect across the whole genome size range; rather it appears that lineages with the largest genome sizes are constrained from finding a way to survive in extreme environments. We encourage further examination of genome size distribution across abiotic gradients, perhaps even including analyses across gradients of elevation and latitude. However, these analyses should include descriptions of mean environmental conditions and habitat types spanning the range of elevations or latitudes considered. A greater range of habitat types is more likely to reveal significant trends—perhaps only after employing quantile regression to examine the effects at the boundaries of these bivariate distributions.
Ultimately the large genome constraints for evolution and ecology must be due to phenotypic variation manifest either directly or indirectly by changes in DNA content. Associations between GS and maximum photosynthetic rate, SLA, seed mass, relative growth rate or generation time are just a few examples of ecologically relevant phenotypic traits that may form a causative link for the large genome constraints outlined above. Results for photosynthetic rate and GS are particularly intriguing. The possibility of arriving at universal scaling laws originating from nucleotypic effects that predict the metabolic efficiency of organisms is exciting. If confirmed, these results may have far-reaching implications for the ecology and evolution of species.
Phenotypic associations with GS also often exhibit non-linear distributions. In general, large-genome species tend to display restricted trait variation, while small-genome species can attain a much wider array of trait states. For example, large-genome species never have small seeds while small-genome species display a much wider range of seed sizes. Similarly, large-genome species have lower photosynthetic rates while small-genome species have a wider range of photosynthetic performance. Also, large-genome species have reduced variation in SLA and tend to have lower SLA in general, compared to small-genome species which have a wider range of SLA.
It is likely that there is strong interdependence of the large genome constraints found at the evolutionary, ecological and functional levels. We can speculate that restricted ecological tolerances may increase probabilities of extinction by reducing population sizes. It may also increase the potential of ‘mutational meltdown’ scenarios to drive populations into extinction (Lynch et al., 1993). In addition, the inability to colonize extreme environments may decrease the chances of long-term isolation and allopatric speciation of large-genome lineages. The latter effect is also amplified by the tendency of large-genome lineages to have large seeds and thus to have lower dispersal abilities.
We emphasize that the endeavour of joining results from plant functional trait databases with estimates of GS has considerable promise to provide new insight into the phenotypic and physiological consequences of variation in plant GS. Combining GIS analyses with these results will also add clues in the search for putative abiotic selection pressures operating on GS. The development of these holistic databases will perhaps allow us to progress beyond pairwise correlations to partial correlation and path analyses. Such analyses should help us finally arrive closer to direct rather than correlated statistical effects. This is a goal worth striving for.
Acknowledgments
We thank Benjamin Carter, Joe Mello, Jonathan Wilson, Alison Chamberlain, Katherine Gordon, Jennifer Moonjian, Gregory Wilvert, Ian Robbins, Boris Igic and two anonymous reviewers for helpful comments that improved the manuscript. C.K. thanks Thomas Mitchell-Olds and the Max Planck Institute for Chemical Ecology in Jena, Germany for financial support in the early stages of this project. D.P. thanks Stanford University for a Terman Award and the Alfred P. Sloan Foundation for a Research Fellowship in Computational Molecular Biology.
