Abstract
• Background and Aims The genus Hordeum exists at three ploidy levels (2x, 4x and 6x) and presents excellent material for investigating the patterns of polyploid evolution in plants. Here the aim was to clarify the ancestry of American polyploid species with the I genome.
• Methods Chromosomal locations of 5S and 18S–25S ribosomal RNA genes were determined by fluorescence in situ hybridization (FISH). In both polyploid and diploid species, variation in 18S–25S rDNA repeated sequences was analysed by the RFLP technique.
• Key Results Six American tetraploid species were divided into two types that differed in the number of rDNA sites and RFLP profiles. Four hexaploid species were similar in number and location of both types of rDNA sites, but the RFLP profiles of 18S–25S rDNA revealed one species, H. arizonicum, with a different ancestry.
• Conclusions Five American perennial tetraploid species appear to be alloploids having the genomes of an Asian diploid H. roshevitzii and an American diploid species. The North American annual tetraploid H. depressum is probably a segmental alloploid combining the two closely related genomes of American diploid species. A hexaploid species, H. arizonicum, involves a diploid species, H. pusillum, in its ancestry; both species share the annual growth habit and are distributed in North America. Polymorphisms of rDNA sites detected by FISH and RFLP analyses provide useful information to infer the phylogenetic relationships of I-genome Hordeum species because of their highly conserved nature during polyploid evolution.
Key words: Chromosome, FISH, genome, Hordeum, in situ hybridization, karyotype evolution, phylogeny, RFLP, ribosomal genes, wild barley
INTRODUCTION
The genus Hordeum (Triticeae) is classified into 31 species (45 taxa in total, including subspecies and cytotypes) and 51 cytotypes exist at the diploid, tetraploid and hexaploid levels with a basic chromosome number of x = 7 (Baden and von Bothmer, 1994; von Bothmer et al., 1995). Phylogenetic relationships of the species in this genus have been studied intensively. Characters examined include morphology, metaphase I (MI) chromosome pairing in interspecific hybrids (von Bothmer et al., 1986, 1987, 1988), C-banding patterns and morphology of satellited (SAT) chromosomes (Linde-Laursen et al., 1995), isozymes (Jørgensen, 1986), repetitive DNA sequences (Molnar et al., 1989; Svitashev et al., 1994), nuclear and chloroplast DNA sequences (Doebley et al., 1992; Komatsuda et al., 1999; Nishikawa et al., 2002; El-Rabey et al., 2002; Petersen and Seberg, 2003), and chromosomal distribution of cloned repetitive DNA sequences (de Bustos et al., 1996; Taketa et al., 1999a, b, 2001). Analysis of MI chromosome pairing (von Bothmer et al., 1995) revealed the presence of four basic genomes (H, I, X and Y). Other studies have confirmed the division of the genus into four groups corresponding to the above four basic genomes, but detailed species relationships, especially those within a group, have not been fully resolved owing to the complicated evolution. A recent molecular cytogenetic study (Taketa et al., 1999a) revealed that three polyploid species/cytotypes, H. secalinum, H. capense and H. brachyantherum ssp. brachyantherum 6x, were allopolyploids having combinations of the X and I genomes. Thus, the genus is classified into five genome groups, namely H, I, X, Y and XI (Taketa et al., 1999a). In this paper, genome designation follows that of Taketa et al. (2001), namely, H. vulgare and H. bulbosum both carry the H genome, H. marinum carries the X genome, H. murinum has the Y genome, and the remaining species share variants of the I genome. In the present genome nomenclature, the genome symbols H and I have been swapped relative to the designation by von Bothmer et al. (1995). This is because the symbol H was allocated to the genomes of H. vulgare and H. bulbosum at the 7th International Barley Genetics Symposium (Linde-Laursen et al., 1997).
The I-genome group is the largest of the five groups and includes 25 species. Fourteen species are diploid, five are tetraploid and four are hexaploid; the remaining two species exists at two and three ploidy levels. The I-genome species are distributed from Central Asia to the American Continents (von Bothmer et al., 1995). The I-genome group contains many morphologically similar species, and the nature of polyploidy and species relationships of this group remain largely uncertain despite the many studies cited above.
Fluorescence in situ hybridization (FISH) is a powerful molecular cytogenetic technique to reveal genome constitution and species' relationships in plants (Heslop-Harrison, 2000). In the genus Hordeum, the technique has been applied to restricted species (de Bustos et al., 1996; Taketa et al., 1999b, 2001). Previously we analysed the physical location of two types of ribosomal DNA (5S rDNA and 18S–5·8S–25S rDNA, hereafter referred to as 18S–25S rDNA) in 15 of the 16 I-genome diploid species (Taketa et al., 1999b, 2001). These studies demonstrated the usefulness of rDNA sites as chromosome landmarks for investigating karyotype evolution and phylogeny in the genus Hordeum. Similar approaches have been utilized successfully in several other plant genera and species (Aegilops, Bedaeva et al., 1996; Gossypium, Hanson et al., 1996; Brachyscome linearloba, Adachi et al., 1997; Hypochaeris, Cerbah et al., 1998; Trifolium, Ansari et al., 1999; Arachis, Raina and Mukai, 1999; Sanguisorba, Mishima et al., 2002). In the present study, research was extended to nine I-genome polyploid species of American origin, providing new information on their origin. Moreover, the study was complemented by Southern hybridization analyses with the wheat 18S–25S rDNA probe pTa71, to detect rDNA sequence variation.
MATERIALS AND METHODS
Plant materials
Five tetraploid and four hexaploid species of the I-genome group, which are distributed in the American Continents, were analysed by FISH and Southern hybridization (Table 1). A South American I-genome diploid species H. comosum, which was not available in our previous study, was also included. For reference, 18 taxa (in total 21 accessions, Table 1) were selected for Southern hybridization analyses with the 18S–25S rDNA probe. Detailed information on collection sites of the materials is given in the Appendix. Seeds are preserved at both Kagawa University, Japan and the Nordic Gene Bank, Sweden, and are available upon request from the senior author.
Table 1.
