Abstract
• Aims To outline the current state of knowledge and discuss the evolution of various viewpoints put forth to explain the mechanism of cellulose biosynthesis.
• Scope Understanding the mechanism of cellulose biosynthesis is one of the major challenges in plant biology. The simplicity in the chemical structure of cellulose belies the complexities that are associated with the synthesis and assembly of this polysaccharide. Assembly of cellulose microfibrils in most organisms is visualized as a multi-step process involving a number of proteins with the key protein being the cellulose synthase catalytic sub-unit. Although genes encoding this protein have been identified in almost all cellulose synthesizing organisms, it has been a challenge in general, and more specifically in vascular plants, to demonstrate cellulose synthase activity in vitro. The assembly of glucan chains into cellulose microfibrils of specific dimensions, viewed as a spontaneous process, necessitates the assembly of synthesizing sites unique to most groups of organisms. The steps of polymerization (requiring the specific arrangement and activity of the cellulose synthase catalytic sub-units) and crystallization (directed self-assembly of glucan chains) are certainly interlinked in the formation of cellulose microfibrils. Mutants affected in cellulose biosynthesis have been identified in vascular plants. Studies on these mutants and herbicide-treated plants suggest an interesting link between the steps of polymerization and crystallization during cellulose biosynthesis.
• Conclusions With the identification of a large number of genes encoding cellulose synthases and cellulose synthase-like proteins in vascular plants and the supposed role of a number of other proteins in cellulose biosynthesis, a complete understanding of this process will necessitate a wider variety of research tools and approaches than was thought to be required a few years back.
Key words: Cellulose, plant cell wall, cotton, Arabidopsis thaliana, Acetobacter xylinum, cellulose synthase, cellulose synthase-like, glycosyltransferases, terminal complex, polymerization, crystallization
INTRODUCTION
Cellulose is often referred to as the most abundant macromolecule on earth (Brown, 2004) and most of the cellulose is produced by vascular plants. Apart from these plants, cellulose synthesis also occurs in most groups of algae, the slime mold Dictyostelium, a number of bacterial species (including the cyanobacteria), and tunicates in the animal kingdom. Cellulose is an extracellular polysaccharide and, with the exception of bacteria and the tunicates, it is part of the cell wall in plants, algae and Dictyostelium. The function of cellulose in these different groups of organisms reflects the diverse roles associated with this simple structural polysaccharide. Whereas it is possible for some of these organisms, specifically bacteria, to survive in the absence of cellulose synthesis, it may not be true for most vascular plant cells to survive in the absence of cellulose synthesis. As such, the importance of cellulose in the life of a plant cannot be overemphasized since it not only provides the necessary strength to resist the turgor pressure in plant cells but also has a distinct role in maintaining the size, shape and division/differentiation potential of most plant cells and ultimately the direction of plant growth (Fig. 1). In the authors' view, deposition of cellulose microfibrils in a specific orientation for determining the direction of plant cell elongation, in a sense, is a stage of commitment akin to the S phase and M phase in eukaryotic cell cycle. Once the cellulose microfibrils are ordered in a specific orientation, the direction of cell elongation is essentially fixed. There are a very large number of questions related to cellulose biosynthesis that need to be addressed; however, at this point it is important to recognize that after a long hiatus there is an exponential increase in the number of research articles that discuss the molecular aspects of cellulose biosynthesis, and many of these advances have been made with the identification of genes, specifically for cellulose synthases, and cellulose-deficient mutants in plants. A comprehensive view of cellulose synthesis and the plant cell wall is provided in reviews by Delmer (1999), Doblin et al. (2002), and Somerville et al. (2004). Excellent articles on individual topics related to cellulose biosynthesis are provided in a special recent issue of the journal Cellulose (Vol. 11, no. 3/4, September/December 2004). In this review, selective topics in cellulose biosynthesis will be discussed with the goal of providing a timely and unique view of this rather exciting field of study from the authors' perspective.
Fig. 1.
