Abstract
• Background and Aims The nuclear DNA of certain varieties of flax (Linum usitatissimum) can vary within a single generation when the plants are grown under specific environmental conditions. This review details the genomic variations that have been identified and associated with this environmental response.
• Conclusions The variation occurs across the whole spectrum of sequence repetition and has been shown to occur in the highly repeated, middle repetitive and low copy number sequences. Although the variation has been shown to be spread throughout the genome it does not occur at random, as similar molecular events have been shown to occur repeatedly. The changes in two labile regions in the nucleus, the ribosomal RNA genes and a site-specific insertion event, have been shown to occur within the period of vegetative growth and over a relatively short period of that growth. The gradual change in total nuclear DNA that has been described would then need to have arisen through an accumulation of changes occurring over the whole, or most of the, period of growth prior to flowering. The polymorphisms that result from these rapidly occurring genomic events have also been observed in many other flax and linseed varieties as well as in the wild progenitors of flax.
Keywords: Linum usitatissimum, flax, insertion sequence, rDNA, nuclear DNA content, rapid genomic variation
INTRODUCTION
The nuclear DNA content varies widely among flowering plants and even between closely related species. The differences within a species are much less than those between species. However, since most of the variation appears to be a historical legacy it is difficult to determine the sequence of events by which DNA variation occurs. In one plant species, Linum usitatissimum (flax) rapid changes in the genome occur that are associated with the environment in which the plant is growing, and these nuclear DNA variants can be stably or unstably inherited. In addition, the magnitude of the variation that occurs in flax has enabled the phenomenon to be identified.
Rapid modifications of the genome, correlated with changes in gene expression, have been observed during plant development and under stress conditions. Several mechanisms, such as quantitative modification of repetitive DNA, DNA methylation, excision and insertion of transposable elements, gene amplification or deletion and histone acetylation have been suggested as points of control for these changes in gene expression (Fedoroff, 1989; Bassi, 1990, 1991; Cullis, 1990; Natali et al., 1993, 1995; Smulders et al., 1995; Johnston et al., 1996; Richards, 1997; Grandbastien, 1998; Kalendar et al., 2000). Speculation on the role of the miniature inverted-repeat transposable element (MITE ) mPing that is actively mobile in rice has concluded that this element might have been responsible for diversification of cultivars and hastened their domestication (Jiang et al., 2003). Thus, in many organisms, the genome may reorganize itself on facing adversity, for which it is unprepared, in order to ensure the organism's survival (McClintock, 1978). The extensive genomic variations that occur in flax within a generation provide the opportunity to study the mechanism(s) by which such variations occur, and the possible phenotypic consequences of these variations.
ORGANIZATION OF THE FLAX GENOME
The organization of the flax genome itself may also have contributed to the phenomenon being observed. The fractions of the total nuclear DNA that are found in highly repetitive tandem arrays, intermediately dispersed and low copy number sequences are somewhat different from most other plants. The highly repetitive tandemly arrayed sequences are made up of a small number of families and comprise about 35 % of the total nuclear DNA. These regions are probably responsible for the blocks of heterochromatin visible in the interphase nucleus. The middle repetitive fraction is only about 15 % of the total nuclear DNA, somewhat less than in most higher plants, while the low copy number fraction is the remaining 50 % of the genome. Therefore flax appears to have a larger fraction of its total genome present in these lower copy number sequences than other plants of similar genome size. An additional difference between flax and other higher plants is the long period interspersion pattern of these classes of sequences (Cullis, 1981). This pattern is consistent with a much lower activity of mobile elements in the genome, or of a restriction of the available sites for insertion of such elements.
INDUCED HERITABLE CHANCES IN FLAX
Genome alterations that occur in response to specific defined environments have been described in the inbred flax variety ‘Stormont Cirrus’ (Pl). Growth of Pl in different fertilizer combinations, or under different temperature regimes, can result in phenotypic and genotypic differences in the first generation progeny. Certain growth conditions can result in the ‘stable’ inheritance of these differences in subsequent generations obtained by self-fertilization (Durrant, 1962; Cullis, 1977, 1981) under many different, but not all, growth conditions tested in these subsequent generations. Therefore stability for the genotrophs is a relative concept and probably needs to be defined for each of the characteristics individually.
