Abstract
• Background and Aims Rice (Oryza sativa) is one of the most important cereal plants in the world. Wild-abortive (WA) and Honglian (HL) cytoplasmic male sterility (CMS) have been used extensively in the production of hybrid seeds. Although a variable number of fertility-restorer genes (Rf) for WA and HL-CMS have been identified in various cultivars, information on Rf in Oryza species with the AA-genome is sparse. Therefore the distribution and heredity of Rf for WA and HL-CMS in wild rice species of Oryza with the AA-genome were investigated.
• Methods Fertility-restorer genes for WA and HL-CMS in wild rice species with the AA-genome were investigated by following the fertility of microspores identified by I2–KI staining and by following the seed-setting rate of spikelets. A genetic model of Rf in some selected restorer accessions was analysed based on the fertility segregation of BC1F1 populations.
• Key Results Fertility analysis showed that 21 out of 35 HL-type F1s, and 13 out of 31 WA-type F1s were scored as fertile. The frequency of Rf in wild rice was 60 % for HL-CMS and 41·9 % for WA-CMS, respectively. The fertility-restorer accessions, especially those with complete restoring ability, aggregated mainly in two species of O. rufipogon and O. nivara. The wild rice accessions with Rf for HL-CMS were distributed in Asia, Oceania, Latin American and Africa, but were centered mainly in Asia, whilst the wild restorer accessions for WA-CMS were limited only to Asia and Africa. Apart from one restorer accession that possessed two pairs of Rf for WA-CMS, all of the other nine tested wild restorer accessions each contained only a single Rf for WA-CMS or HL-CMS. Allele analysis indicated that there existed at least three Rf loci for the WA and HL-CMS systems.
• Conclusions These data support the hypothesis that fertility-restorer genes exist widely in Oryza species with the AA-genome, and that Rf in Oryza sativa originated from the Oryza rufipogon/Oryza nivara complex, the ancestor of cultivated rice in Asia. The origin and evolution of Rf is tightly linked to that of CMS in wild rice, and fertility of a given CMS type is controlled by several Rf alleles in various wild restorer accessions.
Keywords: AA genome, Oryza, cytoplasmic male sterility, fertility-restorer genes, distribution, heredity, Honglian, wild-abortive
INTRODUCTION
Cytoplasmic male sterility (CMS), which causes the production of non-functional pollen and is inherited maternally, is important in commercial hybrid seed production (Kaul, 1988) and breeding programmes. A number of studies on the relationship between CMS and fertility-restorer genes (Rf) have been conducted in various plants and may enable a better understanding of genetic differentiation and the interaction between cytoplasmic and nuclear genomes in plants (Budar and Pelletier, 2001).
The wild-abortive (WA) and Honglian (HL) are two genetically different types of CMS in rice (Oryza sativa) (Li and Zhu, 1988). The former has been used extensively in commercial production, and its fertility is sporophytically restored by the dominant restorer genes (Shen et al., 1998; Jing et al., 2001): fertility-restorer genes are important in the production of hybrid rice. Although a variable number of restorer genes have been proposed in various restorer lines, one or two dominant restorer alleles (Rf3 and Rf4) are usually suggested to be responsible for the fertility (Yao et al., 1997; Tan et al., 1998). HL-CMS is a gametophytic type as is BT-CMS in rice. The Honglian-type hybrid rice has also been widely cultivated in China. It has been suggested that HL-CMS is restored by only one dominant restorer gene, Rf5 or Rf6, in various cultivated lines (Liu et al., 2004).
The maintenance and transference of CMS within natural population of wild rice cannot be separated from Rf and it is easy to assume that the restorer genes exist in wild rice. However, the information about the origin, evolutionary relationships and distribution of the fertility-restorer genes for WA-CMS and HL-CMS systems is fragmentary. To be able to recognize the Rf in wild rice would facilitate not only the exploitation of new Rf alleles but also give a better understanding of the origin and evolution of the fertility-restorer genes. In the present study, the distribution of the Rf for WA-CMS and HL-CMS in wild rice with the AA genome was investigated.
MATERIALS AND METHODS
Materials
Thirty-seven wild rice (Oryza sativa L.) accessions with the AA genome from the International Rice Research Institute (IRRI) (Table 1), a typical Honglian CMS line, i.e. Yuetai A (YtA) and the corresponding maintainer, i.e. Yuetai B (YtB), a typical WA-CMS line, i.e. Zhenshan 97A (ZsA) and the corresponding maintainer, i.e. Zhenshan 97B (ZsB), were used in this study. Plants were grown in the experimental fields within Wuhan University campus, Wuhan, China in the summer and Hainan Island, Hainan, China in the winter during 2001–2003. All of the wild rice and their derived progenies were given a 10-h short photoperiod (0800–1800 h) after they were grown for about 2 months at Wuhan.
Table 1.