Hordeum species analysed in this study and the rDNA fragments obtained from EcoRV digestion of total genomic DNA
|
EcoRV digestion (kb) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species |
Accession |
Ploidy |
Collection site |
FISH image in Fig. 1 |
12–10 |
9·5–7·5 |
4–3 |
2·5 |
Lane number in Fig. 3 |
|||||||||
| Species used for FISH analysis | ||||||||||||||||||
| H. depressum | H2089 | 4x | USA | C | 7·5 | 2·5 | 1 | |||||||||||
| H2005 | 4x | USA | Not shown | 7·5 | 2·5 | 2 | ||||||||||||
| H. jubatum | H1922 | 4x | USA | A | 11·7, 11·5 | 9 | 2·5 | 3 | ||||||||||
| H1976 | 4x | USA | Not shown | 11·7, 11·5 | 9 | 2·5 | 4 | |||||||||||
| H. guatemalense | H2299 | 4x | Guatemala | B | 11·5 | 9 | 2·5 | 6 | ||||||||||
| H. fuegianum | H1418 | 4x | Argentina | Not shown | 11·5 | 8·2, 8 | 2·5 | 7 | ||||||||||
| H. tetraploidum | H6198 | 4x | Argentina | D | - | - | - | - | - | |||||||||
| H. arizonicum | H3254 | 6x | USA | E | 11·7, 11·5 | 9, 8·7 | 4, 3·2 | 2·5 | 18 | |||||||||
| H. lechleri | H6104 | 6x | Argentina | F | 11·7 | 9, 8·5 | 2·5 | 19 | ||||||||||
| H. parodii | H6294 | 6x | Argentina | G | 11·5 | 9, 7·5 | 2·5 | 20 | ||||||||||
| H. procerum | H1789 | 6x | Argentina | H | 11·7 | 9, 8·5 | 2·5 | 21 | ||||||||||
| H. comosum | H10608 | 2x | Argentina | Not shown | - | - | - | - | - | |||||||||
| Species only used for RFLP analysis | ||||||||||||||||||
| H. brevisubulatum ssp. violaceum | H315 | 2x | Iran | * | 11 | 8 | ||||||||||||
| H. bogdanii | H4014 | 2x | Pakistan | * | 10 | 9 | ||||||||||||
| H. roshevitzii | H9152 | 2x | China | * | 11·5 | 10 | ||||||||||||
| H. brachyantherum ssp. californicum | H3317 | 2x | USA | * | 7·5 | 2·5 | 11 | |||||||||||
| H1954 | 2x | USA | * | 9 | 2·5 | 12 | ||||||||||||
| H2419 | 2x | USA | - | 9, 8·5 | 2·5 | 31 | ||||||||||||
| ssp. brachyantherum | JIC line 2 | 4x | unknown | ** | 11 | 5 | ||||||||||||
| H2420 | 4x | USA | - | 11·7 | 9 | 2·5 | 17 | |||||||||||
| H. pusillum | H722 | 2x | USA | * | 8·7 | 4, 3·5 | 2·5 | 13 | ||||||||||
| H. intercedens | H2310 | 2x | USA | * | 8·5 | 2·5 | 14 | |||||||||||
| H. chilense | JIC line 1 | 2x | unknown | ** | 8·5 | 2·5 | 15 | |||||||||||
| H. muticum | H6479 | 2x | Argentina | * | 9 | 2·5 | 16 | |||||||||||
| H. cordobense | H6429 | 2x | Argentina | * | 8·5 | 2·5 | 22 | |||||||||||
| H. flexuosum | H1116 | 2x | Argentina | * | 8 | 2·5 | 23 | |||||||||||
| H. patagonicum | ||||||||||||||||||
| ssp. musterisii | H1358 | 2x | Argentina | * | 7·5 | 2·5 | 24 | |||||||||||
| ssp. patagonicum | H6052 | 2x | Argentina | * | 9 | 2·5 | 25 | |||||||||||
| ssp. setifolium | H1366 | 2x | Argentina | * | 7·5 | 2·5 | 26 | |||||||||||
| ssp. santacrucense | H1353 | 2x | Argentina | * | 7·5 | 2·5 | 27 | |||||||||||
| H. pubiflorum | ||||||||||||||||||
| ssp. halophilum | H1348 | 2x | Argentina | * | 9·5, 7·5 | 2·5 | 28 | |||||||||||
| ssp. pubiflorum | H1296 | 2x | Argentina | * | 9 | 2·5 | 29 | |||||||||||
| H. stenostachys | H1108 | 2x | Argentina | * | 8 | 2·5 | 30 | |||||||||||
FISH images were reported in Taketa et al. (2001)
reported in Taketa et al. (1999b).
–, Not analysed.
Fluorescence in situ hybridization
Actively growing root-tips from germinating seeds or hydrocultured plants were treated in ice water at 0 °C for 16–24 h to accumulate metaphases and fixed with 3 : 1 (v/v) 100 % ethanol : acetic acid. Root tips were stained with a 2 % acetocarmine solution and squashed in a drop of 45 % acetic acid according to the acetocarmine squash technique (see Fukui, 1996). Two DNA clones, pTa794 and pTa71 were used as probes. Clone pTa794 is a BamHI fragment of the 5S rDNA, having a 120-bp coding sequence for the 5S rRNA gene and the intergenic spacers isolated from common wheat, Triticum aestivum (Gerlach and Dyer, 1980). For clone pTa71, the 18S–25S rDNA is a 9-kb EcoRI fragment from common wheat, containing the coding sequences for the 18S, 5·8S and 25S rRNA genes and the intergenic spacer sequences (Gerlach and Bedbrook, 1979). The in situ hybridization procedure described by Taketa et al. (1999b) was adopted. Slides were examined with an Olympus BX-50 epifluorescence microscope with appropriate filter sets (U-MWU for UV, U-MWIB for FITC, U-DM-Cy3 for Cy3 and U-DM-DA/FI/TX for simultaneous visualization of all fluorochromes). Photographs were taken on Fujicolor Super HG400 color print film. Photographs were scanned and processed in Adobe Photoshop using only cropping and processing functions that affect all pixels in the image equally. The number of rDNA sites was determined from observation of at least five metaphases with clear signals.
rDNA–RFLP
For Southern analysis, the total genomic DNA (0·5 µg) of two North American tetraploid species, H. depressum H2089 and H. jubatum H1922 were digested with eight restriction enzymes, BamHI, BglII, DraI, EcoRI, EcoRV, HindIII, XbaI and XhoI separately, electrophoresed in 0·8 % agarose gels, and transferred to a positively charged nylon membrane. The blot was hybridized with the purified rDNA probe from the clone pTa71 according to a standard protocol of ECL chemiluminescence (Amersham). Hybridization and washing stringencies were 78 % and 86 %, respectively. Hybridization sites were detected using ECL detection reagents and recorded directly on Hyperfilm-ECL (Amersham). From this preliminary survey, EcoRV was selected as the most informative restriction enzyme for detecting RFLP in wild Hordeum and used in the subsequent analysis. Exposure times for luminographs (shown in Fig. 3) ranged from 15–60 min.
Fig. 3.
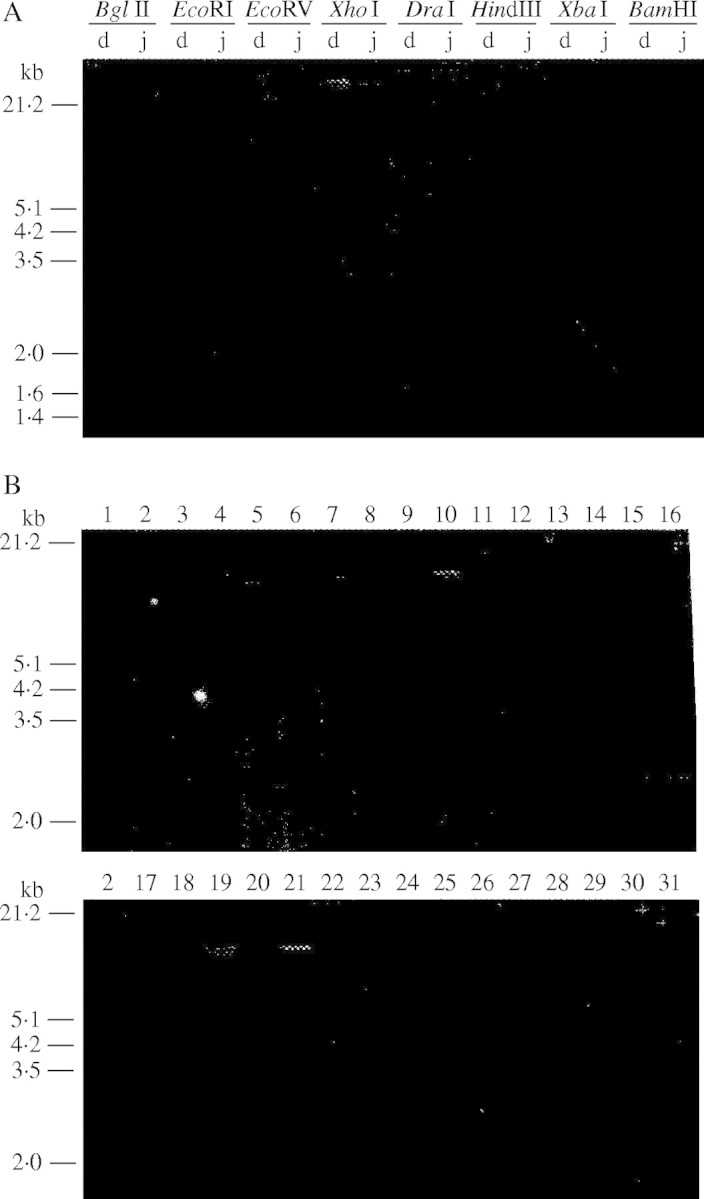
Luminographs showing Southern hybridization of labelled pTa71 probe to digests of genomic DNA from (A) H. depressum H2089 and H. jubatum H1922, and (B) to EcoRV digests of genomic DNA from various I-genome Hordeum species. In (A) eight restriction enzymes were used, and ‘d’ and ‘j’ stand for DNAs of H. depressum and H. jubatum, respectively. In (B) species/accession names are indicated with numbers, which correspond to those given in Table 1.