The lines in which ‘stable’ changes were observed were termed genotrophs (Durrant, 1962). All the individuals of a given genotroph, generally all the progeny of inbred plants that had been subjected to a particular set of growth conditions, were identical to each other. However the ‘stable’ types have been divided into two groups, the large and small genotrophs that differed extensively from each other and Pl, the line from which they were derived. However, not all of the characteristics are identical in all the genotrophs within these groups.
There are four established aspects about the induction process in the flax variety ‘Stormont Cirrus’. First, ‘Stormont Cirrus’ is a predominantly self-fertilizing plant since anther dehiscence and pollination usually occur during flower opening. Second, nearly all of the seeds planted grow under the inducing conditions and can contribute to the next generation (Durrant, 1962). Third, all of the self-fertilized progeny from all the individuals growing in a specific environment were identical to each other, but different from all the progeny of individuals grown in a different environment. Fourth, the induction of the changes has been repeated, resulting in the appearance of similar phenotypic, biochemical and molecular changes (Durrant, 1962; Cullis, 1977, 1981). Thus it is extremely unlikely that any form of selection, in the conventional sense, from a heterogeneous population of plants is the causative agent for the observed change.
The response of the genome to growth in various environments has also been observed in other flax varieties, namely ‘Rembrandt’, ‘Hollandia’ and ‘Liral Monarch’, so that it is not a property unique to the variety ‘Stormont Cirrus’. It is also clear that many varieties do not appear to respond in this way, or at least not to the extent observed in ‘Stormont Cirrus’. However, until all the possible genomic sites that are found to vary have been tested, it cannot be confirmed that any varieties are completely stable. Crosses between ‘stable’ and responding lines has indicated that the ability to respond is under genetic control, but the basis of this control is currently unknown.
CHARACTERISTICS OF FLAX GENOTROPHS
The large and small genotrophs were so described on the basis of a difference in plant weight at maturity when the plants were grown under optimal conditions (Durrant, 1962). A difference in plant height is another characteristic that varies, with both the original small and large genotrophs being shorter than the original line, and the large genotrophs being taller that the small ones. The plant height differences are a more robust character for differentiating between the genotrophs, as it is manifest under all growth conditions, whereas the plant weight at maturity characteristic is dependent on particular growth conditions. However, the use of height does not generate the same classes as the plant weight classification since there was not always a co-appearance of all of the possible phenotypic and molecular changes. Thus LH, a large genotroph, and Sh, a small genotroph, were both as tall as the original line (Durrant, 1971). Additional characteristics that varied between some of the genotroph classes and Pl were the peroxidase isozyme pattern and the seed capsule septa hair number. These two characteristics appear to be stable under many growth conditions and their inheritance obeys the rules of normal Mendelian genetics.
The characteristic that has been most extensively investigated is the change in nuclear DNA amount, as measured by Feulgen staining or by the quantitative characterization of various nuclear DNA sequences. These genomic changes occur within all three fractions of the flax genome (highly repetitive, middle repetitive and low copy number) and appear to occur through the whole chromosome complement (Cullis and Creissen, 1987). These genomic changes will be described in greater detail.