LITERATURE CITED
- Anderson LK, Stack SM, Fox MH, Zhang CS. 1985. The relationship between genome size and synaptonemal complex length in higher plants. Experimental Cell Research 156: 367–378. [DOI] [PubMed] [Google Scholar]
- Armstrong DP, Westoby M. 1993. Seedlings from large seeds tolerate defoliation better: a test using phylogenetically independent contrasts. Ecology 74: 1092–1100. [Google Scholar]
- Austin RB, Morgan CL, Ford MA, Bhagwat SG. 1982. Flag leaf photosynthesis in Triticum aestivum and related diploid and tetraploid species. Annals of Botany 49: 177–189. [Google Scholar]
- Avdulov NP. 1931. Karyo-systematishce untersuchungen der familie Gramineen. Bulletin of Applied Botany Genetics and Plant Breeding 44: 1–428. [Google Scholar]
- Baetcke KP, Sparrow AH, Nauman CH, Schwemme SS. 1967. The relationship of DNA content to nuclear and chromosome volumes and to radiosensitivity (LD50). Proceedings of the National Academy of Sciences of the USA 58: 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranyi M, Greilhuber J. 1999. Genome size in Allium: in quest of reproducible data. Annals of Botany 83: 687–695. [Google Scholar]
- Bennett MD. 1971. The duration of meiosis. Proceedings of the Royal Society of London, Series B 178: 259–275. [Google Scholar]
- Bennett MD. 1972. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London, Series B 181: 109–135. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1973. Nuclear characters in plants. Brookhaven Symposia in Biology 25: 344–366. [Google Scholar]
- Bennett MD. 1976. DNA amount, latitude, and crop plant distribution. Environmental and Experimental Botany 16: 93–108. [Google Scholar]
- Bennett MD. 1976. DNA amount, latitude, and crop plant distribution. In: Jones K, Brandham PE, eds. Current chromosome research. Amsterdam: Elsevier North Holland Biomed. Press, 151–158. [Google Scholar]
- Bennett MD. 1977. The time and duration of meiosis. Philosophical Transactions of the Royal Society of London, Series B 277: 201–226. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200. [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. 2000. Nuclear DNA amounts in angiosperms and their modern uses—807 new estimates. Annals of Botany 86: 859–909. [Google Scholar]
- Bennett MD, Cox AV, Leitch IJ. 2001. Angiosperm DNA C-values database. http://www.rbgkew.org.uk/cval/database1.html. Accessed: 9/1/2003. [Google Scholar]
- Bennett MD, Heslop-Harrison JS, Smith JB, Ward JP. 1983. DNA density in mitotic and meiotic metaphase chromosomes of plants and animals. Journal of Cell Science 63: 173–179. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L. 1998. DNA amounts in two samples of angiosperm weeds. Annals of Botany 82: 121–134. [Google Scholar]
- Bennett MD, Smith JB, Lewis Smith RI. 1982. DNA amounts of angiosperms from the Antartic and South Georgia. Environmental and Experimental Botany 22: 307–318. [Google Scholar]
- Bennetzen J, Ma J, Devos KM. 2005. Mechanisms of recent genome size variation in flowering plants. Annals of Botany 95: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biradar DP, Bullock DG, Rayburn AL. 1994. Nuclear DNA amount, growth, and yield parameters in maize. Theoretical and Applied Genetics 88: 557–560. [DOI] [PubMed] [Google Scholar]
- Black JN. 1958. Competition between plants of different initial seed sizes in swards of subterranean clover (Trifolium subterraneum L.) with particular reference to leaf area and the light microclimate. Australian Journal of Agricultural Research 9: 299–318. [Google Scholar]
- Bottini MCJ, Greizerstein EJ, Aulicino MB, Poggio L. 2000. Relationship among genome size, environmental conditions and geographical distribution in natural populations of NW Patagonian species of Berberis L. (Berberidaceae). Annals of Botany 86: 565–573. [Google Scholar]
- Bretagnolle F, Thompson JD. 1996. An experimental study of ecological differences in winter growth between sympatric diploid and autotetraploid Dactylis glomerata. Journal of Ecology 84: 343–351. [Google Scholar]
