Abstract
• Background The phytohormone auxin is critical for plant growth and orchestrates many developmental processes.
• Scope This review considers the complex array of mechanisms plants use to control auxin levels, the movement of auxin through the plant, the emerging view of auxin-signalling mechanisms, and several interactions between auxin and other phytohormones. Though many natural and synthetic compounds exhibit auxin-like activity in bioassays, indole-3-acetic acid (IAA) is recognized as the key auxin in most plants. IAA is synthesized both from tryptophan (Trp) using Trp-dependent pathways and from an indolic Trp precursor via Trp-independent pathways; none of these pathways is fully elucidated. Plants can also obtain IAA by β-oxidation of indole-3-butyric acid (IBA), a second endogenous auxin, or by hydrolysing IAA conjugates, in which IAA is linked to amino acids, sugars or peptides. To permanently inactivate IAA, plants can employ conjugation and direct oxidation. Consistent with its definition as a hormone, IAA can be transported the length of the plant from the shoot to the root; this transport is necessary for normal development, and more localized transport is needed for tropic responses. Auxin signalling is mediated, at least in large part, by an SCFTIR1 E3 ubiquitin ligase complex that accelerates Aux/IAA repressor degradation in response to IAA, thereby altering gene expression. Two classes of auxin-induced genes encode negatively acting products (the Aux/IAA transcriptional repressors and GH3 family of IAA conjugating enzymes), suggesting that timely termination of the auxin signal is crucial. Auxin interaction with other hormone signals adds further challenges to understanding auxin response.
• Conclusions Nearly six decades after the structural elucidation of IAA, many aspects of auxin metabolism, transport and signalling are well established; however, more than a few fundamental questions and innumerable details remain unresolved.
Keywords: Auxin; IAA; indole-3-acetic acid; 2,4-D; IBA; phytohormone; hormone signalling; proteasome; auxin biosynthesis; auxin conjugate; auxin transport; Arabidopsis thaliana
INTRODUCTION
To fully understand auxin regulation, action, and interactions will be to understand many aspects of plant growth and development. As a critical plant hormone, auxin modulates such diverse processes as tropic responses to light and gravity, general root and shoot architecture, organ patterning, vascular development and growth in tissue culture (Davies, 1995). The importance of auxin for human sustenance is both vital and readily apparent: auxin is required for plant growth. Anthropogenic manipulation of auxin physiology has assisted plant propagation, and, through the blind pressure of artificial selection, the development of modern crop varieties (Multani et al., 2003; Salamini, 2003).
Auxin biology is among the oldest fields of experimental plant research. Charles Darwin performed early auxin experiments, observing the effects of a hypothetical substance modulating plant shoot elongation to allow tropic growth toward light (Darwin, 1880). Darwin's experiments expanded upon Theophil Ciesielski's research examining roots bending toward gravity (Ciesielski, 1872). The term auxin was coined by scientists examining plant growth-modulating substances in human urine named auxins A and B (Kögl and Haagen Smit, 1931). A structurally distinct compound with auxin activity isolated from fungi was called heteroauxin; auxins A and B were gradually abandoned for the reproducibly bioactive heteroauxin, which was later determined to be indole-3-acetic acid (IAA) (Thimann, 1977).
COMPOUNDS WITH AUXIN ACTIVITY
Because auxins influence virtually every aspect of plant growth and development, numerous bioassays for auxin response have been described. These assays have proven useful in the isolation of endogenous auxins, the identification of auxin precursors, and the development of synthetic auxin-like compounds (Thimann, 1977). One of the earliest noted auxin effects was in phototropism, the curvature of stems toward a light source (Darwin, 1880). Application of auxin to decapitated shoots can induce such bending in the absence of a light stimulus (Went, 1926), and several nonphototropic mutants are deficient in auxin signalling components (Harper et al., 2000; Tatematsu et al., 2004).
The pea curvature test also employs auxin-regulated differential growth: dark-grown (etiolated) Pisum sativum stems are decapitated, sliced along part of their length, and floated in solution containing compounds being tested (Wain and Wightman, 1954; Fawcett et al., 1960). In auxin solution, stem segments bend inward, while in water they curl outward (Went and Thimann, 1937). Other tests to establish whether a given compound exerts auxin-like effects include spraying tomato plants and application to wheat coleoptiles, where auxin causes characteristic stem bending and elongation, respectively (Wain and Wightman, 1954; Fawcett et al., 1960).
Another early assay for auxin activity was in tissue culture, where auxins promote rooting from undifferentiated callus (Skoog and Miller, 1957). Along with the phytohormone cytokinin, which induces shoot formation, auxin allows regeneration of plants from cultured callus (Krikorian, 1995).
Current assays for auxin response in the model plant Arabidopsis thaliana often involve growth of seedlings on medium supplemented with the compound of interest. Auxins profoundly influence root morphology, inhibiting root elongation, increasing lateral root production (Fig. 1), and inducing adventitious roots (Zimmerman and Hitchcock, 1942). The relevance of these bioassays to normal plant physiology is supported by the observation that mutants that overproduce auxin tend to have abundant lateral and adventitious roots, along with long hypocotyls and petioles, and epinastic leaves and cotyledons (Boerjan et al., 1995; King et al., 1995; Delarue et al., 1998; Zhao et al., 2001). Conversely, mutants deficient in auxin responses are often characterized by long primary roots, few lateral roots, and short hypocotyls when grown on unsupplemented medium in the light, in addition to reduced auxin responses in the bioassays described above (Estelle and Somerville, 1987; Hobbie and Estelle, 1995; Monroe-Augustus et al., 2003).
Fig. 1.
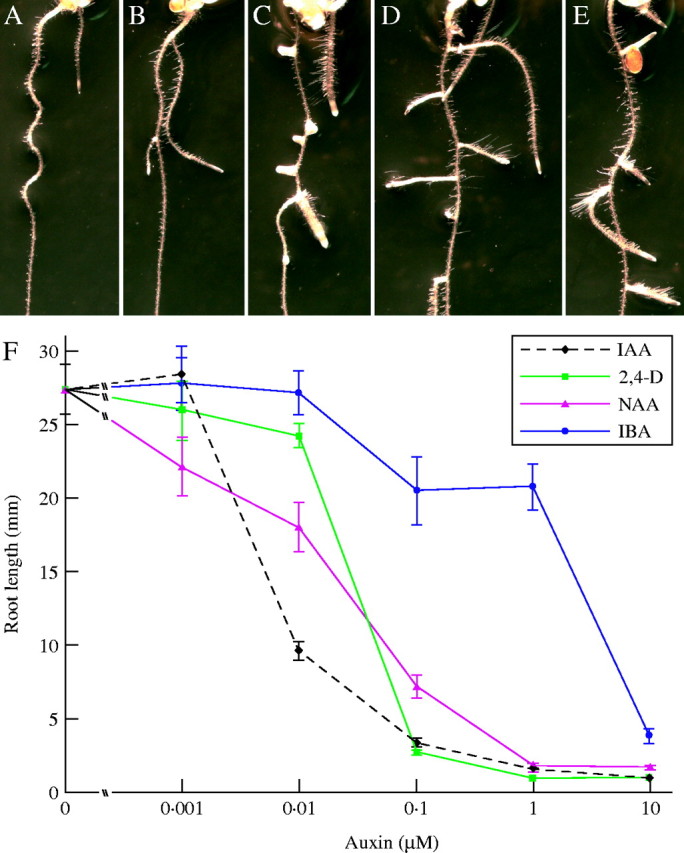
Auxins promote lateral root formation and inhibit root elongation. Arabidopsis thaliana Col-0 ecotype plants were grown on unsupplemented medium (Haughn and Somerville, 1986) for 6 d, then transferred to unsupplemented medium (A) or medium supplemented with 10 nm IAA (B), 100 nM 2,4-D (C), 100 nM NAA (D) or 10 μM IBA (E) and grown for 6 additional days. (F) Plants were grown on various concentrations of natural and synthetic auxins for 8 d. Points represent means ± standard error, n ≥ 8. All plants were grown at 22 °C under yellow light.
Many naturally occurring compounds that exert auxin-like effects have been revealed by these bioassays (Fig. 1). IAA, an extensively studied endogenous auxin, is active in all bioassays described above and is often potent at nanomolar concentrations (Fig. 1). A chlorinated form of IAA with high auxin activity, 4-Cl-IAA, is found in several plants (Slovin et al., 1999). In addition to the indolic auxins, phenylacetic acid (PAA) has been identified in plants and is an active auxin (Wightman, 1977; Ludwig-Müller and Cohen, 2002).
Certain IAA precursors, such as indole-3-acetonitrile and indole-3-pyruvic acid, are also active in bioassays, presumably because of conversion in the tissue to IAA (Thimann, 1977). Similarly, indole-3-butyric acid (IBA), identical to IAA except for two additional methylene groups in the side chain, is effective in bioassays. Like IAA, exogenous IBA inhibits arabidopsis root elongation (Zolman et al., 2000) and induces lateral (Zolman et al., 2000) and adventitious (King and Stimart, 1998) root formation. IBA, originally classified as a synthetic auxin, is in fact an endogenous plant compound (Epstein and Ludwig-Müller, 1993; Ludwig-Müller, 2000; Bartel et al., 2001). IBA is more effective than IAA at lateral root induction, perhaps because, unlike IAA, IBA efficiently induces lateral roots at concentrations that only minimally inhibit root elongation (Zolman et al., 2000); IBA is employed commercially for this purpose (Hartmann et al., 1990). Biochemical analyses in a variety of plants and genetic studies in arabidopsis indicate that IBA acts primarily through conversion to IAA in a process resembling peroxisomal fatty acid β-oxidation (Bartel et al., 2001), though roles for IBA independent of conversion to IAA have been proposed (Ludwig-Müller, 2000; Poupart and Waddell, 2000).
Two main types of synthetic plant growth regulators with auxin-like activity have been described: 1-naphthalacetic acid (NAA) and 2,4-D-related compounds. Both compounds exert auxin-like influences, including root elongation inhibition and lateral root promotion (Fig. 1). The NAA isomer 2-NAA has little activity in bioassays (Thimann, 1977) and provides a weak acid control for auxin experiments employing the active 1-NAA. 2,4-Dichlorophenoxybutyric acid (2,4-DB) is a 2,4-D derivative with two additional methylene groups in the side chain (analogous to the structural relationship between IBA and IAA) that elicits similar responses to those observed after 2,4-D treatment. In general, 2,4-dichlorophenoxyacetic acid (2,4-D) and IAA derivatives with even-numbered carbon side chains have more activity than derivatives with odd-numbered carbon side chains (Wain and Wightman, 1954; Fawcett et al., 1960). This result suggests that a process such as β-oxidation could remove two-carbon units from the side chains, arriving at the active acetate form if the substrate started with an even carbon number (Wain and Wightman, 1954; Fawcett et al., 1960). 2,4,5-Tricholorphenoxybutyric acid (2,4,5-TB) also exerts auxin-like activity; the infamous defoliant herbicide Agent Orange was a mixture of 2,4-D and 2,4,5-TB (Fallon et al., 1994). Agent Orange was particularly toxic because of dioxin produced as a by-product of 2,4,5-TB synthesis (Courtney et al., 1970; Schwetz et al., 1973). Today, 2,4-D alone is a widely used herbicide. In addition to NAA and 2,4-D, several alkylated and halogenated forms of IAA elicit auxin-like growth responses in various bioassays (Antolić et al., 1996; Nigović et al., 2000). Though IAA, 2,4-D, NAA, and other synthetic compounds can cause similar physiological responses in bioassays, the molecules cause distinct but overlapping changes in gene expression (Pufky et al., 2003), perhaps reflecting differences in metabolism, transport, or interaction with the signalling machinery.
IAA BIOSYNTHETIC PATHWAYS
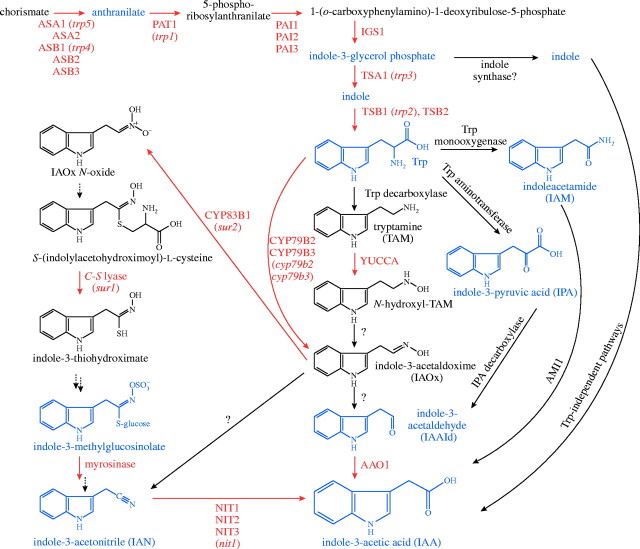
Arabidopsis seedlings can synthesize IAA in leaves, cotyledons and roots; young leaves have the highest biosynthetic capacity (Ljung et al., 2001). Although it is widely accepted that plants use several pathways to synthesize IAA, none of the pathways is yet defined to the level of knowing each relevant gene, enzyme, and intermediate. Plant genes implicated in IAA biosynthesis are listed in Table 1, and the reactions catalysed by the encoded enzymes are illustrated in Fig. 2. Plants use both tryptophan (Trp)-dependent and Trp-independent routes to synthesize IAA; several Trp-dependent pathways have been suggested. Multiple IAA biosynthetic pathways may contribute to regulation of IAA production, but the paucity of informative loss-of-function mutations in IAA biosynthetic enzymes, coupled with functional redundancy, has limited analysis of pathway control and prevented definitive determination of the importance of each pathway. For example, arabidopsis seedlings grown at high temperature accumulate free IAA (Gray et al., 1998) and display high-auxin phenotypes (Gray et al., 1998; Rogg et al., 2001), but the source of the excess IAA is unknown.
Table 1.
Plant genes implicated in de novo IAA biosynthesis
| Gene |
Product* |
Putative localization |
Loss-of-function (LOF) or overexpression (OE) phenotype |
Reference |
|---|---|---|---|---|
| AAO1 | IAAld oxidase | Cytoplasm | OE in sur1 | Sekimoto et al. (1998); Seo et al. (1998) |
| AMI1 | IAM hydrolase | Not reported | Not reported | Pollmann et al. (2003) |
| CYP79B2, CYP79B3 | P450 monooxygenases | Chloroplast | LOF: cyp79B2 cyp79B3: low glucosinolates, IAN, and IAA OE: resistant to Trp analogues; high indolic glucosinolate, IAN, and IAA–X levels | Zhao et al. (2002); Hull et al. (2000); Mikkelsen et al. (2000); |
| CYP83B1/SUR2 | P450 monooxygenase | Cytoplasm | LOF: high IAA, IAAld, and IAA–Asp levels; normal IAN levels; low indolic glucosinolate levels; altered Trp biosynthetic gene expression; defective photomorphogenesis in red light | Delarue et al. (1998); Barlier et al. (2000); Bak et al. (2001); Smolen and Bender (2002); Hoecker et al. (2004) |
| OE: high indolic glucosinolate levels, reduced apical dominance | Bak et al. (2001) | |||
| NIT1 | Nitrilase | Not reported | LOF: IAN resistant, normal IAA levels | Normanly et al. (1997) |
| NIT2 | Nitrilase | Not reported | OE: increased sensitivity to IAN, normal IAA levels | Normanly et al. (1997) |
| ZmNIT2 | Nitrilase (maize) | Not reported | Not reported | Park et al. (2003) |
| SUR1/RTY/ALF1/HLS3 | C-S lyase | Not reported | LOF: high IAA and IAA–X levels, low glucosinolates | Boerjan et al. (1995); Celenza et al. (1995); King et al. (1995); Golparaj et al. (1996); Lehman et al. (1996); Mikkelsen et al. (2004) |
| TDC | Trp decarboxylase (C. roseus) | Cytoplasm | OE: enhanced root curling | De Luca et al. (1989); Guillet et al. (2000) |
| TSA1/TRP3 | Trp synthase α | Chloroplast | LOF: high IAA–X, IAN, indole-glycerol phosphate, and indolic glucosinolate levels; normal free IAA; low Trp; Trp auxotroph | Normanly et al. (1993); Müller and Weiler (2000a); Ouyang et al. (2000) |
| TSB1/TRP2 | Trp synthase β | Chloroplast | LOF: high IAA–X and IAN levels, normal free IAA, low Trp, Trp auxotroph | Normanly et al. (1993); Ouyang et al. (2000) |
| ORP | Trp synthase β (maize) | Chloroplast | LOF: high IAA–X, normal free IAA, Trp auxotroph | Wright et al. (1991, 1992) |
| YUCCA, YUCCA2 | FMO-like | Cytoplasm | LOF: no phenotype OE: high IAA levels | Zhao et al. (2001) |
| FLOOZY | FMO-like (petunia) | Not reported | LOF: defective leaf venation and apical dominance OE: high IAA levels | Tobeña-Santamaria et al. (2002) |
Listed genes are from arabidopsis unless otherwise noted.
Fig. 2.
Potential pathways of IAA biosynthesis in arabidopsis. De novo IAA biosynthetic pathways initiate from Trp or Trp precursors. Compounds quantified in arabidopsis are in blue, enzymes for which the arabidopsis genes are identified are in red, and arabidopsis mutants are in lower-case italics. Suggested conversions for which genes are not identified are indicated with question marks. Trp biosynthesis and the P450-catalysed conversion of Trp to IAOx are chloroplastic, whereas many Trp-dependent IAA biosynthetic enzymes are apparently cytoplasmic. See Table 1 for references.
Trp-dependent IAA biosynthesis
Several Trp-dependent pathways, which are generally named after an intermediate, have been proposed: the indole-3-pyruvic acid (IPA) pathway, the indole-3-acetamide (IAM) pathway, the tryptamine pathway, and the indole-3-acetaldoxime (IAOx) pathway. An arabidopsis enzymatic complex that converts Trp to IAA in vitro has been partially purified (Müller and Weiler, 2000b), and future biochemical and genetic dissection of the process is likely to reveal the relative importance of the pathways discussed below.
The IPA pathway [Trp → IPA → indole-3-acetaldehyde (IAAld) → IAA] is important in some IAA-synthesizing microorganisms (Koga, 1995) and may operate in plants as well (Cooney and Nonhebel, 1991). IPA is found in arabidopsis seedlings (Tam and Normanly, 1998), but genes encoding a Trp aminotransferase that oxidatively transaminates Trp to IPA or an IPA decarboxylase that converts IPA to IAAld have not been identified in plants. The final enzyme in the proposed IPA pathway is an IAAld-specific aldehyde oxidase protein (AAO1) that has increased activity in the IAA-overproducing superroot1 (sur1) mutant (Seo et al., 1998). The identification of arabidopsis AAO1 does not verify the existence of the IPA pathway, however, as IAAld may be an intermediate in other IAA biosynthetic pathways (see below).