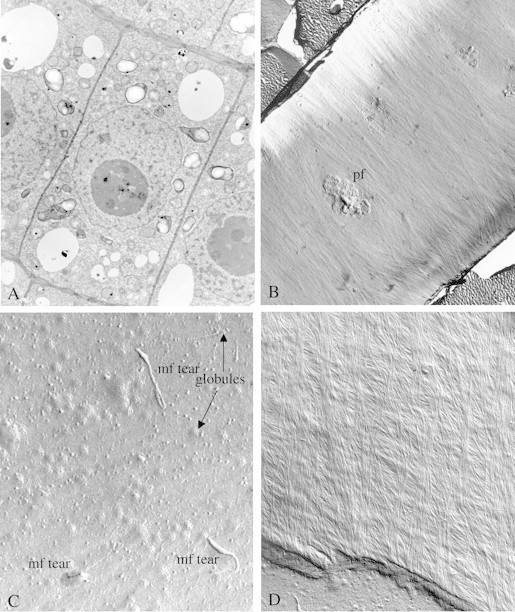
(A) Ultrathin section of recently divided cells just below the meristem of a Zea mays root tip. Note the recently synthesized transverse walls (thinner). The elongation axis will be perpendicular to this direction. (Unpublished micrograph, courtesy of Susette Mueller and R. Malcolm Brown, Jr.) (B) Freeze fracture showing the E fracture face (EF) of a large area of an elongating cell in the root of Zea mays. The direction of microfibril impressions and, hence, the direction of the orientation of the microfibrils themselves, is perpendicular to the axis of elongation. Note also a prominent pit field (pf) in the centre of the micrograph. Microfibril synthesis around this pit field gives clues that suggest a membrane flow mechanism in the plane of the fluid membrane may underlie and direct cellulose microfibril synthesis (see Mueller and Brown, 1982a, b). Evidence to support this hypothesis is based on the direction of microfibrillar tears through the plasma membrane where the terminal globules and direction of synthesis is revealed (see C). In addition, parallel cortical microtubules provide the general ‘channels’ for the membrane flow. Actin microfilaments are found perpendicular to the cortical microtubules and may be the source of motion to propel the directional motions of the fluid membrane. (Unpublished micrograph, courtesy of Susette Mueller and R. Malcolm Brown, Jr.) (C) E fracture face of the plasma membrane of an actively elongating cell in the root of Zea mays showing three prominent tears of microfibrils back through the outer leaflet of the plasma membrane (mf tear). Note that the ‘rip’ terminates at a hole where the microfibril is associated with the rosette TC. In this fracture face, only the globular regions of the tips are shown associated with the TCs (globules). Many other TCs which have not been torn through the plasma membrane are revealed, some in clusters. (Unpublished micrograph, courtesy of Susette Mueller and R. Malcolm Brown, Jr.) (D) Freeze fracture through the innermost layer of a growth wall from an elongating cell in the root of Zea mays. Note the change in pitch of the transverse walls, suggesting that during elongation, the general pitch of the direction of microfibril synthesis is gradually changing from transverse to longitudinal. (Unpublished micrograph, courtesy of Susette Mueller and R. Malcolm Brown, Jr.)
CELLULOSE IS A POLYMORPHIC MOLECULE: THE MANY FORMS OF CELLULOSE DIFFER IN THEIR ARRANGEMENT OF GLUCAN CHAINS
Cellulose is composed of linear polymer chains of β-1,4-linked glucose residues. Depending on the source from which cellulose is obtained, the physical properties such as the crystalline state, degree of crystallinity, and molecular weight may be highly variable. The crystalline state of cellulose is determined by the arrangement of the glucan chains with respect to each other in a unit cell. In nature, most cellulose is produced as crystalline cellulose and is defined as cellulose I. The glucan chains in cellulose I are parallel to each other and are packed side by side to form microfibrils that in most plants are 3 nm thick, but which reach widths of 20 nm in certain algae (Jarvis, 2003). Interestingly, the microfibrillar width in the red alga Erythrocladia subintegra has been shown to vary from 10 to 68 nm (Tsekos et al., 1999). Differing amounts of two crystalline sub-allomorphs of cellulose I, namely Iα and Iβ are found to occur in the cellulose obtained from natural sources (Attala and VanderHart, 1984). Cellulose Iα and cellulose Iβ differ with respect to their crystal packing, molecular conformation and hydrogen bonding and these differences may influence the physical properties of the cellulose (Nishiyama et al., 2003). Cellulose from some algae and bacteria is found to be Iα rich, while cellulose from cotton, wood, ramie and tunicates is Iβ rich (Sugiyama et al., 1991). Since a cellulose microfibril may contain both types of cellulose, some of the physical properties of cellulose fibres will be dependant on the ratio of these two allomorphs. Cellulose Iα is metastable and can be converted to Iβ by annealing.