- Caceres ME, De Pace C, Scarascia Mugnozza GT, Kotsonis P, Ceccarelli M, Cionini PG. 1998. Genome size variation within Dasypyrum villosum: correlation with chromosomal traits, environmental factors and plant phenotypic characteristics and behaviour in reproduction. Theoretical and Applied Genetics 96: 559–567. [Google Scholar]
- Cade BS, Richards JD. 1996. Permutation test for least absolute deviation regression. Biometrics 52: 886–902. [Google Scholar]
- Cade BS, Terrell JW, Schroeder RL. 1999. Estimating effects of limiting factors with regression quantiles. Ecology 80: 311–323. [Google Scholar]
- Cade BS, Guo Q. 2000. Estimating effects of constraints on plant performance with regression quantiles. Oikios 91: 245–254. [Google Scholar]
- Cade BS, Noon BR. 2003. A gentile introduction to quantile regression for ecologists. Frontiers in Ecology and the Environment 1: 412–420. [Google Scholar]
- Campbell BD, Caradus JR, Hunt CL. 1999. Temperature responses and nuclear DNA amounts of seven white clover populations which differ in early spring growth rates. New Zealand Journal of Agricultural Research 42: 9–17. [Google Scholar]
- Castro-Jimenez Y, Newton RJ, Price HJ, Halliwell RS. 1989. Drought stress response of Microseris species differing in nuclear DNA content. American Journal of Botany 76: 789–795. [Google Scholar]
- Cavalier-Smith T. 1985.The evolution of genome size. New York: John Wiley and Sons. [Google Scholar]
- Cavalier-Smith T. 2005. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany 95: 147–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallini A, Natali L, Cionini G, Gennai D. 1993. Nuclear DNA availability within Pisum sativum (Leguminosae): nucleotypic effects on plant growth. Heredity 70: 561–565. [Google Scholar]
- Ceccarelli M, Falisfocco E, Cionini PG. 1992. Variation in genome size and organization within hexaploid Festuca arundinaceae Theoretical and Applied Genetics 83: 273–278. [DOI] [PubMed] [Google Scholar]
- Ceccarelli M, Minelli S, Falcinelli M, Cionini PG. 1993. Genome size and plant development in hexaploid Festuca arundinaceae Heredity 71: 555–560. [Google Scholar]
- Cerbah MJ, Coulaud, Brown SC, Siljak-Yakovlev S. 1999. Evolutionary DNA variation in the genus Hypochaeris. Heredity 82: 261–266. [DOI] [PubMed] [Google Scholar]
- Chooi WY. 1971. Variation in nuclear DNA content in the genus Vicia Genetics 68: 195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Lee JH, Arumuganathan K, Graef GL, Specht JE. 1998. Relationships between nuclear DNA content and seed and leaf size in Soybean. Theoretical and Applied Genetics 96: 1064–1068. [Google Scholar]
- Cornelissen JHC, Lavorel S, Garnier E, Diaz S, Buchmann N, Gurvich DE, Reich PB, ter Steege H, Morgan HD, van der Heijden MGA, Pausas JG, Poorter H. 2003. A handbook of protocols for standardized and easy measrement of plant functional traits worldwide. Australian Journal of Botany 51: 335–380. [Google Scholar]
- Creber HMC, Davies MS, Francis D, Walker HD. 1994. Variation in DNA C value in natural populations of Dactylis glomerata New Phytologist 128: 555–561. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603. [DOI] [PubMed] [Google Scholar]
- Edwards GA, Endrizzi JL. 1975. Cell size nuclear size and DNA content relationships in Gossypium Canadian Journal of Genetics and Cytology 17: 181–186. [Google Scholar]
- Evans GM, Rees H, Snell CL, Sun S. 1972. The relation between nuclear DNA amount and the duration of the mitotic cycle. Chromosomes Today 3: 24–31. [Google Scholar]
- Garrett MK. 1978. Control of photorespiration at RuBP carboxylase/oxygenase level in ryegrass cultivars. Nature 274: 913–915. [Google Scholar]
- Godelle B, Cartier D, Marie D, Brown SC, Siljak-Yakovlev S. 1993. Heterochromatin study demonstrating the non-linearity of fluorometry useful for calculating genomic base composition. Cytometry 14: 618–626. [DOI] [PubMed] [Google Scholar]