The IAM pathway [Trp → IAM → IAA] is a second microbial pathway that also may act in plants. In Agrobacterium tumifaciens and Pseudomonas syringae, for example, Trp monooxygenase (IaaM) converts Trp to IAM, and an IAM hydrolase (IaaH) converts IAM to IAA (Patten and Glick, 1996). IAM lacks auxin activity in arabidopsis, which allows the iaaH gene to be used as a screenable marker that confers IAM sensitivity (Brusslan et al., 1993). Intriguingly, IAM is found in arabidopsis seedlings at levels similar to free IAA (Pollmann et al., 2002), and an arabidopsis amidohydrolase (AMI1) converts IAM to IAA in vitro (Pollmann et al., 2003). It will be interesting to learn whether disruption of AMI1 or AAO1 decreases IAA levels.
YUCCA may catalyse a rate-limiting step in a tryptamine pathway
A tryptamine (TAM) pathway [Trp → TAM → N-hydroxyl-TAM → indole-3-acetaldoxime (IAOx) → IAAld → IAA] could also convert Trp to IAA (Fig. 2). Trp decarboxylase converts Trp to tryptamine in the first committed step in the biosynthesis of Catharanthus roseus monoterpenoid indole alkaloids (Facchini et al., 2000). The arabidopsis genome contains potential Trp decarboxylase genes, but the encoded enzymes have not been characterized, and tryptamine has not been identified in arabidopsis.
The identification of yucca, an IAA-accumulating mutant with classic high-auxin phenotypes (Zhao et al., 2001), suggests that a tryptamine IAA biosynthetic pathway may operate in some plants. yucca is resistant to toxic Trp analogues, suggesting that the accumulating IAA is Trp-derived (Zhao et al., 2001). The yucca phenotype derives from overexpression of a flavin monooxygenase (FMO)-like enzyme that oxidizes tryptamine to N-hydroxyl-tryptamine in vitro (Zhao et al., 2001). The homologous Petunia × hybrida enzyme FLOOZY is defective in a mutant deficient in leaf venation and apical dominance (Tobeña-Santamaria et al., 2002). Although the loss-of-function floozy mutant has wild-type IAA levels, overexpressing FLOOZY results in increased IAA levels in shoot apices and young leaves (Tobeña-Santamaria et al., 2002). YUCCA may be a rate-limiting enzyme in the tryptamine pathway, but a test of this hypothesis is hampered by genetic redundancy. Arabidopsis has a family of ten YUCCA-like enzymes, and insertional mutations in YUCCA and YUCCA2 confer no morphological phenotypes (Zhao et al., 2001). The N-hydroxyl-tryptamine produced by YUCCA could be dehydrogenated to IAOx or dehydrogenated and hydrolysed to IAAld (Fig. 2). Enzymes that catalyse these conversions have not been identified.
Indole-3-acetaldoxime is a precursor to indolic glucosinolates that can be converted to IAA
The IAOx pathway [Trp → IAOx → IAN or IAAld → IAA] is of particular interest in plants like arabidopsis that make indolic glucosinolate secondary metabolites (Fahey et al., 2001), because IAOx is the branch-point between indole-3-methylglucosinolate and IAA biosynthesis (Fig. 2). Two arabidopsis P450 monooxygenases, CYP79B2 and CYP79B3, oxidize Trp to IAOx in vitro (Hull et al., 2000; Mikkelsen et al., 2000). CYP79B2 overexpressors have increased IAA, IAN (Zhao et al., 2002) and indolic glucosinolate levels (Mikkelsen et al., 2000). Conversely, the cyp79B2 cyp79B3 double mutant has morphological phenotypes suggestive of low auxin, reduced IAA in certain growth conditions, lowered IAN levels, and no detectable indolic glucosinolates (Zhao et al., 2002). Taken together, these results are consistent with IAOx serving as a precursor that can be shunted to either auxin or indolic glucosinolates.
A third P450 monooxygenase, CYP83B1, converts IAOx to its N-oxide, the first committed step in indole-3-methylglucosinolate biosynthesis (Fig. 2; Bak et al., 2001). Loss-of-function cyp83b1 alleles were independently isolated in screens for high-auxin seedling phenotypes (superroot2 or sur2; Delarue et al., 1998), altered resistance to toxic Trp analogues (Smolen and Bender, 2002), defective photomorphogenesis in red light (Hoecker et al., 2004), and P450 monooxygenase insertional disruptions (Winkler et al., 1998). The sur2/cyp83B1 mutant accumulates free IAA (Delarue et al., 1998; Barlier et al., 2000) and the IAA precursor IAAld (Barlier et al., 2000). This phenotypic analysis, along with the nature of the defective gene, suggests that IAOx accumulates in the mutant and is converted to IAAld, which is oxidized to IAA (Fig. 2).
The sur1 mutant (Boerjan et al., 1995), also isolated as rooty (King et al., 1995), alf1 (Celenza et al., 1995) and hookless3 (Lehman et al., 1996), provides another link between high auxin and defects in glucosinolate production. This mutant has high-auxin phenotypes resembling sur2 and yucca, and accumulates free IAA and IAA conjugates (Boerjan et al., 1995; King et al., 1995; Lehman et al., 1996). sur1 is defective in a C-S lyase that apparently cleaves S-(indolylacetohydroximoyl)-l-cysteine to indole-3-thiohydroximate, the third step in glucosinolate production from IAOx (Golparaj et al., 1996; Mikkelsen et al., 2004). Indeed, indolic glucosinolates are undetectable in sur1 (Mikkelsen et al., 2004). Given the multiplicity of available pathways to modulate IAA levels, it is intriguing that arabidopsis plants cannot adequately compensate for the increased IAA precursor levels that result when indolic glucosinolate production is dampened.
Indole-3-acetonitrile and nitrilases in IAA biosynthesis
Nitrilases that can hydrolyse IAN to IAA are found in several plant families, including crucifers and grasses (Thimann and Mahadevan, 1964). These enzymes are encoded by the arabidopsis NIT genes (Bartling et al., 1992, 1994; Bartel and Fink, 1994) and Zea mays (maize) ZmNIT2 (Park et al., 2003). NIT1 and NIT2 can hydrolyse IAN applied to plants (Schmidt et al., 1996; Normanly et al., 1997), and an enzymatic complex with nitrilase immunoreactivity converts Trp to IAA in vitro (Müller and Weiler, 2000b). IAN is present in arabidopsis (Normanly et al., 1993; Ilić et al., 1996) and maize (Park et al., 2003), suggesting that this conversion could contribute to IAA homeostasis. In the brassica, IAN is formed following myrosinase-catalysed indole-3-methylglucosinolate hydrolysis, and IAN levels tend to track with indolic glucosinolate levels in arabidopsis mutants (Normanly et al., 1993; Mikkelsen et al., 2000; Müller and Weiler, 2000a; Reintanz et al., 2001; Zhao et al., 2002), consistent with nitrilases acting downstream of glucosinolates in arabidopsis. However, it has also been suggested that IAN is an intermediate in IAOx to IAA conversion (Fig. 2), although enzymes catalysing the conversion of IAOx to IAN have not been isolated, and the source of IAN in maize, which lacks indolic glucosinolates, is unknown.
NIT1 is the most highly expressed of the four arabidopsis NIT genes (Bartel and Fink, 1994). nit1 mutants are resistant to exogenous IAN (Normanly et al., 1997), but lack obvious low-auxin phenotypes, indicating that any role played by NIT1 in IAA biosynthesis is redundant. The NIT2 gene is normally expressed at a low level, but is induced by a bacterial pathogen (Bartel and Fink, 1994), by Plasmodiophora (Grsic-Rausch et al., 2000), during arabidopsis leaf senescence (Quirino et al., 1999), and in response to IAN treatment (Grsic et al., 1998). NIT2 induction correlates with decreased IAN levels and increased IAA levels during senescence (Quirino et al., 1999), increased IAA levels in Plasmodiophora-infected roots (Grsic-Rausch et al., 2000) and higher nitrilase immunoreactivity (Müller and Weiler, 2000a) in the IAN-accumulating trp3 mutant (Normanly et al., 1993). NIT3 expression is induced by sulfur starvation, and is correlated with reduced indolic glucosinolate levels and lateral root proliferation (Kutz et al., 2002). Expression of maize nitrilase ZmNIT2 is elevated in embryonic tissue (Park et al., 2003). Upgrading these correlations between expression and IAA levels to causal relationships awaits the analysis of additional nit family mutants and would be aided by an arabidopsis nit1 nit2 nit3 triple mutant.
Analyses of trp mutants reveal Trp-independent IAA biosynthesis
In addition to the proposed Trp-dependent IAA biosynthetic pathways (Fig. 2), analyses of Trp biosynthetic mutants demonstrate that plants also can synthesize IAA without using a Trp intermediate. The arabidopsis trp3-1 and trp2-1 mutants are defective in Trp synthase α and β, respectively (Last et al., 1991; Radwanski et al., 1996). These mutants accumulate amide- and ester-linked IAA conjugates (Normanly et al., 1993; Ouyang et al., 2000), despite having low soluble Trp levels (Müller and Weiler, 2000a; Ouyang et al., 2000). Similarly, the maize orange pericarp Trp synthase β mutant accumulates IAA conjugates (Wright et al., 1991, 1992). Unlike trp2 and trp3, plants blocked earlier in the Trp pathway, such as trp1 (Last and Fink, 1988) and antisense plants with decreased indole-3-glycerol phosphate synthase (IGS) levels, do not accumulate IAA conjugates (Normanly et al., 1993; Ouyang et al., 2000).
Analyses of the trp mutants imply that a Trp-independent IAA biosynthetic pathway branches from indole-3-glycerol phosphate or indole (Fig. 2). Trp synthase α and β normally channel indole-3-glycerol phosphate to Trp without indole release. In maize, however, Trp synthase α-like enzymes can act without β subunits to produce indole released as a volatile or converted into certain defense compounds (Frey et al., 1997, 2000; Melanson et al., 1997) or possibly IAA. Arabidopsis contains two apparent Trp synthase α genes: TSA1, the gene defective in the trp3 mutant (Radwanski et al., 1996), and a second uncharacterized gene (At4g02610).
Because IAA conjugates are hydrolysed under alkaline conditions (Bialek and Cohen, 1986; Baldi et al., 1989), total (free plus conjugated) IAA is often inferred without knowledge of the conjugates present by quantifying free IAA after alkaline hydrolysis. The specificity of the alkaline hydrolysis evidence used to support the importance of the Trp-independent pathway has been questioned (Müller and Weiler, 2000a). Application of this technique requires accommodation for the indolic biochemistry of the plant under study. For example, IAN, which is present in arabidopsis, is hydrolysed to IAA under alkaline conditions, so IAN must be separately quantified and subtracted from apparent total IAA values (Ilić et al., 1996). As the individual conjugates of arabidopsis are identified and quantified, it will be interesting to learn the precise conjugate profiles in the various trp mutants, and to reinvestigate alkaline-releasable IAA in mutant plants that lack indolic glucosinolates, for example.
An independent method to clarify biosynthetic pathways involves feeding plants isotopically labelled substrates, which, in a linear pathway, will result in isotopic enrichment of a precursor relative to its product. Intact arabidopsis seedlings do not efficiently convert [2H5]Trp into IAA, but the Trp precursor [15N]anthranilate labels IAA more completely than Trp (Normanly et al., 1993), confirming the importance of Trp-independent IAA biosynthesis during normal growth. Arabidopsis shoot and root explants, however, do efficiently convert [2H5]Trp to IAA (Müller et al., 1998b; Müller and Weiler, 2000a). Because the explant process may damage tissue, this result suggests that Trp-dependent IAA biosynthesis may be wound-induced in arabidopsis, as it is in bean (Sztein et al., 2002). Plants may switch from basal Trp-independent IAA biosynthesis to Trp-dependent pathways during stress, when more IAA may be needed (Ribnicky et al., 2002; Sztein et al., 2002). Studies examining metabolism of a recently synthesized, isotopically labelled indole may allow dissection of Trp-independent IAA biosynthesis (Ilić and Cohen, 2004).
IAA STORAGE: CONJUGATES AND INDOLE-3-BUTYRIC ACID
Higher plants can store IAA in the form of IAA conjugates and indole-3-butyric acid (IBA), which can provide free IAA upon hydrolysis or β-oxidation, respectively (Fig. 3). IAA can be ester-linked to sugars or amide-linked to amino acids and peptides. Proposed functions for these conjugates include storage, transport, compartmentalization, excess IAA detoxification, and protection against peroxidative degradation (Cohen and Bandurski, 1982). Certain IAA conjugates are active in auxin bioassays, and several plants store IAA conjugates in seeds that are hydrolysed during germination to provide free IAA to developing seedlings. In contrast, biologically inactive conjugates present in plants are probably intermediates in IAA degradation. Analyses of arabidopsis mutants defective in various facets of IAA homeostasis are revealing the roles of the diverse IAA sources during plant growth and development.
Fig. 3.
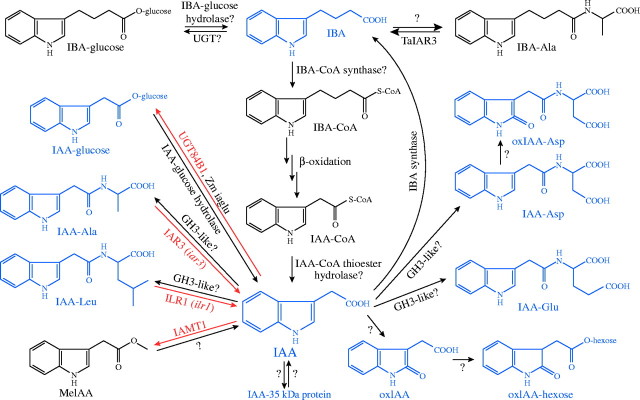
Potential pathways of IAA metabolism. Compounds quantified in arabidopsis are in blue, enzymes for which the arabidopsis genes are cloned are in red, and arabidopsis mutants are in lower-case italics. Suggested conversions for which plant genes are not identified are indicated with question marks. A family of amidohydrolases that apparently resides in the ER lumen can release IAA from IAA conjugates. ILR1 has specificity for IAA–Leu (Bartel and Fink, 1995), whereas IAR3 prefers IAA–Ala (Davies et al., 1999). Maize (Zm) iaglu and arabidopsis UGT84B1 esterify IAA to glucose (Szerszen et al., 1994; Jackson et al., 2001); the enzymes that form and hydrolyse IAA–peptides have not been identified. IBA is likely to be converted to IAA–CoA in a peroxisomal process that parallels fatty acid β-oxidation to acetyl-CoA (Bartel et al., 2001). IAA can be inactivated by oxidation (oxIAA) or by formation of non-hydrolysable conjugates (IAA–Asp and IAA–Glu). IAA–amino acid conjugates can be formed by members of the GH3/JAR1 family (Staswick et al., 2002, 2005). OxIAA can be conjugated to hexose, and IAA–Asp can be further oxidized (Östin et al., 1998). IAMT1 can methylate IAA (Zubieta et al., 2003), but whether this activates or inactivates IAA is not known. IBA and hydrolysable IAA conjugates are presumably derived from IAA; biosynthesis of these compounds may contribute to IAA inactivation. Formation and hydrolysis of IBA conjugates may also contribute to IAA homeostasis; the wheat (Ta) enzyme TaIAR3 hydrolyses IBA–Ala (Campanella et al., 2004).
IAA conjugate identification and functions
Different plant species have distinct IAA conjugate profiles (Cohen and Bandurski, 1982; Slovin et al., 1999). Experiments using alkaline hydrolysis to release free IAA from conjugates indicate that arabidopsis maintains approx. 90 % of IAA in amide linkages, with an additional approx. 10 % as ester-linked conjugates and approx. 1 % as free IAA (Normanly et al., 1993; Tam et al., 2000). Low levels of IAA–Ala, IAA–Asp, IAA–Glu and IAA–Leu are present in arabidopsis seeds (Rampey et al., 2004) and seedlings (Tam et al., 2000; Kowalczyk and Sandberg, 2001; Rampey et al., 2004). However, most of the amide-linked conjugates in arabidopsis seeds are solvent insoluble (Ljung et al., 2002), suggesting that single-amino acid conjugates constitute only part of the amide fraction in this tissue. A 35-kDa IAA–peptide is present in arabidopsis seeds; the large size of this conjugate may contribute to the solvent insolubility of amide conjugates (Ljung et al., 2002). Although genes encoding arabidopsis IAA–peptides have not been identified, an IAA-modified bean protein is similar to a soybean late seed maturation protein (Walz et al., 2002), suggesting that certain seed storage proteins may function in both amino acid and phytohormone storage. In addition to amide conjugates, the ester conjugate IAA–glucose has also been quantified in several dicotyledonous plants (including arabidopsis) and the monocot maize (Tam et al., 2000; Jakubowska and Kowalczyk, 2004).
Among divergent plant phyla, endogenous IAA, IAA–amide and IAA–ester levels are quite variable (Sztein et al., 1999). The lycophyte Selaginella kraussiana accumulates large quantities of conjugates, particularly IAA–amide compounds (Sztein et al., 1999). After feeding labelled IAA to the lycophyte S. kraussiana, the fern Ceratopteris richardii and various mosses and liverworts, varied species-specific conjugate profiles become apparent; the conjugates formed include both previously identified and unknown IAA conjugates (Sztein et al., 1999). These results suggest ancient roles for conjugates in plant biology.
IAA–amino acid conjugates found in plants can be classified into two groups based on bioassay activity and susceptibility to hydrolysis in planta or by plant enzymes. IAA–Ala and IAA–Leu efficiently inhibit arabidopsis root elongation and are substrates of arabidopsis amidohydrolases (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002; Campanella et al., 2003; Rampey et al., 2004). In arabidopsis, IAA–Ala is present at highest levels in shoots, whereas IAA–Leu accumulates in roots (Kowalczyk and Sandberg, 2001), but neither conjugate is formed at detectable levels following IAA application to seedlings or leaves (Östin et al., 1998; Barratt et al., 1999). These results suggest that IAA–Ala and IAA–Leu function to supply free IAA.
In contrast, although IAA–Asp and IAA–Glu also are present in arabidopsis (Tam et al., 2000; Kowalczyk and Sandberg, 2001), they are not appreciably hydrolysed by arabidopsis seedlings (Östin et al., 1998), and are inefficient inhibitors of root elongation (Campanella et al., 1996; LeClere et al., 2002). Tissues such as expanding leaves and roots that contain the highest free IAA levels also contain the highest levels of IAA–Asp and IAA–Glu (Kowalczyk and Sandberg, 2001). These results are consistent with an IAA catabolic role for IAA–Asp and IAA–Glu (see ‘IAA inactivation’ section).