NUCLEAR DNA VARIATION
Detection by Feulgen staining
The described phenotypic variations among the genotrophs (height, weight, capsule hair septa number and isozyme mobility) are but the tip of the iceberg when compared to the extent of variation seen at the DNA level. The original extreme types, L and S, differed by 15 % of their total DNA as determined by Feulgen staining (Evans et al., 1966). However, this nuclear DNA difference could erode with generations grown out-of-doors (essentially at lower temperatures) so that the lines S3 and L3 (the ‘3’ denoting three generations growth out-of-doors) had essentially the same value for the Feulgen stained nuclear DNA content, which was equivalent to that of the original line (Durrant and Jones, 1971). One of the interesting observations was that the suite of phenotypic characteristics that differentiated L and S were still present in L3 and S3, such that phenotypically L3 was indistinguishable from L, and S3 indistinguishable from S. The qualitative variation that had occurred in the genome during the generations between L and L3 and S and S3 indicate that this nuclear DNA amount reversion was not simply a reversal of the original modifications (Cullis et al., 1999) The lines L6 and S6 (again the ‘6’ denoting six generations growth out-of-doors) also had the same Feulgen-based nuclear DNA value, but this value was significantly lower than that for the original line, and similar to that for S. Both of these lines were indistinguishable from S for the other phenotypic characteristics measured. Therefore it is clear that the nuclear DNA estimates by Feulgen staining can vary in flax, and the quantitative variation in DNA as measured this way was not directly correlated with the other phenotypic characteristics observed to vary in the genotrophs.
Since the changes in nuclear DNA content of genotrophs over a number of generations are not necessarily associated with measured phenotypic changes, these DNA variations may not be just a reversal of earlier changes, but may represent the equivalent of a new induction process. Obviously these nuclear DNA measurements are relatively inexact estimates of variation, with very large changes, in terms of the number of base pairs of DNA, necessary for significant variation in staining to be identified. Therefore the characterization of specific sequences would lead to more reliable measures of DNA variation in this system.
Flax genome organization
DNA variation between the genotrophs has been confirmed using renaturation analysis and the characterization of cloned DNA sequences. Renaturation analysis has demonstrated that DNA sequences across the whole spectrum of redundancy (from highly repetitive to low copy numbers) were involved in the variation observed (Cullis, 1973, 1980). The cloning and characterization of flax genomic sequences, including highly repetitive tandemly arrayed families, dispersed repetitive sequences and low copy number sequences has confirmed this initial observation. Notable by their absence in these analyses are the families of transposable elements that are responsible for much of the DNA variation in other species. The flax genome has a lower proportion of middle repetitive sequences (∼15 %, Cullis, 1980) than other plants with an equivalent genome size and so may have a smaller complement of transposable elements. Only one family of transposable elements (dLute) has been isolated from flax and that was isolated through its association with a disease resistance gene (Luck et al., 1998).
Specific DNA sequence variation
The cloning and quantification of families of repetitive sequences (Cullis and Cleary, 1986a, b) and the detailed characterization of the families of large (25S + 18S) and small (5S) ribosomal RNA genes (Cullis, 1976, 1979; Goldsbrough and Cullis, 1981; Schneeberger and Cullis, 1991) are particularly important in the demonstration of significant nuclear DNA variation since these sequences comprise about 35 % of the total nuclear DNA in flax. Supplementing these observations on the repetitive families are many low copy number variable sequences. These have mainly been identified from Random Amplified Polymorphic DNA (RAPD) analysis (Cullis et al., 1999) and representational difference analysis (RDA) (Oh and Cullis, 2003). The RAPD analysis only samples a very small fraction of the genome and so the extent of the changes that are occurring appear to be substantial. Some of these isolated regions have homology to known genes (cytochrome P450, 10-formyltetrahydrofolate dehydrogenase, NADH-ubiquinone oxidoreductase C and 40S ribosomal protein S17). Other regions do not have any similarities to sequences in the current databases in spite of the reporting of the complete genome sequences from Arabidopsis and rice.
Most of the characterizations described above have been using DNA isolated from plant material that had been grown for a number of generations following the Feulgen determination of the nuclear DNA content. Therefore the relationship between these two types of measurement has to be inferential. However, the direct comparison between the original line and stable genotrophs is available for the molecular analysis involving the three lines C1, C2 and C3. DNA isolated from the first generation of plants grown after the induction has been used for comparisons (Cullis, 1981). However, Feulgen-based measurements are not available for these particular lines so that direct comparisons between the specific sequence comparisons and the total nuclear DNA contents have not been made.