- Gregory TR. 2002. A bird's-eye view of the C-value enigma: genome size, cell size, and metabolic rate in the class aves. 56: 121–130. [DOI] [PubMed] [Google Scholar]
- Grime JP, Jeffrey DW. 1965. Seedling establishment in vertical gradients of sunlight. Journal of Ecology 53: 621–642. [Google Scholar]
- Grime JP, Mowforth MA. 1982. Variation in genome size — an ecological interpretation. Nature 299: 151–153. [Google Scholar]
- Grime JP, Shacklock ML, Band SR. 1985. Nuclear DNA contents, shoot phenology and species co-existence in a limestone grassland community. The New Phytologist 100: 435–445. [Google Scholar]
- Grime JP, Thompson K, Hunt R, Hodgson JG, Cornelissen JHC, Rorison IH, Hendry GAF, Ashenden TW, Askew AP, Band SR, et al. 1997. Integrated screening validates primary axes of specialization in plants. Oikos 79: 259–281. [Google Scholar]
- Gross KL, Werner PA. 1982. Colonizing abilities of ‘biennial’ plant species in relation to ground cover: implications for their distributions in a successional series. Ecology 63: 921–931. [Google Scholar]
- Grotkopp E, Stoltenberg R, Rejmanek M, Rost TL. 1998. The effect of genome size on invasiveness. American Journal of Botany 85: 34. [Google Scholar]
- Haber AH, Luippold HJ. 1960. Effects of gibberellin on gamma radiated wheat. American Journal of Botany 47: 140–144. [Google Scholar]
- Hall SE, Dvorak WS, Johnston JS, Price HJ, Williams CG. 2000. Flow cytometric analysis of DNA content for tropical and temperate new world pines. Annals of Botany 86: 1081–1086. [Google Scholar]
- Harms KE, Dalling JW. 1997. Damage and herbivory tolerance through resprouting as an advantage of large seed size in tropical trees and lianas. Journal of Tropical Ecology 13: 617–621. [Google Scholar]
- Husband BC, Schemske DW. 1997. The effect of inbreeding in diploid and tetraploid populations of Epilobium angustifolium (Onagraceae): implications for the genetic basis of inbreeding depression. Evolution 51: 737–745. [DOI] [PubMed] [Google Scholar]
- Jones RN, Brown LM. 1976. Chromosome evolution and DNA variation in Crepis Heredity 36: 91–104. [Google Scholar]
- Joseph MC, Randall DD, Nelson CJ. 1981. Photosynthesis in polyploidy tall fescue. Plant Physiology 68: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner KL, Xiao-Ru W, Johnston JS, Price HJ, Williams CG. 2001. DNA content for Asian pines parallels New World relatives. Canadian Journal of Botany 79: 192–196. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. 2000. Genome evolution of wild barley (Horedeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimate divergence. Proceedings of the National Academy of Sciences of the USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Salomon S, Puchta H. 2000. Species-specific double-strand break repair and genome evolution in plants. The EMBO Journal 19: 5562–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD. 2002. Variation in nuclear DNA content across environmental gradients: a quantile regression analysis. Ecology Letters 5: 66–76. [Google Scholar]
- Koenker R, Bassett G. 1978. Regression quantiles. Econometrica 46: 33–50. [Google Scholar]
- Koenker R, Hallock KF. 2001. Quantile regression: an introduction. Journal of Economic Perspectives 15: 143–156. [Google Scholar]
- Labani RM, Elkingtion TT. 1987. Nuclear DNA variation in the genus Allium L. (Lilaceae). Heredity 59: 119–128. [Google Scholar]
- Lande R, Schemske DW. 1985. The evolution of self-infertility and inbreeding depression in plants. I. Genetic models. Evolution 39: 24–31. [DOI] [PubMed] [Google Scholar]
- Laurie DA, Bennett MD. 1985. Nuclear content in the genera Zea and Sorghum Intergeneric, interspecic and intraspecic variation. Heredity 55: 307–313. [Google Scholar]
- Lawrence ME. 1985.Senecio L. (Asteraceae) in Australia: nuclear DNA amounts. Australian Journal of Botany 33: 221–232. [Google Scholar]