Accessions of wild rice used in the study (from the IRRI)
| Series number |
Accession number |
Species |
Source (country) |
|---|---|---|---|
| w06 | 100219 | O. rufipogon | Thailand |
| w07 | 100968 | O. glumaepatula | Suriname |
| w08 | 100970 | O. glumaepatula | Brazil |
| w09 | 101213 | O. longistaminata | Ivory coast |
| w10 | 101255 | O. barthii | Cameroon |
| w11 | 101411 | O. meridionalis | Australia |
| w12 | 101791 | O. glaberrima | Senegal |
| w13 | 101855 | O. glaberrima–Saria 480 | Berkina Faso |
| w14 | 101959 | O. barthii | Senegal |
| w15 | 101971 | O. nivara | India |
| w16 | 101974 | O. rufipogon | India |
| w17 | 102452 | O. glaberrima | Mali |
| w18 | 102641 | O. glaberrima | Liberia |
| w19 | 103580 | O. barthii | Chad |
| w20 | 103836 | O. nivara | Bangladesh |
| w21 | 104078 | O. barthii | Nigeria |
| w22 | 104081 | O. barthii | Nigeria |
| w23 | 104085 | O. meridionalis | Australia |
| w24 | 104127 | O. longistaminata | Chad |
| w25 | 104147 | O. longistaminata | Cameroon |
| w26 | 104540 | O. glaberrima—Ex Kano | Nigeria |
| w27 | 104599 | O. rufipogon—Uru Wee | Sri Lanka |
| w28 | 104680 | O. nivara | India |
| w29 | 104705 | O. nivara | India |
| w30 | 105204 | O. longistaminata— Zurha/Sukimia | Ethiopia |
| w31 | 105283 | O. meridionalis | Australia |
| w32 | 105293 | O. meridionalis | Australia |
| w33 | 105303 | O. meridionalis | Australia |
| w34 | 105419 | O. nivara–Uru Wee | Sri Lanka |
| w35 | 105561 | O. glumaepatula | Colombia |
| w36 | 105661 | O. glumaepatula– Arroz Bravo | Brazil |
| w37 | 105704 | O. nivara | Nepal |
| w38 | 105736 | O. nivara–Srange | Cambodia |
| w39 | 105887 | O. rufipogon–Jhora | Bangladesh |
| w40 | 106036 | O. rufipogon–Padi Hantu | Malaysia |
| w41 | 106083 | O. rufipogon | India |
| w42 | 106158 | O. rufipogon | Laos |
| w43 | 106194 | O. barthii | Guinea |
| w44 | 106260 | O. rufipogon | Papua New Guinea |
| w45 | 106309 | O. nivara | Cambodia |
| w46 | 106321 | O. rufipogon | Cambodia |
| w47 | 106344 | O. nivara | Myanmar |
| w-Guilin | O. rufipogon | China | |
| w-Dongxiang-1 | O. rufipogon | China | |
| w-Dongxiang-2 | O. rufipogon | China |
Field scores of the fertility of the plants
Fertility evaluation was conducted using two different criteria: (1) does the pollen stain in a 1 % I2-KI solution? and (2) at what rate is the seed set on a spikelet? (Dalmacio et al., 1992). Plants were considered completely fertile if >40 % (HL-type) or >80 % (WA-type) of their pollen stained darkly and the seed-setting rate of a bagged spikelet was >30 %. If the proportion of darkly stained pollen ranged from 10 % to 40 % (HL-type) or to 80 % (WA-type), and the seed-setting rate of a bagged spikelet ranged from 5 % to 30 %, the plants were considered partially fertile. Otherwise, the plants were scored sterile.
Genetic analysis
All the wild rice with the ability to restore fertility was crossed as the male parent with Zhenshan 97B and Yuetai B. Zhenshan 97A and Yuetai A were test-crossed as female parents with a fertile hybrid F1. The fertility segregation of the populations derived from the BC1F1 was evaluated for genetic analysis of the restorer genes.
Hypothesis
If there are no sterile plants observed in the population derived from the test-cross of A//Rf/Rf', the restoring loci between two different restorer lines (Rf and Rf') are thought to be allelic. Otherwise, the two restoring loci are considered non-allelic.