DNA comparison between Pl and C3
Highly repetitive sequences
The fraction of the genome that was contained in the highly repetitive tandemly arrayed sequences in the two lines Pl and C3 was measured using slot blots (Cullis and Cleary, 1986a, b). These data are shown in Table 1. The results demonstrate that the differences in just these sequences results in a reduction in total nuclear DNA content of C3 relative to the progenitor line Pl by 6·8 %. Since these comparisons contain representatives from all the highly repetitive families in the flax genome, compensatory increases cannot occur in C3 in other highly repetitive regions to reverse this difference. The data from RAPDs does not indicate that there is a compensatory increase anywhere else in the C3 genome. The data in Cullis and Cleary (1986a, b) also indicates that the two genotrophs that represented the extremes for the numbers of the ribosomal RNA genes alone (25S, 18S and 5S) would result in a nuclear DNA variation of 3·5 %.
Table 1.
Fraction of the genome (%) comprised of various highly repeated sequences in the lines Pl and C3 (data from Cullis and Cleary, 1986a)
| Sequence ID |
rDNA |
5S |
21 |
13 |
53 |
2 |
8 |
87 |
7 |
|---|---|---|---|---|---|---|---|---|---|
| Pl | 1·5 | 3·0 | 5·6 | 2·2 | 2·3 | 4·2 | 2·6 | 2·5 | 3·2 |
| C3 | 0·8 | 2·2 | 5·6 | 1·2 | 1·6 | 2·5 | 2·1 | 1·9 | 1·7 |
Only one of the highly repetitive sequence families, the light satellite sequence that comprised about 15 % of the total nuclear DNA, did not vary either within the genotrophs or within the sampled varieties. Overall, the variation observed in comparisons between other flax varieties mirrored the variation observed in the genotrophs. The potential difference in nuclear DNA content between two lines that had all the highest or all the lowest measured values for the highly repetitive families is 12·5 %. Given the fact that the light satellite DNA does not vary, this difference represents a more than two-fold difference in the quantity of the remaining highly repetitive DNA families.
Most of these highly repetitive tandemly repeated sequences are scattered throughout the genome, with the exception of the large ribosomal RNA genes. Therefore it has not been possible to identify any specific regions of the chromosomes that might be the selective targets of the genomic variation. However, with the magnitude of the variation and the fact that the flax genome is packaged into fifteen approximately equally sized chromosomes, the variation must be spread throughout the chromosome complement in order for chromosome size differences not to be observable (Cullis and Creissen, 1987).
Specific polymorphic sequences
The identification of specific polymorphisms has allowed the characterization of the reproducibility of the genomic changes and also facilitated the characterization of these changes through development.
The DNAs isolated from genotrophs have been compared using RAPDs (Cullis et al., 1999) and by representational difference analysis (RDA; Oh and Cullis, 2003). These data have again confirmed the large extent of variation between the closely related lines (Cullis et al., 1999). The comparison between Pl and C3 using RAPDs identified 94 polymorphisms. The RAPD bands amplified represented approximately 1·5 % of the total flax genome, so if the RAPD reactions were randomly sampling the flax genome about 6000 variable sites should be present in the whole genome. The molecular bases for these polymorphisms have not been identified, but if these involved significant insertions or deletions then additional quantitative nuclear DNA variation would result. Many fewer polymorphic regions have been identified using RDA (Oh and Cullis, 2003). However, all of the polymorphisms that have been characterized result from amplifications or deletions, and therefore would contribute to the variation in the total nuclear DNA.
One specific polymorphism resulting from a complex insertion event (termed LIS-1, for Linum Insertion Sequence 1) has been identified (Schneeberger, 1992; Chen, 1999) and the sequence determined (GenBank Accession numbers AJ131991, AJ131992, AJ131993, AJ131994). This insertion event occurs at a specific single copy site in a number of the genotrophs, with Pl being homozygous for the uninserted site. The presence of LIS-1 has also been shown in other flax and linseed varieties. The relative positions of four primers that can be used to identify inserted and uninserted sites are shown in Fig. 1. This particular polymorphism is a clear example of the reproducibility of the genomic variation that is associated with the induced changes.