- Leishman MR. 1999. How well do plant traits correlate with establishment ability? Evidence from a study of 16 calcareous grassland species. New Phytologist 141: 487–496. [Google Scholar]
- Leishman MR, Westoby M. 1994. The role of large seed size in shaded conditions: experimental evidence. Functional Ecology 8: 205–214. [Google Scholar]
- Leishman MR, Westoby M. 1994. The role of large seeds in seedling establishment in dry soil conditions±experimental evidence from semi-arid species. Journal of Ecology 82: 249–258. [Google Scholar]
- Leishman MR, Wright IJ, Moles AT, Westoby M. 2000. The evolutionary ecology of seed size. In: Fenner M, ed. Seeds: the ecology of regeneration in plant communitues. Wallingford, UK: CAB International, 31–57. [Google Scholar]
- Levin DA, Funderburg SW. 1979. Genome size in angiosperms: temperate versus tropical species. American Naturalist 114: 784–795. [Google Scholar]
- Longley AE, Kato TAY. 1965.Chromosome morphology of certain races of maize in Latin America Chapingo, Mexico: International Center for the Improvement of Maize and Wheat, Mexico Research Bulletin 1. [Google Scholar]
- Lynch M, Bürger R, Butcher D, Gabriel W. 1993. Mutational meltdowns in asexual populations. Journal of Heredity 84: 339–344. [DOI] [PubMed] [Google Scholar]
- Lysak M, Rostkova A, Dixon J, Rossi G, Dolezel J. 2000. Limited genome size variation in Sesleria albicans. Annals of Botany 86: 399–403. [Google Scholar]
- Mabberley DJ. 1999.The plant book : a portable dictionary of the vascular plants, 2nd edn. Cambridge, UK: Cambridge University Press. [Google Scholar]
- MacGillivray CW, Grime JP. 1995. Genome size predicts frost resistance in British herbaceous plants: implications for rates of vegetation response to global warming. Functional Ecology 9: 320–325. [Google Scholar]
- Magallon S, Sanderson MJ. 2001. Absolute diversification rates in angiosperm clades. Evolution 55: 1762–1780. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf PC, Cameron JW. 1942. Western Guatemala: a secondary center of origin of cultivated maize varieties. Harvard University, Botanical Museum Leaflets 10: 217–252. [Google Scholar]
- Maranon T, Grubb PJ. 1993. Physiological basis and ecological significance of the seed size and relative growth rate relationship in Mediterranean annuals. Functional Ecology 7: 591–599. [Google Scholar]
- Masterson J. 1994. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–24. [DOI] [PubMed] [Google Scholar]
- McMurphy LM, Rayburn AL. 1991. Genome size variation in maize populations selected for cold tolerance. Plant Breeding 106: 190–195. [Google Scholar]
- Miksche JP. 1967. Variation in DNA content in several gymnosperms. Canadian Journal of Genetics and Cytology 9: 717–722. [Google Scholar]
- Miksche JP. 1971. Intraspecific variation of DNA per cell between picea sitchensis (Bong.) Carr. provenances. Chromosoma 32: 343–352. [DOI] [PubMed] [Google Scholar]
- Milberg P, Perez-Fernandez MA, Lamont BB. 1998. Seedling growth response to added nutrients depends on seed size in three woody genera. Journal of Ecology 86: 624–632. [Google Scholar]
- Minelli S, Moscariello P, Ceccarelli M, Cioniini PG. 1996. Nucleotype and phenotype in Vicia faba Heredity 76: 524–530. [Google Scholar]
- Mowforth MA. 1985.Variation in nuclear DNA amounts in flowering plants: an ecological analysis. PhD Thesis. University of Sheffield, UK. [Google Scholar]
- Mowforth MA, Grime JP. 1989. Intra-population variation in nuclear DNA amount, cell size and growth rate in Poa annua L. Functional Ecology 3: 289–295. [Google Scholar]
- Nandini AV, Murray BG. 1997. Intra- and interspecific variation in genome size in Lathyrus (Leguminosae). Botanical Journal of the Linnean Society 125: 359–366. [Google Scholar]
- Natali L, Cavallini A, Cionini G, Sassoli O, Cionini PG, Durante M. 1993. Nuclear changes within Helianthus annuus L.: changes within single progenies and their relationships with plant development. Theoretical and Applied Genetics 85: 506–512. [DOI] [PubMed] [Google Scholar]