RESULTS
Distribution of the fertility restorer genes in wild rice with the AA genome
Thirty-seven wild rice accessions with the AA genome collected from IRRI were test-crossed with HL-CMS and WA-CMS lines, from which 35 HL-type F1 and 31 WA-type F1 plants were obtained, respectively. The F1 fertility evaluation showed that 13 out of 35 HL-type F1 plants and five out of 31 WA-type F1 plants were scored completely fertile, and eight out of 35 HL-type F1 plants and eight out of 31 WA-type F1 plants were scored partially fertile. The frequency of the complete-restoration Rf in wild rice was 37·1 % for HL-CMS and 16·1 % for WA-CMS and the frequency of the partial-restoration Rf in wild rice was 22·9 % for HL-CMS and 25·8 % for WA-CMS, while the frequency of the Rf for HL-CMS was relatively higher than that for WA-CMS. Further analysis showed the following differences between HL-CMS and WA-CMS in the distribution of the Rf. (a) Apart from O. longistaminata, the Rf was found in all of the other six wild rice species with the AA genome. The Rf aggregated mainly in the two species, O. rufipogon and O. nivara, and only one or two accessions in the other four species possessed Rf (Table 2). (b) The fertility-restoring ability differed among the wild restorer accessions in the rice species. The majority of the wild accessions in O. rufipogon and O. nivara could restore the fertility of HL-CMS and WA-CMS, but w46 in O. rufipogon, w13 in O. glaberrima, w35 in O. glumaepatula and w20 and w37 in O. nivara possessed only the Rf for HL-CMS, whereas the w6 in O. rufipogon had the restoring ability for WA-CMS (Table 3). (c) The difference between the restoring ability for HL-CMS and WA-CMS was also observed within the same wild accession; w38 in O. nivara and w39 in O. rufipogon were complete restorers for HL-CMS, but partial restorers for WA-CMS. The complete-restorer accessions aggregated mainly in the two species of O. nivara and O. rufipogon.
Table 2.
Frequency of the fertility-restorer genes in the wild rice species with the AA genome
| Species |
No. of test-crossed wild rice accessions (HL/WA) |
No. of accessions with Rf for HL-CMS |
No. of accessions with Rf for WA-CMS |
No. of accessions with Rf for HL and WA-CMS |
|---|---|---|---|---|
| O. barthii | 6/5 | 1 | 1 | 1 |
| O. glaberrima | 3/3 | 1 | 0 | 0 |
| O. glumaepatula | 5/5 | 1 | 0 | 0 |
| O. meridionalis | 4/2 | 2 | 0 | 0 |
| O. nivara | 8/8 | 8 | 5 | 5 |
| O. rufipogon | 9/8 | 8 | 7 | 6 |
| Total | 35/31 | 21 | 13 | 12 |
Table 3.
Fertility analysis of the HL- and WA-type hybrid F1s
| Wild-abortive type |
Honglian type |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Series no. |
Species |
Fertile pollen (%) |
Bagged seed-setting rate (%) |
Natural seed-setting rate (%) |
Fertile pollen (%) |
Bagged seed-setting rate (%) |
Natural seed-setting rate (%) |
||||
| w06 | O. rufi | 53·3 ± 3·7 | 55·2 ± 4·1 | 67·3 ± 4·4 | 0 | 0 | 4·8 ± 0·7 | ||||
| w07 | O. glum | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w10 | O. bart | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w11 | O. meri | _ | _ | _ | 25·3 ± 2·0 | 10·3 ± 1·1 | 35·7 ± 3·3 | ||||
| w12 | O. glab | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w13 | O. glab | 52·7 ± 2·6 | 0·1 ± 0 | 0·5 ± 0 | 33·7 ± 2·5 | 22·7 ± 1·8 | 28·1 ± 1·4 | ||||
| w14 | O. bart | 98·4 ± 1·4 | 60·4 ± 2·9 | 74·7 ± 3·3 | 99·1 ± 3·1 | 38·6 ± 2·7 | 66·8 ± 3·5 | ||||
| w15 | O. niva | 99·1 ± 0·5 | 23·7 ± 1·1 | 57·3 ± 1·5 | 98·3 ± 5·6 | 24·7 ± 1·1 | 56·0 ± 2·2 | ||||