A few organisms produce crystalline cellulose II naturally, and this form also is produced by mutants of Acetobacter xylinum, a bacterium that normally produces cellulose I. The glucan chain arrangement in cellulose II is antiparallel, and this may take place as a result of chain folding during synthesis as demonstrated in A. xylinum (Kuga et al., 1993). An additional hydrogen bond per glucose residue in cellulose II makes this allomorph as the most thermodynamically stable form. Apart from the crystalline states, cellulose also occurs in a non-crystalline state, and this form of cellulose has been observed to be present along with the cellulose I crystallites in cellulose microfibrils. A large number of crystalline forms of cellulose are obtained by physical and chemical treatments of cellulose post-synthesis. Many of these crystalline forms are characterized using physical techniques. A new form of derived cellulose referred to as nematic ordered cellulose (NOC) is obtained by specific drawing of glucan chains from water-swollen cellulose (Kondo et al., 2001). The structure of NOC is highly ordered but not crystalline, and films obtained from this cellulose exhibit properties different from conventional cellulose films. In a majority of cases, cellulose modified after synthesis has properties not found in the native cellulose that is obtained from living organisms.
Although cellulose is one of the simplest known polysaccharides, non-enzymatic chemical synthesis of this polysaccharide has not been very successful. One can ascribe a large number of reasons for the difficulty in synthesizing cellulose chemically, including the difficulty in realizing regio- and stereo-control at each step of addition of a monosaccharide sub-unit (Kobayashi and Shoda, 1995) as well as the insolubility and folding of β-1,4-linked glucan chains with increasing degree of polymerization. However, in spite of the limitations just mentioned, chemical synthesis of cellulose II has been obtained in vitro using β-cellobiosyl fluoride (a synthetic substrate) and a crude preparation of cellulases from various sources (Kobayashi et al., 1991) and cellulose I from purified preparations (Lee et al., 1994). The mechanism by which cellulases catalyse the synthesis of cellulose from β-cellobiosyl fluoride in an organic solvent is not very well understood, but it does highlight the concept that an ordered assembly of catalytic sites is essential for the parallel orientation of the glucan chains during the crystallization into cellulose I. It is important to consider that in the β-1,4-linked backbone in cellulose, every glucose residue is rotated or inverted 180° with respect to its neighbouring residue. This structure of the backbone implies that the repeating unit in the backbone is cellobiose as opposed to a glucose residue, and the glucan chain itself is relatively straight. Moreover, this aspect of the backbone structure has had a major influence in understanding the biosynthesis of cellulose, especially since the natural substrate is UDP-glucose and not cellobiose. In nature, synthesis of cellulose requires the enzyme cellulose synthase that uses UDP-glucose as the substrate. These two features of cellulose synthesis are now certain for all known organisms even though the mechanism by which cellulose is synthesized in different organisms is still being debated.
THE CELLULOSE-SYNTHESIZING COMPLEX: AN ELEGANT NANOMACHINE
Living cells employ a sophisticated membrane complex for synthesis of cellulose I microfibrils. Not unlike the DNA-replication machinery in cells (Baker and Bell, 1998), the cellulose-synthesizing machinery may be composed of a number of proteins arranged in a very specific manner. During DNA replication, proteins assemble at the replication fork, and synthesis of two polynucleotide chains takes place simultaneously. In cellulose synthesis, a large number of glucan chains are synthesized simultaneously from a large membrane-localized complex that has been visualized by microscopy (Brown and Montezinos, 1976; Mueller and Brown, 1980; Itoh and Brown, 1984; Tsekos and Reiss, 1992). The association of organized membrane complexes to one end of the cellulose microfibril impression in freeze-fracture replicas suggested that these complexes are the sites of synthesis of cellulose. Unlike the DNA-synthesizing machinery, only a single component, the cellulose synthase, has been identified in the cellulose-synthesizing machinery (Kimura et al., 1999). In vascular plants, this complex appears as a ‘rosette’ with a six-fold symmetry and a diameter of 25–30 nm (Mueller and Brown, 1980). Structures analogous to the rosette (e.g. some form of linear synthesizing complexes) have been observed in all cellulose-synthesizing organisms and in general have been referred to as terminal complexes (TCs) (Brown, 1985; Tsekos, 1999; Okuda, 2002).
