Abstract
• Background and Aims The aim of this study was to develop species-specific molecular markers for Bambusa balcooa and B. tulda to allow for their proper identification, in order to avoid unintentional adulteration that affects the quality and quantity of paper pulp production.
• Methods Two putative, species-specific RAPD markers, Bb836 for B. balcooa and Bt609 for B. tulda were generated using a PCR-based RAPD technique. Species-specificity of these two markers was confirmed through Southern hybridization in which RAPD gels were blotted and hybridized with radiolabelled cloned RAPD markers. Southern hybridization analyses were also performed to validate homology of the co-migrating Bb836 and Bt609 marker bands amplified from 16 different populations of B. balcooa and B. tulda, respectively. Sequence-characterized amplified region (SCAR) markers were developed from Bb836 and Bt609 sequences, using 20-mer oligonucleotide primers designed from both the flanking ends of the respective RAPD primers.
• Key Results As anticipated, Bb836 hybridized with an amplified band from B. balcooa and Bt609 hybridized only with an amplified product from B. tulda; the two markers did not hybridize with the amplified products of any of the other 14 bamboo species studied. The two pairs of SCAR primers amplified the target sequences only in the respective species. The species-specific SCAR fragments were named as ‘Balco836’ for B. balcooa and ‘Tuldo609’ for B. tulda. The species-specific ‘Balco836’ was amplified from the genomic DNA of 80 individuals of 16 populations of B. balcooa studied. Similarly, the presence of ‘Tuldo609’ was noted in all the 80 individuals representing 16 populations of B. tulda assessed. These SCAR fragments contained no obvious repetitive sequence beyond the primers.
• Conclusion These two molecular markers are potentially useful for regulatory agencies to establish sovereign rights of the germplasms of B. balcooa and B. tulda. In addition, this is the first report of species-specific SCAR marker development in bamboo.
Keywords: Bambusa balcooa, Bambusa tulda, SCAR markers, species characterization, RAPD
INTRODUCTION
India has the second largest reserve of bamboo populations in the world. The fibres of bamboo are mainly used in the pulp, paper and charcoal industries, while the culms have several other uses as ‘poor man's timber’. The total annual bamboo demand in India has been estimated to be approx. 5 million tonnes, of which about 3·5 million tonnes are required by the paper and pulp industry alone (Sharma, 1987). Bambusa balcooa is one of the strongest species and is thus preferred for construction purposes; it is also used for paper pulp production (Bhatt et al., 2003). On the other hand, B. tulda is sturdy, tall and one of the five quick-growing (70 cm d−1) species of bamboo (Dransfield and Widjaja, 1995). The species is suitable for the production of quality paper due to its long fibres (Upreti and Sundriyal, 2001) and also for producing furniture. Among the anatomical characteristics of bamboo culm, fibre length is important for technical evaluation, especially in the pulp and fibre-based industries. Fibre wall thickness of the culm predetermines the pulping characteristics, paper quality, permeability and strength relationships (Mohmod, 2001). Bambusa balcooa and B. tulda are two abundant tropical species that are recognized as priority bamboo species by the FAO (www.unep-wcmc.org, www.inbar.int) amongst eighteen other bamboo species found globally.
Due to the unusually long sexual cycle and unavailability of any other diagnostic tool, identification of bamboo is mainly dependent on vegetative descriptors such as culm morphology, and the morphology of the culm-sheath including ligule and auricle (Ohrnberger and Goerrings, 1986). PCR-based genetic markers are now well documented for species/cultivar identification (Khasa and Dancik, 1996; Samec and Nasinec, 1996; Raina et al., 2001; Johnson et al., 2003). A number of PCR-based methods, including randomly amplified polymorphic DNA (RAPD; Welsh and McClelland, 1990; Williams et al., 1990) and amplified fragment length polymorphism (AFLP; Vos et al., 1995), are available that do not require previous sequence information of the genome to be studied. In a RAPD assay, a short, usually ten nucleotides long, arbitrary primer is used, which generally anneals with multiple sites in different regions of the genome and amplifies several genetic loci simultaneously. This technique is simple, relatively inexpensive and has been employed to analyze the intra-and intergeneric genetic diversity of bamboo (Gielis et al., 1997; Ding, 1998; Wu et al., 1998; Nayak et al., 2003). In contrast to RAPD, the AFLP technique generates relatively complex patterns.
To overcome the reproducibility problem associated with the RAPD technique, RAPD markers have been converted into sequence-characterized amplified regions (SCAR; Paran and Michelmore, 1993). SCAR markers have been developed for several crops, including lettuce (Paran and Michelmore, 1993), common bean (Adam-Blondon et al., 1994), raspberry (Parent and Page, 1995), grape (Reisch et al., 1996), rice (Naqvi and Chattoo, 1996), Brassica (Barret et al., 1998) and wheat (Hernandez et al., 1999). Although molecular genotyping through SCAR primers has been documented in a few woody species, e.g. olive (Hernandez et al., 2001), papaya (Deputy et al., 2002), apple (Evans and James, 2003) and Salix (Gunter et al., 2003), there is no published report on development of species-specific markers in any of the bamboo species.
The identification of species-specific DNA markers in bamboo would be of immense importance for regulatory agencies to establish sovereign rights of the species in the country of its origin, as well as for the industry to avoid unintentional adulteration that affects the quality and quantity of merchandise production.