Genetic analysis of IAA conjugate hydrolysis
Several mutant screens using different bioactive IAA–amino acid conjugates have been conducted. If conjugates with auxin activity function solely through free IAA release, then conjugate-resistant mutants that retain wild-type sensitivity to IAA may have defects in conjugate uptake or hydrolysis. If bioactive conjugates play additional roles, these also may be uncovered through mutant analyses. ilr1 was isolated as an IAA–Leu resistant mutant with reduced sensitivity to root elongation inhibition caused by exogenous IAA–Leu. ilr1 is defective in an amidohydrolase that cleaves IAA–Leu and IAA–Phe (Bartel and Fink, 1995). Similarly, iar3 is IAA–Ala resistant and is defective in an amidohydrolase homologous to ILR1 that specifically hydrolyses IAA–Ala (Davies et al., 1999). The ILR1-like protein ILL2 is the most active IAA amidohydrolase in vitro (LeClere et al., 2002); however, no ill2 alleles were isolated in genetic screens for conjugate-resistant root elongation. Though ILR1 and IAR3 are expressed in seedling roots, ILL2 appears to be expressed predominantly in the shoot (Rampey et al., 2004). An ill2 T-DNA allele is sensitive to IAA–Leu, IAA–Phe and IAA–Ala, but, when combined in double and triple mutants with ilr1 and iar3, ill2 contributes to IAA–Phe resistance in roots and hypocotyls and IAA–Ala resistance in hypocotyls (Rampey et al., 2004).
Interestingly, ilr1 iar3 ill2 triple mutant seedlings display reductions in lateral root number, hypocotyl elongation in the light, sensitivity to exogenous IAA, and free IAA levels (Rampey et al., 2004). These results suggest that the endogenous IAA conjugate substrates of these hydrolases (IAA–Ala and IAA–Leu) are physiologically relevant sources of free IAA. The IAA–Leu insensitivity of the ilr1 iar3 ill2 mutant implies that at least some IAA conjugates with auxin activity act solely via their hydrolysis to free IAA. However, the triple hydrolase mutant retains partial responsiveness to IAA–Ala (Rampey et al., 2004), suggesting that IAA–Ala has some hydrolysis-independent activity or that additional enzymes hydrolysing IAA–Ala remain to be discovered.
The iar1 mutant is resistant to the known substrates of the ILR1 and IAR3 amidohydrolases and is defective in a membrane protein (Lasswell et al., 2000) that weakly resembles the ZIP family of metal transporters (Guerinot, 2000). Although the substrate and membrane localization of IAR1 are unknown, the fact that the amidohydrolases require divalent cations such as Mn2+, Co2+ or Cu2+ for activity in vitro (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002) suggests that metal homeostasis could impact conjugate hydrolysis by modulating amidohydrolase activity. Further supporting a role for metal homeostasis in IAA conjugate metabolism, the IAA–Leu and IAA–Phe resistant ilr2 mutant is also resistant to exogenous Co2+ and Mn2+ (Magidin et al., 2003). Because the novel ILR2 protein appears to influence metal transport and the ilr2 mutant has a resistance profile similar to ilr1, ILR2 may indirectly affect IAA-conjugate metabolism by negatively regulating transport of metals that influence ILR1 activity (Magidin et al., 2003).
The IAA–Ala resistant mutant iar4 harbours a defective mitochondrial-type pyruvate dehydrogenase E1α (LeClere et al., 2004). iar4 is generally defective in root elongation, but is resistant to several IAA–amino acid conjugates. Although a direct role for pyruvate dehydrogenase in IAA-conjugate hydrolysis is difficult to envision, the slight resistance of iar4 to the synthetic auxin 2,4-D implies that the mutant may be generally deficient in auxin metabolism or response. It is possible that pyruvic acid itself, or an anabolic or catabolic product, influences IAA homeostasis. Alternatively, a complex including IAR4 may function directly in IAA biosynthesis, catalysing indole-3-pyruvic acid dehydrogenation to yield IAA–CoA, a hypothetical precursor of IAA or IAA conjugates (LeClere et al., 2004).
The genes defective in the icr1 (IAA-conjugate resistant), icr2 (Campanella et al., 1996), and ilr3 (R. A. Rampey, M. Tierney and B. Bartel, unpubl. res.) mutants have not been reported. Genes currently implicated in IAA-conjugate responses are listed in Table 2. Because ilr2, ilr3, iar4, icr1 and icr2 are each represented by a single allele isolated in forward genetic screens, it is likely that conjugate resistance screens are not yet saturated. Sequence analysis suggests that the IAA–amino acid conjugate hydrolases reside in the ER (endoplasmic reticulum) lumen (Bartel and Fink, 1995; Davies et al., 1999). Interestingly, the essential auxin binding protein ABP1 (Chen et al., 2001) is also predominantly ER-localized (Jones, 1994), reinforcing the possibility of a role for this compartment in auxin biology. Analysis of additional mutants may reveal genes required for conjugate import into or IAA efflux from the ER, amidohydrolase transcript accumulation, or amidohydrolase localization, activity or stability. In theory, conjugate-resistant mutants that fail to import conjugates from the medium might be isolated as well (see ‘Auxin transport’ section).
Table 2.
Plant genes implicated in IAA conjugate metabolism
| Gene |
Product* |
Putative localization |
Loss-of-function (LOF) or overexpression (OE) phenotype |
Reference |
|---|---|---|---|---|
| ILR1 | IAA–amino acid amidohydrolase | ER lumen | LOF: IAA–Leu resistant | Bartel and Fink (1995) |
| IAR3 | IAA–amino acid amidohydrolase | ER lumen | LOF: IAA–Ala resistant | Davies et al. (1999) |
| TaIAR3 | IBA–Ala amidohydrolase (wheat) | Not reported | Not reported | Campanella et al. (2004) |
| ILL1 | IAA–amino acid amidohydrolase | ER lumen | Not reported | Bartel and Fink (1995) |
| ILL2 | IAA–amino acid amidohydrolase | ER lumen | LOF: enhances IAA–amino acid conjugate resistance of ilr1 and iar3 | Bartel and Fink (1995); Rampey et al. (2004) |
| GH3.1 | Putative IAA–amino acid synthase | Not reported | LOF: IAA hypersensitive | Staswick et al. (2005) |
| YDK1/GH3.2 | IAA–amino acid synthase | Not reported | OE: dwarf, few lateral roots, de-etiolated | Takase et al. (2004); Staswick et al. (2005) |
| LOF: IAA hypersensitive | ||||
| GH3.3 | IAA–amino acid synthase | Not reported | None reported | Staswick et al. (2005) |
| GH3.4 | IAA–amino acid synthase | Not reported | None reported | Staswick et al. (2005) |
| GH3.5 | IAA–amino acid synthase | Not reported | LOF: IAA hypersensitive | Staswick et al. (2005) |
| DFL1/GH3.6 | IAA–amino acid synthase | Not reported | OE: IAA resistant, few lateral roots, dwarf, short hypocotyl in light, accumulates IAA–Asp | Nakazawa et al. (2001); Staswick et al. (2005) |
| GH3.17 | IAA–amino acid synthase | Not reported | LOF: IAA hypersensitive | Staswick et al. (2005) |
| IAR1 | Putative ZIP family transporter | Membrane | LOF: IAA–amino acid conjugate resistant | Lasswell et al. (2000) |
| ILR2 | Novel protein | Not reported | LOF: IAA–Leu resistant | Magidin et al. (2003) |
| IAR4 | Putative pyruvate dehydrogenase E1α subunit | Mitochondrion | LOF: IAA–Ala resistant | LeClere et al. (2004) |
| ICR1 | Not reported | Not reported | IAA–Phe resistant | Campanella et al. (1996) |
| ICR2 | Not reported | Not reported | IAA–Phe resistant | Campanella et al. (1996) |
| ILR3 | Not reported | Not reported | IAA–Leu resistant | R. A. Rampey, M. Tierney, and B. Bartel (unpubl. res.) |
| UGT84B1 | IAA glucosyl-transferase | Not reported | OE: IAA resistant, reduced apical dominance | Jackson et al. (2001, 2002) |
| iaglu | IAA glucosyl-transferase (maize) | Not reported | Not reported | Szerszen et al. (1994) |
| IAMT1 | IAA–methyl transferase | Not reported | Not reported | Zubieta et al. (2003) |
Listed genes are from arabidopsis unless otherwise noted.
The endogenous auxin IBA is converted to IAA in peroxisomes
IBA is a naturally occurring auxin in a variety of plants (Epstein and Ludwig-Müller, 1993; Ludwig-Müller, 2000; Bartel et al., 2001). Arabidopsis seedlings contain somewhat less free IBA than IAA (Ludwig-Müller et al., 1993), although detailed studies indicating whether this trend holds at all developmental stages have not been completed. Conditions that change IAA levels tend to similarly alter IBA levels (Ludwig-Müller et al., 1993), suggesting that IAA and IBA metabolism are linked. Indeed, arabidopsis seedlings fed labelled IAA make labelled IBA, suggesting that IBA is synthesized from IAA (Ludwig-Müller and Epstein, 1994). Because IBA also acts as an IAA precursor (see below), IBA could function similarly to bioactive IAA conjugates in IAA homeostasis (Bartel et al., 2001).
The auxin activity of IBA results, at least in part, from its conversion to IAA (Fig. 3). Isolation and characterization of arabidopsis mutants with IBA-resistant, IAA-sensitive root elongation are clarifying our understanding of IBA action (Poupart and Waddell, 2000; Zolman et al., 2000, 2001a, b; Zolman and Bartel, 2004; Woodward and Bartel, 2005). Mutants with specific β-oxidation defects are IBA resistant, suggesting that IBA is converted to IAA in a process paralleling fatty acid β-oxidation. Because plants β-oxidize fatty acids solely in peroxisomes (Gerhardt, 1992; Kindl, 1993), and several IBA-response mutants also have peroxisomal defects, IBA to IAA conversion is likely peroxisomal.
Peroxisomal β-oxidation of seed storage lipids provides energy to germinating seedlings in oil-seed plants like arabidopsis. As a result, arabidopsis fatty acid utilization mutants require supplemental sucrose after germination to prevent developmental arrest (Hayashi et al., 1998). Similarly, many IBA-response mutants are sucrose-dependent during seedling development, have reduced rates of seed storage lipid utilization, and are IBA resistant in both root elongation and lateral root initiation (Zolman et al., 2000, 2001a, b; Zolman and Bartel, 2004; Woodward and Bartel, 2005). These phenotypes suggest defects in the peroxisomal β-oxidation of long-chain fatty acids and IBA. Other IBA-response mutants appear to metabolize long-chain fatty acids normally during germination (Zolman et al., 2000; Adham et al., 2005), but may still have defects in IBA β-oxidation, perhaps due to lesions in isozymes specific to short-chain substrates and IBA.
Cloning the genes defective in several IBA-response mutants (Table 3) has substantiated the essential role of peroxisomal β-oxidation in IBA activity. In addition to the proteins required directly in peroxisomal metabolism, more than 20 proteins are required for peroxisome biogenesis and import of peroxisomal matrix proteins from the cytoplasm (Olsen, 1998; Subramani, 1998; Tabak et al., 1999; Mullen et al., 2001). Mutations in PEX5 or PEX7, receptors that bind and transport proteins into the peroxisomal matrix (Olsen, 1998; Subramani, 1998), confer IBA-response defects (Zolman et al., 2000; Woodward and Bartel, 2005). pex5 and pex7 are likely to have defects importing β-oxidation enzymes from the cytoplasm, slowing β-oxidation and causing IBA resistance. Another IBA-response mutant is defective in the peroxisome biogenesis gene PEX6 and has abnormal peroxisome morphology (Zolman and Bartel, 2004). PXA1, a membrane protein that is approx. 30 % identical to human and yeast ATP-binding cassette transporters implicated in importing long-chain fatty acids into peroxisomes (Dubois-Dalcq et al., 1999; Holland and Blight, 1999), is defective in another IBA-response mutant (Zolman et al., 2001b). Because pxa1 is resistant to IBA and is sucrose-dependent during seedling development, PXA1 is probably necessary for the import of both IBA and fatty acids (or the corresponding CoA esters) into peroxisomes (Zolman et al., 2001b; Footitt et al., 2002; Hayashi et al., 2002).
Table 3.
Arabidopsis genes implicated in IBA metabolism
| Gene |
Product |
Putative localization |
Mutant phenotypes* |
Reference |
|---|---|---|---|---|
| ACX1 | Long chain acyl-CoA oxidase | Peroxisome | IBA resistant, sucrose independent | Hooks et al. (1999); Adham et al. (2005) |
| ACX3 | Medium chain acyl-CoA oxidase | Peroxisome | IBA resistant, 2,4-DB resistant, sucrose independent | Eastmond et al. (2000); Adham et al. (2005) |
| ACX4 | Short chain acyl-CoA oxidase | Peroxisome | IBA resistant, 2,4-DB resistant, sucrose independent | Rylott et al. (2003); Adham et al. (2005) |
| AIM1 | Multifunctional protein | Peroxisome | abnormal inflorescence meristems, IBA resistant, sucrose dependent | Richmond and Bleecker (1999); Zolman et al. (2000) |
| PED1 | Thiolase | Peroxisome | IBA resistant, 2,4-DB resistant, sucrose dependent | Hayashi et al. (1998); Zolman et al. (2000) |
| PXA1/PED3/CMT | ABC transporter-like | Peroxisome | IBA resistant, 2,4-DB resistant, sucrose dependent, fewer lateral roots | Zolman et al. (2001b); Footitt et al. (2002); Hayashi et al. (2002) |
| PEX5 | Receptor for peroxisomal matrix protein import | Peroxisome | IBA resistant, weak sucrose dependence | Brickner et al. (1998); Zolman et al. (2000) |
| PEX6 | ATPase | Peroxisome | IBA resistant, sucrose dependent, fewer lateral roots | Zolman and Bartel (2004) |
| PEX7 | Receptor for peroxisomal matrix protein import | Peroxisome | IBA resistant, sucrose independent; strong sucrose dependence and fewer lateral roots in pex7-1 pex5-1 double mutant | Woodward and Bartel (2005) |
| PEX14/PED2 | Docking protein for PEX5 | Peroxisome | IBA resistant, sucrose dependent | Hayashi et al. (1998); Hayashi et al. (2000); Monroe-Augustus (2004) |
| IBR1 | Not reported | Not reported | IBA resistant, sucrose independent | Zolman et al. (2000) |
| IBR3 | Not reported | Not reported | IBA resistant, sucrose independent | Zolman et al. (2000) |
All mutants are presumed loss-of-function alleles.
Defects in β-oxidation enzymes can also lead to IBA resistance (Table 3). Several arabidopsis peroxisomal β-oxidation defective mutants have been isolated using resistance to the IBA analogue 2,4-dichlorophenoxybutyric acid (2,4-DB) (Hayashi et al., 1998), which is converted to the active synthetic auxin 2,4-D similarly to IBA β-oxidation (Wain and Wightman, 1954). 2,4-DB-resistant mutants include acx3 (Eastmond et al., 2000), acx4 (Rylott et al., 2003), aim1 (Richmond and Bleecker, 1999) and ped1 (Hayashi et al., 1998), which are also IBA resistant (Zolman et al., 2000; Adham et al., 2005). acx mutants have defects in acyl-CoA oxidases catalysing the second step of fatty acid β-oxidation, aim1 (abnormal inflorescence meristem) is a mutant in a multifunctional protein acting in the third and fourth steps of fatty acid β-oxidation (Richmond and Bleecker, 1999), and peroxisome defective l (ped1) is defective in a thiolase catalysing the final step of β-oxidation (Hayashi et al., 1998). Moreover, mutations in the gene encoding PEX14/PED2, which docks PEX5 at the peroxisome membrane, confer resistance to 2,4-DB (Hayashi et al., 1998, 2000) and IBA (Monroe-Augustus, 2004).
Because arabidopsis mutants defective in fatty acid β-oxidation enzymes and peroxisome biogenesis proteins are IBA resistant, IBA is likely to be converted to IAA in peroxisomes. It remains to be determined whether enzymes that catalyse fatty acid β-oxidation also directly catalyse IBA β-oxidation, or whether there are peroxisomal enzymes dedicated to IBA β-oxidation. At least some fatty acid β-oxidation enzymes appear not to act on IBA, as evidenced by the normal IBA and 2,4-DB responses of the lacs6 lacs7 double mutant, which is sucrose dependent due to defects in peroxisomal acyl-CoA synthetases catalysing the first step of fatty acid β-oxidation (Fulda et al., 2004). If IBA to IAA conversion requires dedicated enzymes, one would expect to recover IBA-response mutants defective in these isozymes that retain normal fatty acid β-oxidation. Moreover, the inferred peroxisomal localization of IBA to IAA conversion implies the existence of a hydrolase that releases IAA from the CoA ester (Fig. 3), unless this thioester is efficiently hydrolysed nonenzymatically, and a transporter that effluxes IAA or IAA–CoA out of the peroxisome. Indeed, several sucrose-independent IBA-response mutants, including ibr1 and ibr3, are candidates for having defects in such functions (Zolman et al., 2000).
Several peroxisome defective IBA-response mutants have reduced lateral root initiation not only following IBA exposure (Zolman et al., 2000), but also in the absence of exogenous auxin (Zolman et al., 2001b; Zolman and Bartel, 2004; Woodward and Bartel, 2005). Similarly, certain Pyrus communis (pear) plants with adventitious root formation defects apparently do not convert IBA to IAA (Baraldi et al., 1993). These defects imply that the IAA formed from endogenous IBA β-oxidation during seedling development is important for lateral root initiation. The lateral rooting defects in the peroxisome defective IBA-response mutants (Zolman et al., 2001b; Zolman and Bartel, 2004; Woodward and Bartel, 2005) are more severe than those of the conjugate hydrolase triple mutant (Rampey et al., 2004), suggesting that conjugate hydrolysis does not fully compensate for a lack of IBA β-oxidation, and vice versa.
A few IBA-response mutants with apparently normal fatty acid β-oxidation are less sensitive than wild type to the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) and auxin transport inhibitors (Zolman et al., 2000). The rib1 (resistant to IBA) mutant is in this class (Poupart and Waddell, 2000). Moreover, the Lateral rootless (Lrt1) Oryza sativa (rice) mutant is resistant to IAA, IBA and 2,4-D in terms of root elongation, but only IBA can restore lateral root initiation to the mutant (Chhun et al., 2003). Identifying the genes defective in these IBA-response mutants may reveal IAA-independent roles for IBA or unique features of IBA biology, such as factors differentially mediating IBA and IAA transport (Rashotte et al., 2003).
Like IAA, much of the IBA in plants is conjugated to other moieties through amide- and ester-linkages (Epstein and Ludwig-Müller, 1993; Ludwig-Müller, 2000). A wheat homologue of the arabidopsis IAR3 IAA–Ala hydrolase is inactive on IAA conjugates, but rather hydrolyses amino acid conjugates of IBA including IBA–Ala, which is present in wheat extracts (Campanella et al., 2004). It will be interesting to learn whether the other members of the monocot amidohydrolase family have specificity for IAA– or IBA–amino acid conjugates. In arabidopsis, IBA is largely ester linked (Ludwig-Müller et al., 1993), suggesting that a different family of enzymes will catalyse IBA release. Although the complete IAA and IBA conjugate profiles have not been reported for any plant, it is likely that these profiles will be complex and reflect the diversity and specificities of the corresponding conjugate hydrolases and synthases.