RESULTS
Chromosomal in situ mapping of the ribosomal genes
First, a South American diploid, H. comosum, was analysed. This species was found to be ‘chilense type’ (Taketa et al., 2001), having two SAT chromosome pairs (one submetacentric with double 5S rDNA sites and one metacentric), typical of American I-genome diploid species (data not shown). Figure 1A–H shows the somatic metaphase chromosomes of eight I-genome polyploid species after in situ hybridization with 5S and 18S–25S rDNA probes and counterstaining with 4′, 6-diamidino-2-phenylindole (DAPI). Idiograms of the haploid complement of the chromosomes with rDNA sites in all nine species are presented in Fig. 2. The present in situ hybridization experiment detected 5S and major and minor 18S–25S rDNA sites and achieved detection sensitivity similar to that of our previous studies (Taketa et al., 1999b, 2001). In this paper, we define SAT chromosomes as chromosomes with a major 18S–25S rDNA site because secondary constrictions were hard to observe in polyploids.
Fig. 1.
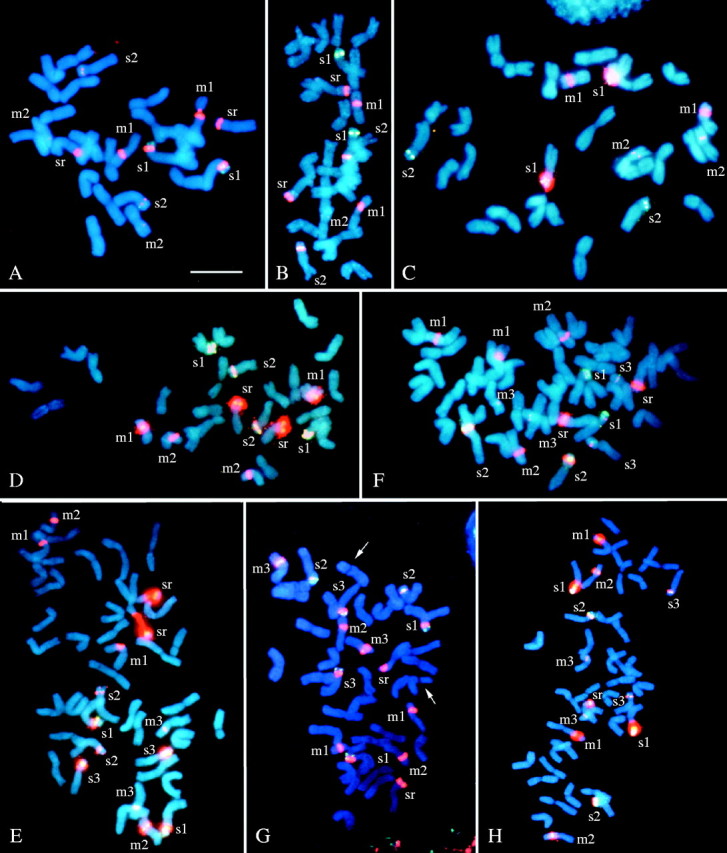
Double-target in situ hybridization to root-tip metaphase cells in polyploid Hordeum species with the I genome. The micrographs were taken with a triple band filter allowing the simultaneous visualization of the DAPI-stained chromosomes (blue), the hybridization sites of the 5S rDNA (green) and the 18S–25S rDNA (red). (A) H. jubatum H1922; (B) H. guatemalense H2299; (C) H. depressum H2089; (D) H. tetraploidum H6198; (E) H. arizonicum H3254; (F) H. lechleri H6104; (G) H. parodii H6294; (H) H. procerum H1789. Chromosomes with rDNA site(s) are labelled: the first letter indicates the morphology of chromosomes, where ‘s’ is a submetacentric and ‘m’ is a metacentric. The chromosome marked with ‘sr’ indicates the H. roshevitzii-specific submetacentric SAT chromosome (‘sr SAT chromosome’). In (B) one m2 chromosome can not be identified due to overlapping of chromosomes. In (G) arrows indicate a tertiary constriction. The metaphase plate shown in (H) is incomplete (2n = 41) and one ‘sr SAT chromosome’ is missing. The scale bar represents 10 μm in (A) and (C); 12 μm in (B), (D–F); and 16 μm in (G) and (H).
Fig. 2.
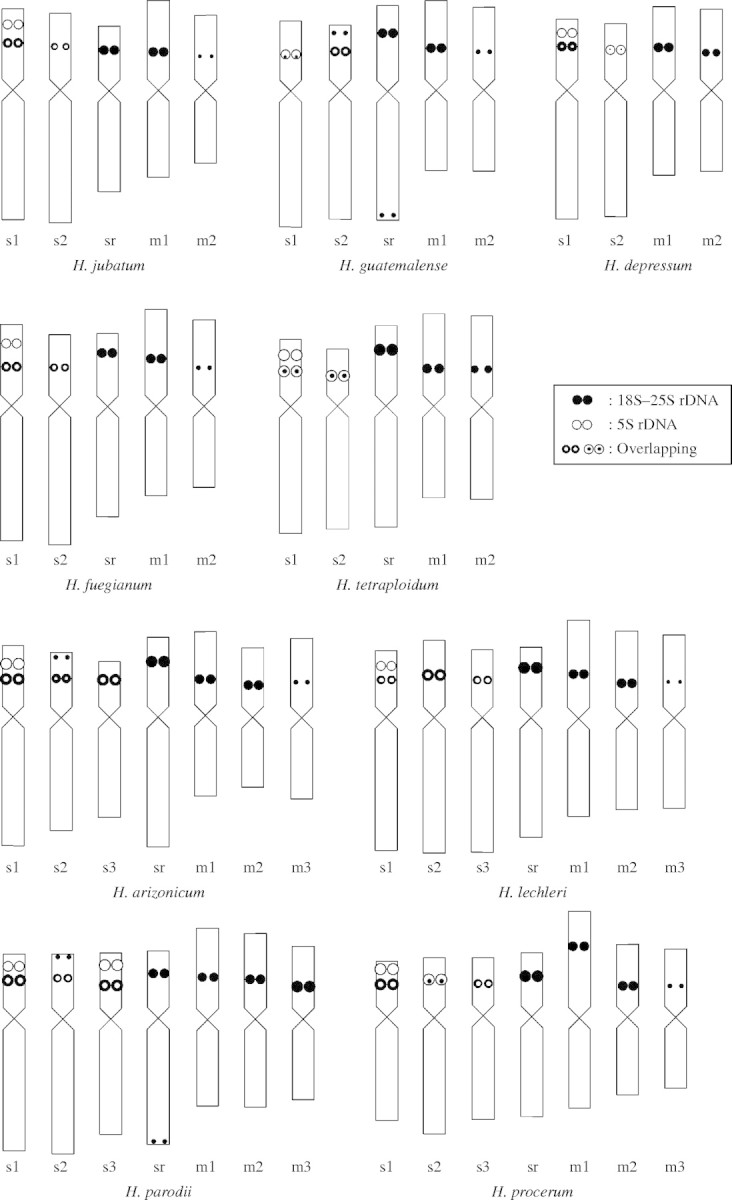
Idiograms of the haploid complement of the chromosomes carrying 5S and 18S–25S rDNA in nine polyploid Hordeum species with the I genome investigated in the study. Letters under the chromosomes are the same as in Fig. 1.