THE ORGANIZATION OF THE TERMINAL COMPLEX DETERMINES THE DIMENSION OF THE CELLULOSE MICROFIBRIL
In general, TCs are observed as particle arrays by freeze-fracture electron microscopy, and the arrangement of these particles can be in the form of a solitary rosette (as observed in charophycean green algae and land plants) or as linear row(s) of rosettes (as in certain algae) (Kiermayer and Sleytr, 1979; Giddings et al., 1980). A great variation is observed in organisms that have linear TCs (Tsekos, 1999). A single row of particles is observed in prokaryotes (as in A. xylinum), brown algae and some red algae. Multiple rows are observed in the glaucophycean algae (Willison and Brown, 1978), some red algae (Tsekos and Reiss, 1992), chlorophycean and ulvophycean green algae (Itoh and Brown, 1984), the slime mold Dictyostelium (Grimson et al., 1996) and the tunicates (Kimura and Itoh, 1996). Diagonal rows of particles are observed in the xanthophycean algae such as Vaucheria hamata (Mizuta and Brown, 1992). Surveying all the different cellulose-synthesizing organisms, it is clear that the greatest TC diversity is observed in different groups of algae. Based on a number of studies, a strong relationship is observed between the TC structure and the dimensions of the cellulose microfibril (Brown, 1996; Tsekos, 1999; Okuda et al., 2004). The rosette TCs of land plants and some green algae synthesize cellulose microfibrils 3·5–10 nm in thickness, consisting of 36–90 glucan chains (Herth, 1983; Ha et al., 1998), while the large linear TCs of the green alga Valonia macrophysa produces microfibrils of up to 1400 glucan chains (Sugiyama et al., 1985).
TERMINAL COMPLEXES MAY BE ASSEMBLED AT THE PLASMA MEMBRANE OR TRANSPORTED PREASSEMBLED VIA THE ER–GOLGI–VESICLE PATHWAY
The plasma membrane is the site of synthesis and assembly of the cellulose microfibril. If the dimension of the cellulose microfibril is determined by the arrangement of cellulose-synthesizing sites in a TC, how and when are these sites organized on the plasma membrane? Two major views for the assembly of TCs have been proposed from ultrastructural studies. In the first case, TCs are assembled prior to their insertion in the plasma membrane and are obtained from Golgi-derived vesicles (Haigler and Brown, 1986). Alternatively, TCs are assembled directly on the plasma membrane from particulate precursors which are supplied by Golgi-derived vesicles (Itoh and Brown, 1988; Tsekos et al., 1996). More recently, Okuda et al. (2004) observed TC-like structures in the membrane of large, dense cytoplasmic vesicles that were distinct from Golgi vesicles in the xanthophycean alga Botrydiopsis intercedens. These authors propose that groups of TC precursors, which consist of diagonal rows of particles, are loaded in the plasma membrane through the fusion of large, cytoplasmic vesicles in this alga. Once present in the plasma membrane, the TC precursors adjust the arrangement of diagonal rows of particles to form a functional TC. Yet another possibility is that, although the arrangement of sub-units in a TC may be determined by the interaction of proteins in the TC, the TC structure may tighten and appears to be much more distinct when it produces the cellulose microfibril. A tight interaction is observed between the glucan chains and cellulose synthases when cellulose is synthesized in vitro, and this interaction may very well exist in vivo. As mentioned earlier, the only component identified in a rosette TC from plants is the cellulose synthase. Based on mutant and molecular analysis, a model for assembly of distinct cellulose synthases in the rosette TC of land plants has been proposed and this will be discussed in a later section.
GENES ENCODING CELLULOSE SYNTHASES IN PLANTS HAVE BEEN IDENTIFIED BY RANDOM SEQUENCING AND SEQUENCE COMPARISONS WITH BACTERIAL CELLULOSE SYNTHASE AND OTHER β-GLYCOSYLTRANSFERASES
One of the most interesting features of cellulose biosynthesis to be discovered in the past few years has been the identification of a large number of genes that encode cellulose synthases with possibly non-redundant functions in vascular plants. DNA sequences encoding cellulose synthases in plants were first identified following sequencing of random clones from a cotton fibre cDNA library (Pear et al., 1996). Derived protein sequences of two cDNA clones (GhCesA1 and GhCesA2) from this library showed similarity to the amino acid sequence of bacterial cellulose synthase (Saxena et al., 1990; Wong et al., 1990) and the D,D,D,QXXRW motif found to be conserved in processive β-glycosyltransferases (Saxena et al., 1995) was identified in these sequences. The expression pattern of the GhCesA1 gene in the developing cotton fibre, and the ability to bind UDP-glucose by a region of the protein synthesized in Escherichia coli further confirmed that the clones obtained encoded cellulose synthase (Pear et al., 1996). Features of plant cellulose synthases, determined using the DNA sequence of the cotton GhCesA1 cDNA clone, revealed that they were larger than the bacterial cellulose synthase and contained regions that were not present in their bacterial counterparts. Bacterial cellulose synthases are