IAA INACTIVATION
Pathways that inactivate IAA (Fig. 3) counteract the inputs to the IAA pool. As discussed above, IAA conjugates that accumulate following exposure of arabidopsis to IAA apparently differ from those used for IAA storage, consistent with the conjugated moiety dictating the fate of the attached IAA (Cohen and Bandurski, 1982). Arabidopsis permanently inactivates applied IAA by ring oxidation to oxIAA (Fig. 3), which can then be conjugated to hexose (Östin et al., 1998). In addition, IAA is conjugated to Asp and Glu after applying 5 μm IAA (Östin et al., 1998), and to Asp, Glu, Gln and glucose in response to 500 μm IAA (Barratt et al., 1999). Arabidopsis seedlings do not appreciably hydrolyse IAA–Asp and IAA–Glu, and IAA–Asp can be further oxidized to oxIAA–Asp (Östin et al., 1998), reinforcing the catabolic nature of Asp conjugation. The catabolic conjugation system is probably present during normal growth, because IAA–Asp and IAA–Glu are present at low levels in arabidopsis seedlings (Tam et al., 2000; Kowalczyk and Sandberg, 2001; Rampey et al., 2004).
In response to elevated IAA levels, catabolic conjugation pathways may be up-regulated and storage conjugation pathways down-regulated. For example, the sur2 mutant accumulates free IAA (see above) and IAA–Asp (Barlier et al., 2000), an intermediate in permanent IAA inactivation (Normanly, 1997; Slovin et al., 1999). However, sur2 plants inefficiently form the putative arabidopsis IAA storage compound IAA–Leu (Barlier et al., 2000). The high-auxin phenotype in the yucca mutant is suppressed by expressing iaaL (Zhao et al., 2001), a microbial IAA-conjugating enzyme (Glass and Kosuge, 1986), suggesting that IAA-conjugating activities are insufficient in yucca. In contrast, the trp2 and trp3 mutants apparently accumulate IAA conjugates but not free IAA (Normanly et al., 1993), implying that the conjugation pathways are sufficient to accommodate the accumulating IAA in this case.
A screen for mutants accumulating new conjugates following exposure to prolonged high-IAA challenge revealed that the photorespiration mutant gluS accumulates IAA–Gln at the expense of IAA–Asp following IAA treatment (Barratt et al., 1999). Because the gluS mutant, which is defective in chloroplastic glutamate synthase, has increased soluble Gln levels (Somerville and Ogren, 1980), this altered conjugate profile implies that conjugates formed following IAA challenge are, in part, dependent on amino acid pool sizes.
Remarkably, the enzymes that conjugate IAA to amino acids are encoded by members of the GH3 family of auxin-induced genes (see ‘Auxin-induced transcripts’ section). These enzymes are in the luciferase superfamily (Staswick et al., 2002) and are related to the JAR1 enzyme that conjugates the hormone jasmonic acid to amino acids (Staswick and Tiryaki, 2004). Two members of the arabidopsis GH3 family have been uncovered as genes overexpressed in dwarf mutants with reduced apical dominance, dfl1-D (Nakazawa et al., 2001) and ydk1-D (Takase et al., 2004). dfl1-D is resistant to applied IAA, and both dfl1-D and ydk1-D have reduced lateral rooting and hypocotyl elongation (Nakazawa et al., 2001; Takase et al., 2004). These phenotypes are consistent with decreased free auxin levels, which would be expected when overexpressing an IAA-conjugating enzyme; indeed, IAA–Asp levels are elevated in dfl1-D (Staswick et al., 2005). Further, disruption of certain GH3 genes confers hypersensitivity to specific forms of auxin conjugated by the encoded GH3 (Staswick et al., 2005). The characterized GH3-like enzymes apparently prefer to synthesize inactivating (IAA–Asp and –Glu) over hydrolysable (IAA–Ala and -Leu) conjugates in vitro (Staswick et al., 2005); it will be interesting to learn which enzymes are responsible for synthesizing hydrolysable conjugates in vivo.
In addition to IAA–amino acid conjugates, the esterified conjugate IAA–glucose is found in plants (Chisnell and Bandurski, 1988; Tam et al., 2000; Jakubowska and Kowalczyk, 2004) and plant proteins with IAA glucosyltransferase activity have been identified (Leznicki and Bandurski, 1988; Szerszen et al., 1994; Jackson et al., 2001; Jakubowska and Kowalczyk, 2004). Maize iaglu (Szerszen et al., 1994) and arabidopsis UGT84B1A (Jackson et al., 2001) encode glucosyl transferases that conjugate IAA to glucose. Overexpressing UGT84B1 renders plants resistant to exogenous IAA and disrupts gravitropism (Jackson et al., 2002), consistent with a role in IAA inactivation. Paradoxically, free IAA levels are actually increased in UGT84B1-overexpressing lines (Jackson et al., 2002). Because UGT84B1 also has considerable activity on IBA (Jackson et al., 2001), it or a related glucosyl transferase may be responsible for the IBA–glucose formed from labelled IBA fed to arabidopsis seedlings (Ludwig-Müller and Epstein, 1993).
An enzyme that methylates the carboxyl side chain of IAA has recently been described (Zubieta et al., 2003). This enzyme, IAMT1, is a member of a family of carboxyl methyltransferases; other members methylate plant hormones such as jasmonic acid (JA) and salicylate (Zubieta et al., 2003). Methylation will increase the volatility of IAA, but it is not clear whether this modification activates or inactivates the hormone. In the case of JA, overexpressing the gene encoding jasmonate methyltransferase JMT increases resistance to a fungal pathogen (Seo et al., 2001), implying that methyl jasmonate is an active signalling molecule. Understanding of the in vivo roles of methylated IAA awaits analysis of plants with increased and decreased accumulation of this derivative.
Finally, as arabidopsis can convert IAA to IBA (Ludwig-Müller and Epstein, 1994), IBA synthase might contribute to IAA inactivation (Fig. 3). An IBA synthase regulated by a variety of biotic and abiotic stresses (Ludwig-Müller, 2000) has been partially purified from maize (Ludwig-Müller and Hilgenberg, 1995), but the specific components or its effects on the free IAA pool have not been determined.
AUXIN TRANSPORT
Although many tissues can synthesize auxin (Ljung et al., 2001), auxin transport is complex and highly regulated, involving many identified proteins. Chemical and genetic studies have revealed that transport of auxin to distant sites is clearly required for normal development. For example, IAA transport is necessary for proper lateral root development (R.C. Reed et al., 1998; Bhalerao et al., 2002), vascular development (Mattsson et al., 1999), phyllotaxis (Reinhardt et al., 2003), embryonic axis development (Friml et al., 2003) and tropisms (Friml et al., 2002).
Auxin is produced largely in shoot apical regions, historically identified as the shoot apical meristem. However, application of auxin transport inhibitors blocks IAA accumulation in the shoot apex, suggesting that apical auxin is transported from other regions, probably young leaves and developing leaf primordia (Ljung et al., 2001; Avsian-Kretchmer et al., 2002). IAA is transported basipetally in shoots (Lomax et al., 1995) and suppresses lateral shoot growth (Thimann and Skoog, 1934). Both acropetal (Scott and Wilkins, 1968) and basipetal (Davies and Mitchell, 1972) transport occurs in roots. Opposing directions of auxin transport in roots is achieved by spatial separation, with acropetal transport in the central cylinder and basipetal transport in the epidermis (Tsurumi and Ohwaki, 1978).
Several arabidopsis mutants are defective in proteins mediating polar auxin transport. Among the earliest genes cloned that were defective in auxin-resistant mutants was AUX1, which encodes a transmembrane protein similar to amino acid permeases (Bennett et al., 1996). AUX1, and possibly other closely related proteins, mediates influx of IAA into cells (Marchant et al., 1999). AUX1 is localized asymmetrically in the plasma membrane of certain cell files, facilitating directional auxin transport (Swarup et al., 2001, 2004). Interestingly, IAA and the synthetic auxin 2,4-D appear to be AUX1 substrates, but aux1 remains sensitive to the synthetic compound NAA; indeed, NAA can restore aux1 gravitropism (Yamamoto and Yamamoto, 1998).
Once IAA has entered a cell via AUX1, several factors regulate efflux. The pin-formed (pin1) arabidopsis mutant is characterized by shoot meristem defects causing inflorescences to terminate in pin-shaped points generally lacking lateral organs (Okada et al., 1991). PIN1 is a member of a multigene family (including EIR1/AGR1/PIN2, another gene implicated in polar auxin transport) that encodes transmembrane auxin efflux facilitator proteins with homology to bacterial efflux carriers (Chen et al., 1998; Gälweiler et al., 1998; Luschnig et al., 1998; Müller et al., 1998a; Utsuno et al., 1998). Like AUX1, PIN1 is asymmetrically localized in the cell, consistent with a role in polar auxin efflux (Gälweiler et al., 1998).
The polar localization of PIN proteins is complex and dynamic. After a change in the gravity vector, PIN3 quickly moves from all lateral sides of columella cells specifically to the lateral side newly oriented toward gravity (Friml et al., 2002). Both PIN1 (Geldner et al., 2001) and PIN3 (Friml et al., 2002) rapidly cycle between the plasma membrane and unidentified endosomal compartments. PIN1 cycling is inhibited by the auxin transport inhibitor TIBA; movement from endosomal compartments to the plasma membrane is likewise inhibited by the vesicular trafficing inhibitor BFA (Geldner et al., 2001). PIN localization responds to cues from the PINOID serine-threonine kinase; PINOID overexpression or disruption alters the polar localization of PIN proteins in the cell (Friml et al., 2004). PIN cycling is actin-dependent (Geldner et al., 2001), and links between actin, polar auxin transport and gravitropism have been reported in several plants (Hou et al., 2003, 2004; Sun et al., 2004).
Flavonoids, compounds that accumulate in specific locations in light-grown arabidopsis (Buer and Muday, 2004), negatively regulate auxin transport (Brown et al., 2001) and PIN1 cycling (Peer et al., 2004). PIN protein expression and localization are altered in flavonoid mutants (Peer et al., 2004) and the arabidopsis transparent testa4 (tt4) mutant lacks flavonoids and has increased root basipetal and acropetal IAA transport and delayed gravitropism (Buer and Muday, 2004).
In addition to PIN proteins, certain MULTIDRUG RESISTANCE-like (MDR) proteins similar to mammalian MDR proteins are necessary for polar auxin transport in arabidopsis (Noh et al., 2001), maize and Sorghum bicolor (Multani et al., 2003). The arabidopsis MDR proteins MDR1 and P-GLYCOPROTEIN 1 (PGP1) bind the auxin transport inhibitor NPA (Noh et al., 2001). Basal PIN1 localization is disrupted in the mdr1 pgp1 double mutant (Noh et al., 2003). PXA1, the peroxisomal membrane transporter necessary for IBA β-oxidation (Zolman et al., 2001b), is also an MDR-like protein.
The arabidopsis mutant transport inhibitor response 3 (tir3, allelic to doc1, big and umb1; see ‘Auxin interactions with other hormones’ section) has reduced auxin transport (Ruegger et al., 1997). The mutant, resistant to root elongation inhibition by NPA, has reduced NPA binding to microsomal fractions (Ruegger et al., 1997). Another mutant with pleiotropic phenotypes, roots curl in NPA (rcn1, allelic to eer1; see ‘Auxin interactions with other hormones’ section), is defective in a protein phosphatase 2A subunit (Deruère et al., 1999) and actually has increased basipetal auxin transport (Rashotte et al., 2001).
Differential transport of auxin precursors and storage forms offers a potential point of auxin regulation. Little is known about the transport of IAA conjugates. The aux1 auxin influx carrier mutant (Bennett et al., 1996) is resistant to IAA–Ala and IAA–Leu (B. Bartel, unpubl. res.), as well as to IAA (Pickett et al., 1990), so it is possible that at least these conjugates enter cells similarly to free IAA. Alternatively, the conjugate resistance of aux1 could reflect failure to take up IAA freed by conjugate hydrolysis.
Similarly, the aux1 mutant (Bennett et al., 1996) is less sensitive than wild type to root elongation inhibition by exogenous IBA (Zolman et al., 2000) and IAA competes effectively for labelled IBA uptake into arabidopsis seedlings (Ludwig-Müller et al., 1995), consistent with a shared importer. However, experiments with labelled IBA suggest that it is transported more efficiently than IAA in roots, and labelled IBA transport is unaffected in the aux1 mutant (Rashotte et al., 2003). Thus, the IBA-resistance of aux1 root elongation may reflect resistance to IAA derived from IBA.
IBA appears not to be a good substrate of the IAA efflux carrier. Unlike wild type, roots of the eir1/agr1/pin2 auxin transport mutant (Chen et al., 1998; Luschnig et al., 1998; Müller et al., 1998a; Utsuno et al., 1998) bend and enter vertically oriented medium containing substrates of the efflux carrier, such as IAA or NAA (Utsuno et al., 1998). However, eir1 roots fail to bend and enter media containing 2,4-D (Utsuno et al., 1998) or IBA (Poupart and Waddell, 2000; Zolman et al., 2000), suggesting that IBA, like 2,4-D, does not use the IAA efflux carrier. In support of this observation, labelled IBA transport is unaffected in eir1 roots and is NPA-insensitive in wild-type roots (Rashotte et al., 2003). Proteins specifically mediating IBA transport remain to be identified.
AUXIN SIGNALLING
Auxin-induced transcripts
IAA biosynthesis, metabolism, and transport together ensure that appropriate auxin levels are in place to orchestrate plant development. How the signalling between auxin and downstream effectors occurs is the subject of much research. Auxin rapidly and transiently induces accumulation of at least three families of transcripts: SMALL AUXIN-UP RNAs (SAURs), GH3-related transcripts and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) family members.
SAUR transcripts accumulate rapidly after auxin exposure in soybean (Walker and Key, 1982) and many other species, including arabidopsis (Gil et al., 1994). Maize ZmSAUR2 is a small nuclear protein that, like the encoding transcript, is rapidly degraded (Knauss et al., 2003). The short half-lives of SAUR mRNAs appear to be conferred by downstream elements (DSTs) in the 3′ untranslated region of the messages (Sullivan and Green, 1996). Arabidopsis mutants that stabilize DST-containing RNAs, and thus stabilize SAUR transcripts, have no reported morphological phenotype (Johnson et al., 2000), and the function of these small RNAs remains unknown.
GH3 transcript accumulation is also induced by auxin (Hagen et al., 1984) and numerous plants have auxin-responsive GH3-like genes. At least some IAA-induced GH3 genes encode IAA–amino acid conjugating enzymes (Staswick et al., 2005), whereas several GH3-related proteins that are not auxin regulated function to adenylate or conjugate amino acids to molecules other than IAA, including jasmonic acid (Staswick et al., 2002; Staswick and Tiryaki, 2004). Thus, the auxin induction of GH3 genes likely serves to dampen the auxin signal by inactivating IAA via conjugation (see ‘IAA inactivation’ section).
Like the GH3 family, Aux/IAA transcripts accumulate following auxin exposure, and the encoded proteins also apparently serve to dampen auxin signalling. Induction of some Aux/IAA genes occurs within minutes of auxin application and does not require new protein synthesis (Abel et al., 1994; Abel and Theologis, 1996). The Aux/IAA family includes 28 proteins in arabidopsis (Liscum and Reed, 2002; Remington et al., 2004), and homologous genes, some of which are also auxin-induced, are present in other plants. The encoded proteins share extensive sequence identity in four conserved domains. Domain I is a transcriptional repressor (Tiwari et al., 2004). Domain II is critical for Aux/IAA instability; several mutations in this domain have been isolated as gain-of-function alleles that stabilize the proteins and confer auxin-resistant phenotypes (Kim et al., 1996; Rouse et al., 1998; Soh et al., 1999; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001; Fukaki et al., 2002; Hamann et al., 2002; Tatematsu et al., 2004; Yang et al., 2004). In fact, a 13-amino acid fragment of domain II from IAA7/AXR2 is sufficient to confer auxin-mediated degradation to a fusion protein (Ramos et al., 2001). Domains III and IV are involved in homodimerization and heterodimerization with other Aux/IAA proteins and with AUXIN REPONSE FACTORs (ARFs) (Kim et al., 1997; Ulmasov et al., 1999a; Hardtke et al., 2004).
Many genes with auxin-induced expression, including most SAUR, GH3 and Aux/IAA genes, share a common sequence in their upstream regulatory regions, TGTCTC or variants, first identified from the promoter region of the pea PS-IAA4/5 gene (Ballas et al., 1993). Regions including this sequence, known as the Auxin-Responsive Element, or AuxRE, confer auxin-induced gene expression in synthetic constructs (Ulmasov et al., 1995, 1997b). More recently, genome-wide profiling experiments have revealed a wealth of auxin-induced genes (Sawa et al., 2002; Pufky et al., 2003; Cluis et al., 2004; Himanen et al., 2004), many of which contain AuxREs in putative regulatory regions (Pufky et al., 2003; Nemhauser et al., 2004).
Auxin response factors mediate auxin-induced changes in gene expression
Identification of the AuxRE led to isolation of ARF1, the founding member of the AuxRE-binding protein family, using a yeast one-hybrid screen (Ulmasov et al., 1997a). ARF proteins can either activate or repress target gene transcription, depending on the nature of a central domain (Ulmasov et al., 1999b; Tiwari et al., 2003). ARFs can bind tandem repeat AuxRE sequences as homodimers, dimers with other ARFs, or dimers with repressive Aux/IAA proteins (Ulmasov et al., 1997a, 1999b). Auxin responsiveness depends on ARF motifs similar to Aux/IAA protein domains III and IV and is mediated through dimerization with Aux/IAA proteins (Tiwari et al., 2003). Because the dimerization domains are dispensable for ARF activation of reporter genes in Daucus carota protoplast assays, it is likely that ARFs can function as monomers (Tiwari et al., 2003).
Mutations in several arabidopsis ARF genes confer gene-specific developmental defects. Mutations in ETTIN/ARF3 (ETT) lead to floral abnormalities (Sessions and Zambryski, 1995; Sessions et al., 1997) that can be phenocopied by NPA application to flowers (Nemhauser et al., 2000). Genetic interaction between ETT and the SEUSS gene is necessary for proper floral development (Pfluger and Zambryski, 2004). Further, seuss mutants are defective not only in flower morphology, but are generally defective in auxin responses (Pfluger and Zambryski, 2004).
A screen for suppressors of a hookless1 (hls1) mutant, which fails to form an ethylene-mediated apical hook in darkness (Lehman et al., 1996), identified hooklessl suppressor 1/arf2 (Li et al., 2004). Ethylene acts through HLS1 to negatively regulate ARF2 levels and achieve an apical hook; ARF2 overexpression, like HLS1 deficiency, inhibits apical hook formation (Lehman et al., 1996; Li et al., 2004). Loss of arf2 function has no observable effect on apical hook formation when not in combination with hls1, but does cause various defects in adult morphology (Li et al., 2004).