| w17 | O. glum | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w18 | O. glum | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w19 | O. bart | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w20 | O. niva | 8·6 ± 0·7 | 0·6 ± 0·02 | 1·6 ± 0·02 | 71·1 ± 3·4 | 24·1 ± 1·6 | 52·8 ± 2·7 | ||||
| w21 | O. bart | _ | _ | _ | 6·3 ± 0·1 | 1·2 ± 0 | 2·9 ± 0·1 | ||||
| w22 | O. bart | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w23 | O. meri | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w26 | O. glab | 0 | 0 | 0 | 1·0 | 0 | 0 | ||||
| w28 | O. niva | 86·7 ± 4·7 | 50·3 ± 1·4 | 81 ± 2·1 | 97·1 ± 4·6 | 64·7 ± 1·6 | 80·4 ± 2·8 | ||||
| w29 | O. niva | 98·6 ± 0·8 | 37·4 ± 1·3 | 82·2 ± 3·3 | 93·6 ± 1·4 | 66·1 ± 3·1 | 72·3 ± 2·8 | ||||
| w32 | O. meri | 0 | 0 | 0 | 43·5 ± 0·8 | 37·9 ± 1·3 | 53·3 ± 2·0 | ||||
| w33 | O. meri | _ | _ | _ | 90·7 ± 1·9 | 0 | 0 | ||||
| w34 | O. niva | 44 ± 1·6 | 0·1 ± 0 | 4·8 ± 0·3 | 45·1 ± 1·6 | 50·3 ± 2·7 | 67·3 ± 2·3 | ||||
| w35 | O. glum | 0 | 0 | 0 | 46·3 ± 0·9 | 34·8 ± 1·1 | 73·7 ± 3·4 | ||||
| w36 | O. glum | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w37 | O. niva | 0 | 0 | 0 | 74·4 ± 2·7 | 44·8 ± 3·8 | 77·5 ± 2·9 | ||||
| w38 | O. niva | 50·4 ± 2·5 | 14·7 ± 2·3 | 32·5 ± 1·8 | 50·9 ± 2·6 | 41·5 ± 3·3 | 46·9 ± 2·8 | ||||
| w39 | O. rufi | 51·0 ± 4·4 | 55·8 ± 3·6 | 69·9 ± 4·0 | 50·3 ± 0·6 | 49·2 ± 2·9 | 55·9 ± 1·7 | ||||
| w40 | O. rufi | _ | _ | _ | 86·6 ± 3·4 | 65·3 ± 2·7 | 70·2 ± 2·4 | ||||
| w41 | O. rufi | 97·7 ± 1·7 | 40·2 ± 2·8 | 46 ± 2·3 | 67·4 ± 1·5 | 62·7 ± 3·8 | 74·3 ± 3·0 | ||||
| w42 | O. rufi | 71·4 ± 2·5 | 37·9 ± 3·9 | 52·6 ± 2·4 | 48·3 ± 2·6 | 20·5 ± 1·4 | 55·7 ± 1·9 | ||||
| w43 | O. bart | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| w45 | O. niva | 98 ± 0·4 | 79·3 ± 3·8 | 77·1 ± 3·3 | 84·8 ± 2·9 | 75·6 ± 2·5 | 79·6 ± 3·7 | ||||
| w46 | O. rufi | 0 | 0 | 0 | 81·2 ± 3·7 | 57·4 ± 2·2 | 84·1 ± 2·4 | ||||
| w-GL | O. rufi | 48 ± 1·5 | 11·4 ± 0·7 | 38·6 ± 2·1 | 81·0 ± 1·8 | 17·3 ± 0·4 | 63·7 ± 1·7 | ||||
| w-DX1 | O. rufi | 62·8 ± 2·0 | 22·7 ± 1·1 | 53·4 ± 0·9 | 44·6 ± 0·5 | 27·6 ± 1·3 | 54·3 ± 1·4 | ||||
| w-DX2 | O. rufi | 31·6 ± 2·7 | 19·6 ± 0·4 | 40·7 ± 1·1 | 50·9 ± 1·1 | 21·3 ± 0·9 | 34·4 ± 1·7 | ||||
All the experiments were performed with triplicate; values reported are means of three replicates ± standard deviation.
O. bart, O. barthii; O. glab, O. glaberrima; O. glum, O. glumaepatula; O. long, O. longistaminata; O. niva, O. nivara; O. rufi, O. rufipogon; GL, Guilin; DX, Dongxiang.
Genetic analysis of the fertility-restorer genes in wild rice
To investigate the genetic mode of the fertility-restorer genes in wild rice, a series of backcrosses were carried out. The populations for Rf analysis were derived mainly from backcrosses and were based mainly on the following two cases: (1) the HL-CMS was genetically a gametophytic-restoration CMS type with all the F2 plants fertile; (2) the easier shattering of the F2 plants derived from the test-crosses decreased the reliability of the seed-setting rate of the spikelets.
To analyse the genetic mode of the Rf for Honglian CMS in wild rice, the fertility of the plants in ten backcrosses derived from various wild-rice accessions were investigated. The size of all the populations was about 100 plants. The ratios between fertile and sterile plants were all equal to 1 : 1, and fit to the genetic mode of one pair of genes (Table 4), indicating that all of the ten wild-rice accessions each contained only one pair of fertility-restorer genes for HL-CMS.
Table 4.