transmembrane proteins that have a large globular region in which the conserved β-glycosyltransferases residues D,D,D, QXXRW are present. The globular region is predicted to be present in the cytoplasm with transmembrane segments present at the N-terminal and C-terminal regions. The cotton cellulose synthase was shown to have a similar arrangement of the globular and transmembrane regions, but containing a zinc-binding domain at the N-terminus and variable regions within the globular region. Genetic identification of cellulose synthase genes in vascular plants came about following the analysis of the rsw1 conditional mutant in arabidopsis (Arioli et al., 1998). This mutant exhibits a normal phenotype when grown at 21°C, but shows swelling of roots and stunted growth at 31°C. Furthermore it produces reduced amounts of crystalline cellulose at the non-permissive temperature but increased amounts of a product characterized as non-crystalline cellulose. Using positional cloning, the mutation in the rsw1 mutant was found to be within a gene (rsw1/AtCesA1) that encoded a protein similar to the cotton cellulose synthase. Moreover, the mutation in the rsw1 mutant was corrected upon transfer of a wild-type rsw1 gene, confirming that the mutant phenotype resulted from a mutation in the rsw1 gene. Characterization of a number of other mutants led to the identification of a number of other genes encoding cellulose synthases in arabidopsis. The genome sequence of arabidopsis has now made it possible to obtain information on the complete set of cellulose synthases in this plant. In arabidopsis and maize, at least ten distinct CesA genes have been identified (Holland et al., 2000; Richmond and Somerville, 2000). Genes encoding cellulose synthase (CesA) and cellulose synthase-like (Csl) proteins have now been identified in almost 170 species of plants (http://cellwall.stanford.edu; see also http://128.83.195.51/cen/library/tree/default.htm).
ASSEMBLY OF A CELLULOSE MICROFIBRIL IN VASCULAR PLANTS REQUIRES ASSEMBLY OF THREE DIFFERENT CELLULOSE SYNTHASES IN THE ROSETTE TC
A number of arabidopsis mutants altered in their growth and development have been characterized, and changes in some of them are related to the decreased amount of cellulose produced in these mutants (see Robert et al., 2004). In these strains, mutations are observed in genes predicted to have a role in cellulose biosynthesis, including those that encode cellulose synthase. Gene expression of CesA genes in different tissues, developmental stages and under different environmental conditions has been analysed in a number of plants including arabidopsis (Hamann et al., 2004), maize (Appenzeller et al., 2004) and hybrid aspen (Djerbi et al., 2004). In most cases, no significant differences have been observed in the expression of the different CesA genes in different tissues. However, different groups of genes are co-expressed in cells that synthesize cellulose in the primary cell wall versus those that are active in the synthesis of cellulose in the secondary cell wall. A relationship between these genes has also been obtained from mutant analysis as well as phylogenetic analysis. In arabidopsis, AtCesA1, AtCesA3 and AtCesA6 are proposed to be required for primary cell wall cellulose synthesis (Fagard et al., 2000; Scheible et al., 2001; Burn et al., 2002) and AtCesA4, AtCesA7 and AtCesA8 are proposed to be required for secondary cell wall cellulose synthesis (Taylor et al., 2003). Similar sets of genes have also been identified in other plants (Tanaka et al., 2003). These observations have led to the suggestion that three different CesA gene products may be required for the formation of a functional rosette TC in plants (Doblin et al., 2002). Although the three different CesA gene products encode cellulose synthase, they are non-redundant. A mutation in any one results in the loss of cellulose microfibril formation. Hypothetical models showing the arrangement of the different CesA sub-units have been proposed, but as yet there is no experimental evidence as to how the different CesA sub-units are arranged in the rosette TC (Perrin, 2001). Rosettes associated with cellulose microfibrils have a six-fold symmetry and each particle in the rosette is believed to contain six CesA sub-units allowing for an assembly of 36 CesA sub-units in a rosette. The number of CesA sub-units in a rosette is predicted from the number of glucan chains present in a cellulose microfibril. Interaction between the three cellulose synthases (AtCesA4, AtCesA7 and AtCesA8) that are required for cellulose synthesis in the secondary cell wall has been demonstrated (Taylor et al., 2003). Moreover, the interaction between the different cellulose synthase sub-units to give rise to a multimeric rosette structure has been suggested to take place via intermolecular disulfide bridges formed in the N-terminal zinc finger regions of cellulose synthases (Kurek et al., 2002). At this point it is important to consider that only a part of the rosette structure is exposed to the extracellular side of the plasma membrane with a significantly larger proportion of this complex being present in the cytoplasm (Kudlicka et al., 1987).