In this study, a number of species-specific RAPD markers were scored in an investigation on the phylogenetic relationships amongst 15 tropical/sub-tropical bamboo species. Two potential species-specific RAPD markers, Bb836 for B. balcooa and Bt609 for B. tulda were identified. To validate this finding, 16 different populations of each of these two species were collected from isolated climatic regions of West Bengal, India. The presence of these two putative species-specific RAPD markers in all the populations studied led us to design species-specific SCAR primers, allowing the amplification of a unique DNA fragment only from the expected species following PCR reaction under more stringent conditions. To the best of our knowledge, this study is the first to document the conversion of species-specific RAPD markers into dependable SCAR markers for the identification of B. balcooa and B. tulda.
MATERIALS AND METHODS
Plant material
Leaf samples from healthy plants of 15 bamboo species were collected from the germplasm stock maintained at the garden of Botanical Survey of India, Howrah, West-Bengal. The samples included 11 species of the genus Bambusa (B. affinis Munro., B. arundinacea Retz., B. atra Lindl., B. auriculata Kurz., B. balcooa Roxb., B. multiplex ‘riviereorum’ R. Maire, B. oliveriana Gamble, B. striata Lodd. Ex Wendl., B. tulda, Roxb., B. vulgaris Schrad. Ex Wendl., B. wamin, Camus), two species of the genus Dendrocalamus (D. giganteus Munro., D. strictus Roxb. Nees), one species of the genus Gigantochloa (G. atroviolacea Widjaja), and one species of the genus Pseudobambusa (P. kurzii Munro Ohrnberger). In addition, healthy leaves of B. balcooa and B. tulda populations were collected from 16 geographically isolated populations. The geographical details are given in Table 1 and Fig. 1. Five independent stands were sampled randomly for each population, resulting in a total of 80 plants for each species employed in this study.
Table 1.
Principal geographical features of the collection sites
| Species |
Collection number* |
Place of collection |
Latitude |
Longitude |
Altitude (m) |
Max. temp. (°C) |
Min. temp. (°C) |
Average annual rainfall (mm) |
Soil type |
|---|---|---|---|---|---|---|---|---|---|
| Bambusa balcooa | MD/ARA/02/024 | Arambagh | 22°52′N | 87°46′E | 14·0 | 36·0 | 15·0 | 1605·0 | Alluvial |
| B. tulda | MD/ARA/02/025 | ||||||||
| B. balcooa | MD/BAD/02/037 | Badu | 22°41′N | 88°27′E | 14·0 | 30·5 | 19·2 | 1625·0 | Alluvial |
| B. tulda | MD/BAD/02/038 | ||||||||
| B. balcooa | AP/BAR/99/181 | Baromile | 26°46′N | 88°26′E | 280·0 | 29·0 | 8·0 | 2160·0 | Alluvial red |
| B. tulda | AP/BAR/99/194 | ||||||||
| B. balcooa | SB/BHA/03/023 | Bhadreswar | 22°49′N | 88°21′E | 11·6 | 36·0 | 17·5 | 1523·0 | Deltic Alluvial |
| B. tulda | SB/BHA/03/032 | ||||||||
| B. balcooa | MD/CON/01/007 | Contai | 21°46′N | 87°45′E | 5·0 | 36·3 | 19·4 | 1610·0 | Alluvium transported buried laterite |
| B. tulda | MD/CON/01/008 | ||||||||
| B. balcooa | MD/DIA/03/069 | Diamond Harbour | 22°11′N | 88°11′E | 0 | 35·0 | 19·0 | 1480·0 | Deltic Alluvial |
| B. tulda | MD/DIA/03/071 | ||||||||
| B. balcooa | MD/DIG/03/084 | Digberia | 22°43′N | 88°31′E | 13·0 | 30·7 | 19·6 | 1600·8 | Alluvial |
| B. tulda | MD/DIG/03/088 | ||||||||
| B. balcooa | MD/HAS/02/064 | Hasnabad | 22°34′N | 88°55′E | 6·0 | 32·5 | 19·1 | 1630·0 | Deltic Alluvial |
| B. tulda | MD/HAS/02/066 | ||||||||
| B. balcooa | AP/KAL/99/121 | Kalimpong | 27°40′N | 88°28′E | 787·0 | 25·0 | 7·0 | 2200·0 | Alluvial red |
| B. tulda | AP/KAL/99/149 | ||||||||
| B. balcooa | SB/MEM/03/067 | Memari | 23°11′N | 88°7′E | 24·0 | 36·0 | 13·2 | 1621·0 | Alluvial |
| B. tulda | SB/MEM/03/069 | ||||||||
| B. balcooa | MD/NIL/02/049 | Nilganj | 22°45′N | 88°22′E | 14·0 | 36·0 | 20·0 | 1500·0 | Deltic Alluvial |
| B. tulda | MD/NIL/02/050 | ||||||||
| B. balcooa | MD/PUR/03/077 | Purulia | 23°19′N | 86°22′E | 227·0 | 42 | 5·0 | 1250·0 | Laterite |
| B. tulda | MD/PUR/03/081 | ||||||||
| B. balcooa | AP/SAL/99/157 | Salugaura | 26°51′N | 88°16′E | 134·0 | 20·1 | 6·0 | 2500·0 | Alluvial red |
| B. tulda | AP/SAL/99/173 | ||||||||
| B. balcooa | MD/SIB/02/057 | Sibpur | 22°34′N | 88°19′E | 10·0 | 30·7 | 19·8 | 1633·6 | Deltic Alluvial |
| B. tulda | SB/SIB/02/015 | ||||||||
| B. balcooa | SB/SIN/03/056 | Singur | 22°48′N | 88°13′E | 13·0 | 36·0 | 16·8 | 1445·0 | Alluvial |
| B. tulda | SB/SIN/03/058 | ||||||||
| B. balcooa | SB/SRE/03/046 | Srerampore | 22°53′N | 88°24′E | 14·2 | 35·0 | 18·0 | 1580·0 | Deltic Alluvial |
| B. tulda | SB/SRE/03/047 |
Collection number of representative individual of each population is given.