Fertility segregation in the BC1F1 crosses between Yuetai and wild rice
| Backcrosses |
Fertile : sterile |
Expected ratio |
χ2 value |
|---|---|---|---|
| YtA//YtB/w-Dongxiang-2 | 73 : 65 | 1 : 1 | 0·464 |
| YtA//YtB/w-Guilin | 59 : 67 | 1 : 1 | 0·508 |
| YtA//YtB/w13 | 61 : 74 | 1 : 1 | 0·910 |
| YtA//YtB/w14 | 48 : 42 | 1 : 1 | 0·40 |
| YtA//YtB/w29 | 37 : 48 | 1 : 1 | 1·423 |
| YtA//YtB/w34 | 57 : 63 | 1 : 1 | 0·20 |
| YtA//YtB/w37 | 48 : 41 | 1 : 1 | 0·551 |
| YtA//YtB/w39 | 51 : 56 | 1 : 1 | 0·234 |
| YtA//YtB/w40 | 83 : 71 | 1 : 1 | 0·935 |
| YtA//YtB/w45 | 66 : 75 | 1 : 1 | 0·578 |
In the genetic analysis of the Rf for WA-CMS in wild rice, eight wild rice accessions were randomly selected, and ten populations were investigated, which included eight derived from backcrosses and two from the F2 generation. Fertility analysis showed that the ratios among fertile, semi-fertile and sterile plants in the F2 generation of w37 and w-Guilin were all equal to 1 : 2 : 1; while the ratios between the fertile and sterile plants derived from backcrosses of w37 and w-Guilin almost fit to 1 : 1, indicating that these two wild-rice accessions each contained only one pair of Rf. Whereas, in the backcross population of w15, there were 25 sterile plants and 104 fertile plants, the ratio was equal to 1 : 3 and fitted to the action mode of two pairs of genes, indicating that w15 contains two pairs of restorer genes for WA-CMS. Further analysis showed that the segregation ratio of fertility in the other five BC1F1 populations of wild rice, including w6, w29, w38, w42 and w45, all fitted to the heredity mode of one pair of genes (Table 5), indicating that, apart from w15 which possessed two pair of Rf, the other seven wild-rice accessions with the AA genome all contained only one pair of Rf for WA-CMS.
Table 5.
Fertility segregation in the BC1F1 crosses derived from Zhenshan 97 and wild rice
| Backcrosses |
Fertile : (semi-fertile) : sterile |
Expected ratio |
χ2 value |
|---|---|---|---|
| Zs97A//Zs97B/w-Guilin | 86 : 71 | 1 : 1 | 1·433 |
| Zs97A/w-Guilin(F2) | 74 : 131 : 56 | 1 : 2 : 1 | 2·488 |
| Zs97A//Zs97B/w6 | 55 : 48 | 1 : 1 | 0·746 |
| Zs97A//Zs97B/w15 | 25 : 104 | 1 : 3 | 1·811 |
| Zs97A//Zs97B/w29 | 58 : 50 | 1 : 1 | 0·64 |
| Zs97A//Zs97B/w34 | 60 : 71 | 1 : 1 | 0·924 |
| Zs97A/w34 (F2) | 59 : 98 : 61 | 1 : 2 : 1 | 3·367 |
| Zs97A//Zs97B/w38 | 88 : 79 | 1 : 1 | 0·485 |
| Zs97A//Zs97B/w41 | 115 : 96 | 1 : 1 | 1·711 |
| Zs97A//Zs97B/w45 | 80 : 69 | 1 : 1 | 0·812 |
Allelic analysis of the fertility-restorer genes in wild rice
It is necessary to analyse the relationship of the fertility-restoring loci among the wild-rice accessions with the AA genome to understand better the evolution and transference of Rf in the natural populations of wild rice. To evade the reproductive barrier of the F1 hybrids between wild-rice accessions, 9311 and Milyang 23 (My23), two cultivars with Rf, were employed as bridge parents. Milyang 23 was suggested to restore HL-CMS and WA-CMS systems, and their restoring loci for HL-CMS are non-allelic (Liu et al., 2004). 9311 is a restorer line only for HL-CMS.
To compare the relationship of restoring loci among wild rice, eight wild-rice accessions were selected, of which w-Dongxiang-2, w38, w40 and w45 were hybridized with Milyang 23 and 9311 and w20, w15, w29 and w34 were hybridized with Milyang 23 or 9311 only. A fertility assay showed that sterile plants were observed in the population derived from test-crosses of YtA//Milyang23/Dongxiang-2, YtA//9311/Dongxiang-2, YtA//Milyang23/w38 and YtA//9311/w38, suggesting that the restoring loci among Dongxiang-2, w38, 9311 and Milyang 23 were all non-allelic.
The recombination frequencies between w-Dongxiang-2 and 9311 and Milyang 23 were 26·97 % and 17·58 %, respectively, and the corresponding frequencies between w38 and 9311 and Milyang 23 were 6·61 % and 17·31 %, respectively. The restoring loci among w20, w39, w40 and 9311 were all allelic, whereas the restoring loci between w45 and 9311 were non-allelic. Interestingly, on the contrary, the restoring loci among Milyang23, w29, w34 and w40 were non-allelic, but between w45 and Milyang 23 the locus was allelic, and the recombination frequency was 17·72 %. Furthermore, it was also found that there were sterile plants in the population derived from Yuetai A//w34/w15 (Table 6), indicating that these two restoring loci were also non-allelic. From the data above, it is concluded that at least three restoring loci for HL-CMS existed in wild rice because Dongxiang-2 and w38 were non-allelic to those of 9311 and Milyang 23.
Table 6.