Tetraploid species
In American tetraploid species, the number of chromosome pairs with rDNA sites was five for H. jubatum (Fig. 1A), H. guatemalense (Fig. 1B), H. fuegianum and H. tetraploidum (Fig. 1D), but was four for H. depressum (Fig. 1C). The former four species show a perennial growth habit, but the latter is an annual species of the western USA (von Bothmer et al., 1995). The four perennial species, H. jubatum (North America and also in eastern Siberia), H. guatemalense (Central America) and H. fuegianum and H. tetraploidum (both from South America), had essentially the same rDNA pattern as that previously reported for the tetraploid cytotype of H. bracyantherum ssp. brachyantherum (Taketa et al., 1999b). Hordeum bracyantherum ssp. brachyantherum is a perennial from North America and Kamchatka (hereafter called H. brachyantherum 4x). In the consensus FISH pattern of perennial tetraploid species, rDNA sites are located on three pairs of submetacentric chromosomes and two pairs of metacentric chromosomes. Minor differences in the number and intensity of rDNA sites were found among the species. Except for H. guatemalense, all perennial tetraploid species had double 5S rDNA sites on the short arm of a submetacentric chromosome pair. Similar double 5S rDNA sites were observed in the ‘chilense-type’ American diploid species (Taketa et al., 1999b, 2001). We will refer to the FISH pattern found in the five American perennial tetraploid species as the ‘jubatum type’.
Both accessions of H. depressum (H2089 and H2005) had three pairs of 5S rDNA sites and four (two major and two minor) pairs of 18S–25S rDNA sites on four pairs of chromosomes (two submetacentrics and two metacentrics). Compared to the ‘jubatum type’, H. depressum lacked a submetacentric chromosome pair with a major 18S–25S rDNA site in its short arm, but other rDNA-carrying chromosome pairs in this species can find their possible counterparts in the ‘jubatum type’ (Fig. 2). We designate the unique FISH pattern of H. depressum the ‘depressum type’. Linde-Laursen et al. (1995) pointed out that the unique submetacentric SAT chromosome pair that is absent in H. depressum but is present in other American polyploid species was derived from the Asian diploid I-genome species, H. roshevitzii. In this paper, we will call it the ‘sr SAT chromosome’.
Hexaploid species
All four American hexaploid species had rDNA sites on seven pairs of chromosomes and showed more-or-less similar FISH patterns (Figs. 1E–H, 2). The seven pairs of chromosomes with rDNA sites consisted of four submetacentrics and three metacentrics. The four submetacentric pairs included three pairs with both 5S and 18S–25S rDNA sites in their short arms and the ‘sr SAT chromosome’ pair. The three metacentric pairs had an 18S–25S rDNA site in one arm. Of the seven pairs of chromosomes with rDNA sites in hexaploid species, five can find their possible counterparts in the ‘jubatum-type’ tetraploid species, and the remaining two pairs were one submetacentric and one metacentric. These results indicate that American hexaploid species were formed from hybridization between a ‘jubatum-type’ tetraploid species and an American diploid species.
There was variation in the number and intensity of 5S and 18S–25S rDNA sites among hexaploid species. A North American annual/biannual species H. arizonicum (Fig. 1E) and a South American perennial species H. lechleri (Fig. 1F) were similar in FISH pattern, except that the former had an extra minor 18S–25S rDNA site in the short arm of one submetacentric SAT chromosome pair (marked s2). A South American perennial species H. parodii (Fig. 1G) was unique in having two pairs of submetacentric chromosomes with double 5S rDNA sites (marked s1 and s3), while the three other hexaploid species had only one pair of similar chromosomes. In addition, H. parodii had an extra minor 18S–25S rDNA site in the long arm terminal of the ‘sr SAT chromosome’ pair, and also had a metacentric chromosome pair with a tertiary constriction. In H. procerum (Fig. 1H), one metacentric SAT chromosome pair showed an extended chromosome arm with a more distally located 18S–25S rDNA site (marked m1), as reported by Linde-Laursen et al. (1990). A similar metacentric SAT chromosome pair was reported in the South American diploid H. cordobense (Linde-Laursen et al., 1989). Because the present study analysed only one accession in each hexaploid species, it is not known whether these variations are species-specific or not.
rDNA–RFLP
Two representative tetraploid species, H. depressum and H. jubatum, were analyzed by Southern hybridization using eight restriction enzymes and the pTa71 clone as a probe (Fig. 3A). Two restriction enzymes (HindIII and XhoI) gave no clear pattern in either species, indicating the absence of restriction sites in the pTa71 repeat unit. The other five restriction enzymes detected one or more solid fragments and detected a clear RFLP between them. EcoRI and XbaI had a single restriction site in both species and detected a pTa71 repeat unit length polymorphism. DraI detected the same polymorphism, but a weak high-molecular-weight fragment was observed only in H. depressum. The other three enzymes, BglII, BamHI and EcoRV, produced two or more fragments in both species and detected RFLP. In the BglII and EcoRV profiles, smaller fragments sum up to the repeat unit length of the respective species. The BamHI profiles showed only a minor difference between the two species, and their overall patterns were similar to those reported by Molnar et al. (1989). Available data from different restriction enzymes suggest the presence of at least two kinds of pTa71 repeat unit sequences in each species. On the basis of these results, we selected EcoRV for subsequent analyses. This restriction enzyme was not used in previous rDNA–RFLP analyses of the genus Hordeum (Molnar and Fedak, 1989; Molnar et al., 1989, 1992).
In the present study, except for H. tetraploidum, all American I-genome polyploid species were analysed by Southern hybridization of EcoRV digests using the pTa71 probe. Figure 3B shows RFLP profiles and Table 1 summarizes the results. In tetraploid species, most species/accessions shared similar profiles consisting of three classes of fragments: 12–10 kb, 9·5–7·5 kb and 2·5 kb (lanes 3–7 and 17), but both accessions of H. depressum (lanes 1 and 2) and a H. brachyantherum 4x accession JIC line 2 (lane 5) showed a unique profile. Hordeum depressum had only two fragments: 7·5 kb and 2·5 kb. Hordeum brachyantherum 4x accession JIC line 2 had only a single fragment (11 kb), but another accession of this cytotype (H2420, lane 17) showed a profile representative of tetraploid species. In hexaploid species (lanes 18–21), all except H. arizonicum (lane 18) showed a profile similar to that of representative tetraploid species, but the 12–10 kb and 9·5–7·5 kb fragment classes consisted of complicated multiple bands with size and intensity variation among species. In H. arizonicum, faint double fragments of a new class range (4–3 kb) were detected.