- Orel N, Kyryk A, Puchta H. 2003. Different pathways of homologous recombination are used for the repair of double-strand breaks within tandemly arranged sequences in the plant genome. The Plant Journal 35: 604–612. [DOI] [PubMed] [Google Scholar]
- Orgel LE, Crick FHC. 1980. Selfish DNA: the ultimate parasite. Nature 284: 604–607. [DOI] [PubMed] [Google Scholar]
- Palomino G. 1993. DNA content for seven diploid species and populations of Echeandia (Lilliaceae). American Journal of Botany, 80: 77. [Google Scholar]
- Palomino G, Sousa SM. 2000. Variation of nuclear DNA content in the biflorus species of Lonchocarpus (Leguminosae). Annals of Botany 85: 69–76. [Google Scholar]
- Petrov DA. 2002. Mutational equilibrium model of genome size evolution. Theoretical Population Biology 61: 531–543. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Lozovskaya ER, Hartl DL. 1996. High Intrinsic rate of DNA loss in Drosophila Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Poggio L, Rosato M, Chiavarino AM, Naranjo C. 1998. Genome size and environmental correlations in Maize (Zea mays ssp. mays, Poaceae). Annals of Botany 82 (Suppl. A): 107–115. [Google Scholar]
- Price HJ, Chambers KL, Bachmann K. 1981. Geographic and ecological distribution of genomic DNA content variation in Microseris douglasii (Asteraceae). Botanical Gazette 142: 415–426. [Google Scholar]
- Randall DD, Nelson CJ, Asay KH. 1977. Ribulose bisphosphate carboxylase. Altered genetic expression in tall fescue. Plant Physiology 59: 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayburn AL. 1990. Genome size variation in Southwestern United States Indian maize adapted to various altitudes. Evolutionary Trends in Plants 4: 53–57. [Google Scholar]
- Rayburn AL, Auger JA. 1990. Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theoretical and Applied Genetics 79: 470–474. [DOI] [PubMed] [Google Scholar]
- Rayburn AL, Price HJ, Smith JD, Gold JR. 1985. C-band heterochromatin and DNA content in Zea mays American Journal of Botany 72: 1610–1617. [Google Scholar]
- Rayburn AL, Dudley JW, Biradar DP. 1994. Selection for early flowering results in simultaneous selection for reduced nuclear DNA content in maize. Plant Breeding 112: 318–322. [Google Scholar]
- Reeves G, Francis D, Davies MS, Rogers HJ, Hodkinson TR. 1998. Genome size is negatively correlated with altitude in natural populations of Dactylis glomerata Annals of Botany 82(Suppl. A): 99–105. [Google Scholar]
- Reader RJ. 1993. Control of seedling emergence by ground cover and seed predation in relation to seed size for some old-field species. Journal of Ecology 81: 169–175. [Google Scholar]
- Rees H, Cameron FM, Hazarika MH, Jones GH. 1966. Nuclear variation between diploid angiosperms. Nature 211: 828–830. [DOI] [PubMed] [Google Scholar]
- Reich PB, Walters MB, Ellensworth DS. 1997. From tropics to tundra: Global convergence in plant functioning. Proceedings of the National Academy of Sciences of the USA 94: 13730–13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich PB, Walters MB, Ellsworth DS, Vose JM, Volin JC, Gresham C, Bowman WD. 1998. Relationships of leaf dark respiration to leaf nitrogen, specific leaf area and leaf life-span: a test across biomes and functional groups. Oecologia 114: 471–482. [DOI] [PubMed] [Google Scholar]
- Richards JA. 1997.Plant breeding systems. London: Chapman and Hall. [Google Scholar]
- Saverimuttu T, Westoby M. 1996. Seedling longevity under deep shade in relation to seed size. Journal of Ecology 84: 681–689. [Google Scholar]
- Setter TL, Schrader LE, Bingham ET. 1978. Carbon dioxide exchange rates, transpiration in genetically equivalent ploidy levels in alfalfa. Crop Science 18: 327–332. [Google Scholar]
- Sims LE, Price HJ. 1985. Nuclear DNA content variation in Helianthus (Asteraceae). American Journal of Botany 72: 1213–1219. [Google Scholar]