THE ROSETTE STRUCTURE REVISITED
Since the discovery of the rosette TC in vascular plants (Mueller and Brown, 1980), the concept of this multi-enzyme complex has centred upon the freeze fracture image of a six-fold symmetry of particle sub-unit found on the P fracture face of the plasma membrane. It was only later from sectioned material that the cross-section of a linear TC (Kudlicka et al., 1987) indicated that most of the structure was deeply embedded in the cytoplasm of the cell (Fig. 2B). As a result, it became clear that the ‘linear or rosette’ TC morphology is based only on a small fraction of the structural unit, and this view has been supported by the purification of an intact rosette TC and its activation to synthesize cellulose I microfibril in vitro (Fig. 2A; W. Laosinchai and R. M. Brown, Jr, unpubl. res.). This evidence, as well as that obtained from recent molecular, biochemical and structural data, provides impetus for the current model of the rosette TC to be revised. The revised model of the rosette takes into consideration two levels of assembly of the cellulose synthases (Fig. 3). In the first level, assembly and processing of three different homodimers (each dimer being composed of a unique cellulose synthase) occurs to form a linear array with six particles, presumably deep within the cytoplasmic base of the TC structure. In the next level, the linear arrays are arranged in a rosette with a six-fold symmetry. The assembly and processing of the linear arrays and their assembly into the complete rosette TC complex presumably occurs in the endoplasmic reticulum and the Golgi apparatus. The assembled rosette TC is then transported to the plasma membrane for activation and cellulose microfibril synthesis. In the revised model, the linear rows within each rosette allow formation of glucan sheets by van der Waals forces. This has been experimentally confirmed from studies of cellulose biosynthesis in A. xylinum (Cousins and Brown, 1995, 1997a, b). Formation of monomolecular glucan sheets is the first of two steps in cellulose crystallization. In the next stage, six separate glucan chain sheets are directed into the exit channel of the TC complex, where they pass through the rosette aperture and are then hydrogen-bonded into the crystalline cellulose I microfibril. While not fully understood, this model is very attractive in that it seems to explain all of the available evidence thus far discovered, from understanding the requirement for more than a single CesA gene product for cellulose I microfibril assembly to the two-step crystallization model. In the absence of any one specific CesA gene product, assembly of the rosette would be affected. At the same time, a mutant CesA may be incorporated in the rosette but would not allow synthesis of cellulose I microfibrils.
Fig. 2.
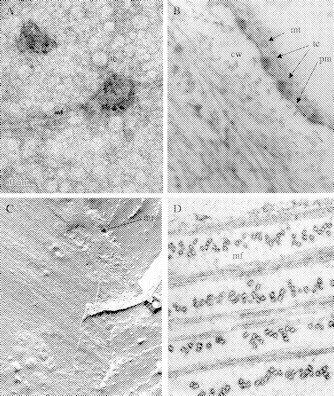
(A) Negative staining of immunity-purified cellulose synthase from Gossypium hirsutum showing synthesis of cellulose I microfibrils in vitro. The identity of the isolated components of the rosette TC is demonstrated by immunolabelling using antibodies to CesA that are coupled with colloidal gold. The TC complex (tc) attached to a cellulose I microfibril is labelled with the antibody. When cellulose synthases are isolated using specific detergents and purified by immunoaffinity methods, they remain sufficiently intact to synthesize microfibrils (mf) as they would in vivo. This unpublished micrograph, courtesy of Walairat Laosinchai and R. Malcolm Brown, Jr, shows that the TC structure at the ‘business end’ is very different from the classical view of a rosette with a six-fold symmetry. (B) Ultrathin section through the plasma membrane of Boergesenia forbesii which has characteristic linear TCs, each with three rows of TC sub-units (see Kudlicka et al., 1987). In thin sections, these linear TCs can be observed in cross-section (tc), revealing structures never revealed by freeze fracture. In this case, a very large cytoplasmic component is imaged just beneath the plasma membrane (pm), and this proves that the typical TC structures revealed by freeze fractures show only ‘the tip of the iceberg’. These observations are consistent with the isolated functional TCs from Gossypium hirsutum (A) and form the basis for the revised model of TC structure/function (see Fig. 3). Note a single cortical microtubule (mt) adjacent to the plasma membrane and the cell wall (cw). (Unpublished micrograph, courtesy of Krystyna Kudlicka and R. Malcolm Brown, Jr.) (C) A multiple fracture through the cytoplasm and inner leaflet of the plasma membrane of an expanding cell in the root tip of Zea mays. This very unusual micrograph reveals the longitudinal fractures through cortical microtubules (mt) which parallel the underlying innermost layer of active microfibril synthesis. (Unpublished micrograph, courtesy of Susette Mueller and R. Malcolm Brown, Jr.) (D) Ultrathin section through the cell wall of the alga, Glaucocystis nostocherinum revealing the ordered arrangement of giant microfibrils (mf) synthesized by linear TCs. The alga synthesizes nearly pure cellulose Iα (Nishiyama et al., 2003). The microfibrils are synthesized in a complex helical pattern over the cell surface to reveal a precise rectangular shape. The microfibrils are coated with non-cellulose materials which stain well with a tannic acid post stain. These microfibrils are proposed to have more than 500 glucan chains per microfibril. While not identical to vascular plant cell walls, the Glaucocystis cell wall is perhaps one of the most beautiful examples to demonstrate the relationship between microfibril deposition and orientation to produce an ellipsoidal single cell. (Unpublished micrograph, courtesy of J. H. Martin Willison and R. Malcolm Brown, Jr.)