Fig. 1.
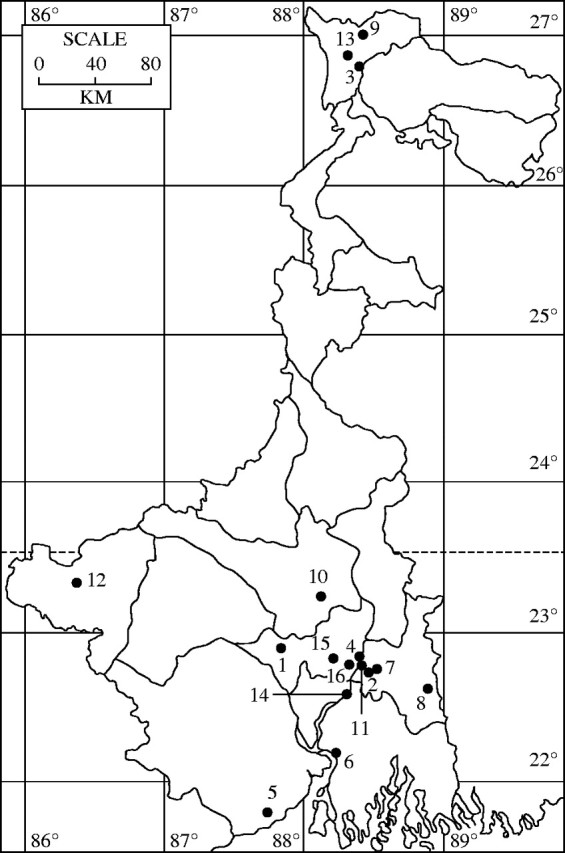
Map of West Bengal (India) depicting 16 different geographical locations of the collection sites of B. balcooa and B. tulda populations: 1, Arambagh; 2, Badu; 3, Baromile; 4, Bhadreswar; 5, Contai; 6, Diamond Harbour; 7, Digberia; 8, Hasnabad; 9, Kalimpong; 10, Memari; 11, Nilganj; 12, Purulia; 13, Salugaura; 14, Sibpur; 15, Singur; 16, Srerampore.
Isolation of PCR-compatible genomic DNA
Bamboo leaves were surface-sterilized following a modified procedure of Schultz et al. (1993) and genomic DNA was isolated from 200 mg of tissue using a protocol modified from Dellaporta et al. (1983). Briefly, tissues were ground to a fine powder and extracted for 30 min at 60 °C using a high-salt extraction buffer (50 mm Tris pH 8·0, 100 mM EDTA pH 8·0, 150 mm NaCl, 1·8 % SDS, 1·0 % PVP and PVPP). The supernatant was extracted 3–4 times with chloroform-isoamyl alcohol (24 : 1), 2·5× volume of chilled alcohol added, stored at −20 °C overnight and centrifuged at 10 000 g for 10 min. The pellet was suspended in TE buffer (10 mm Tris and 1 mm EDTA, pH 8·0) and total RNA was removed by RNase treatment. The concentration of DNA samples was determined by comparing band intensity with known concentrations of lambda DNA digest after 0·8 % agarose gel electrophoresis and ethidium bromide staining.
RAPD analysis and marker selection
Thirty RAPD primers were initially screened to detect species-specific markers. The PCR amplifications were carried out in a 50 μL reaction mixture containing 100 ng of genomic DNA, 100 ng of RAPD primer, 1× PCR buffer containing 1·5 mm MgCl2, 250 μm of each dNTPs and 1 unit of Taq DNA polymerase (Bangalore Genei, India). The amplifications were carried out in a Perkin Elmer Cetus 2400 thermal cycler using the following programme: 4 min at 95 °C followed by 35 amplification cycles (45 sec at 94 °C, 45 sec at 35 °C, 1 min at 72 °C) and finally 10 min at 72 °C for elongation. The amplified PCR products were resolved by electrophoresis on 1·5 % agarose (Sigma, USA) gel and TAE buffer. Gels were visualized by ethidium bromide staining and recorded with a gel doc 1000 camera using the programme Molecular Analyst version 1·5 (Bio Rad, USA). Two bright and highly reproducible RAPD bands—putative markers—were selected for both the species to develop species-specific SCAR markers.