Fig. 1.
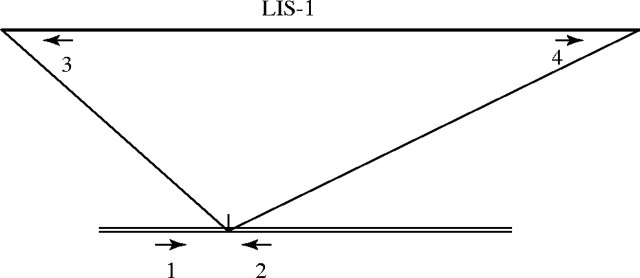
The relative positions of the primers used to follow the appearance of LIS-1 during growth of Pl under inducing conditions. Primers 1 and 2 only amplify a fragment if LIS-1 is absent, while primers 1 and 3, and 2 and 4 only amplify from sites where LIS-1 has been inserted.
Reproducibility of the sites of variation
A notable characteristic of all the DNA comparisons is the specificity of the changes between Pl and the genotrophs. Thus independent induction events have resulted in identical new structures at many loci, confirming the precise nature of the induced changes. For example, specific polymorphisms have been repeatedly generated, in independently induced genotrophs, in subsets of the 5SRNA gene family (Schneeberger and Cullis, 1991; Schneeberger, 1992), in the repeated appearance of specific RAPD polymorphisms (Cullis et al., 1999), and in the generation of specific polymorphisms in plants growing under inducing conditions (Cullis and Charlton, 1981; Y. Chen, R. Lowenfeld and C. Cullis, unpubl. res.).
The use of these polymorphic sequences in comparisons with other flax and linseed varieties has shown that the variation seen in Pl is a reflection of the type and extent of the variation seen in natural populations. Every one of the sequences that have been shown to be different between the genotrophs and subsequently tested on a wider range of flax varieties has also highlighted polymorphisms among these varieties. Where different-sized bands are seen, rather than the presence or absence of a band, the same-sized fragments are seen as those observed in the genotrophs. Therefore this set of sequences also appears to be highly variable in natural populations of flax, but with a limited range of events leading to the polymorphisms.
Induction of changes
The majority of the variation that has been described has resulted from comparisons between ‘stable’ lines. Except for the comparisons between Pl and the three lines C1, C2 and C3, all the DNA samples have been isolated from plants that have been grown for a number of generations after the changes were induced. Therefore the question that arises is whether the changes occurred during that ‘inducing’ treatment or in subsequent generations. Three sets of data address this question. The first is the measurement of the nuclear DNA by Feulgen staining during the growth of Pl under inducing conditions (Evans et al., 1966). The second is the determination of the ribosomal RNA gene number at various points along the stem during growth (Cullis and Charlton, 1981). The third is the identification of a unique complex insertion event (LIS-1) that can be followed during growth of the plants. A combination of samples of these data is shown in Fig. 2. Each of the growth conditions that were used to generate the data shown in Fig. 2 result in the appearance of a small genotroph. The total nuclear DNA had a steady decline during growth prior to flowering (data from Evans et al., 1966). The rDNA measurements indicated that the reduction in gene number occurred over a very short period of the growth prior to flowering. The complex insertion event, LIS-1, also appeared over a short period of growth prior to flowering. Measurements of the variation in all the other repetitive sequence families would indicate if there was a particular stage in development when a particular sequence family varied, and the accumulation of these variations were responsible for the apparent gradual change in the total nuclear DNA content.
Fig. 2.