For reference, representative I-genome diploid species were examined for rDNA–RFLP (Fig. 3B, Table 1). Southern analyses showed clear differentiation between Asian and American diploid species, supporting our previous FISH results (Taketa et al., 2001). All three Asian diploid species, H. brevisubulatum 2x, H. bogdanii and H. roshevitzii (lanes 8–10) had only a single fragment of 12–10 kb. In contrast, all American diploid species had fragments at two size classes: 9·5–7·5 kb and 2·5 kb. Among American species, H. pusillum (lane 13) was unique in having extra double fragments in the 3–4 kb class.
By comparing the RFLP patterns of diploids with those of polyploids, we obtained interesting findings. The 12–10 kb fragment, which is specific to Asian diploids, was detected in all American polyploid species except for H. depressum. The 2·5 kb fragment was conserved across all American species except for one H. brachyantherum 4x accession, but was absent in all Asian diploids. The fragments in the 9·5–7·5 kb class were highly polymorphic both between and within American species. For example, all three accessions of H. brachyantherum ssp. californicum differed in fragment patterns of this class (lanes 11, 12 and 31).
DISCUSSION
Ancestry of the perennial tetraploid species
I-genome polyploid species have not previously been analysed for their rDNA FISH patterns to any significant extent. Only two species/cytotypes, H. brevisubulatum 4x and H. brachyantherum 4x, have been described so far (Taketa et al., 1999b). Our previous FISH studies on American I-genome diploid species revealed a low level of polymorphism (Taketa et al., 1999b, 2001). Therefore, in the present study we employed both FISH and RFLP techniques for more sensitive detection of rDNA polymorphisms in the American I-genome polyploid species. Previous rDNA–RFLP analyses on Hordeum (Molnar and Fedak 1989; Molnar et al., 1989, 1992) failed to reveal a detailed relationship among I-genome species owing to a low level of polymorphisms. However, the present study revealed that the restriction enzyme EcoRV is very informative in rDNA–RFLP analysis of Hordeum.
The similar FISH pattern in the five perennial tetraploid species (the ‘jubatum type’) suggests their close relationships and common origin. Because the FISH pattern of the ‘jubatum type’ deviates significantly from a set of four homologous chromosomes, these species are considered alloploids. Similar RFLP profiles were observed in all five perennial tetraploid species except for one H. brachyantherum 4x accession. The presence of both the ‘sr SAT chromosome’ pair and the Asian species-specific rDNA fragment in all ‘jubatum-type’ species indicates that H. roshevitzii is one of their diploid ancestors. The other diploid ancestor is probably a North American ‘chilense-type’ species because the summed FISH signals of H. roshevitzii and the ‘chilense type’ match those of the ‘jubatum type’. If we consider the current geographical distribution of the Hordeum species (von Bothmer et al., 1995), it is reasonable to assume that the ‘jubatum-type’ species were probably formed somewhere in Asia or North America, in which the supposed ancestral diploid species coexisted. Because H. roshevitzii-like diploid species are absent in Central and South America, tetraploid species in these areas are probably descendents of North American tetraploids that migrated southward.
The ‘chilense-type’ diploid ancestor of respective ‘jubatum-type’ species may be inferred from FISH and RFLP patterns. Hordeum jubatum, H. guatemalense and the standard accession of H. brachyantherum 4x seem to have an ancestor identical to H. brachyantherum ssp. californicum accession H1954, owing to the common RFLP fragments. Hordeum brachyantherum ssp. californicum includes two types of karyotypes, namely those having one or two pairs of SAT chromosomes (Linde-Laursen et al., 1986, 1995; Taketa et al., 2001). The accession H1954 carries two SAT chromosome pairs and shows the ‘chilense-type’ FISH pattern (Taketa et al., 2001); therefore this accession could be an ancestor of the three North American tetraploid species. On the basis of karyotypes, Linde-Laursen et al. (1995) reached a similar conclusion. Studies on chloroplast DNA also suggested that H. brachyantherum ssp. californicum is the maternal parent of these three tetraploid species (Nishikawa et al., 2002). On the other hand, a South American tetraploid, H. fuegianum, has a slightly different RFLP profile in the 9·5–7·5 kb fragment class. This may be due to a mutation or an introgression from South American diploid species. On the basis of C-banding pattern, Linde-Laursen et al. (1990) proposed an introgression into South American tetraploids from a South American diploid, H. patagonicum. We could not verify this from the RFLP pattern.
As mentioned above, H. brachyantherum 4x included a novel accession that has only a single 11-kb rDNA fragment similar to that found in Asian diploids. We suppose that this accession was probably derived from hybridization between H. roshevitzii and a H. brachyantherum ssp. californicum accession having an RFLP profile identical to that of H. roshevitzii. Such a special H. brachyantherum ssp. californicum accession has not been identified yet and might have become extinct by now. But, if present, it may represent a transitional form from Asian to American diploid species. On the basis of MI chromosome pairing, von Bothmer et al. (1986) concluded that, of the three North American diploids, the genome of H. brachyantherum ssp. californicum is very similar to that of the Asian diploid H. roshevitzii, but that the genomes of the two annuals, H. interecedens and H. pusillum, are very similar to those of the South American diploids. This may support derivation of H. brachyantherum ssp. californicum from H. roshevitzii. The presence of two distinct forms in H. brachyantherum 4x that differ in rDNA–RFLP profile may suggest multiple origins of this cytotype. Wide morphological variation of H. brachyantherum 4x has been reported (von Bothmer et al., 1993).
Ancestry of H. depressum
The FISH and RFLP patterns of the North American annual species H. depressum were clearly different from those of the ‘jubatum-type’ species. The absence of the ‘sr SAT chromosome’ pair in H. depressum may be explained by the deletion of the 18S–25S rDNA site in the ‘jubatum-type’, but the unique rDNA–RFLP profile of this species rules out this possibility. The absence of the Asian diploid species-specific rDNA fragment in H. depressum indicates that this species originated in North America independently of the ‘jubatum-type’ species. The nature of polyploidy in H. depressum has been a matter of debate. An autoploid origin of this species was suggested from a high autosyndetic pairing between the two H. depressum genomes in intergeneric hybrids (Sakamoto, 1974; Petersen, 1991). On the other hand, H. depressum was proposed to be an alloploid involving the diploid H. brachyantherum ssp. californicum as one parent and either H. interecedens (Baum and Bailey, 1988) or H. pusillum (Covas, 1949) as the other. On the basis of crossing experiments, Salomon and von Bothmer (1998) concluded that H. depressum arose from hybridization between H. intercendens and H. brachyantherum ssp. californicum. Studies on the chloroplast DNA suggested H. brachyantherum ssp. californicum as the maternal parent of H. depressum (Doebley et al., 1992; Nishikawa et al., 2002). The present study indicates that H. depressum is not a simple autoploid because its FISH pattern slightly deviates from a set of four homologous chromosomes. Hordeum brachyantherum ssp. californicum, H. pusillum and H. intercedens had a FISH pattern that is expected for a diploid ancestor of H. depressum (see Taketa et al., 2001). However, the RFLP analysis revealed that only one accession (H3317) of H. brachyantherum ssp. californicum showed a RFLP profile identical to that of H. depressum, and that the other species/accessions had fragment(s) not found in H. depressum and therefore were unsuitable as diploid ancestors. Thus, available results suggest that H. depressum is a segmental alloploid having two closely related genomes, one of which was derived from a specific accession of H. brachyantherum ssp. californicum. A study of repetitive DNA sequences (Svitashev et al., 1994) also indicated a close relationship between these two taxa. The donor of the second genome of H. depressum remains unknown. As suggested by Linde-Laursen et al. (1995), the second genome could have been derived from another accession of H. brachyantherum ssp. californicum. Hordeum depressum is assumed to have a strong diplodizing mechanism because it shows exclusive bivalent formation at meiosis and good seed fertility (von Bothmer et al., 1987). Such a mechanism ensures stable seed propagation and must have been a prerequisite for its successful establishment as an annual (Knutsson and von Bothmer, 1993). Further detailed studies on intra- and inter-specific variation on North American species are required to elucidate the ancestry of H. depressum.