Cloning and sequencing of RAPD products
Two species-specific RAPD markers [Bb836 derived from the primer PW-02 (5′-TCGTCGGCGT-3′) and Bt609 derived from the primer OPA-08 (5′-GTGACGTAGG-3′)] were excised from 1·5 % agarose gels with a sterile gel slicer and the DNA was purified using the MinElute Gel Extraction kit (QIAGEN, USA). The eluted fragments were ligated into pGEM T Easy Vector (Promega, USA) following the supplier's instructions, transformed into competent Escherichia coli strain, DH5oc, and the plasmid DNA purified from the white colonies as described by Sambrook et al. (1989). Selected transformed clones were screened by PCR analysis with corresponding RAPD primers and the sizes of the DNA inserts were checked by EcoRI (Roche, Germany) restriction digestion. The inserted DNA fragments were sequenced at the University of Delhi, South Campus, India, using an ABI Prism 3100 automated DNA sequencer.
Hybridization analysis
The RAPD products of the PW-02 primer from the 15 bamboo species and 16 randomly selected individuals from 16 populations of B. balcooa were transferred to two positively charged nylon membranes (Roche) and probed with αP32 labelled EcoRI excised Bb836 marker. Probe labelling was performed using the Prime-a-gene labelling system kit (Promega). Approximately 25 ng of probe DNA was labelled with αP32 dCTP (BARC, India). Transfer of DNA to a nylon membrane, UV cross-linking, prehybridization and hybridization at 68 °C followed by repeated washing for 20 min each at 25 °C, 55 °C and 65 °C were performed following the methods of Sambrook et al. (1989). Signals were detected by exposing the membranes to X-ray film (Kodak). The marker fragment was used as a positive control. The objective of the first hybridization experiment was to check the species-specificity of the Bb836 marker, and that of the second experiment was to validate the homology of the co-migrating Bb836 marker band amplified from different populations of B. balcooa.
Similarly, PCR amplified products of 15 bamboo species and 16 representative populations of B. tulda with the OPA-08 primer were transferred to two positively charged membranes and hybridized with the excised Bt609 marker. The marker fragment was used as a positive control.
Designing SCAR primers and amplification of genomic regions
Based on the sequences of the cloned RAPD markers, Bb836 and Bt609, two pairs of oligonucleotide primers of 20 nucleotides each, were designed by extending the original ten bases of the RAPD primer with the next ten nucleotides of the DNA sequences at the 3′ ends. The SCAR primer sequences (Balco836F and Balco836R for B. balcooa, and Tuldo609F and Tuldo609R for B. tulda) were custom-synthesized from Integrated DNA Technologies Inc, USA (Table 2). The two species-specific SCAR primer pairs were used for PCR amplifications of genomic DNA of the 15 bamboo species, including one individual representative of the 16 populations of B. balcooa and B. tulda. PCR conditions for genomic DNA amplification with SCAR primers were the same as for RAPD, except for the annealing temperatures. The optimal annealing temperatures to achieve species-specific amplification were determined (Table 2).
Table 2.
Bamboo species-specific SCAR primer sequences derived from cloned RAPD fragments and optimal annealing temperature for each set of reactions
| RAPD primer |
SCAR primer* |
Sequence (5′-3′)† |
Annealing temperature |
|---|---|---|---|
| PW-02 | Balco800F | TCGTCGGCGTAGACGGAGAG | 60 °C |
| Balco800R | TCGTCGGCGTTCGAGCTTAT | ||
| OPA-08 | Tuldo600F | GTGACGTAGGCGAACATGGC | 60 °C |
| Tuldo600R | GTGACGTAGGGCATACCTTG |
The numbers preceding the R (reverse) and F (forward) refer to the approximate size of the SCAR band (in bp), ‘Balco’ and ‘Tuldo’ indicate the two source genomes, B. balcooa and B. tulda, respectively.
The nucleotides underlined represent the sequence of the RAPD primers used.
Analysis of sequence data
The DNA sequences of Bb836 and Bt609 were named as ‘Balco836’ and ‘Tuldo609’ and submitted to GenBank (Accession numbers AY653073, AY684298). Homology searches were performed within GenBank's non-redundant Viridiplantae database using the BLAST 2.2.8 (Basic Local Alignment Search Tool; Altschul et al., 1997) algorithm at http://www.ncbi.nlm.nih.gov/BLAST/ of the National Center for Biotechnology Information (NCBI), with the program BLASTN. The ORF signatures within the two sequences were detected using the NCBI ORF finder tool.
RESULTS
Identification of species-specific RAPD markers in bamboo
Of the 30 RAPD primers screened, 22 primers produced distinct, reproducible, polymorphic profiles within the 15 bamboo species surveyed. The approximate size range of the RAPD products was 200 bp to 2 kb. Reproducibility of the amplification pattern was checked by repeating each reaction at least twice without deliberate alteration in the protocol. Alhough a number of species-diagnostic RAPD bands were noted, most of them were either rather faint or not repeatedly found in all the representative individuals of the 15 species. Thus, a large number of potentially species-specific, informative RAPD bands were eliminated from consideration. In contrast, the primer PW-02 amplified a single, bright band of approx. 800 bp from B. balcooa, which was absent in the rest of the 14 bamboo species (Fig. 2). This band was named Bb836, and was amplified from all the 80 individuals representing 16 different populations of B. balcooa without exception. Similarly, the primer OPA-08 amplified a distinct, reproducible band of approx. 600 bp, termed Bt609, from B. tulda, which was not found in the other 14 bamboo species (Fig. 3). This band was consistently present in all the 80 plants collected from the 16 different populations of B. tulda. These two bands, Bb836 and Bt609, were selected as putative species-specific markers.