Fig. 3.
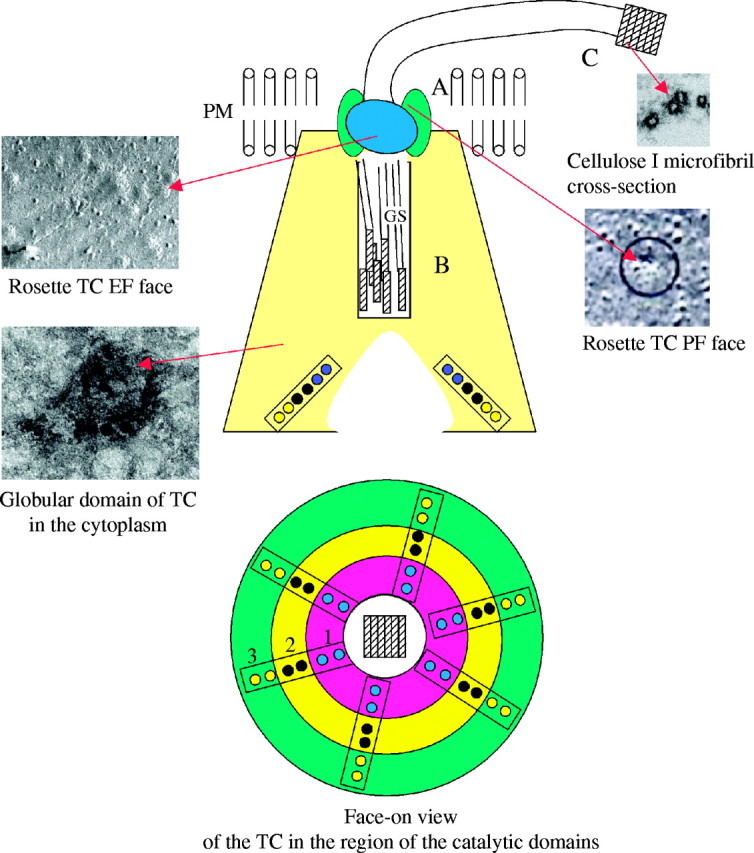
A revised model for the structure and function of the rosette TC in cellulose I microfibril biosynthesis. The 25-nm rosette portion of the TC (A) is shown in green where the six sub-units are largely localized to the innermost leaflet of the plasma membrane. The cytoplasmic portion of the TC is shown in yellow (B) and it contains the globular region of the catalytic sub-units. In this model, two identical sub-units of at least three different gene products form homodimers, all of which are required for cellulose I biosynthesis. Interestingly, the linear rows, each comprised of the three different cellulose synthases, are positioned such that the glucan chains produced by each sub-unit can rapidly associate by van der Waals interactions to produce the first stage of the crystalline cellulose product, namely a glucan chain sheet. Six separate glucan chain sheets are directed into the exit channel of the TC complex (B) where they pass through the rosette aperture and are then H-bonded into the crystalline cellulose I microfibril (C) that passes through this region to the surface of the cell. The face-on view of the cytoplasmic domain shows three different cellulose synthases, indicated as 1, 2 and 3, that are assembled as homodimers and organized in a linear row.