Hexaploid species
In hexaploid species, only H. arizonicum is an annual/biannual and is distributed in North America, while the other three are all perennials and grow in South America. The RFLP analysis unequivocally shows that a North American annual, H. pusillum, is the diploid ancestor of H. arizonicum. The tetraploid ancestor is probably H. jubatum. On the basis of crossing experiments, Rajhathy and Symko (1966) concluded that H. arizonicum is an amphiploid between H. pusillum (female parent) and H. jubatum. Nishikawa et al. (2002) reported that H. arizonicum and H. pusillum have very similar chloroplast sequences. The present results provide the first firm evidence from nuclear genomes for the involvement of H. pusillum in the ancestry of H. arizonicum. Among diploid Hordeum species, H. pusillum is known to have shorter chromosomes and smaller DNA content (Kankanpää et al., 1996). Two short chromosome pairs with rDNA sites in H. arizonicum (marked s3 and m2 in Fig. 1E) were probably derived from H. pusillum.
A South American hexaploid species, H. lechleri, has a FISH pattern very similar to that of H. arizonicum, but the RFLP data indicate their different origins. The present data and morphological observation (von Bothmer et al., 1995) suggest that the tetraploid ancestor of H. lechleri is probably H. jubatum. The diploid ancestor could not be identified because among the South American diploids there were no species matching in the FISH or RFLP pattern of rDNA.
On the basis of C-banding pattern and morphology of marker SAT chromosomes, Linde-Laursen et al. (1990) proposed that H. parodii is an alloploid between H. tetraploidum and H. muticum, and that H. procerum is an alloploid between H. jubatum and H. cordobense. The FISH patterns indicate that H. parodii and H. procerum have a ‘jubatum-type’ species, probably H. fuegianum or H. tetraploidum, as the tetraploid ancestor. It is impossible to infer their diploid ancestor based solely on the FISH pattern owing to a lack of diagnostic polymorphism (Taketa et al., 2001; present study). In the RFLP profile of these species, different rDNA fragments in the 9·5–7·5 kb class were intensified; H. parodii had a strong 7·5 kb fragment, while H. procerum had a strong 8·5 kb fragment. The RFLP data suggest that the diploid ancestors of H. parodii and H. procerum may be a H. patagonicum subspecies with the 7·5 kb fragment and H. cordobense, respectively.
rDNA evolution in I-genome polyploid species
In I-genome Hordeum species, FISH patterns of both 5S and 18S–25S rDNA sites allowed estimation of lower-ploidy-level ancestors of most polyploid species. This is because rDNA sites are highly conserved between polyploid species and their putative ancestors, generally showing additive relationships. Although the FISH signal intensities differed between corresponding rDNA sites of polyploids and their estimated ancestors, such discrepancies are probably caused by nucleolar dominance (Cermeno and Lacadena, 1985) and amplification or reduction of rDNA repeat unit sequences (Heslop-Harrison, 2000). Some I-genome polyploid species had one or two minor 18S–25S rDNA sites that were not observed in their putative ancestors. These extra minor sites were probably formed by dispersion of rDNA sites, as proposed by Dubcovsky and Dvorák (1995). This highly conserved nature of both 5S and 18S–25S rDNA sites during polyploid evolution in I-genome Hordeum species sharply contrasts with observations made in other plant species. In Gossypium, the cultivated tetraploid species G. hirstum had as many as four extra minor 18S–28S rDNA loci that were not found in either of the two diploid ancestral species, while 5S rDNA sites were the sum of its supposed ancestors in both number and location. In polyploid series of the Brachyscome lineariloba complex, 5S rDNA sites progressively increased as the ploidy level elevated, but 18S–26S rDNA was restricted to a single major locus (Adachi et al., 1997). In polyploids of Sanguisorba, however, the reverse tendency was reported (Mishima et al., 2002). Thus, during polyploid evolution, plant species differ in the degree of the stability of rDNA sites, and different species show different trends in rDNA site-number change.
rDNA–RFLP analysis detected rDNA polymorphisms more sensitively and corroborated the estimation of ancestry based on the FISH pattern. RFLP analysis showed that I-genome polyploid species of Hordeum generally retain variants of 18S–25S rDNA repeat sequences contributed by their putative ancestral species, although quantitative changes in their copy numbers after polyploidization were apparent in some species. A completely different situation was reported in Gossypium, where all rDNA repeats in allopolyploids were converted to one of the ancestral diploid types by concerted evolution (Wendel et al., 1995). In I-genome Hordeum species, intra- and inter-locus concerted evolution of rDNA sequences appears to be operating at a much slower rate than that reported in Gossypium (Wendel et al., 1995). The variants of 18S–25S rDNA repeat units detected by EcoRV digestion in I-genome Hordeum species probably involve changes in intergenic spacer regions, but this must be confirmed by constructing restriction site maps. Moreover, chromosomal distribution of rDNA repeat unit variants needs to be clarified. Phylogenic studies of Triticeae species have utilized 5S rDNA spacer sequences (Baum and Johnson, 1998) and internal transcribed spacer (ITS) sequences of 18S–25S rDNA (Hsiao et al., 1995) with some success. However, application of these parameters to the phylogenetic analysis of I-genome polyploid Hordeum species may require some caution because the materials may violate the basic assumption of homogeneity between the repeated units of rDNA. Due to the complications associated with polyploidy, only a few polyploid Hordeum species have been analysed by other molecular markers and comparative sequencing approaches, mostly failing to reveal their ancestors (Komatsuda et al., 2001; de Bustos et al., 2002; El-Rabey et al., 2002). In conclusion, the highly conserved nature of rDNA sites during polyploid evolution in the I-genome group of Hordeum has enabled the estimation of the ancestry of polyploid species based on the FISH and RFLP data. The genus Hordeum is an excellent material to study the patterns of plant evolution because of availability of many species and accessions with a variety of ploidy levels, life forms, reproduction modes and adaptability. Large amounts of rDNA FISH and cytological data have been accumulated in this genus (Leitch and Heslop-Harrison, 1992, 1993; Pedersen and Linde-Laursen, 1994; Linde-Laursen et al., 1995; de Bustos et al., 1996; Taketa et al., 1999b, 2001, 2003; present study). Future elucidation of homoeology of wild barley chromosomes would allow comparative analysis of karyotype evolution not only within the genus but also with other genera in the grass family. Moreover, such analysis may help solve an important question: why polyploid series were developed only in wild Hordeum species, but not in cultivated barley (H. vulgare ssp. vulgare, 2n = 2n = 14), an important cereal crop.