Allelism analysis of the fertility-restorer genes for Honglian-CMS among wild rice accessions
| Test crosses |
Sterile plants |
Population size |
Recombination frequency (%) |
Allelism |
|---|---|---|---|---|
| YtA//My23/Dongxiang2 | 8 | 91 | 17·58 | Non-allelic |
| YtA//9311/Dongxiang2 | 12 | 89 | 26·97 | Non-allelic |
| YtA//9311/w20 | 0 | 140 | 0 | Allelic |
| YtA//My23/w29 | 14 | 103 | 27·18 | Non-allelic |
| YtA//My23/w34 | 17 | 129 | 26·35 | Non-allelic |
| YtA//w34/w15 | 5 | 155 | 6·45 | Non-allelic |
| YtA//My23/w38 | 9 | 104 | 17·31 | Non-allelic |
| YtA//9311/w38 | 4 | 121 | 6·61 | Non-allelic |
| YtA//9311/w39 | 0 | 107 | 0 | Allelic |
| YtA//My23/w40 | 3 | 90 | 6·67 | Non-allelic |
| YtA//9311/w40 | 0 | 110 | 0 | Allelic |
| YtA//My23/w45 | 0 | 143 | 0 | Allelic |
| YtA//9311/w45 | 7 | 79 | 17·72 | Non-allelic |
Because 9311 possesses no fertility-restorer gene for WA-CMS, only Milyang was used as a bridge parent to compare the genetic relationship of the Rf loci among wild-rice accessions. Six crosses of Milyang/wild rice were carried out using Dongxing-2, w15, w29, w34, w38 and w40. A fertility survey showed that the restoring loci among Dongxiang-2, w38, w40 and Milyang23 were allelic, whereas w15, w29, w34 and Milyang 23 were non-allelic. Furthermore, seven out of 133 plants were found to be fertile in the population derived from the test-cross of Zhenshan 97A//w34/w15 (Table 7), indicating that the restoring loci between these two wild-rice accessions were also non-allelic. Therefore, it is concluded also that there were at least three restoring loci for WA-CMS in the wild rice with the AA genome.
Table 7.
Allelism analysis of the fertility-restorer genes for WA-CMS between wild rice accessions and Milyang 23
| Test crosses |
Sterile plants |
Population size |
Recombination frequency (%) |
Allelism |
|---|---|---|---|---|
| Zs97A//My23/DX-2 | 0 | 138 | 0 | Allelic |
| Zs97A//My23/w15 | 3 | 109 | 5·50 | Non-allelic |
| Zs97A//My23/w29 | 9 | 95 | 18·95 | Non-allelic |
| Zs97A//My23/w34 | 5 | 126 | 7·94 | Non-allelic |
| Zs97A//My23/w38 | 0 | 146 | 0 | Allelic |
| Zs97A//My23/w40 | 0 | 142 | 0 | Allelic |
| Zs97A//w34/w15 | 7 | 133 | 10·52 | Non-allelic |
DISCUSSION
Universality and disequilibrium of the Rf in the species of Oryza with the AA genome
The few reports about the identification of the Rf in wild rice with the AA genome are mostly limited to a few species, usually O. sativa and O. rufipogon, and there is no systematic analysis of all the species of Oryza with the AA genome (Chen et al., 1995; Song et al., 1998; Fu, 2002). Test-cross analysis showed that apart from O. glumaepatula, the fertility restorer genes were observed in the other six wild rice species. The frequency of the Rf differed depending on species; the Rf congregate mainly in the species of O. rufipogon and O. nivara, and only one or two restorer accessions were identified in the other four wild rice species. Most of the restorer accessions within O. rufipogon and O. nivara can restore HL and WA-CMS systems.
It is easy to understand such a characteristic of the Rf in wild rice based on a popular evolution theory about the Rf and CMS. It has been suggested that the Rf is the precondition for the existence and transfer of CMS in the natural population of plants (De Haan et al., 1997a, b). The HL-CMS and WA-CMS were all derived from a common wild rice in China. It is certain that the Rf exist in a common wild rice or the complex O. rufipogon/O. nivara (it is difficult of discriminate O. rufipogon from O. nivara at the molecular level) (Ren et al., 2003) so as to keep the spread of the cytoplasmic sterility factors in the natural populations of wild rice. It has been reported that almost all of the common wild rice which originated in Hainan, Jiangxi and Guangdong provinces in China have the ability to restore the fertility of the CMS line, and the seed-setting rate of spikelets of the hybrids reached over 70 % (Li and Zhu, 1988). This is consistent with the present results.