Fig. 2.
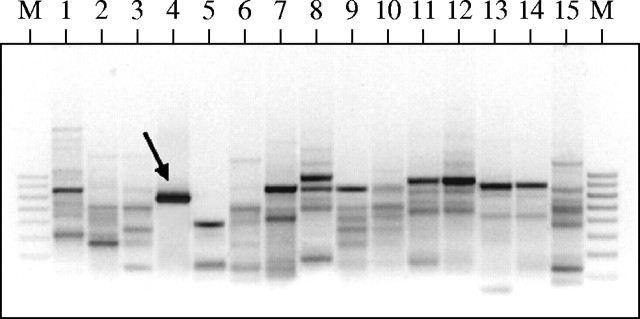
RAPD profiles of 15 bamboo species amplified with PW-02 primer. Lane 1, Bambusa atra; lane 2, B. auriculata; lane 3, B. arundinacea; lane 4, B. balcooa; lane 5, B. multiplex; lane 6, B. oliverina; lane 7, B. striata; lane 8, B. tulda; lane 9, B. vulgaris; lane 10, B. wamin; lane 11, B. affinis; lane 12, Pseudobambusa kurzii; lane 13, Gigantochloa atroviolacea; lane 14, Dendrocalamus giganteus; lane 15, D. strictus. M, molecular marker (100 bp ladder), the arrow indicates migration of Bb836 band.
Fig. 3.
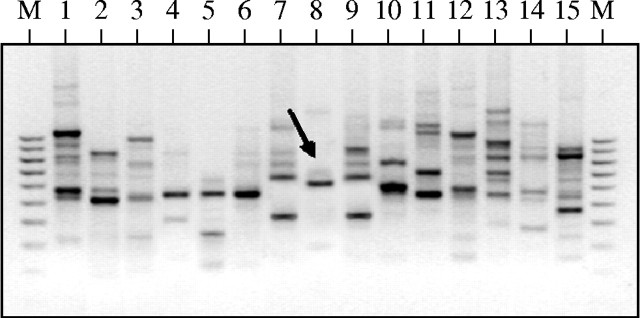
RAPD profiles of 15 bamboo species amplified with OPA-08 primer. The arrow indicates the migration of Bt609 band. Lane numbers represent different bamboo species as shown in Fig. 2.
Cloning and sequencing of the two putative species-specific markers
The two putative species-specific RAPD markers were successfully cloned and sequenced. The size of the inserted DNA fragments were confirmed by both PCR and restriction digestion analysis. The first ten nucleotides of the sequences obtained matched completely with the corresponding RAPD primers used.
Hybridization analysis
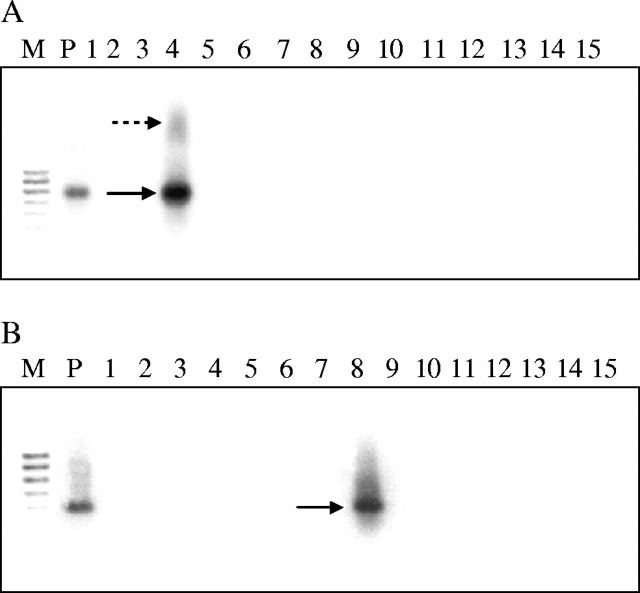
The cloned Bb836 band was used to probe the Southern blots of PW-02-generated RAPD products of the 15 species (Fig. 2). A hybridization signal was obtained only with B. balcooa and the positive control (Fig. 4A). In addition to the expected RAPD band, the probe cross-hybridized weakly to another slow-migrating band, which could not be resolved by ethidium bromide staining. Similarly, the hybridization signal was obtained only with the lane containing OPA-08-generated PCR products of B. tulda and the positive control (Fig. 4B) when probed with the cloned Bt609 band. As the position of each hybridization signal corresponded to that of the expected RAPD band, it can be inferred that the cloned fragments were derived from the amplified RAPD products of the genomic DNA of the bamboo species, and not from contaminants of the same size present at concentrations below the resolution of ethidium bromide staining. No non-specific signal was found, indicating the absence of any homologous sequence in the amplified products of the other 14 species studied in both cases.
Fig. 4.
Hybridization of (A) Bb836 and (B) Bt609 bands with two Southern blots of corresponding agarose gels containing RAPD products of 15 bamboo species amplified with PW-02 and OPA-08 primers, respectively. P, positive control; the solid arrow indicates the hybridization signal of the target band and the dotted arrow indicates the false positive band.