Defects in MONOPTEROS/ARF5 (MP), a transcriptionally activating ARF, result in aberrant seedling morphology, often with a single cotyledon and a loss of basal structures (Hardtke and Berleth, 1998). Mutations in a second activating ARF, NON-PHOTOTROPIC HYPOCOTYL4 (NPH4/TIR5/MSG1/ARF7), confer deficient shoot phototropism, an auxin-mediated process (Harper et al., 2000). While the phenotypes of mp/arf5 and nph4/arf7 initially appeared unrelated, combining a weak allele of mp with nph4 in a double mutant enhances the mp fused cotyledon phenotype (Hardtke et al., 2004), suggesting some functional overlap between the two ARFs.
Thus, mutation of several ARF genes confers developmental phenotypes, and abundant evidence from studies employing reporter constructs in protoplast assays implicates ARFs in auxin responsive transcription. Studies in intact plants are beginning to directly link ARFs with auxin-responsive transcription at native promoters. For example, nph4/arf7 mutant seedlings display dramatically reduced levels of several auxin-induced transcripts both before and after auxin application (Stowe-Evans et al., 1998) and expression of several Aux/IAA genes responds to MP/ARF5 levels (Mattsson et al., 2003).
The diversity of arf mutant phenotypes makes it clear that the rules governing the interactions between ARFs and AuxREs will be complex; the fact that only a few arf mutants have been reported indicates that much of this complexity remains to be uncovered. ARF8 disruption leads to mild but significant high-auxin phenotypes such as stronger apical dominance, increased lateral root number, and a longer hypocotyl in the light; conversely, ARF8 overexpression results in opposite phenotypes, suggesting reduced auxin response (Tian et al., 2004). These observations are initially counterintuitive, because ARF8 is an activating ARF (Ulmasov et al., 1999b). However, these results can be neatly explained by the observation that ARF8 induces expression of several GH3 genes (Tian et al., 2004) involved in auxin inactivation (Staswick et al., 2005). Indeed, free IAA levels are reduced in hypocotyls of ARF8 overexpression lines (Tian et al., 2004), demonstrating the intimate connection between auxin responses and auxin levels.
Aux/IAA proteins repress ARF function
Aux/IAA proteins interact with ARF proteins via C-terminal domains III and IV conserved between the Aux/IAA family and most ARF proteins (Ulmasov et al., 1999a). At least some Aux/IAA proteins can directly repress transcription (Tiwari et al., 2004), and the interaction of Aux/IAA proteins with activating ARF proteins can prevent transcriptional activation in protoplast assays (Tiwari et al., 2003).
It may seem counterintuitive that expression of some Aux/IAA genes, which function to repress auxin signalling, is auxin-induced. However, Aux/IAA protein levels plummet immediately following auxin exposure (see below). Increased transcription of Aux/IAA genes after an auxin stimulus is likely to be mediated by ARF proteins via AuxREs in Aux/IAA promoter regions. Thus, Aux/IAA accumulation is subject to negative feedback; Aux/IAA protein levels decline after auxin exposure, allowing increased transcription of Aux/IAA genes and thereby ensuring a transient auxin response.
Gain-of-function Aux/IAA mutations generally reduce auxin sensitivity in root elongation assays and confer dramatic auxin-related developmental defects, including altered gravitropism and apical dominance in axr2/iaa7 (Wilson et al., 1990; Nagpal et al., 2000), axr3/iaa17 (Rouse et al., 1998) and axr5/iaa1 (Yang et al., 2004), severe lateral root defects in iaa28 (Rogg et al., 2001) and slr/iaa14 (Fukaki et al., 2002), photomorphogenic defects in shy2/iaa3 (Soh et al., 1999; Tian and Reed, 1999), hypocotyl tropism defects in msg2/iaa19 (Tatematsu et al., 2004), and embryonic patterning defects in bdl/iaa12 (Hamann et al., 2002). Remarkably, these dominant missense mutations all map to a small region of domain II and several have been shown to stabilize the encoded Aux/IAA proteins (Worley et al., 2000; Gray et al., 2001; Ramos et al., 2001; Tiwari et al., 2001; Zenser et al., 2001), underscoring the importance of this region in vivo. In contrast to the dramatic defects conferred by stabilizing Aux/IAA proteins, the few reported loss-of-function Aux/IAA alleles confer only subtle phenotypes. Most were isolated as suppressors of gain-of-function alleles mutated in the same gene (Rouse et al., 1998; Tian and Reed, 1999; Nagpal et al., 2000). Loss-of-function shy2/iaa3 mutants have large cotyledons and short hypocotyls (Tian and Reed, 1999), and null axr2/iaa7 mutants have slightly longer hypocotyls than wild type (Nagpal et al., 2000). The dramatic phenotypes of the gain-of-function Aux/IAA mutants coupled with the subtle effects of losing individual Aux/IAA genes suggests that these genes play important but largely overlapping roles in wild-type plants.
Aux/IAA degradation is mediated by the SCFTIR1 ubiquitin ligase
The Aux/IAA proteins, which inhibit auxin responses, are unstable even in the absence of a stimulus (Abel et al., 1994). Auxin application further destabilizes Aux/IAA proteins (Gray et al., 2001; Zenser et al., 2001), which is presumed to free activating ARF proteins from repression and thereby allow auxin-induced gene expression (Fig. 4). The transient nature of auxin-induced transcription is likely to result in part from many of the Aux/IAA genes themselves being transcriptionally induced by auxin (Abel et al., 1995b).
Fig. 4.
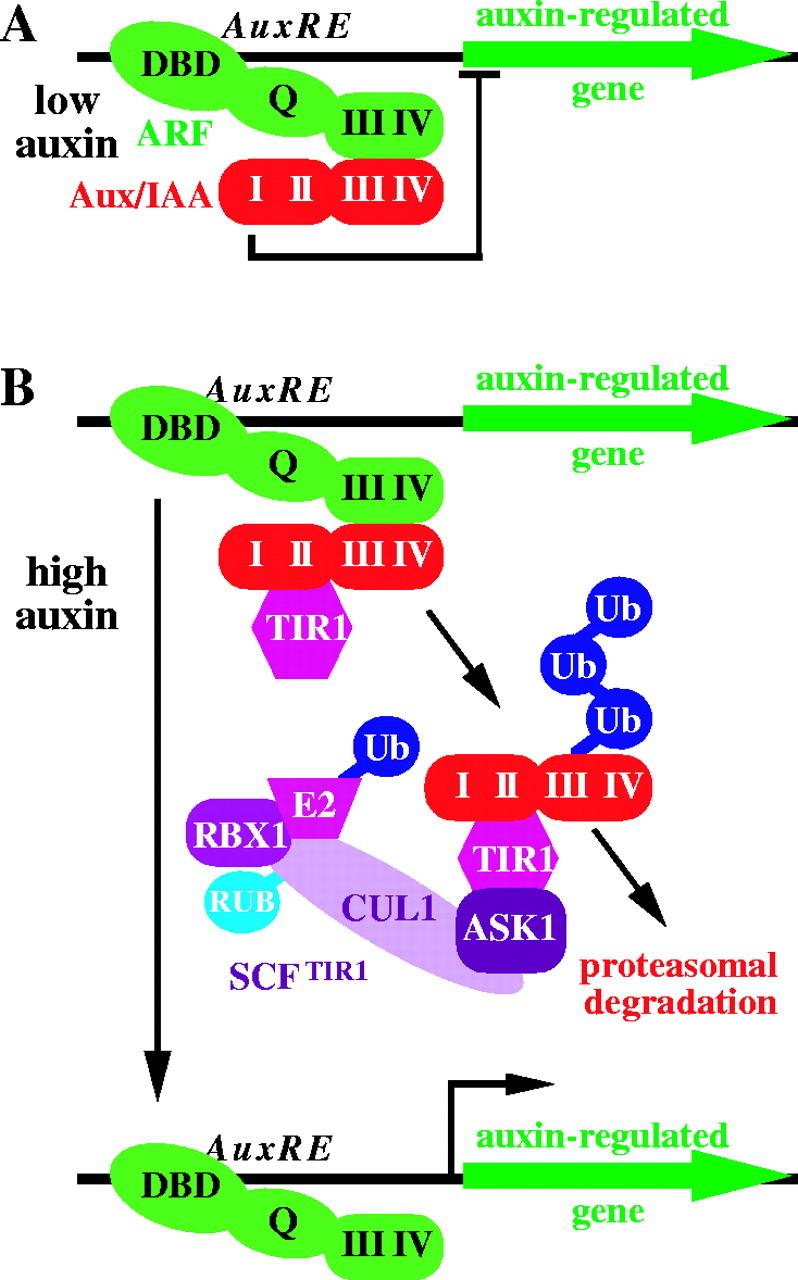
The SCFTIR1 relieves Aux/IAA repression of activating ARFs. (A) An activating ARF protein (green) binds an AuxRE promoter element via an N-terminal DNA binding domain (DBD). Under low-auxin conditions, an Aux/IAA repressor (red) binds the activating ARF via heterodimerization between Aux/IAA and ARF domains III and IV. (B) Auxin promotes Aux/IAA domain II-TIR1 association, bringing the Aux/IAA protein to the SCFTIR1 complex (purple) for ubiquitination (Ub) and subsequent destruction by the 26S proteasome. The activating ARF, with a Gln-rich (Q) middle domain, is then freed to promote auxin-induced gene expression.
Aux/IAA proteins are unstable because they are targets of ubiquitin-mediated degradation. Ubiquitin is covalently attached to substrate proteins in a three-step process that begins with activation of the ubiquitin C-terminus by an E1 enzyme, followed by ubiquitin transfer from the E1 to an E2 intermediary protein. Finally, the E2–ubiquitin complex is brought into proximity to the target protein via a specificity-providing E3 protein or protein complex, which mediates the ubiquitination of target proteins. Once targets are multiply ubiquitinated, they are substrates for degradation by the 26S proteasome. One type of E3 complex is the Skp1-Cullin-F-box (SCF) class, named for the defining components. SCF complexes are comprised of mostly general subunits, while one of numerous F-box proteins confers target specificity. The F-box protein associates with a cullin (CUL1) via a SKP adapter protein named ASK (from arabidopsis SKP1-like). CUL1 also binds to the ubiquitin E2-interacting protein RBX1 (also known as HRT1 or ROC1). Thus, the SCF complex recruits an E2 protein bearing ubiquitin to a specific protein target to facilitate ubiquitination of the target protein.
Several auxin-resistant mutants stabilize Aux/IAA proteins by disrupting components of the SCFTIR1 ubiquitin ligase or proteins that regulate the SCF (Table 4). tir1 mutants are resistant to auxins, auxin transport inhibitors and the auxin mimic sirtinol (Ruegger et al., 1998; Zhao et al., 2003b). TIR1 encodes a leucine-rich repeat-containing F-box protein (Ruegger et al., 1998) that lends Aux/IAA ubiquitination specificity to the SCFTIR1 complex. The auxin-resistant axr6 mutant is defective in CUL1 (Hellmann et al., 2003), and misexpressing RBX1 results in auxin resistance (Gray et al., 2002). Also, ASK1 is necessary for proper auxin response, and ASK1 and ASK2, members of a 19-member family (Farrás et al., 2001), interact with the TIR1 F-box (Gray et al., 1999).
Table 4.
Auxin-related SCF components and SCF-regulatory genes from arabidopsis
 |
Red, auxin-related phenotypes; blue, ethylene-related phenotypes; purple, jasmonate-related phenotypes; green, floral development phenotypes.
SCFTIR1 regulation is complex, and auxin resistance mutant screens have revealed regulatory components as well. AXR1 encodes one subunit of a ubiquitin E1-like enzyme complex (Leyser et al., 1993) with the partner protein E1-LIKE CONJUGATING ENZYME-RELATED 1 (ECR1) (del Pozo et al., 1998, 2002). The AXR1-ECR1 dimer activates and transfers RELATED TO UBIQUITIN 1 (RUB1, known as NEDD8 in animals) to the RUB1-CONJUGATING ENZYME RCE1 (S. Dharmasiri et al., 2003). RUB is subsequently transferred to a specific lysine on CUL1 (del Pozo and Estelle, 1999), apparently using RBX1 as the RUB E3 ligase to facilitate RUB transfer from RCE1 to CUL1 (Gray et al., 2002). Although mutants defective in ECR1 have not been reported, overexpression of ECR1 with the active site cysteine mutated confers developmental defects in the shoot (del Pozo et al., 2002). Defects in AXR1 (Estelle and Somerville, 1987; Lincoln et al., 1990), RUB (Bostick et al., 2004) or RCE1 (S. Dharmasiri et al., 2003; Larsen and Cancel, 2004) confer auxin resistance, and axr1 rce1 double mutants have embryonic defects similar to mp/arf5 (S. Dharmasiri et al., 2003), prompting the hypothesis that RUB modification positively regulates SCFTIR1 activity.
Recent results suggest a more complicated situation. CONSTITUTIVE PHOTOMORPHOGENESIS 9 (COP9) is a component of the COP9 signalosome complex, which resembles the 26S proteasome lid (Li and Deng, 2003). COP9 signalosome activity is necessary both for RUB removal from CUL1 and for proper auxin response (Schwechheimer et al., 2001). In addition, overexpressing RBX1 results in increased RUB-CUL1 levels and 2,4-D resistance (Gray et al., 2002). Thus, it appears that RUB addition to, and removal from, CUL1 are both required for SCFTIR1 function, implying a necessary RUB modification cycle. However, RUB deconjugation is separable from at least some developmental roles of the COP9 signalosome (Wang et al., 2002). Evidence that the COP9 signalosome interacts with proteasome components indicates that the COP9 signalosome may be a bona fide proteasome lid (Schwechheimer and Deng, 2001; Li and Deng, 2003; Peng et al., 2003) that could function directly in both RUB removal and ubiquitin-mediated substrate degradation.
A screen for mutations that enhance the 2,4-D resistance of tir1 uncovered several previously identified SCFTIR1 components as well as enhancer of tir1-1 auxin resistance 3 (eta3) (Gray et al., 2003). The eta3 single mutant phenotype is similar to tir1, and when combined in tir1 eta3, enhances the tir1 lateral root deficit, short hypocotyl and 2,4-D resistance phenotypes (Gray et al., 2003). eta3 is deficient in SGT1b (Gray et al., 2003), a homologue of yeast SGT1, which interacts with SCF complexes (Kitagawa et al., 1999). While the role of SGT1b remains obscure, eta3 is not deficient in SCFTIR1 assembly or auxin-responsive interaction of TIR1 with an Aux/IAA protein in vitro (Gray et al., 2003).
Further modulation of SCF is achieved by a plant orthologue of the human Cullin-Associated and Neddylation-Dissociated (CAND) protein, which is predicted to regulate SCF assembly (Liu et al., 2002; Zheng et al., 2002; Oshikawa et al., 2003). Defects in arabidopsis CAND1/ETA2 result in auxin resistance (Cheng et al., 2004; Chuang et al., 2004; Feng et al., 2004). Though CUL1 and RUB-CUL1 levels appear wild type in cand1 mutants (Chuang et al., 2004; Feng et al., 2004), pulldowns with the SCFTIR1-interacting domain of AXR2/IAA7 reveal increased CUL1, but decreased ASK1, association (Chuang et al., 2004). Further experiments examining SCF component and substrate interactions in cand1 are needed to understand these results, but it is clear that CAND1 is essential for proper SCFTIR1 function.
Auxin induction of Aux/IAA–TIR1 interaction
Auxin application to plants (Gray et al., 2001) or addition to plant cell lysate (N. Dharmasiri et al., 2003) can quickly induce Aux/IAA domain II interaction with TIR1. Thus, auxin could destabilize Aux/IAA proteins in planta by promoting association with the SCFTIR1 E3 ubiquitin ligase complex. The in vitro dissection of this TIR1–Aux/IAA interaction holds promise to reveal at least one of the long-sought auxin receptors.
Many substrates of SCF complexes require modification prior to ubiquitination (Deshaies, 1999). Several observations led to the early hypothesis that alterations to proline residues in domain II could provide a mechanism for auxin promotion of TIR1–Aux/IAA domain II interaction. First, several dominant auxin-resistant mutants harbour mutations altering one of two adjacent prolines within domain II (Rouse et al., 1998; Soh et al., 1999; Tian and Reed, 1999; Nagpal et al., 2000; Rogg et al., 2001; Fukaki et al., 2002; Hamann et al., 2002; Tatematsu et al., 2004; Yang et al., 2004); many of these mutations are known to stabilize the mutant Aux/IAA protein (Worley et al., 2000; Ouellet et al., 2001). In addition, a mutant resistant to the auxin-like effects of sirtinol is defective in SIR1, a protein containing a domain present in certain Peptidyl Prolyl cis-trans Isomerases (PPIases; Zhao et al., 2003b), and the parvulin-class PPIase inhibitor juglone inhibits Aux/IAA–TIR1 interaction (N. Dharmasiri et al., 2003; Tian et al., 2003).
However, although proline modification in Aux/IAA domain II may be relevant to TIR1 interaction, it is not the change induced by auxin. Mass spectroscopic analysis of AXR3/IAA17 domain II reveals possible proline hydroxylation, but this state is not affected by auxin addition (Kepinski and Leyser, 2004). Indeed, auxin does not appear to cause any covalent modification of domain II (Kepinski and Leyser, 2004). In addition, sirtinol does not promote Aux/IAA–TIR1 interaction (Kepinski and Leyser, 2004); therefore, sirtinol (and SIR1) influence Aux/IAA turnover in some other way, perhaps by regulating SCFTIR1 or proteasome activity. SIR1 is in fact more closely related to E1 ligases than to PPIases, suggesting yet another ubiquitin-like modification (in addition to the known roles of ubiquitin and RUB) important for auxin responsiveness. Finally, juglone affects TIR1 or an associated protein, not Aux/IAA domain II, to repress Aux/IAA–TIR1 association (Kepinski and Leyser, 2004).
The signal by which auxin promotes Aux/IAA–TIR1 interaction remains unclear. Tantalizing results show that in vitro treatment of extracts with NAA induces modification of TIR1 or a TIR1-associated protein rather than the Aux/IAA proteins (Kepinski and Leyser, 2004). Thus the most direct hypothesis, that TIR1 is itself an auxin receptor and that association of auxin with TIR1 promotes its binding and ubiquitination of substrate Aux/IAA proteins, will soon be testable.
AUXIN INTERACTIONS WITH OTHER HORMONES
Auxin modulates hormone levels
Physiological studies suggest many links between phytohormones. Several other hormones modulate or are modulated by auxin levels and responses. One of the most-told stories in plant biology is the relationship between auxin and cytokinin, which can be employed in vitro to induce root and shoot development, respectively (Skoog and Miller, 1957). Auxin and cytokinin levels are inversely correlated in vivo (Eklöf et al., 2000) and auxin treatment can rapidly inhibit cytokinin biosynthesis (Nordström et al., 2004).