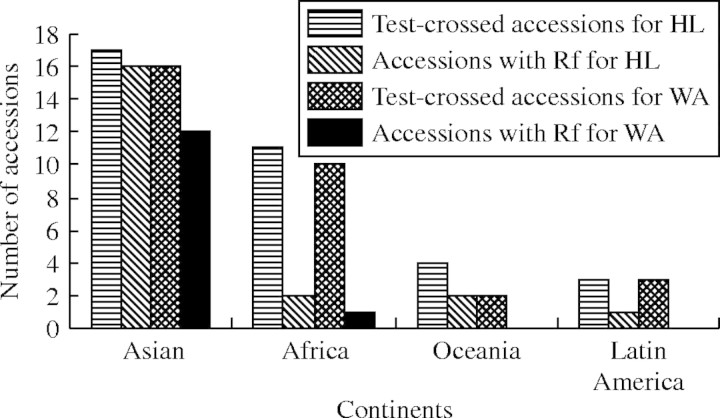
Further analysis showed that there was great variation in the geographical origin of the wild rice accessions with the Rf. The wild-rice accessions with the Rf for HL-CMS occur in all four continents but are centred mainly in Asia (Fig. 1). For WA-CMS, with the exception of two wild-rice accessions from Africa, no other wild-rice accessions from Oceania and Latin Africa were found to have the Rf. Li and Zhu (1988) have reported that the Rf exist mainly in the varieties from southern Asia and south-eastern Asia, and the varieties from North America, Latin America and Africa have no restoring ability. Zhu (1986) has also reported that the Rf occur mainly in the native varieties from southern China and the valley of the Yangtze river in China. It is suggested that rice originated in southern and south-eastern Asia and southern China, and these regions overlap geographically with the distribution of the wild rice species O. rufipogon and O. nivara. This indicates that the Rf in modern cultivars are inherited from their wild ancestors, O. rufipogon and O. nivara. Interestingly, Besides Asia and Africa, the Rf for HL-CMS was also found in Oceania and Latin America. The restoring spectrum for HL-CMS is distinctively wider than that of the WA-CMS system. This seems to be consistent with the distribution of HL-cytoplasm in wild rice. It has been reported that the HL-CMS-related gene, orfH79, was found not only in O. rufipogon and O. nivara, but also in O. meridionalis and O. barthii. This indicates that the Rf is tightly linked to the origin of CMS and the evolution of rice cultivars (Frank, 1989).
Fig. 1.
Geographical distribution of the fertility-restorer accessions for HL-CMS and WA-CMS in wild rice with the AA genome.
Relationship between the restorer gene and CMS in wild rice
Wide hybridization and inter-species and inter-subspecies crossing are commonly used approaches to produce CMS lines in a breeding programme. It has been reported that a series of CMS lines was obtained from 132 hybrid crosses derived from inter-species crosses between one of the four wild relatives, O. rufipogon, O. nivara, O. barthii, O. longistaminata, and one of the two species of O. sativa and O. glaberrima. And a few of the CMS lines are suggested to be different from WA-CMS for the variant restoring–maintaining relationship (Hoan et al., 1998). In the same manner, a great number of CMS lines were also produced via hybrid/cultivar crossing by IRRI, and some of these were found to have no fertility restorer lines in the cultivars, because of the cytoplasm in the CMS lines was derived from wild relatives of rice beyond the AA genome (Subudhi et al., 1998). Fu (2002) once reported that no restorer lines were found in the cultivars of the two CMS lines from IRRI that share the nuclear background of IR64, a typical restorer line for WA-CMS, of which IR54755A carries the cytoplasm from O. perenis and IR67700A from O. glumaepatula (Dalmacio et al., 1992). Chen et al. (1995) have also reported that no restorer lines were found in cultivars for the CMS line with the cytoplasm of Dongxiang wild rice (Jiangxi province, China). On the contrary, the lines from the natural populations of Dongxian wild rice and the Chaling wild rice all possessed the ability to restore the fertility of the Dongxiang CMS line. This indicates that the Rf and the CMS are dependent on each other in evolution, the lines with CMS in the wild rice population being the best resources for the corresponding restorer genes. Many types of CMS may coexist in the wild rice population, and each has various restorer genes.
Origin and diversity of the restorer genes in wild rice
Allelism analysis of the Rf alleles showed that there are at least three Rf loci for each of the HL-CMS and WA-CMS systems. It appears that there are three types of CMS in Plantago lanceolata (de Haan et al., 1997a), and multiple restorer alleles, which work independently or interactively, are responsible for the fertility of each of the CMS systems (de Haan et al., 1997b). Six variations of orfH79, a mitochondrial gene related to HL-CMS, have been identified in the wild-rice accessions with the AA genome; the different types of CMS genes may correspond to various restorer alleles. If comparison of the fertility of microspores in an F1 hybrid revealed 10–90 % fertile microspores among various crosses, then variation in the restoring ability of the restorer alleles in wild-rice accessions would be indicated. However, the HL-CMS is genetically a gametophytic system, with the restorer allele acting in a gametophytic mode, so in theory only 50 % of the microspores can be fertile. It is therefore perplexing that pollen fertility in the test crosses between YtA and w14, w15, w28 and w29 reached up to 90 %, w14, w15 and w29 each possessed only one pair of Rf allele, the traditional Rf-CMS theory seems beyond explaining the cause.