Confirmatory tests were performed to check the homology of the co-migrating bands generated from different representative individuals of 16 populations of the two species concerned. The two sets of blotted RAPD gels for the two species were probed with the respective cloned marker bands and the position of the hybridization signal was compared with the position of the species-specific RAPD band. In both the cases, the hybridization signal corresponded with the position of the species-specific RAPD marker (data not shown). The results suggest that the bands of the same size that amplified in different individuals of a species using the same primer do indeed represent the same sequence.
Sequence data analysis
The BLAST results revealed that both the sequences have homology with known plant nucleotide sequences at different sequence-similarity levels (data not shown). Repeats were not found within any of the sequences. The length of the Bb836 marker sequence obtained was 836 bp with 46 % G+C (A = 226; C = 236; G = 145; T = 229). The NCBI ORF finder revealed that the derived amino acid sequence of Bb836 contains an ORF signature from amino acid 85 to 201. The BLAST analysis of this ORF showed considerable similarity (33·9 bits, E-value = 0·95) with an unknown protein of Oryza sativa ‘japonica’ (AAT38047).
The length of the Bt609 sequence obtained was 609 bp with 52 % G+C (A = 151; C = 173; G = 157; T = 154). An ORF was also detected within the in silico translated product of Bt609 from 93 to 184, which showed similarity (35·0 bits and E-value = 0·43) with a hypothetical protein of Caenorhabditis briggsae (CAE65839).
Amplification using SCAR primers
The designed putatively species-specific SCAR primer pairs were used to amplify genomic DNAs of the 15 bamboo species. A single, distinct and brightly resolved band of the same size as the original RAPD fragment was obtained in both B. balcooa and B. tulda and no non-specific amplification was observed in the other 14 species (Figs 5, 6). When 80 individuals of B. balcooa and B. tulda were screened with the corresponding SCAR primer pairs, single bands of the expected length were amplified from each of the samples (data not shown). Reduction of the annealing temperatures did not generate an alternate or extra allele other than the SCAR, confirming the specificity of the SCAR primers.
Fig. 5.
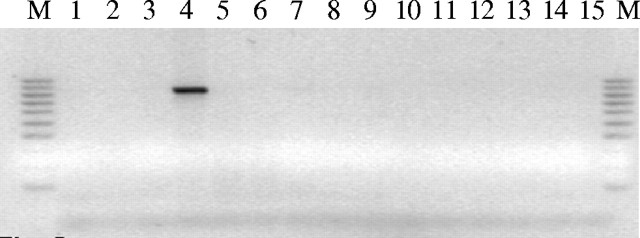
Electrophorogram of agarose gel (1·5 %) showing the amplification product from the genomic DNA of B. balcooa only using ‘Balco836F’ and ‘Balco836R’ SCAR-primer-pairs, and the absence of the markers in 14 other bamboo species.
Fig. 6.
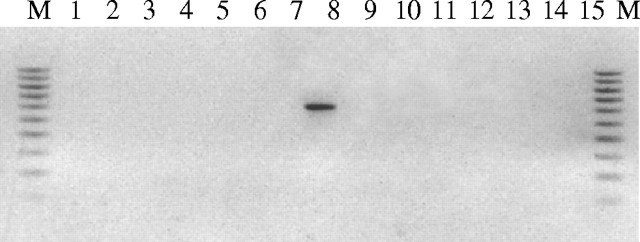
Electrophorogram of agarose gel (1·5 %) showing the amplification product only from the genomic DNA of B. tulda using ‘Tuldo609F’ and ‘Tuldo609R’ SCAR-primer-pairs, and the absence of the markers in 14 other bamboo species.
DISCUSSION
In this investigation two species-specific SCAR markers, ‘Balco836’ for Bambusa balcooa and ‘Tuldo609’ for B. tulda, were developed by designing primers from sequenced, putatively species-specific, RAPD bands. Inconsistencies and lack of reproducibility for amplified product profiles of RAPD reactions in independent laboratories have made RAPD markers less useful than anticipated. Species-specific marker development for plant taxa require reliable, reproducible, amplified genomic sequences, especially when they are to be used in germplasm patent disclosures and in legal issues of varietal infringement. The sensitivity of the RAPD reaction to a number of reaction parameters at a low annealing temperature has failed to generate consistent profiles even under laboratory conditions, which necessitates designing SCAR primers from polymorphic RAPD bands.
The Southern analysis using putatively species-specific RAPD bands, Bb836 and Bt609, showed homology with the target amplified RAPD band of the same size, but did not show any hybridization signal with the amplified products of the other 14 species studied (compare Fig. 2 with Fig. 4A, and Fig. 3 with Fig. 4B). These results predicted that the two putative RAPD markers, Bb836 and Bt609, might be specific for B. balcooa and B. tulda, respectively. Weeden et al. (1992) have affirmed that without DNA hybridization, or DNA sequence analysis, it is extremely difficult to establish that RAPD bands amplified in individuals of undefined relationships are homologous. This was also considered as a major impediment to use of RAPD amplification with distantly related plant species. In the case of B. balcooa, an additional faint signal was found in the autoradiogram, which was not detected by ethidium bromide staining (compare Fig. 2 with Fig. 4A). Previously, Vidal et al. (2000) have reported that partial sequence homology was found amongst bands of different molecular weights in the Vitis genome.