Auxin and the gaseous hormone ethylene are also intimately linked. Exogenous auxin exposure stimulates ethylene production (Morgan and Hall, 1962) through induction of a gene encoding the rate-limiting enzyme in ethylene biosynthesis (Abel et al., 1995a). Conversely, ethylene inhibits lateral (Burg and Burg, 1966) and basipetal (Suttle, 1988) auxin transport.
As with ethylene, auxin elicits increased gibberellic acid (GA) production (Ross et al., 2000), and basipetally transported auxin is necessary for the production of the active gibberellins GA1 and GA3 in barley (Wolbang et al., 2004). GAs act, at least in part, through promoting degradation of DELLA repressors (Silverstone et al., 2001); disrupting auxin transport precludes GA-mediated DELLA protein degradation (Fu and Harberd, 2003).
Auxin response is also connected to brassinosteroids (BRs), which act in concert with auxin to promote root gravitropic curvature in maize (Kim et al., 2000). BR and auxin treatments induce accumulation of many of the same transcripts (Goda et al., 2004; Nemhauser et al., 2004). Exposure to the hormone abscisic acid (ABA) decreases free IAA levels while increasing esterified IAA conjugates in muskmelon ovaries (Dunlap and Robacker, 1990). Antagonistic to auxin, exogenous ABA inhibits lateral root formation (De Smet et al., 2003).
The SCF links auxin signalling with other stimuli
Several mutants with defects in auxin response are also defective in responses to other phytohormones, suggesting communication or cross-talk between phytohormone response pathways. For example, mutations in the general components comprising or regulating the SCF cause deficient responses to multiple hormonal and environmental stimuli (Schwechheimer et al., 2002). Though the full phenotypic complexity of mutants compromised in multiple pathways is likely to be revealed in years to come, pleiotropic defects have already been reported for many mutants with compromised auxin response (Table 4).
axr1 mutants fail to efficiently modify CUL1 with RUB (del Pozo et al., 2002) and are resistant to the root elongation inhibition caused by auxins (Estelle and Somerville, 1987), cytokinin, ethylene (Timpte et al., 1995), methyl jasmonate (Schwechheimer et al., 2002) and epi-brassinolide (Tiryaki and Staswick, 2002). These data suggest that RUB modification is necessary for active SCF complexes and hormone responses in addition to SCFTIR1 and auxin.
Indeed, SCFCOI1 is necessary for jasmonate responses (Xie et al., 1998), and the RUB E2 enzyme-defective mutant rce1 is auxin and methyl-jasmonate resistant (S. Dharmasiri et al., 2003) and overproduces ethylene (Larsen and Cancel, 2004). In addition, reduced expression of the CSN5 subunit of the COP9 signalosome that removes RUB from CUL1, or of the SCF component RBX1 that adds RUB to CUL1, or defects in the putative CUL1-regulatory protein SGT1b, each cause deficient auxin and methyl-jasmonate responses (Schwechheimer et al., 2001; Gray et al., 2003).
axr1 and cand1 are also resistant to hypocotyl elongation inhibition by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) in darkness (Cheng et al., 2004). Apical hook formation in darkness, an auxin-mediated differential growth response promoted by ethylene, is reduced in cand1 (Chuang et al., 2004). However, the RUB1 E2 conjugating enzyme mutant rce1, though defective in auxin response (S. Dharmasiri et al., 2003), overproduces ethylene and has an exaggerated triple response (Larsen and Cancel, 2004). Likewise, reducing RUB levels causes ethylene overproduction and enhanced triple response (Bostick et al., 2004). Indeed, SCF complexes with the F-box proteins EBF1 and EBF2 repress ethylene response by targeting the ethylene response transcriptional activator EIN3 for degradation; ebf1 ebf2 mutants show constitutive ethylene responses (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004).
Gibberellins promote degradation of repressor DELLA proteins mediated by SCFSLY1 (McGinnis et al., 2003; Dill et al., 2004) and the functionally redundant SCFSNE (Strader et al., 2004). Though no altered morphological response to GA has been reported in general SCF mutants, a DELLA protein–GFP fusion is more stable in axr1-12 than wild type following GA application (Fu and Harberd, 2003).
In addition to hormone responses, SCF complexes can modulate development and environmental responses. For example, the F-box protein UFO is necessary for floral development (Samach et al., 1999). axr1 plants and transgenic plants overexpressing an inactive ECR1 both bear abnormal flowers; introducing the inactive ECR1 transgene into axr1 enhances the severity of axr1 floral defects (del Pozo et al., 2002). Also, though a role for an F-box complex in cold response has not been demonstrated, axr1, csn5 and rbx1 each display delayed cold-regulated gene induction (Schwechheimer et al., 2002).
The full impact of many response pathways feeding through a single type of protein degradation apparatus, the SCF, remains to be determined. However, it is tempting to imagine competition for shared SCF components among hundreds of specificity-determining F-box proteins. Additionally, it is possible that regulatory mechanisms such as RUB modification may differentially regulate specific SCF complexes. Further, different phytohormones modulate SCF activity in opposing ways: repressive proteins are degraded in response to auxin (Rogg and Bartel, 2001), gibberellin (Itoh et al., 2003) and presumably jasmonate (Xu et al., 2002), whereas ethylene impedes destruction of the transcriptional activator EIN3 by SCFEBF1/EBF2 (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004).
Other genes bridge auxin response with other stimuli
In addition to factors affecting SCF function, defects in several proteins with less-defined roles in auxin response display altered response to other hormones and environmental conditions. For example, several lines of evidence suggest that protein phosphorylation influences response to various stimuli including auxin exposure. First, auxin, salicylic acid, wounding and salt treatments induce MAPK (mitogen activated protein kinase) activity in arabidopsis roots, whereas kinase inhibitors block auxin inducibility of a reporter gene (Mockaitis and Howell, 2000).
Second, expression of constitutively active forms of the ANP family members of arabidopsis mitogen-activated protein kinase kinase kinases (MAPKKKs) dampens responsiveness of an auxin-inducible reporter gene construct in protoplasts (Kovtun et al., 2000). The same kinase family positively regulates responses to oxidative stress, and, like constitutively active ANPs, H2O2 exposure inhibits auxin-responsive reporter gene induction (Kovtun et al., 2000). Likewise, a constitutively active form of the related tobacco MAPKKK NPK1 dampens auxin responsiveness of a reporter gene (Kovtun et al., 1998) and confers resistance to freezing, heat and salt stresses (Kovtun et al., 2000). Based on these results and auxin activation of kinase activity, there is both a positive and negative modulation of auxin responsiveness by kinases.
Third, a mutant isolated because of altered response to an auxin transport inhibitor, roots curl in NPA 1 (rcn1) is deficient in a protein phosphatase 2A subunit (Garbers et al., 1996; Deruère et al., 1999). In addition to having increased IAA transport (Rashotte et al., 2001) and overproducing ethylene (Larsen and Chang, 2001; Larsen and Cancel, 2003), rcn1 is defective in NPA, gravity (Rashotte et al., 2001) and ABA (Kwak et al., 2002) responses. Likewise, protein phosphatase inhibitors phenocopy rcn1 gravitropism and IAA transport defects when applied to wild-type arabidopsis (Rashotte et al., 2001).
Finally, the indole-3-butyric acid-response mutant ibr5 is deficient in an apparent dual-specificity phosphatase (Monroe-Augustus et al., 2003). ibr5 is resistant to auxins and ABA (Monroe-Augustus et al., 2003). In addition, ibr5 makes fewer lateral roots, a longer primary root, and a shorter hypocotyl than wild type, phenotypes consistent with generally deficient auxin response (Monroe-Augustus et al., 2003).
In addition to phosphorylation-related genes, genes classically involved in auxin signalling are involved in response to other environmental cues. For example, Aux/IAA and ARF proteins are involved in light responses. Stabilizing gain-of-function mutations in shy2/iaa3, axr2/iaa7 and axr3/iaa17 cause de-etiolation in darkness (Kim et al., 1996; J.W. Reed et al., 1998; Nagpal et al., 2000). Moreover, the constitutive photomorphogenic mutant long hypocotyl 5 (hy5) (Koornneef et al., 1980) can be partially rescued by overexpressing AXR2/IAA7 (Cluis et al., 2004). Further, hookless1 (hls1) fails to make an apical hook in darkness (Lehman et al., 1996), and this phenotype is suppressed by mutation of the auxin response factor ARF2 (Li et al., 2004).
The arabidopsis mutant abscisic acid insensitive 3 (abi3) was isolated because of ABA-insensitive germination and is deficient in a transcription factor (Giraudat et al., 1992). In addition to ABA resistance, abi3 is resistant to lateral root proliferation induced by auxin and lateral root repression by the auxin transport inhibitor NPA, and ABI3:GUS reporter accumulation is induced by auxin exposure (Brady et al., 2003).
Additionally, certain mutants defective in auxin transport exhibit altered responses to other stimuli. The auxin influx mediator aux1 is resistant to specific auxins and to ethylene (Pickett et al., 1990). rcn1 has increased auxin transport and numerous developmental and phytohormone phenotypes (see above). Further, mutations in the auxin transport facilitator TRANSPORT INHIBITOR RESPONSE 3 (TIR3) (Ruegger et al., 1997) cause not only low-auxin phenotypes, but also altered responses to light, ethylene, cytokinin and gibberellin (Kanyuka et al., 2003). tir3 alleles have been isolated as dark overexpression of CAB1 (doc1; Li et al., 1994), the cytokinin-resistant mutant umbrella1, and attenuated shade avoidance1 (Kanyuka et al., 2003); the gene has been renamed BIG (Gil et al., 2001).
CONCLUSIONS AND FUTURE PROSPECTS
Auxin is a critical phytohormone. Complex and redundant regulation of IAA abundance, transport, and response allow an intricate system of auxin utilization that achieves a variety of purposes in plant development. As a result, the study of auxin biology is making an impact on our understanding of an astounding variety of processes, from regulated protein degradation to signal transduction cascades, from organelle biogenesis to plant morphogenesis. Despite prodigious historical and ongoing auxin research, many of the most fundamental original questions remain incompletely answered.
Redundancy is a key theme in auxin metabolism. IAA is produced both from the amino acid tryptophan and from an independent pathway utilizing a Trp precursor (Normanly et al., 1993). IAA can be stored in certain conjugated forms, which can be hydrolysed (Bartel and Fink, 1995), or as IBA, which can be β-oxidized (Zolman et al., 2000) to regenerate free IAA. Further, the isolation of a protein bearing covalently attached IAA (Walz et al., 2002) suggests an exciting new area for auxin storage research. A variety of auxin degradation products are found in plants (Östin et al., 1998). Future work will establish the relative contributions to auxin homeostasis from each of these redundant mechanisms.
In addition, tantalizing clues are emerging about the subcellular compartmentalization of auxin metabolism and response. Differential compartmentalization and controlled entry and exit from organelles may provide regulatory points for auxin and its precursors, although it is unknown where free IAA and its storage forms accumulate within the cell. Trp biosynthetic enzymes are plastidic (Radwanski and Last, 1995), whereas several potential downstream Trp-dependent IAA biosynthetic enzymes, including YUCCA and AAO1, are apparently cytoplasmic, and the subcellular location of Trp-independent IAA biosynthesis remains a mystery. Interestingly, CYP79B2 and CYP79B3 have chloroplast targeting signals (Hull et al., 2000); perhaps compartmentalization aids channelling common intermediates to either IAA or indolic glucosinolates. Directly comparing IAA and Trp precursor and metabolite levels in yucca, sur1 and sur2 mutants may reveal the importance of this potential compartmentalization. The subcellular location of IAA-conjugate biosynthesis and IAA degradation are also unknown. IAA-conjugate hydrolase sequence are consistent with ER retention (Bartel and Fink, 1995; Davies et al., 1999). Also, β-oxidation of IBA to yield IAA apparently takes place in peroxisomes (Zolman et al., 2000). Finally, it is unclear how many different sites in the cell can perceive auxin, though at least one auxin signalling apparatus is apparently entirely soluble (N. Dharmasiri et al., 2003).
The effectors through which auxin signalling influences growth and development are beginning to be elucidated. These will include the direct targets of ARFs and downstream effectors of these genes, combined with any non-transcriptional auxin responses. Transcript changes associated with lateral root production in response to auxin are beginning to be identified (Himanen et al., 2004); some of these are likely to be ARF targets. For example, lateral root proliferation in response to auxin is mediated by the transcription factor encoded by the auxin-induced NAC1 gene, which acts downstream of TIR1 (Xie et al., 2000).
In addition to auxin effectors downstream of TIR1, some processes appear to be mediated by auxin independent of the TIR1 signalling pathway. For example, auxin leads to rapid acidification of the extracellular space by enhancing plasma membrane ATPase activity, allowing cell wall loosening and thereby enabling cell growth (Hager, 2003). This process may be regulated by auxin, at least in part, via a signal transduction pathway acting in parallel to the SCFTIR1 pathway (Hager, 2003).
Auxin is also proving useful in the study of various cellular processes of general interest. For example, IBA and 2,4-DB response mutants are revealing key steps in peroxisome biogenesis and function, and the search for genes influencing IAA–amino acid conjugate responses has revealed new players in metal homeostasis (Lasswell et al., 2000; Magidin et al., 2003). Further, studies of disrupted auxin transport reveal that the actin cytoskeleton, unidentified endosomal compartments, and a rapid vesicular trafficking are all involved (Surpin and Raikhel, 2004). Finally, the study of auxin-response mutants has provided multiple new insights into the roles of SCF complexes in ubiquitin-mediated proteolysis (Dharmasiri and Estelle, 2002).
A tremendous area of future research will be the characterization and understanding of interactions between different phytohormones and environmental stimuli. The channelling of several hormone signalling pathways through specific SCF complexes with shared components is one possible arena for cross-talk. In addition, many mutants with altered response to auxin and other stimuli remain to be fully characterized or understood.
The recent identification of microRNAs and their targets in plants (Bartel and Bartel, 2003) has revealed intriguing links to auxin signalling. Several of these tiny riboregulators, which direct negative regulation of complementary mRNAs (Bartel, 2004), target messages implicated in auxin responsiveness. For example, miR167 targets ARF6 and ARF8 mRNAs (Rhoades et al., 2002; Kasschau et al., 2003), which encode activating ARFs (Ulmasov et al., 1999b), and miR160 targets ARF10, ARF16, and ARF17 mRNAs (Rhoades et al., 2002; Kasschau et al., 2003), which have not been functionally characterized but resemble repressing ARFs. Moreover, miR393 targets mRNAs encoding TIR1 and the three most closely related F-box proteins (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). In addition to microRNAs influencing auxin signalling, microRNAs are likely to aid in signal interpretation. For example, miR164 targets CUC1 and CUC2 (Laufs et al., 2004; Mallory et al., 2004), which establish organ boundaries in embryos and flowers (Aida et al., 1997) and are negatively regulated by PIN1 and PINOID (Furutani et al., 2004). Thus microRNAs, like ubiquitin-mediated protein degradation, are likely to be post-transcriptional regulators defining the extent of auxin signalling and response during development and in response to environmental cues.
The single most pressing question of auxin biology remains the identity of the auxin receptors. Tremendous progress has been made in this area, starting with the identification of SCFTIR1 as a critical conduit of auxin response (Leyser et al., 1993; Gray et al., 1999). Recent results suggest that auxin causes modification of the TIR1 F-box, or a tightly associated protein, to increase TIR1 interaction with Aux/IAA repressors, bringing them to the SCF to be targeted for destruction (Kepinski and Leyser, 2004). However, the nature of the modification, as well as the proximity to the auxin receptor, remain undetermined. While it is likely that SCFTIR1 mediates many transcriptional responses to auxin, it remains to be seen whether there are SCFTIR1-independent transcriptional auxin responses, or whether this signalling complex also mediates rapid responses to auxin that are not transcriptional.
Acknowledgments
We are grateful to our colleagues for sharing results prior to publication and to A. Raquel Adham, Diana Dugas, Dereth Phillips, Rebekah Rampey, Jeanne Rasbery, Lucia Strader, Bethany Zolman and an anonymous reviewer for critical comments on the manuscript. Auxin research in the authors' laboratory is supported by grants from the National Science Foundation and the Robert A. Welch Foundation. A.W.W. is a recipient of a Houston Livestock Show and Rodeo Scholarship.