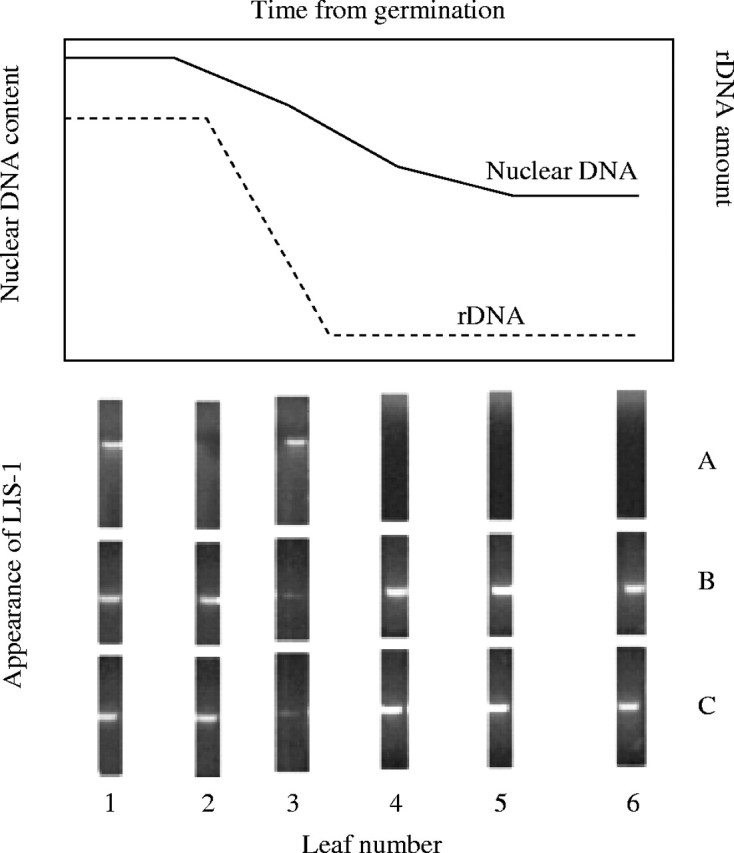
Total nuclear DNA, rDNA and LIS-1 variation during growth. These data are a compilation of the nuclear DNA amount from Evans et al. (1966), rDNA variation from Cullis and Charlton (1981), and unpublished data for LIS-1 from Y. Chen and C. A. Cullis. The LIS-1 data represents the DNA isolated from six leaves equally spaced along the stem (1 being the lowest leaf and 6 being a leaf from the top of the stem at flowering) of a plant grown under inducing conditions that resulted in a small genotroph. As can be seen, by the fourth sample the leaves have become homozygous for the presence of LIS-1. A, Amplification using primers 1 and 2; B, amplification using primers 1 and 3; C, amplification using primers 2 and 4.
The variation in LIS-1 demonstrates another of the primary characteristics of the induced changes, namely the genotrophs are homozygous for all the DNA alterations that have been characterized and that this homozygosity occurs immediately within the inducing generation.
All these studies confirm that the DNA changes occur during the vegetative growth of the plants before flowering. The data for the total nuclear DNA indicates that there was a progressive decrease in DNA amount that spanned the whole of the pre-flowering period. The data for the two specific sequences are somewhat different in that the changes in each of the specific sequences occurred over a relatively short, but possibly different, period of the vegetative growth. These two observation can be reconciled if each of the variable sites within the genome change independently, but at a specific time during development. The accumulation of the altered sites results in the progressive change in total nuclear DNA amount.
The total nuclear DNA estimations were made using apical meristematic cells. Therefore the DNA can be observed to change in the apical meristem during vegetative growth. The changes seen in the rDNA and LIS-1 are both consistent with the changes occurring in the meristem during a short period of growth, and becoming homozygous almost immediately. With both LIS-1 and the rDNA, tissue samples were collected from the lower regions of the plant both early and late in the growth stage. No differences were observed in these two sets of samples, which is consistent with the idea that the DNA variations are only occurring in the meristem and not in differentiated tissue. Thus all the subsequent organs laid down following the change have the altered regions. That the variation only occurs in the meristematic cells is also supported by the fact that even at flowering the plants are still chimeric, with the lower parts of the plant having the characteristic genome constitution of the previous generation, while the upper regions of the plant have the altered constitution. The characterization of two varying sequences reinforces the observations on the total nuclear DNA amount that the variation takes place during vegetative growth. The identification has not been performed with both probes on the same experimental material. Therefore it is not certain that each of the sequences varies independently and at a different stage of the life cycle. However, in both cases the variation does take place over a short period of the life cycle.