Xu et al. (1995) have reported that in four Vitis rootstocks, in which no original RAPD band was revealed by ethidium bromide-stained agarose gels, hybridization signals were achieved with a RAPD-derived probe. It was concluded that, although some amount of DNA amplification took place in the rootstocks, the concentrations were too low to be detected by ethidium bromide staining. In two other rootstocks, positive RAPD bands showed no detectable hybridization signals, perhaps due to generation of amplified bands by a mismatch at the priming sites. These results suggest that Southern hybridization experiments are absolutely necessary for analysing the molecular identity of the RAPD markers, especially prior to the designing of SCAR primers.
The sequence analysis of ‘Balco836’ and ‘Tuldo609’ indicated the absence of repeated sequence motifs. Previously, Paran and Michelmore (1993) reported that RAPD products were not ideal for use as hybridization probes because they often contain repetitive DNA sequences. Clearly, in the present study this factor was not a problem. Additionally, the in silico translated 255–605 nucleotide sequence of ‘Balco836’ showed considerable homology with an as yet unassigned open reading frame coding for a protein of Oryza sativa ‘japonica’ (AAT 38047). This is probable as bamboo and rice both belong to the family Poaceae. On the other hand, the derived amino acid sequence from 279–554 nucleotides of ‘Tuldo609’ showed the highest homology with a hypothetical protein of Caenorhabditis briggsae (CAE65839). This is not improbable as such trans-kingdom homology between ancient proteins is quite common. For example, the amino-terminal domain of the nucleotide-binding site of plant disease resistance proteins shows high homology to both the ‘Toll’ protein of Drosophila and ‘interleukin receptor like’ proteins of mammals (Hammond-Kosack and Jones, 1997). It may be assumed that the genome of B. tulda has retained some proteins of ancient origin, which might have been lost or modified substantially during evolution in the other 14 bamboo species investigated. The polyphyletic nature of the genus Bambusa was demonstrated by Loh et al. (2000) during an investigation on genetic variation and relationships among bamboo species using AFLP markers. In an independent study, we have analyzed the phylogenetic relationships among 15 bamboo species using 120 polymorphic loci, generated by RAPD primers. The cladistic analysis is in gross agreement with respect to the phylogeny of the species of Bambusa. The study revealed that the dissimilarity coefficient between B. balcooa and B. tulda is 0·607 (unpublished data).
Taxonomists have been relying solely on vegetative characters for the identification of all the bamboo species in the absence of reproductive characters. Bamboos flower once in a lifetime and then die: the vegetative phase lasts from 1–120 years depending on the species. Usually, genera are identified on the basis of culm, rhizome and branching characters, while species characterization is mainly dependent on culm sheath, ligule and auricle characters (Ohrnberger and Goerrings, 1986). Species identification based only on morphological characters may be misleading as vegetative characters are often influenced by environmental factors (Wu, 1962). In spite of the serious, outstanding need to develop a method to identify bamboo species at the molecular level, attention has been lacking. To the best of our knowledge, this is the first report of identification of B. balcooa and B. tulda using species-specific SCAR markers developed from RAPD markers. We hope that our approach of developing species-diagnostic SCAR markers may pave the way for the unambiguous identification of the bamboo species.
Acknowledgments
This investigation was supported by a research grant from the Council of Scientific and Industrial Research [Grant No. 38 (1062)/03/EMR-II], New Delhi, India. The authors are thankful to Dr T. K. Ghose for his comments and valuable suggestions for the improvement of the manuscript.
LITERATURE CITED
- Adam-Blondon AF, Sevignac M, Bannerot H, Dron M. 1994. SCAR, RAPD and RFLP markers linked to a dominant gene (Are) conferring resistance to anthracnose in common bean. Theoretical and Applied Genetics 88: 865–870. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret P, Delourme R, Foisset N, Renard M. 1998. Development of a SCAR sequence characterized amplified region) marker for molecular tagging of the dwarf BREIZH (Bzh) gene in Brassica napus L. Theoretical and Applied Genetics 97: 828–833. [DOI] [PubMed] [Google Scholar]
- Bhatt BP, Singha LB, Singh K, Sachan MS. 2003. Some commercial edible bamboo species of North East India: production, indigenous uses, cost–benefit and management strategies. Bamboo Science and Culture 17: 4–20. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter 1: 19–21. [Google Scholar]
- Deputy JC, Ming R, Ma H, Liu Z, Fitch MMM, Wang M, Manshardt R, Stiles JI. 2002. Molecular markers for sex determination in papaya (Carica papaya L.). Theoretical and Applied Genetics 106: 107–111. [DOI] [PubMed] [Google Scholar]
- Ding Y. 1998.Systematic studies on Phyllostachys. Ph.D. Dissertation, Nanjing University, China. [Google Scholar]
- Dransfield S, Widjaja EA. 1995.Plant resources of South-East Asia, No 7. Bamboos. Leiden; Backhuys Publishers. [Google Scholar]
- Evans KM, James CM. 2003. Identification of SCAR markers linked to Pl-w mildew resistance in apple. Theoretical and Applied Genetics 106: 1178–1183. [DOI] [PubMed] [Google Scholar]