APPENDIX
Collection sites of Hordeum species with the I genome analysed in this study
| Species |
Ploidy |
Accession |
Collection site |
|||
|---|---|---|---|---|---|---|
| FISH | ||||||
| H. jubatum L. | 4x | H1922 | USA: New Mexico, E Santa Fe | |||
| 4x | H1976 | USA: California, Solano co, | ||||
| H. guatemalense Both.& al. | 4x | H2299 | Guatemala: dep. Huehuetenango | |||
| H. depressum (Scribn.&Sm.) Rydb. | 4x | H2089 | USA: California, San Joaquin co. | |||
| 4x | H2005 | USA: California, San Luis Obispo co. | ||||
| H. fuegianum Both.& al. | 4x | H1418 | Argentina: Tierra del Fuego | |||
| H. tetraploidum Covas | 4x | H6198 | Argentina: Santa Cruz, Rio Pelke | |||
| H. arizonicum Cov. | 6x | H3254 | USA: Arizona, Cochise co. | |||
| H. lechleri (Steud.) Schenck | 6x | H6104 | Argentina: Tierra del Fuego | |||
| H. parodii Covas | 6x | H6294 | Argentina: prov. Chubut | |||
| H. procerum Nevski | 6x | H1789 | Argentina: prov. San Juan | |||
| H. comosum Presl | 2x | H10608 | Argentina | |||
| Species only used for Southern hybridizaiton | ||||||
| H. brevisubulatum (Trin.) Link | ||||||
| ssp. violaceum (Boiss. & Hohen.) Tzvel | 2x | H315 | Iran: prov. Mazanderan | |||
| H. bogdanii Wil. | 2x | H4014 | Pakistan: Gilgit, Nagar valley | |||
| H. roshevitzii Bowden | 2x | H9152 | China: Gansu | |||
| H. brachyantherum Nevski | ||||||
| ssp. californicum (Cov. & Steb.) Both. & et al. | 2x | H3317 | USA: California, Ventura co | |||
| 2x | H1954 | USA: California, Carmel vy, Haystack Hill | ||||
| 2x | H2419 | USA: California, San Luis Obispo co | ||||
| ssp. brachyantherum | 4x | JIC line2 | Unknown | |||
| 4x | H2420 | USA: California, San Luis Obispo co | ||||
| H. pusillum Nutt. | 2x | H722 | USA: Texas, Childress co | |||
| H. intercedens Nevski | 2x | H2310 | USA: California, Ventura co | |||
| H. chilense Roem. & Schult. | 2x | JIC line 1 | Unknown | |||
| H. muticum Presl | 2x | H6479 | Argentina: prov. Jujuy | |||
| H. cordobense Both. & al. | 2x | H6429 | Argentina: prov. Mendoza | |||
| H. flexuosum Steud. | 2x | H1116 | Argentina: prov. Buenos Aires | |||
| H. patagonicum (Haum.) Cov. | ||||||
| ssp. musterisii (Nico.) Both. & al. | 2x | H1358 | Argentina: prov. Santa Cruz | |||
| ssp. patagonicum | 2x | H6052 | Argentina: prov. Santa Cruz | |||
| ssp. setifolium (Paro. & Nico.) Both. & al. | 2x | H1366 | Argentina: prov. Santa Cruz | |||
| ssp. santacrucense (Paro. & Nico.) Both. & al. | 2x | H1353 | Argentina: prov. Santa Cruz | |||
| H. pubiflorum Hook. f. | ||||||
| ssp. halophilum Grise. | 2x | H1348 | Argentina: prov. Santa Cruz | |||
| ssp. pubiflorum Hook. f. | 2x | H1296 | Argentina: prov. Santa Cruz | |||
| H. stenostachys Godr. | 2x | H1108 | Argentina: prov. Buenos Aires | |||
Seeds of these accessions are preserved at Kagawa University, Japan and Nordic Gene Bank, Sweden.
Acknowledgments
We are grateful to Professor M. Murata (Okayama University) for advice on FISH, and Miss Y. Sakurai for technical assistance. The work was supported in part by grants from CREST, the Ministry of Education, Science, Sports and Culture, Japan (No.09760006), Kagawa University, and the Scandinavia–Nippon Sasakawa foundation.
LITERATURE CITED
- Adachi J, Watanabe K, Fukui K, Ohmido N, Kosuge K. 1997. Chromosomal location and reorganization of the 45S and 5S rDNA in the Brachyscome lineariloba complex (Asteraceae). Journal of Plant Research 110: 371–377. [Google Scholar]
- Ansari HA, Ellison NW, Reader SM, Bedaeva ED, Friebe B, Miller TE, Williams WM. 1999. Molecular cytogenetic organization of 5S and 18S–25S rDNA loci in white clover (Trifolium repens L.) and related species. Annals of Botany 83: 199–206. [Google Scholar]
- Baden C, von Bothmer R. 1994. A taxonomic revision of Hordeum sect. Critesion. Nordic Journal of Botany 14: 117–136. [Google Scholar]
- Baum BR, Bailey LG. 1988. A taxanomic study of the annual Hordeum depressum and related species. Canadian Journal of Botany 66: 401–408. [Google Scholar]
- Baum BR, Johnson DA. 1998. The 5S rRNA gene in sea barley (Hordeum marinum Hudson sensu lato): sequence variation among repeat units and relationship to the X haplome in barley (Hordeum). Genome 41: 652–661. [PubMed] [Google Scholar]
- Bedaeva ED, Friebe B, Gill BS. 1996. Genome differentiation in Aegilops 2. Physical mapping of 5S and 18S-26S ribosomal RNA gene families in diploid species. Genome 39: 1150–1158. [DOI] [PubMed] [Google Scholar]
- von Bothmer R, Fink J, Landström T. 1986. Meiosis in interspecific Hordeum hybrids. I. Diploid combinations. Canadian Journal of Genetics and Cytology 28: 525–535. [Google Scholar]
- von Bothmer R, Fink J, Landström T. 1987. Meiosis in interspecific Hordeum hybrids. II. Triploid hybrids. Evolutionary Trends in Plants 1: 42–50. [Google Scholar]
- von Bothmer R, Fink J, Landström T. 1988. Meiosis in interspecific Hordeum hybrids. IV. Tetraploid (4x × 4x) hybrids. Genome 30: 479–485. [Google Scholar]
- von Bothmer R, Jacobsen N, Seberg O. 1993. Variation and taxonomy in Hordeum depressum and in the H. brachyantherum complex (Poaceae). Nordic Journal of Botany 13: 3–17. [Google Scholar]
- von Bothmer R, Jacobsen N, Baden C, Jørgensen RB, Linde-Laursen I. 1995.Systematic and ecogeographical studies on crop genepools 7. An ecogeographical study of the genus Hordeum, 2nd edn. Rome: IBPGR. [Google Scholar]
- de Bustos A, Cuadrado A, Soler C, Jouve N. 1996. Physical mapping of repetitive DNA sequences and 5S and 18S–26S rDNA in five wild species of the genus Hordeum Chromosome Research 4: 491–499. [DOI] [PubMed] [Google Scholar]
- de Bustos A, Loarce Y, Jouve N. 2002. Species relationships between antifungal chitinase and nuclear rDNA (internal transcribed spacer) sequences in the genus Hordeum Genome 45: 339–342. [DOI] [PubMed] [Google Scholar]
- Cerbah M, Coulaud J, Siljak-Yakovlev S. 1998. rDNA organization and evolutionary relationships in the genus Hypochaeris (Asteraceae). Journal of Heredity 89: 312–318. [Google Scholar]
- Cermeno MC, Lacadena JR. 1985. Nucleolar organizer competition in Aegilops–rye hybrids. Genome 27: 479–483. [Google Scholar]
- Covas G. 1949. Taxonomic observations on the North American species of Hordeum Madoroño 10: 1–21. [Google Scholar]
- Doebley J, von Bothmer R, Larson S. 1992. Chloroplast DNA variation and the phylogeny of Hordeum (Poaceae). American Journal of Botany 79: 576–584. [Google Scholar]
- Dubcovsky J, Dvorák J. 1995. Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rabey HA, Badr A, Scäfer-Pregl R, Martin W, Salamini F. 2002. Speciation and species separation in Hordeum L. (Poaceae) resolved by discontinuous molecular markers. Plant Biology 4: 567–575. [Google Scholar]