- Singh KP, Raina SN, Singh AK. 1996. Variation in chromosomal DNA associated with the evolution of Arachis species. Genome 39: 890–897. [DOI] [PubMed] [Google Scholar]
- Smith JB, Bennett MD, Gustafson JP. 1976. Variation in DNA amount and heterochromatin patterns in Scecale. In: Jones K, Brandham PE, eds. Current chromosome research. Amsterdam: Elsevier/North Holland Biomed, 232–233. [Google Scholar]
- Suda J, Kyncl T, Freiova R. 2003. Nuclear DNA amounts in Macronesian angiosperms. Annals of Botany 92: 152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh SB, Rees H. 1976. Nuclear DNA amounts in populations of Picea glauca and Pinus species. Heredity 36: 123–137. [Google Scholar]
- Temsch E, Greilhuber J. 2001. Genome size in Arachis duranensis: a critical study. Genome 44: 826–830. [PubMed] [Google Scholar]
- The Angiosperm Phylogeny Group. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society 141: 399–436. [Google Scholar]
- Thibault J. 1998. Nuclear DNA amount in pure species and hybrid willows (Salix): a flow cytometric investigation. Canadian Journal of Botany 76: 157–165. [Google Scholar]
- Thompson K. 1990. Genome size, seed size and germination temperature in herbaceous angiosperms. Evolutionary Trends in Plants 4: 113–116. [Google Scholar]
- Tito CM, Poggio L, Naranjo CA. 1991. Cytogenetic studies in genus Zea 3. DNA content and heterochromatin in species and hybrids. Theoretical and Applied Genetics 83: 58–64. [DOI] [PubMed] [Google Scholar]
- Torrel M, Valles J. 2001. Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): systematic, evolutionary, and ecological implications. Genome 44: 231–238. [PubMed] [Google Scholar]
- Turpeinen T, Kulmala J, Nevo E. 1999. Genome size variation in Hordeum spontaneum populations. Genome 42: 1094–1099. [DOI] [PubMed] [Google Scholar]
- Van't Hof J. 1975. The duration of chromosomal DNA synthesis, of the mitotic cycle, and of meiosis of higher plants. King RC, ed. Handbook of genetics. New York: Plenum Press. [Google Scholar]
- Vilhar B, Vidic T, Jogan N, Dermastia M. 2002. Genome size and the neucleolar number as estimators of ploidy level in Dactylis glomerata in the Slovenian Alps. Plant Systematics and Evolution 234: 1–13. [Google Scholar]
- Vinogradov AE. 1995. Nucleotypic effects in homeotherms: body-mass-corrected basal metabolic rate of mammals is related to genome size. Evolution 49: 1249–1259. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 1997. Nucleotypic effects in homeotherms: body-mass-independent resting metabolic rate of Passerine birds is related to genome size. Evolution 51: 220–225. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 2001. Mirrored genome size distributions in monocot and dicot plants. Acta Biotheoretica 49: 43–51. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. 2003. Selfish DNA is maladaptive: evidence from the plantRed list. Trends in Genetics 19: 609–614. [DOI] [PubMed] [Google Scholar]
- Wakamiya I, Newton RJ, Johnston SJ, Price JH. 1993. Genome size and environmental factors in the genus Pinus American Journal of Botany 80: 1235–1241. [Google Scholar]
- Wakamiya I, Price JH, Messina MG, Newton RJ. 1996. Pine genome size diversity and water relations. Physiologia Plantarum 96: 13–20. [Google Scholar]
- Wellhausen EJ, Roberts LM, Hernandez EX. 1952.Races of maize in Mexico: their origin, characteristics and distribution. Cambridge, MA: Harvard University Press. [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249. [PubMed] [Google Scholar]
- Westoby M, Jurado E, Leishman M. 1992. Comparative evolutionary ecology of seed size. Trends in Ecology and Evolution 7: 368–372. [DOI] [PubMed] [Google Scholar]
- Willson MF. 1983.Plant reproductive ecology. New York: John Wiley and Sons. [Google Scholar]
- Wullschleger SD, Sanderson MA, McLaughlin SB, Biradar DP, Rayburn AL. 1996. Photosynthetic rates and ploidy levels among populations of switchgrass. Crop Science 36: 306–312. [Google Scholar]