LITERATURE CITED
- Abel S, Nguyen MD, Chow W, Theologis A. 1995.ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana Journal of Biological Chemistry 270: 19093–19099. [DOI] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A. 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana Journal of Molecular Biology 251: 533–549. [DOI] [PubMed] [Google Scholar]
- Abel S, Oeller PW, Theologis A. 1994. Early auxin-induced genes encode short-lived nuclear proteins. Proceedings of the National Academy of Science of the USA 91: 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A. 1996. Early genes and auxin action. Plant Physiology 111: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adham AR, Zolman BK, Millius A, Bartel B. 2005. Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β-oxidation. Plant Journal 41: 859–874. [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. 1997. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. The Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolić S, Kojić-Prodić B, Tomić S, Nigović B, Magnus V, Cohen JD. 1996. Structural studies on monofluorinated derivatives of the phytohormone indole-3-acetic acid (auxin). Acta Crystallographica B 52: 651–661. [Google Scholar]
- Avsian-Kretchmer O, Cheng JC, Chen L, Moctezuma E, Sung ZR. 2002. Indole acetic acid distribution coincides with vascular differentiation pattern during Arabidopsis leaf ontogeny. Plant Physiology 130: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. 2001. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. The Plant Cell 13: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi BG, Maher BR, Cohen JD. 1989. Hydrolysis of indole-3-acetic acid esters exposed to mild alkaline conditions. Plant Physiology 91: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Theologis A. 1993. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). Journal of Molecular Biology 233: 580–596. [DOI] [PubMed] [Google Scholar]
- Baraldi R, Bertazza G, Predieri S, Bregoli AM, Cohen JD. 1993. Uptake and metabolism of indole-3-butyric acid during the in vitro rooting phase in pear cultivars (Pyrus communis). Acta Horticulturae 329: 289–291. [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C. 2000. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proceedings of the National Academy of Science of the USA 97: 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt NM, Dong W, Gage DA, Magnus V, Town CD. 1999. Metabolism of exogenous auxin by Arabidopsis thaliana: identification of the conjugate Nα-(indol-3-ylacetyl)-glutamine and initiation of a mutant screen. Physiologia Plantarum 105: 207–217. [Google Scholar]
- Bartel B, Bartel DP. 2003. MicroRNAs: at the root of plant development? Plant Physiology 132: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. 1994. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana Proceedings of the National Academy of Science of the USA 91: 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. 1995. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748. [DOI] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK. 2001. Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. Journal of Plant Growth Regulation 20: 198–216. [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Mithöfer A, Weiler EW. 1992. Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. European Journal of Biochemistry 205: 417–424. [DOI] [PubMed] [Google Scholar]
- Bartling D, Seedorf M, Schmidt RC, Weiler EW. 1994. Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: key enzymes in the biosynthesis of the plant hormone indole-3-acetic acid. Proceedings of the National Academy of Science of the USA 91: 6021–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schultz B, Feldmann KA. 1996.Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklöf J, Ljung K, Marchant A, Bennett M, Sandberg G. 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant Journal 29: 325–332. [DOI] [PubMed] [Google Scholar]
- Bialek K, Cohen JD. 1986. Isolation and partial characterization of the major amide-linked conjugate of indole-3-acetic acid from Phaseolus vulgaris L. Plant Physiology 80: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inzé D. 1995.superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. The Plant Cell 7: 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Lochhead SR, Honda A, Palmer S, Callis J. 2004. Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. The Plant Cell 16: 2418–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis Plant Journal 34: 67–75. [DOI] [PubMed] [Google Scholar]
- Brickner DG, Brickner JH, Olsen LJ. 1998. Sequence analysis of a cDNA encoding Pex5p, a peroxisomal targeting signal type 1 receptor from Arabidopsis. Plant Physiology 118: 330. [Google Scholar]
- Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK. 2001. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiology 126: 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusslan JA, Karlin-Neumann GA, Huang L, Tobin EM. 1993. An Arabidopsis mutant with a reduced level of cab140 RNA is a result of cosuppression. The Plant Cell 5: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK. 2004. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. The Plant Cell 16: 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burg SP, Burg EA. 1966. The interaction between auxin and ethylene and its role in plant growth. Proceedings of the National Academy of Science of the USA 55: 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Ludwig-Mueller J, Town CD. 1996. Isolation and characterization of mutants of Arabidopsis thaliana with increased resistance to growth inhibition by indoleacetic acid-amino acid conjugates. Plant Physiology 112: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanella JJ, Ludwig-Mueller J, Bakllamaja V, Sharma V, Cartier A. 2003. ILR1 and sILR1 amidohydrolase homologs differ in expression pattern and substrate specificity. Plant Growth Regulation 41: 215–223. [Google Scholar]
- Campanella JJ, Olajide AF, Magnus V, Ludwig-Müller J. 2004. A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiology 135: 2230–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Grisafi PL, Fink GR. 1995. A pathway for lateral root formation in Arabidopsis thaliana Genes and Development 9: 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chen J-G, Ullah H, Young JC, Sussman MR, Jones AM. 2001. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes and Development 15: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. 1998. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proceedings of the National Academy of Science of the USA 95: 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2004. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiology 135: 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Taketa S, Tsurumi S, Ichii M. 2003. Interaction between two auxin-resistant mutants and their effects on lateral root formation in rice (Oryza sativa L.). Journal of Experimental Botany 54: 2701–2708. [DOI] [PubMed] [Google Scholar]
- Chisnell JR, Bandurski RS. 1988. Translocation of radiolabeled indole-3-acetic acid and indole-3-acetyl-myo-inositol from kernel to shoot of Zea mays L. Plant Physiology 86: 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HW, Zhang W, Gray WM. 2004. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. The Plant Cell 16: 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski T. 1872. Untersuchungen über die Abwärtskrümmung der Wurzel. Beitrage zur Biologie der Pflanzen 1: 1–30. [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS. 2004. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant Journal 38: 332–347. [DOI] [PubMed] [Google Scholar]
- Cohen JD, Bandurski RS. 1982. Chemistry and physiology of the bound auxins. Annual Review of Plant Physiology 33: 403–430. [Google Scholar]
- Cooney TP, Nonhebel HM. 1991. Biosynthesis of indole-3-acetic acid in tomato shoots: Measurement, mass-spectral identification and incorporation of 2H from 2H2O into indole-3-acetic acid, D- and L-tryptophan, indole-3-pyruvate and tryptamine. Planta 184: 368–376. [DOI] [PubMed] [Google Scholar]
- Courtney KD, Gaylor DW, Hogan MD, Falk HL, Bates RR, Mitchell I. 1970. Teratogenic evaluation of 2,4,5-T. Science 168: 864–866. [DOI] [PubMed] [Google Scholar]
- Darwin C. 1880.The power of movement in plants. London: John Murray. [Google Scholar]
- Davies PJ. 1995.Plant hormones. Dordrecht: Kluwer Academic Publishers. [Google Scholar]
- Davies PJ, Mitchell EK. 1972. Transport of indoleacetic acid in intact roots of Phaseolus coccineus Planta 105: 139–154. [DOI] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. 1999.IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. The Plant Cell 11: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, Marineau C, Brisson N. 1989. Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proceedings of the National Academy of Science of the USA 86: 2582–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. 1999. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proceedings of the National Academy of Science of the USA 96: 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. 2002. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. The Plant Cell 14: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. 1998. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis Science 280: 1760–1763. [DOI] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Van Onckelen H, Caboche M, Bellini C. 1998.Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant Journal 14: 603–611. [DOI] [PubMed] [Google Scholar]
- Deruère J, Jackson K, Garbers C, Söll D, DeLong A. 1999. The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo Plant Journal 20: 389–399. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. 1999. SCF and Cullin/RING H2-based ubiquitin ligases. Annual Review of Cell and Development Biology 15: 435–467. [DOI] [PubMed] [Google Scholar]
- De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H. 2003. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis Plant Journal 33: 543–555. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. 2003. Auxin action in a cell-free system. Current Biology 13: 1418–1422. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. 2003. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis EMBO Journal 22: 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S, Estelle M. 2002. The role of regulated protein degradation in auxin response. Plant Molecular Biology 49: 401–409. [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. 2004. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. The Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois-Dalcq M, Feigenbaum V, Aubourg P. 1999. The neurobiology of X-linked adrenoleukodystrophy, a demyelinating peroxisomal disorder. Trends in Neuroscience 22: 4–12. [DOI] [PubMed] [Google Scholar]
- Dunlap JR, Robacker KM. 1990. Abscisic acid alters the metabolism of indole-3-acetic acid in senescing flowers of Cucumis melo L. Plant Physiology 94: 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Hooks MA, Williams D, Lange P, Bechtold N, Sarrobert C, Nussaume L, Graham IA. 2000. Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. Journal of Biological Chemistry 275: 34375–34381. [DOI] [PubMed] [Google Scholar]
- Eklöf S, Åstot C, Sitbon F, Moritz T, Olsson O, Sandberg G. 2000. Transgenic tobacco plants co-expressing Agrobacterium iaa and ipt genes have wild-type hormone levels but display both auxin- and cytokinin-overproducing phenotypes. Plant Journal 23: 279–284. [DOI] [PubMed] [Google Scholar]
- Epstein E, Ludwig-Müller J. 1993. Indole-3-butyric acid in plants: occurrence, synthesis, metabolism, and transport. Physiologia Plantarum 88: 382–389. [Google Scholar]
- Estelle MA, Somerville C. 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Molecular and General Genetics 206: 200–206. [Google Scholar]
- Facchini PJ, Huber-Allanach KL, Tari LW. 2000. Plant aromatic L-amino acid decarboxylases: Evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry 54: 121–138. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P. 2001. The chemical diversity and distribution of glucosinolates and isothiocynanates among plants. Phytochemistry 56: 5–51. [DOI] [PubMed] [Google Scholar]
- Fallon H, Tollerud D, Breslow N, Berlin J, Bolla K, Colditz G, Goetz C, Kaminski N, Kriebel D, Mottet NK, et al. 1994. The U.S. military and the herbicide program in Vietnam. In Veterans and agent orange: health effects of herbicides used in Vietnam Washington, DC: National Academy Press, 74–110. [Google Scholar]
- Farrás R, Ferrando A, Jásik J, Kleinow T, Ökrész L, Tiburcio A, Salchert K, del Pozo JC, Schell J, Koncz C. 2001. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO Journal 20: 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett CH, Wain RL, Wightman F. 1960. The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proceedings of the Royal Society of London. Series B. Biological Science 152: 231–254. [DOI] [PubMed] [Google Scholar]
- Feng S, Shen Y, Sullivan JA, Rubio V, Xiong Y, Sun TP, Deng XW. 2004. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. The Plant Cell 16: 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. 2002. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO Journal 21: 2912–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, Simcox K, Gierl A. 1997. Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699. [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Paré PW, Schmelz EA, Tumlinson JH, Gierl A. 2000. An herbivore elicitor activates the gene for indole emission in maize. Proceedings of the National Academy of Science of the USA 97: 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis Nature 426: 147–153. [DOI] [PubMed] [Google Scholar]
- Friml J, Wísniewska J, Benková E, Mendgen K, Palme K. 2002. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis Nature 415: 806–809. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PBF, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP. 2003. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421:d40–743. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis Plant Journal 29: 153–168. [DOI] [PubMed] [Google Scholar]
- Fulda M, Schnurr J, Abbadi A, Heinz E, Browse J. 2004. Peroxisomal acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana The Plant Cell 16: 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M, Vernoux T, Traas J, Kato T, Tasaka M, Aida M. 2004.PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131: 5021–5030. [DOI] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. 2004.Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proceedings of the National Academy of Science of the USA 101: 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K. 1998. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282: 2226–2230. [DOI] [PubMed] [Google Scholar]
- Garbers C, DeLong A, Deruére J, Bernasconi P, Söll D. 1996. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis EMBO Journal 15: 2115–2124. [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. 2001. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Gerhardt B. 1992. Fatty acid degradation in plants. Progress in Lipid Research 31: 417–446. [DOI] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J. 2001. BIG: a calossin-like protein required for polar auxin transport in Arabidopsis Genes and Development 15: 1985–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Liu Y, Orbović V, Verkamp E, Poff KL, Green PJ. 1994. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis Plant Physiology 104: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Kosuge T. 1986. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi Journal of Bacteriology 166: 598–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiology 134: 1555–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golparaj M, Tseng T-S, Olszewski N. 1996. The Rooty gene of Arabidopsis encodes a protein with highest similarity to aminotransferases. Plant Physiology 111 (suppl.): 114. [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana Genes and Development 13: 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. 2002. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. The Plant Cell 14: 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE. 2003. Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. The Plant Cell 15: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. 1998. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis Proceedings of the National Academy of Science of the USA 95: 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grsic S, Sauerteig S, Neuhaus K, Albrecht M, Rossiter J, Ludwig-Müller J. 1998. Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in sense or antisense direction. Journal of Plant Physiology 153: 446–456. [Google Scholar]
- Grsic-Rausch S, Kobelt P, Siemens JM, Bischoff M, Ludwig-Müller J. 2000. Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiology 122: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML. 2000. The ZIP family of metal transporters. Biochimica et Biophysica Acta 1465: 190–198. [DOI] [PubMed] [Google Scholar]
- Guillet G, Poupart J, Basurco J, De Luca V. 2000. Expression of tryptophan decarboxylase and tyrosine decarboxylase genes in tobacco results in altered biochemical and physiological phenotypes. Plant Physiology 122: 933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. 2003. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Hagen G, Kleinschmidt A, Guilfoyle T. 1984. Auxin-regulated gene expression in intact soybean hypocotyl and excised hypocotyl sections. Planta 162: 147–153. [DOI] [PubMed] [Google Scholar]
- Hager A. 2003. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. Journal of Plant Research 116: 483–505. [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Bäurle I, Kientz M, Jürgens G. 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes and Development 16: 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO Journal 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. 2004. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4 Development 131: 1089–1100. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. 2000. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. The Plant Cell 12: 757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann HT, Kester DE, Davies FT. 1990. Anatomical and physiological basis of propagation by cuttings. In Plant propagation: principles and practices Englewood Cliffs, NJ: Prentice Hall, 199–245. [Google Scholar]
- Haughn GW, Somerville C. 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana Molecular and General Genetics 204: 430–434. [Google Scholar]
- Hayashi H, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. 2002. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid β-oxidation. Plant Cell Physiology 43: 1–11. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M. 2000.AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO Journal 19: 5701–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 1998. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. The Plant Cell 10: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M. 2003.Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO Journal 22: 3314–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inzé D, Beeckman T. 2004. Transcript profiling of early lateral root initiation. Proceedings of the National Academy of Science of the USA 101: 5146–5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie L, Estelle M. 1995. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant Journal 7: 211–220. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH. 2004. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219: 195–200. [DOI] [PubMed] [Google Scholar]
- Holland IB, Blight MA. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. Journal of Molecular Biology 293: 381–399. [DOI] [PubMed] [Google Scholar]
- Hooks MA, Kellas F, Graham IA. 1999. Long-chain acyl-CoA oxidases of Arabidopsis Plant Journal 20: 1–13. [DOI] [PubMed] [Google Scholar]
- Hou G, Kramer VL, Wang YS, Chen R, Perbal G, Gilroy S, Blancaflor EB. 2004. The promotion of gravitropism in Arabidopsis roots upon actin disruption is coupled with the extended alkalinization of the columella cytoplasm and a persistent lateral auxin gradient. Plant Journal 39: 113–125. [DOI] [PubMed] [Google Scholar]
- Hou G, Mohamalawari DR, Blancaflor EB. 2003. Enhanced gravitropism of roots with a disrupted cap actin cytoskeleton. Plant Physiology 131: 1360–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. 2000.Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proceedings of the National Academy of Science of the USA 97: 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilić N, Cohen JD. 2004. Synthesis of [13C]-isotopomers of indole and tryptophan for use in the analysis of indole-3-acetic acid biosynthesis. Journal of Labelled Compounds and Radiopharmaceuticals 47: 635–646. [Google Scholar]
- Ilić N, Normanly J, Cohen JD. 1996. Quantification of free plus conjugated indoleacetic acid in Arabidopsis requires correction for the nonenzymatic conversion of indolic nitriles. Plant Physiology 111: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM. 2003. A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends in Plant Science 8: 492–497. [DOI] [PubMed] [Google Scholar]
- Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J, Sandberg G, Bowles DJ. 2002. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterization of transgenic lines. Plant Journal 32: 573–583. [DOI] [PubMed] [Google Scholar]
- Jackson RG, Lim E-K, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. 2001. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. Journal of Biological Chemistry 276: 4350–4356. [DOI] [PubMed] [Google Scholar]
- Jakubowska A, Kowalczyk S. 2004. The auxin conjugate 1-O-indole-3-acetyl-β-d-glucose is synthesized in immature legume seeds by IAGlc synthase and may be used for modification of some high molecular weight compounds. Journal of Experimental Botany 55: 791–801. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Pérez-Amador MA, Lidder P, Green PJ. 2000. Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proceedings of the National Academy of Science of the USA 97: 13991–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM. 1994. Auxin-binding proteins. Annual Review of Plant Physiology and Plant Molecular Biology 45: 393–420. [Google Scholar]
- Jones-Rhoades MW, Bartel DP. 2004. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, Halliday KJ. 2003. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant Journal 35: 57–70. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2004. Auxin-induced SCFTIR1–Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proceedings of the National Academy of Science of the USA 101: 12381–12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Kang BJ, Furuya M, Nam HG. 1996. Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2 Plant Journal 9: 441–456. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. 1997. Protein-protein interactions among the Aux/IAA proteins. Proceedings of the National Academy of Science of the USA 94: 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS. 2000. Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiology 123: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindl H. 1993. Fatty acid degradation in plant peroxisomes: function and biosynthesis of the enzymes involved. Biochimie 75: 225–230. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP. 1998. Genetic analysis of variation for auxin-induced adventitious root formation among eighteen ecotypes of Arabidopsis thaliana L. Heynh. Journal of Heredity 89: 481–487. [DOI] [PubMed] [Google Scholar]
- King JJ, Stimart DP, Fisher RH, Bleecker AB. 1995. A mutation altering auxin homeostasis and plant morphology in Arabidopsis. The Plant Cell 7: 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. 1999.SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Molecular Cell 4: 21–33. [DOI] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L. 2003. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. Journal of Biological Chemistry 278: 23936–23943. [DOI] [PubMed] [Google Scholar]
- Koga J. 1995. Structure and function of indolepyruvate decarboxylase, a key enzyme in indole-3-acetic acid biosynthesis. Biochimica et Biophysica Acta 1249: 1–13. [DOI] [PubMed] [Google Scholar]
- Kögl F, Haagen Smit AJ. 1931. Über die Chemie des Wuchsstoffs. K. Akad. Wetenschap. Amsterdam Proceedings. Section Science 34: 1411–1416. [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. 1980. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift fur Pflanzenphysiology. International Journal of Plant Physiology 100S: 147–160. [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J. 2000. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proceedings of the National Academy of Science of the USA 97: 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Zeng W, Sheen J. 1998. Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395: 716–720. [DOI] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. 2001. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiology 127: 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Krikorian AD. 1995. Hormones in tissue culture and micropropagation. In: Davies PJ, ed. Plant hormones. Dordrecht: Kluwer, 774–796. [Google Scholar]
- Kutz A, Müller A, Hennig P, Kaiser WM, Piotrowski M, Weiler EW. 2002. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana Plant Journal 30: 95–106. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI. 2002. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. The Plant Cell 14: 2849–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD. 2003. Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1 Plant Journal 34: 709–718. [DOI] [PubMed] [Google Scholar]
- Larsen PB, Cancel JD. 2004. A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis Plant Journal 38: 626–638. [DOI] [PubMed] [Google Scholar]
- Larsen PB, Chang C. 2001. The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiology 125: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasswell J, Rogg LE, Nelson DC, Rongey C, Bartel B. 2000. Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. The Plant Cell 12: 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR. 1991. Tryptophan mutants in Arabidopsis: The consequences of duplicated tryptophan synthase β genes. The Plant Cell 3: 345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Fink GR. 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana Science 240: 305–310. [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. 2004. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131: 4311–4322. [DOI] [PubMed] [Google Scholar]
- LeClere S, Rampey RA, Bartel B. 2004.IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiology 135: 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. 2002. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis Journal of Biological Chemistry 277: 20446–20452. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. 1996.HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. 1993.Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164. [DOI] [PubMed] [Google Scholar]