- Gielis J, Everaert I, De LM. 1997. Analysis of genetic variability and relationships in Phyllostachys using random amplified polymorphic DNA. In: Chapman G, eds. The bamboos. London: Academic Press, 107–124. [Google Scholar]
- Gunter LE, Roberts GT, Lee K, Larimer FW, Tuskan GA. 2003. The development of two flanking SCAR markers linked to a sex determination locus in Salix viminalis L. Journal of Heredity 94: 185–189. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. 1997. Plant disease resistance genes. Annual Review of Plant Physiology and Plant Molecular Biology 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Hernandez P, Martin A, Dorado G. 1999. Development of SCARs by direct sequencing of RAPD products: A practical tool for the introgression and marker assisted selection of wheat. Molecular Breeding 5: 245–253. [Google Scholar]
- Hernandez P, Rosa RDL, Rallo L, Martin A, Dorado G. 2001. First evidence of retrotransposon like element in olive (Olea europaea): Implications in plant variety identification by SCAR marker development. Theoretical and Applied Genetics 102: 1082–1087. [Google Scholar]
- Johnson EL, Saunders JA, Mischke S, Helling CS, Emche SD. 2003. Identification of Erythroxulum taxa by AFLP DNA analysis. Phytochemistry 64: 187–197. [DOI] [PubMed] [Google Scholar]
- Khasa PD, Dancik BP. 1996. Rapid identification of white Engelmann spruce species by RAPD markers. Theoretical and Applied Genetics 92: 46–52. [DOI] [PubMed] [Google Scholar]
- Loh JP, Kiew R, Set O, Gan LH, Gan YY, 2000. A study of genetic variation and relationships within the bamboo subtribe Bambusinae using amplified fragment length polymorphism. Annals of Botany 85: 607–612. [Google Scholar]
- Mohmod AL. 2001. Anatomical features of Bambusa vulgaris and Gigantochloa scortechnii from four harvesting sites in peninsular Malaysia. Journal of Tropical Forest Products 7: 10–28. [Google Scholar]
- Naqvi NI, Chattoo BB. 1996. Development of a sequence-characterized amplified region (SCAR) based indirect selection method for a dominant blast resistance gene in rice. Genome 39: 26–30. [DOI] [PubMed] [Google Scholar]
- Nayak SGR, Rout GR, Das P. 2003. Evaluation of the genetic variability in bamboo using RAPD markers. Plant Soil and Environment 49: 24–28. [Google Scholar]
- Ohrnberger D, Goerrings J. 1986.The bamboos of the world. Odenthal, Germany. [Google Scholar]
- Paran I, Michelmore RW. 1993. Development of reliable PCR based markers linked to downy mildew resistance genes in lettuce. Theoretical and Applied Genetics 85: 985–993. [DOI] [PubMed] [Google Scholar]
- Parent JG, Page D. 1995. Evaluation of SCAR markers to identify raspberry cultivars. HortScience 30: 856 (Abstract). [Google Scholar]
- Raina SN, Rani V, Kojima T, Ogihara Y, Singh KP, Deyarumath RM. 2001. RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44: 763–772. [PubMed] [Google Scholar]
- Reisch BI, Weeden NF, Lodhi MA, Ye G, Soylemezoglu G. 1996. Linkage map construction in two hybrid grapevine (Vitis sp.) populations. In: Plant genome IV: Proceedings of the Fourth International Conference on the Status of Plant Genome Research. Maryland, USA: USDA, ARS, 26 (Abstract). [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. 1989.Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Samec P, Nasinec V. 1996. The use of RAPD technique for the identification and classification of Pisum sativum L. genotypes. Euphytica 89: 229–234. [Google Scholar]
- Schultz B, Wanke U, Draeger S, Aust HJ. 1993. Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycological Research 97: 1447–1450. [Google Scholar]
- Sharma YML. 1987. Inventory and resource of bamboo. In: Rao AN, Dhanarajan G, Sastry CB, eds. Recent research in bamboo. Proceedings of the International Bamboo Workshop. China. Chinese Academy of Forestry, China and International Development Research Centre, Canada, 14–17. [Google Scholar]
- Upreti TC, Sundriyal RC. 2001. Bamboo and cane resources of Arunachal Pradesh: utilization pattern and implications for management. Bamboo Science and Culture 15: 20–34. [Google Scholar]
- Vidal JR, Delavault P, Coarer M, Defontaine A. 2000. Design of grapevine (Vitis vinifera L.) cultivar-specific SCAR primers for PCR fingerprinting. Theoretical and Applied Genetics 101: 1194–1201. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. 1995. AFLP, a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeden NF, Timmerman GM, Hemmat M, Kneen BE, Lodhi MA. 1992. Inheritance and reliability of RAPD markers. Applications of RAPD Technology to Plant Breeding. Joint Plant Breeding Symposia Series. Madison, WI: Crop Science Society of America, 12–17. [Google Scholar]
- Welsh J, Mc Clelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 19: 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18: 6531–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC-Y. 1962. Classification of Bambuseae based on leaf anatomy. Botanical Bulletin of Academia Sinica 3: 83–107. [Google Scholar]
- Wu Y, Chun Nong H, Jun Hui W. 1998. Primary study on the RAPD fingerprinting of four bamboo species. Journal of Bamboo Research 17: 10–14. [Google Scholar]
- Xu Hong, Wilson DJ, Arulsekar S, Bakalinsky AT. 1995. Sequence specific polymerase chain reaction markers derived from randomly amplified polymorphic DNA markers for fingerprinting grape (Vitis) rootstocks. Journal American Society of Horticultural Science 120: 714–720. [Google Scholar]