The data demonstrate that the genomic variation occurs within the meristematic cells and that in this system the genomic variation becomes homozygous very rapidly. The data cannot differentiate between mechanisms in which either the homozygosity occurs by two independent events at each locus, or by some type of gene conversion event. Since a subset of changes occur in every individual plant under certain growth conditions, it is possible that these have some direct adaptive effect on the individuals in which they occur.
CONCLUDING REMARKS
The accumulation of evidence in flax has demonstrated the response at the genomic level to the growth environment. This response appears to be under genetic control since responsive and non-responsive genotypes have been described. Clearly there are many unanswered questions concerning this system. Foremost is that relating to the adaptive significance of any of the genomic variations. Experiments that are designed to follow the genomic variation, and associated phenotypic characters, under controlled changing growth environments may help answer this question. As genomic variations are identified that are correlated with phenotypic attributes, their occurrence in natural populations may point to their importance and activity in the genomic restructuring system active in natural populations.
The identity of the genes controlling the ability to respond can be identified through genomic mapping and marker-based gene isolation using flax varieties that differ in their responsiveness. Although the genomic resources available in flax are still limited, the small genome, and simple organization, should facilitate the identification and isolation of the genes underlying this important phenomenon. The identification of the genes and the underlying processes by which the genomic restructuring processes proceed in flax will be essential for the search for analogous systems in other plants. A strategy of ab initio screening in an attempt to uncover similar phenomena elsewhere is likely to prove difficult since even in flax, where the changes are large, the variation in the timing and stability of genomic variation would make it difficult for the phenomenon to be rediscovered.
LITERATURE CITED
- Bassi P. 1990. Quantitative variation of nuclear DNA during plant development: A critical analysis. Biological Reviews 65: 185–225. [Google Scholar]
- Bassi P. 1991. Repetitive non-coding DNA: A possible link between environment and gene expression in plants? Biologisches Zentralblatt 110: 1–13. [Google Scholar]
- Chen Y. 1999.An insertion sequence in flax induced by the environment. PhD Thesis, Case Western Reserve University, USA. [Google Scholar]
- Cullis CA. 1973. DNA differences between flax genotypes. Nature 243: 515–516. [DOI] [PubMed] [Google Scholar]
- Cullis CA. 1976. Environmentally induced changes in ribosomal RNA cistron number in flax. Heredity 36: 73–79. [DOI] [PubMed] [Google Scholar]
- Cullis CA. 1977. Molecular aspects of the environmental induction of heritable changes in flax. Heredity 38: 129–154. [Google Scholar]
- Cullis CA. 1979. Quantitative variation of ribosomal RNA genes in flax genotrophs. Heredity 42: 237–246. [Google Scholar]
- Cullis CA. 1980. DNA sequence organization in the flax genome. Biochimica et Biophysica Acta 652: 1–15. [DOI] [PubMed] [Google Scholar]
- Cullis CA. 1981. Environmental induction of heritable changes in flax: defined environments inducing changes in rDNA and peroxidase isozyme band pattern. Heredity 47: 87–94. [Google Scholar]
- Cullis CA. 1990. DNA rearrangements in response to environmental stress. Advances in Genetics 28: 73–97. [DOI] [PubMed] [Google Scholar]
- Cullis CA, Charlton LM. 1981. The induction of ribosomal DNA changes in flax. Plant Science Letters 20: 213–217. [Google Scholar]
- Cullis CA, Cleary W. 1986. Rapidly varying DNA sequences in flax. Canadian Journal of Genetics and Cytology 28: 252–259. [Google Scholar]