- Leznicki AJ, Bandurski RS. 1988. Enzymic synthesis of indole-3-acetyl-1-O-β-D-glucose. I. Partial purification and characterization of the enzyme from Zea mays Plant Physiology 88: 1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. 2004. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis Developmental Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Li H-M, Altschmied L, Chory J. 1994.Arabidopsis mutants define downstream branches in the phototransduction pathway. Genes and Development 8: 339–349. [DOI] [PubMed] [Google Scholar]
- Li L, Deng XW. 2003. The COP9 signalosome: an alternative lid for the 26S proteasome? Trends in Cell Biology 13: 507–509. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. 1990. Growth and development of the axr1 mutants of Arabidopsis The Plant Cell 2: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Molecular Biology 49: 387–400. [PubMed] [Google Scholar]
- Liu J, Furukawa M, Matsumoto T, Xiong Y. 2002. NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Molecular Cell 10: 1511–1518. [DOI] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G. 2001. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant Journal 28: 465–474. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G. 2002. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana Plant Molecular Biology 50: 309–332. [DOI] [PubMed] [Google Scholar]
- Lomax TL, Muday GK, Rubery PH. 1995. Auxin transport. In: Davies PJ, ed. Plant hormones. Dordrecht: Kluwer, 509–530. [Google Scholar]
- Ludwig-Müller J, Epstein E. 1993. Indole-3-butyric acid in Arabidopsis thaliana II. In vivo metabolism. Plant Growth Regulation 13: 189–195. [Google Scholar]
- Ludwig-Müller J. 2000. Indole-3-butyric acid in plant growth and development. Plant Growth Regulation 32: 219–230. [Google Scholar]
- Ludwig-Müller J, Cohen JD. 2002. Identification and quantification of three active auxins in different tissues of Tropaeolum majus Physiologia Plantarum 115: 320–329. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Epstein E. 1994. Indole-3-butyric acid in Arabidopsis thaliana III. In vivo biosynthesis. Plant Growth Regulation 14: 7–14. [Google Scholar]
- Ludwig-Müller J, Hilgenberg W. 1995. Characterization and partial purification of indole-3-butyric acid synthetase from maize (Zea mays). Physiologia Plantarum 94: 651–660. [Google Scholar]
- Ludwig-Müller J, Raisig A, Hilgenberg W. 1995. Uptake and transport of indole-3-butyric acid in Arabidopsis thaliana: comparison with other natural and synthetic auxins. Journal of Plant Physiology 147: 351–354. [Google Scholar]
- Ludwig-Müller J, Sass S, Sutter EG, Wodner M, Epstein E. 1993. Indole-3-butyric acid in Arabidopsis thaliana I. Identification and quantification. Plant Growth Regulation 13: 179–187. [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana Genes and Development 12: 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. The Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidin M, Pittman JK, Hirschi KD, Bartel B. 2003.ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana Plant Journal 35: 523–534. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. 2004. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Current Biology 14: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO Journal 18: 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T. 2003. Auxin signaling in Arabidopsis leaf vascular development. Plant Physiology 131: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T. 1999. Responses of plant vascular systems to auxin transport inhibition. Development 126: 2979–2991. [DOI] [PubMed] [Google Scholar]
- Melanson D, Chilton M-D, Masters-Moore D, Chilton WS. 1997. A deletion in an indole synthase gene is responsible for the DIMBOA-deficient phenotype of bxbx maize. Proceedings of the National Academy of Science of the USA 94: 13345–13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Hansen CH, Wittstock U, Halkier BA. 2000. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. Journal of Biological Chemistry 275: 33712–33717. [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA. 2004.Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant Journal 37: 770–777. [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Howell SH. 2000. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant Journal 24: 785–796. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M. 2004. Genetic approaches to elucidating the mechanisms of indole-3-acetic acid and indole-3-butyric acid function in Arabidopsis thaliana. PhD Thesis, Department of Biochemistry and Cell Biology, Rice University, Houston, TX, 202 pp. [Google Scholar]
- Monroe-Augustus M, Zolman BK, Bartel B. 2003. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. The Plant Cell 15: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PW, Hall WC. 1962. Effect of 2,4-dichlorophenoxyacetic acid on the production of ethylene by cotton and grain sorghum. Physiologia Plantarum 15: 420–427. [Google Scholar]
- Mullen RT, Flynn CR, Trelease RN. 2001. How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends in Plant Science 6: 256–261. [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. 1998.AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO Journal 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Hillebrand H, Weiler EW. 1998. Indole-3-acetic acid is synthesized from L-tryptophan in roots of Arabidopsis thaliana Planta 206: 362–369. [DOI] [PubMed] [Google Scholar]
- Müller A, Weiler EW. 2000. Indolic constituents and indole-3-acetic acid biosynthesis in the wild-type and a tryptophan auxotroph mutant of Arabidopsis thaliana Planta 211: 855–863. [DOI] [PubMed] [Google Scholar]
- Müller A, Weiler EW. 2000. IAA-synthase, an enzyme complex from Arabidopsis thaliana catalyzing the formation of indole-3-acetic acid from (S)-tryptophan. Biology and Chemistry 381: 679–686. [DOI] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302: 81–84. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. 2000.AXR2 encodes a member of the Aux/IAA protein family. Plant Physiology 123: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M. 2001.DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant Journal 25: 213–221. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Feldman LJ, Zambryski PC. 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis PLoS Biology 2: 1460–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigović B, Antolić S, Kojić-Prodić B, Kiralj R, Magnus V, Salopek-Sondi B. 2000. Correlation of structural and physico-chemical parameters with the bioactivity of alkylated derivatives of indole-3-acetic acid, a phytohormone (auxin). Acta Crystallographica B 56: 94–111. [DOI] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. 2003. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002. [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. 2001.Multidrug Resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. The Plant Cell 13: 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Åstot C, Dolezal K, Sandberg G. 2004. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proceedings of the National Academy of Science of the USA 101: 8039–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. 1997. Auxin metabolism. Physiologia Plantarum 100: 431–442. [Google Scholar]
- Normanly J, Cohen JD, Fink GR. 1993.Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proceedings of the National Academy of Science of the USA 90: 10355–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J, Grisafi P, Fink GR, Bartel B. 1997. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. The Plant Cell 9: 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. 1991. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. The Plant Cell 3: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LJ. 1998. The surprising complexity of peroxisome biogenesis. Plant Molecular Biology 38: 163–189. [PubMed] [Google Scholar]
- Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. 2003. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochemical and Biophysical Research Communications 303: 1209–1216. [DOI] [PubMed] [Google Scholar]
- Östin A, Kowalyczk M, Bhalerao RP, Sandberg G. 1998. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiology 118: 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. 2001. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. The Plant Cell 13: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Shao X, Li J. 2000. Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana Plant Journal 24: 327–333. [DOI] [PubMed] [Google Scholar]
- Park WJ, Kriechbaumer V, Müller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E. 2003. The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiology 133: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten CL, Glick BR. 1996. Bacterial biosynthesis of indole-3-acetic acid. Canadian Journal of Microbiology 42: 207–220. [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. 2004. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana The Plant Cell 16: 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Shen Y, Feng S, Wang X, Chitteti BN, Vierstra RD, Deng XW. 2003. Evidence for a physical association of the COP9 signalosome, the proteasome, and specific SCF E3 ligases in vivo Current Biology 13: R504–R505. [DOI] [PubMed] [Google Scholar]
- Pfluger J, Zambryski P. 2004. The role of SEUSS in auxin response and floral organ patterning. Development 131: 4697–4707. [DOI] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. 1990. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiology 94: 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Müller A, Piotrowski M, Weiler EW. 2002. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana Planta 216: 155–161. [DOI] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Weiler EW. 2003. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62: 293–300. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. 2003. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Poupart J, Waddell CS. 2000. The rib1 mutant is resistant to indole-3-butyric acid, an endogenous auxin in Arabidopsis. Plant Physiology 124: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufky J, Qiu Y, Rao MV, Hurban P, Jones AM. 2003. The auxin-induced transcriptome for etiolated Arabidopsis seedlings using a structure/function approach. Functional & Integrative Genomics 3: 135–143. [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RM. 1999. Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Molecular Biology 40: 267–278. [DOI] [PubMed] [Google Scholar]
- Radwanski ER, Barczak AJ, Last RL. 1996. Characterization of tryptophan synthase alpha subunit mutants of Arabidopsis thaliana Molecularand General Genetics 253: 353–361. [DOI] [PubMed] [Google Scholar]
- Radwanski ER, Last RL. 1995. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. The Plant Cell 7: 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. 2001. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. The Plant Cell 13: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. 2004. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiology 135: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK. 2001. Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. The Plant Cell 13: 1683–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Poupart J, Waddell CS, Muday GK. 2003. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiology 133: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Elumalai RP, Chory J. 1998. Suppressors of an Arabidopsis thaliana phyB mutation identify genes that control light signaling and hypocotyl elongation. Genetics 148: 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiology 118: 1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. 2001.bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. The Plant Cell 13: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiology 135: 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Ribnicky DM, Cohen JD, Hu W-S, Cooke TJ. 2002. An auxin surge following fertilization in carrots: a mechanism for regulating plant totipotency. Planta 214: 505–509. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Bleecker AB. 1999. A defect in β-oxidation causes abnormal inflorescence development in Arabidopsis. The Plant Cell 11: 1911–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Bartel B. 2001. Auxin signaling: derepression through regulated proteolysis. Developmental Cell 1: 595–604. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. 2001. A gain-of-function mutation in IAA28 suppresses lateral root development. The Plant Cell 13: 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LHJ, Elliott RC. 2000. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant Journal 21: 547–552. [DOI] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes and Development 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M. 1997. Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. The Plant Cell 9: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. 2003.Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. Journal of Biological Chemistry 278: 21370–21377. [DOI] [PubMed] [Google Scholar]
- Salamini F. 2003. Hormones and the green revolution. Science 302: 71–72. [DOI] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL. 1999. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant Journal 20: 433–445. [DOI] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T. 2002. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis Plant Journal 32: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Schmidt R-C, Müller A, Hain R, Bartling D, Weiler EW. 1996. Transgenic tobacco plants expressing the Arabidopsis thaliana nitrilase II enzyme. Plant Journal 9: 683–691. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW. 2001. COP9 signalosome revisited: a novel mediator of protein degradation. Trends in Cell Biology 11: 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng X-W. 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292: 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Deng XW. 2002. Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. The Plant Cell 14: 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwetz BA, Norris JM, Sparschu GL, Rowe VK, Gehring PJ, Emerson JL, Gerbig CG. 1973. Toxicology of chlorinated dibenzo-p-dioxins. Environmental Health Perspectives 5: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TK, Wilkins MB. 1968. Auxin transport in roots. II. Polar flux of IAA in Zea roots. Planta 83: 323–334. [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T. 1998. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana Plant Cell Physiology 39: 433–442. [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. 2001. Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proceedings of the National Academy of Science of the USA 98: 4788–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Akaba S, Oritani T, Delarue M, Bellini C, Caboche M, Koshiba T. 1998. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana Plant Physiology 116: 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McCall A, Roe JL, Feldmann KA, Zambryski PC. 1997.ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491. [DOI] [PubMed] [Google Scholar]
- Sessions RA, Zambryski PC. 1995.Arabidopsis gynoecium structure in the wild type and in ettin mutants. Development 121: 1519–1532. [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P. 2002. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis Molecular Biology of the Cell 13: 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. 2001. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. The Plant Cell 13: 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F, Miller CO. 1957. Chemical regulation of growth and organ formation in plant tissues cultured in vitro Symposia of the Society for Experimental Biology 11: 118–131. [PubMed] [Google Scholar]
- Slovin JP, Bandurski RS, Cohen JD. 1999. Auxin. In: Hooykaas PJJ, Hall MA, Libbenga KR, eds. Biochemistry and molecular biology of plant hormones. Amsterdam: Elsevier, 115–140. [Google Scholar]
- Smolen G, Bender J. 2002. Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 160: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Kim BC, Vizir I, Park DH, Choi G, Hong MY, Chung Y-Y, Furuya M, Nam HG. 1999. Regulation of both light- and auxin-mediated development by the Arabidopsis IAA3/SHY2 gene. Journal of Plant Biology 42: 239–246. [Google Scholar]
- Somerville CR, Ogren WL. 1980. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286: 257–259. [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell 17: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. 2004. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell 16: 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I, Rowe ML. 2002. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. The Plant Cell 14: 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans EL, Harper RM, Motchoulski AV, Liscum E. 1998. NPH4, a conditional modulator of auxin-dependent differential growth responses in Arabidopsis. Plant Physiology 118: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. 2004. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY Proceedings of the National Academy of Science of the USA 101: 12771–12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. 1998. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiological Reviews 78: 171–188. [DOI] [PubMed] [Google Scholar]
- Sullivan ML, Green PJ. 1996. Mutational analysis of the DST element in tobacco cells and transgenic plants: identification of residues critical for mRNA instability. RNA 2: 308–315. [PMC free article] [PubMed] [Google Scholar]
- Sun H, Basu S, Brady SR, Luciano RL, Muday GK. 2004. Interactions between auxin transport and the actin cytoskeleton in developmental polarity of Fucus distichus embryos in response to light and gravity. Plant Physiology 135: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. 2004. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell 16: 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Raikhel N. 2004. Traffic jams affect plant development and signal transduction. Nature Reviews. Molecular Cell Biology 5: 100–109. [DOI] [PubMed] [Google Scholar]
- Suttle JC. 1988. Effect of ethylene treatment on polar IAA transport, net IAA uptake and specific binding of N-1-naphthylphthalamic acid in tissues and microsomes isolated from etiolated pea epicotyls. Plant Physiology 88: 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M. 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes and Development 15: 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, et al. 2004. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. The Plant Cell 16: 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen JB, Szczyglowski K, Bandurski RS. 1994.iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265: 1699–1701. [DOI] [PubMed] [Google Scholar]
- Sztein AE, Cohen JD, de la Fuente IG, Cooke TJ. 1999. Auxin metabolism in mosses and liverworts. American Journal of Botany 86: 1544–1555. [PubMed] [Google Scholar]
- Sztein AE, Ilic N, Cohen JD, Cooke TJ. 2002. Indole-3-acetic acid biosynthesis in isolated axes from germinating bean seeds: the effect of wounding on the biosynthetic pathway. Plant Growth Regulation 36: 201–207. [Google Scholar]
- Tabak HF, Braakman I, Distel B. 1999. Peroxisomes: simple in function but complex in maintenance. Trends in Cell Biology 9: 447–453. [DOI] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M. 2004.ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant Journal 37: 471–483. [DOI] [PubMed] [Google Scholar]
- Tam YY, Epstein E, Normanly J. 2000. Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiology 123: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Normanly J. 1998. Determination of indole-3-pyruvic acid levels in Arabidopsis thaliana by gas chromatography-selected ion monitoring-mass spectrometry. Journal of Chromatography A 800: 101–108. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. 2004.MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana The Plant Cell 16: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV. 1977.Hormone action in the whole life of plants. Amherst: University of Massachusetts Press. [Google Scholar]
- Thimann KV, Mahadevan S. 1964. Nitrilase. I. Occurrence, preparation, and general properties of the enzyme. Archives of Biochemistry and Biophysics 105: 133–141. [DOI] [PubMed] [Google Scholar]
- Thimann KV, Skoog, F. 1934. On the inhibition of bud development and other functions of growth substance in Vicia faba. Proceedings of the Royal Society of London Series B. Biological Science 114: 317–339. [Google Scholar]
- Tian CE, Muto H, Higuchi K, Matamura T, Tatematsu K, Koshiba T, Yamamoto KT. 2004. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant Journal 40: 333–343. [DOI] [PubMed] [Google Scholar]
- Tian Q, Nagpal P, Reed JW. 2003. Regulation of Arabidopsis SHY2/IAA3 protein turnover. Plant Journal 36: 643–651. [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW. 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721. [DOI] [PubMed] [Google Scholar]
- Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. 1995. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant Journal 8: 561–569. [DOI] [PubMed] [Google Scholar]
- Tiryaki I, Staswick PE. 2002. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1 Plant Physiology 130: 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T. 2003. The roles of auxin response factor domains in auxin-responsive transcription. The Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. 2004. Aux/IAA proteins contain a potent transcriptional repression domain. The Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ. 2001. Aux/IAA proteins are active repressors, and their stability and activity are modulated by auxin. The Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobeña-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM, Souer E, Koes R. 2002. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes and Development 16: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi S, Ohwaki Y. 1978. Transport of 14C-labeled indoleacetic acid in Vicia root segments. The Plant Cell Physiology 19: 1195–1206. [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1997. ARF1, a transcription factor that binds auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. Dimerization and DNA binding of auxin response factors. Plant Journal 19: 309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Science of the USA 96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu Z-B, Hagen G, Guilfoyle TJ. 1995. Composite structure of auxin response elements. The Plant Cell 7: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T. 1998.AGR, an agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant Cell Physiology 39: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Wain RL, Wightman F. 1954. The growth-regulating activity of certain ω-substituted alkyl carboxylic acids in relation to their β-oxidation within the plant. Proceedings of the National Academy of Science of the USA 142: 525–536. [DOI] [PubMed] [Google Scholar]
- Walker JC, Key JL. 1982. Isolation of cloned cDNAs to auxin-responsive poly(A)+ RNAs of elongating soybean hypocotyl. Proceedings of the National Academy of Science of the USA 79: 7185–7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz A, Park S, Slovin JP, Ludwig-Müller J, Momonoki YS, Cohen JD. 2002. A gene encoding a protein modified by the phytohormone indoleacetic acid. Proceedings of the National Academy of Science of the USA 99: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kang D, Feng S, Serino G, Schwechheimer C, Wei N. 2002. CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure-function study of CSN1 subunit of COP9 signalosome. Molecular Biology of the Cell 13: 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went FW. 1926. On growth-accelerating substances in the coleoptile of Avena sativa Proceedings of the Koninklijke Nederlandse Akademie van Wetenschappen. Series C. Biological and Medical Sciences 30: 10–19. [Google Scholar]
- Went FW, Thimann KV. 1937.Phytohormones. New York: The Macmillan Company. [Google Scholar]
- Wightman F. 1977.Plant growth regulation. Berlin: Springer-Verlag. [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. 1990. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Molecular and General Genetics 222: 377–383. [DOI] [PubMed] [Google Scholar]
- Winkler RG, Frank MR, Galbraith DW, Feyereisen R, Feldmann KA. 1998. Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis: isolation of mutations in the cytochrome P450 gene superfamily. Plant Physiology 118: 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Chandler PM, Smith JJ, Ross JJ. 2004. Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiology 134: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Molecular Biology of the Cell 16: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. 2000. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant Journal 21: 553–562. [DOI] [PubMed] [Google Scholar]
- Wright AD, Moehlenkamp CA, Perrot GH, Neuffer MG, Cone KC. 1992. The maize auxotrophic mutant orange pericarp is defective in duplicate genes for tryptophan synthase β. The Plant Cell 4: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD. 1991. Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254: 998–1000. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. 1998.COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua N-H. 2000.Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes and Development 14: 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. 2002. The SCFC°I1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. The Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yamamoto KT. 1998. Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of Arabidopsis, aux1 Plant Cell Physiology 39: 660–664. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M. 2004. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1 Plant Journal 40: 772–782. [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proceedings of the National Academy of Science of the USA 98: 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H. 2003. Members of the Arabidopsis-SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiology 133: 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. 2003. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL. 2002. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes and Development 16: 3100–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. 2002. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular Cell 10: 1519–1526. [DOI] [PubMed] [Google Scholar]
- Zimmerman PW, Hitchcock AE. 1942. Substituted phenoxy and benzoic acid growth substances and the relation of structure to physiological activity. Contributions of the Boyce Thompson Institute 12. [Google Scholar]
- Zolman BK, Bartel B. 2004. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proceedings of the National Academy of Science of the USA 101: 1786–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B. 2001.chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. Journal of Biological Chemistry 276: 31037–31046. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. 2001. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiology 127: 1266–1278. [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. 2000. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Ross JR, Koscheski P, Yang Y, Pichersky E, Noel JP. 2003. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. The Plant Cell 15: 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]