Acknowledgments
The work was supported by National Nature Science Foundation of China (30270149) and Chinese National 973 Program (Grant number: 2001CB108806).
LITERATURE CITED
- Budar F, Pelletier G. 2001. Male sterility in plants: occurrence, determinism, significance and use. Life Science 324: 543–550. [DOI] [PubMed] [Google Scholar]
- Chen DZ, Xiao YQ, Zhao SX, Xiao HJ. 1995. Study on the restoring resource for CMS lines with the cytoplasm of Jiangxi Dongxiang wild rice. Hybrid Rice 6: 4–6. [Google Scholar]
- Dalmacio R, Brar DS, Sitch LA, Virmani SS, Khush GS. 1992.Oryza perennis: a new source of cytoplasmic male sterility in rice. Rice Genetics Newsletter 9: 108–110. [Google Scholar]
- De Haan AA, Luyten RMJM, Bakx-schotman TJMT, van Damme JMM. 1997. The dynamics of gynodioecy in Plantago lanceolata L. I. Frequency of male-steriles and their cytoplasmic male sterility types. Heredity 79: 453–462. [Google Scholar]
- De Haan AA, Luyten RMJM, Bakx-schotman TJMT, van Damme JMM. 1997. The dynamics of gynodioecy in Plantago lanceolata L. II. Mode of action and frequencies of restorer alleles. Genetics 147: 1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. 1989. The evolutionary dynamics of cytoplasmic male sterility. American Naturalist 133: 345–376. [Google Scholar]
- Fu HW. 2002. Fertility restorer lines for the CMS originated from O. perennis Agriculture Science of Anhui 30: 170–174. [Google Scholar]
- Hoan NT, Sarma NP, Siddiq EA. 1998. Wide hybridization for diversification of CMS in rice. International Rice Research Notes 23: 5–6. [Google Scholar]
- Jing RC, Li XM, Yi P, Zhu YG. 2001. Mapping fertility-restoring genes of rice WA cytoplasmic male sterility using SSLP markers. Botanical Bulletin of Academia Sinica 42: 167–171. [Google Scholar]
- Kaul MLH. 1988. Male sterility in high plants. In: Frankel R, Grossman M, Maliga P, Riley R, eds. Monographs of Theoretical Applied Genetics 10. Heidelberg: Springer-Verlag, 775–797. [Google Scholar]
- Li ZB, Zhu YG. 1988. Rice male sterile cytoplasm and fertility restoration. In: Hybrid rice—Proceedings of the International Symposium on Hybrid Rice. Manila: International Rice Research Institute, 85–102. [Google Scholar]
- Liu XQ, Xu X, Tan YP, Li SQ, Yang DC, Li YS, Zhu YG. 2004. Inheritance and molecular mapping of two fertility-restoring loci for Honglian gametophytic cytoplasmic male sterility in rice (Oryza sativa L.). Molecular Genetics and Genomics 271: 586–594. [DOI] [PubMed] [Google Scholar]
- Ren F, Lu B, Li S, Huang J, Zhu Y. 2003. A comparison study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theoretical Applied Genetics 108: 113–120. [DOI] [PubMed] [Google Scholar]
- Shen YW, Guan ZQ, Lu J, Zhuang JY, Zheng KL, Gao MW, Wang XM. 1998. Linkage analysis of a fertility restoring mutant generated from CMS rice. Theoretical and Applied Genetics 97: 261–266. [Google Scholar]
- Song DM, Wang Z, Liu YS, Wang ML, Long TK, Sun JS. 1998. Development of anther-defective cytoplasmic male sterile line from interspecific hybridization between Dongxiang wild rice and cultivated rice and preliminary observations on its derivatives. Acta Botanica Sinica 40: 184–185. [Google Scholar]
- Subudhi PK, Nandi S, Casal C, Virmani SS, Huang N. 1998. Classification of rice germplasm: III. High-resolution fingerprinting of cytoplasmic genetic male-sterile (CMS) lines with AFLP. Theoretical and Applied Genetics 96: 941–949. [Google Scholar]
- Tan XL, Vanavichit A, Amornsilpa S, Trangoonrung S. 1998. Genetic analysis of rice CMS-WA fertility restoration based on QTL mapping. Theoretical and Applied Genetics 96: 994–999. [Google Scholar]
- Yao FY, Xu CG, Yu SB, Li JX, Gao YJ, Li XH, Zhang QF. 1997. Mapping and genetic analysis of two fertility restorer loci in the wild-abortive cytoplasmic male sterility system of rice (Oryza sativa L.). Euphytica 98: 183–187. [Google Scholar]
- Zhu YG. 1986. Geographical distribution of fertility-restoring genes for two male-sterile lines in China. Rice Genetics Newsletter 3: 50–51. [Google Scholar]