- Fukui K. 1996. Plant chromosomes at mitosis. In: Fukui K, Nakayama S, eds. Plant chromosomes: laboratory methods. Tokyo: CRC Press, 1–17. [Google Scholar]
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Research 7: 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Dyer TA. 1980. Sequence organization of the repeated units in the nucleus of wheat which contains 5S-rDNA genes. Nucleic Acids Research 8: 4851–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ. 1996. Distribution of 5S and 18S–28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105: 55–61. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS. 2000. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. The Plant Cell 12: 617–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C, Chatterton NJ, Asay KH, Jensen KB. 1995. Phylogenetic relationship of the monogenomic species of the wheat tribe, Triticeae (Poaceae), inferred from nuclear rDNA (internal transcribed spacer) sequences. Genome 38: 211–223. [DOI] [PubMed] [Google Scholar]
- Jørgensen RB. 1986. Relationships in the barley genus (Hordeum): an electrophoretic examination of proteins. Hereditas 104: 273–291. [Google Scholar]
- Kankanpää J, Mannonen L, Schulman AH. 1996. The genome sizes of Hordeum species show considerable variation. Genome 39: 730–735. [DOI] [PubMed] [Google Scholar]
- Knutsson T, von Bothmer R. 1993. Interspecific hybridization with Hordeum depressum (Poaceae). Nordic Journal of Botany 13: 389–394. [Google Scholar]
- Komatsuda T, Tannno K, Salomon B, Bryngelsson T, von Bothmer R. 1999. Phylogeny in the genus Hordeum based on nucleotide sequences closely linked to the vrs1 locus (row number of spikelets). Genome 42: 973–981. [DOI] [PubMed] [Google Scholar]
- Komatsuda T, Salomon B, Bryngelsson T, von Bothmer R. 2001. Phylogenetic analysis of Hordeum marinum Huds. based on nucleotide sequences linked to the vrs1 locus. Plant Systematics and Evolution 227: 137–144. [Google Scholar]
- Leitch IJ, Heslop-Harrison JS. 1992. Physical mapping of the 18S–5·8S–26S rRNA genes in barley by in situ hybridization. Genome 35: 1013–1018. [Google Scholar]
- Leitch IJ, Heslop-Harrison JS. 1993. Physical mapping of four sites of 5S rDNA sequences and one site of the alpha-amylase-2 gene in barley (Hordeum vulgare). Genome 36: 517–523. [DOI] [PubMed] [Google Scholar]
- Linde-Laursen I, von Bothmer R, Jacobsen N. 1986. Giemsa C-banded karyotypes of Hordeum taxa from North America. Canadian Journal of Genetics and Cytology 28: 42–62. [Google Scholar]
- Linde-Laursen I, von Bothmer R, Jacobsen N. 1989. Giemsa C-banded karyotypes of South American Hordeum (Poaceae). I. 14 diploid taxa. Hereditas 110: 289–305. [Google Scholar]
- Linde-Laursen I, von Bothmer R, Jacobsen N. 1990. Giemsa C-banded karyotypes of South and Central American Hordeum (Poaceae). II. 6 polyploid taxa. Hereditas 112: 93–107. [Google Scholar]
- Linde-Laursen I, von Bothmer R, Jacobsen N. 1995. Karyotype differentiation and evolution in the genus Hordeum (Poaceae). In: Brandham PE, Bennett MD, eds. Kew Chromosome Conference IV. Kew, UK: Royal Botanic Gardens, 233–247. [Google Scholar]
- Linde-Laursen I, Heslop-Harrison JS, Shepherd KW, Taketa S. 1997. The barley genome and its relationship with the wheat genomes. A survey with an internationally agreed recommendation for barley chromosome nomenclature. Hereditas 126: 1–16. [Google Scholar]
- Mishima M, Ohmido N, Fukui K, Yahara T. 2002. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110: 550–558. [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Fedak G. 1989. Polymorphism in ribosomal DNA repeat units of 12 Hordeum species. Genome 32: 1124–1127. [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Gupta PK, Fedak G, 1989. Ribosomal DNA repeat unit polymorphism in 25 Hordeum species. Theoretical and Applied Genetics 78: 387–392. [DOI] [PubMed] [Google Scholar]
- Molnar SJ, Wheatcroft R, Fedak G. 1992. RFLP analysis of Hordeum species relationship. Hereditas 116: 87–91. [Google Scholar]
- Nishikawa T, Salomon B, Komatsuda T, von Bothmer R, Kadowaki K. 2002. Molecular phylogeny of the genus Hordeum using three chloroplast DNA sequences. Genome 45: 1157–1166. [DOI] [PubMed] [Google Scholar]
- Pedersen C, Linde-Laursen I. 1994. Chromosomal locations of four minor rDNA loci and marker microsatellite sequence in barley. Chromosome Research 2: 65–71. [DOI] [PubMed] [Google Scholar]
- Petersen G. 1991. Intergeneric hybridization between Hordeum and Secale II. Analysis of meiosis in hybrids. Hereditas 114: 141–159. [Google Scholar]
- Petersen G, Seberg O. 2003. Phylogenetic analyses of the diploid species of Hordeum (Poaceae) and a revised classification of the genus. Systematic Botany 28: 293–306. [Google Scholar]
- Raina SN, Mukai Y. 1999. Detection of a variable number of 18S–5·8S–26S and 5S ribosomal DNA loci by fluorescent in situ hybridization in diploid and tetraploid Arachis species. Genome 42: 52–59. [Google Scholar]
- Rajhathy T, Symko S. 1966. The synthesis of a species: Hordeum arizonicum Canadian Journal of Botany 44: 1224–1228. [Google Scholar]
- Sakamoto S. 1974. Intergeneric hybridization among three species of Heteranthelium, Eremopyrum and Hordeum, and its significance for the genetic relationships within the tribe Triticeae. New Phytologist 73: 341–350. [Google Scholar]
- Salomon B, von Bothmer R. 1998. The ancestry of Hordeum depressum (Poaceae, Triticeae). Nordic Journal of Botany 18: 257–265. [Google Scholar]
- Svitashev S, Bryngelsson T, Vershinin A, Pederesen C, Säll T, von Bothmer R. 1994. Phylogenetic analysis of the genus Hordeum using repetitive DNA sequences. Theoretical and Applied Genetics 89: 801–810. [DOI] [PubMed] [Google Scholar]
- Taketa S, Ando H, Takeda K, von Bothmer R. 1999. Detection of Hordeum marinum genome in three polyploid Hordeum species and cytotypes by genomic in situ hybridization. Hereditas 130: 185–188. [DOI] [PubMed] [Google Scholar]
- Taketa S, Harrison GE, Heslop-Harrison JS. 1999. Comparative physical mapping of the 5S and 18S–25S rDNA in nine wild Hordeum species and cytotypes. Theoretical and Applied Genetics 98: 1–9. [Google Scholar]
- Taketa S, Ando H, Takeda K, von Bothmer R. 2001. Physical locations of 5S and 18S–25S rDNA in Asian and American diploid Hordeum species with the I genome. Heredity 86: 522–530. [DOI] [PubMed] [Google Scholar]
- Taketa S, Linde-Laursen I, Künzel G. 2003. Cytogenetic diversity in barley (Hordeum vulgare). In: von Bothmer R, Knüpffer H, van Hintum T, Sato K, eds. Diversity in barley. Amsterdam: Elsevier, 97–119. [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proceedings of National Academy of Science, USA 92: 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]