- Cullis CA, Cleary W. 1986. DNA variation in flax tissue culture. Canadian Journal of Genetics and Cytology 28: 247–251. [Google Scholar]
- Cullis CA, Creissen GP. 1987. Genomic variation in plants. Annals of Botany 60 (Supp 4): 103–113. [Google Scholar]
- Cullis CA, Song Y, Swami S. 1999. RAPD polymorphisms in flax genotrophs. Plant Molecular Biology 41: 795–800. [DOI] [PubMed] [Google Scholar]
- Durrant A. 1962. The environmental induction of heritable changes in Linum Heredity 17: 27–61. [Google Scholar]
- Durrant A. 1971. Induction and growth of flax genotrophs. Heredity 27: 277–298. [Google Scholar]
- Durrant A, Jones TWA. 1971. Reversion of induced changes in the amount of nuclear DNA in Linum Heredity 30: 369–379. [Google Scholar]
- Evans GM, Durrant A, Rees H. 1966. Associated nuclear changes in the induction of flax genotrophs. Nature 212: 697–699. [Google Scholar]
- Fedoroff NV. 1989. Maize transposable elements. In: Berg DE, Howe MM, eds. Mobile DNA. Washington DC: Amerian Society of Microbiology, 657–678. [Google Scholar]
- Goldsbrough PB, Cullis CA. 1981. Characterization of the genes for ribosomal RNA in flax. Nucleic Acid Research 9: 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien MA. 1998. Activation of plant retrotransposons under stress conditions. Trends in Plant Science 3: 181–187. [Google Scholar]
- Jiang N, Bao ZR, Zhang XY, Hirochika H, Eddy SR, McCouch SR, Wessler SR. 2003. An active DNA transposon family in rice Nature 421: 163–167. [DOI] [PubMed] [Google Scholar]
- Johnston JS, Jensen A, Czeschin DG, Price HJ. 1996. Environmentally induced nuclear 2C DNA content instability in Helianthus annus (Asteraceae). American Journal of Botany 83: 1113–1120. [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. 2000. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimate divergence. Proceedings of the National Academy of Sciences, USA 97: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck JE, Lawrence GJ, Finnegan EJ, Jones DA, Ellis JG. 1998. A flax transposon identified in two spontaneous mutant alleles of the L6 rust resistance gene. The Plant Journal 16: 365–369. [DOI] [PubMed] [Google Scholar]
- McClintock B. 1978. Mechanisms that rapidly reorganize the genome. Stadler Symp. 10: 25–48. [Google Scholar]
- Natali L, Cavallini A, Cionini G, Sassoli O, Cionini PG, Durante M. 1993. Nuclear DNA changes within Helianthus annus L.: changes within single progenies and their relationships with plant development. Theoretical and Applied Genetics 85: 506–512. [DOI] [PubMed] [Google Scholar]
- Natali L, Cavallini A, Cremonini R, Bassi P, Cionini PG. 1986. Amplification of nuclear DNA sequences during induced plant cell dedifferentiation. Cell Differentiation 18: 157 – 161. [DOI] [PubMed] [Google Scholar]
- Natali L, Giordani T, Cionini G, Pugliesi C, Fambrini M, Cavallini A. 1995. Heterochromatin and repetitive DNA frequency variation in regenerated plants of Helianthus annus L. Theoretical and Applied Genetics 91: 395–400. [DOI] [PubMed] [Google Scholar]
- Oh TJ, Cullis CA. 2003. Labile DNA Sequences in Flax Identified by Combined Sample Representational Difference Analysis (csRDA). Plant Molecular Biology 52: 527–536. [DOI] [PubMed] [Google Scholar]
- Richards EJ. 1997. DNA methylation and plant development. Trends in Genetics 13: 319–322. [DOI] [PubMed] [Google Scholar]
- Schneeberger R. 1992.Characterization of DNA polymorphisms associated with environmentally induced heritable changes in flax. Ph.D. Thesis, Case Western Reserve University, USA. [Google Scholar]
- Schneeberger R, Cullis CA. 1991. Specific DNA alterations associated with the environmental induction of heritable change in flax. Genetics 128: 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders MJM, Rus-Kortekaas W, Vosman, B. 1995. Tissue culture induced DNA methylation polymorphisms in repetitive DNA of tomato calli and regenerated plants. Theoretical and Applied Genetics 91: 1257–1264. [DOI] [PubMed] [Google Scholar]