IN VITRO CELLULOSE SYNTHESIS IS ESSENTIAL FOR UNDERSTANDING CELLULOSE SYNTHESIS IN VIVO
To determine the approximate, if not the precise, conditions under which cellulose synthesis takes place in the cell it is important to demonstrate cellulose synthesis in vitro using purified or partially purified cellular components. This is not unlike any other cellular reaction being mimicked in vitro; however, demonstration of in vitro synthesis of cellulose using extracts from plants has been notoriously difficult. Numerous reasons can be cited for the difficulties in determining cellulose synthase activities from plant extracts, not the least of which is the large amount of callose (β-1,3-glucan) being produced under most reaction conditions (Nakashima et al., 2003). Other reasons for failure in determining synthesis of cellulose in vitro from plant extracts could just be that the proper reaction conditions have not yet been determined and the difficulty in characterizing the in vitro cellulose product. At this point it has to be mentioned that in vitro cellulose synthesis has been routinely achieved using membrane preparations and detergent-solubilized proteins (including partially pure preparations) from the bacterium A. xylinum (Lin and Brown, 1989). Whereas the cellulose produced in vitro generally is obtained as cellulose II, cellulose I microfibrils have also been observed under specific conditions. Although in vitro cellulose products using cell-free preparations of A. xylinum were described in 1958 (Glaser, 1958), conditions for obtaining high rates of cellulose synthesis in vitro were not defined until a much later date (Aloni et al., 1982). As we now know, these conditions allowed the formation of the activator c-di-GMP (Ross et al., 1987). By manipulating the use of detergents and reaction conditions, in vitro cellulose synthesis was demonstrated using extracts from cotton fibres (Okuda et al., 1993; Kudlicka et al., 1995; Peng et al., 2002), mung bean (Kudlicka and Brown, 1997), blackberry (Lai-Kee-Him et al., 2002) and cell suspension cultures of hybrid aspen (Colombani et al., 2004). Although callose still makes a large part of the in vitro product, Kudlicka and Brown (1997) were able to separate the callose synthase activity from cellulose synthase activity by native gel electrophoresis. However, no conclusive evidence regarding the similarity or differences between callose synthase activity and cellulose synthase activity could be obtained by analyzing the polypeptide composition in these two fractions. In certain cases, the cellulose I microfibrils obtained in vitro were shown to have dimensions similar to microfibrils obtained from primary cell walls (Lai-Kee-Him et al., 2002), suggesting that synthesis of native cellulose microfibrils can be mimicked in vitro. Interestingly, the same cellulose product (cellulose I) is obtained when cellobiosyl fluoride and cellulase are used in an in vitro reaction (Lee et al., 1994). As with A. xylinum, no unique effector has so far been identified for modulating in vitro cellulose synthase activity in vascular plants (Li and Brown, 1993). Hopefully, identification of this missing link would allow determination of the optimal conditions under which cellulose synthesis occurs not only in vitro but also in plant cells.
POLYMERIZATION AND CRYSTALLIZATION: LINKED STEPS IN THE ASSEMBLY OF CELLULOSE MICROFIBRILS
The parallel arrangement of glucan chains in the cellulose microfibril requires that the newly synthesized glucan chains align with each other and lock into a specific crystalline arrangement (cellulose I), otherwise they would fold into the more thermodynamically stable cellulose II or simply exist as non-crystalline cellulose. The coordinated synthesis of a large number of glucan chains (polymerization) from ordered sites present in the TC allows these glucan chains to be positioned adjacent to each other before crystallization occurs. Whereas polymerization of glucose residues requires the enzyme cellulose synthase and the substrate UDP-glucose, no proteins have been directly implicated in the crystallization process in vascular plants. Indirectly, proteins associated with the organization of the cellulose-synthesizing sites and for the export of the glucan chains across the plasma membrane probably play a role in the crystallization step. That the cellulose synthases do affect crystallization in vascular plants is clear from studies with the rsw1 mutant in arabidopsis, where there is an increase in the synthesis of non-crystalline cellulose under non-permissive conditions (Arioli et al., 1998). In bacteria, specifically A. xylinum, other proteins (BcsC and BcsD) encoded by genes in the cellulose-synthesizing operon have been implicated in the assembly of the glucan chains and thereby affect crystallization indirectly (Saxena et al., 1994). Although crystallization occurs soon after the glucan chains have been extruded from the cell, it does not occur instantaneously. The glucan chains are able to bind to agents such as Calcofluor (or Tinopal) and Congo Red after synthesis but before crystallization, suggesting stages in the process of crystallization (Haigler et al., 1980). Based on their studies with A. xylinum and incorporating results from molecular modelling, Cousins and Brown (1997a, b) have proposed a two-step model for cellulose I crystallization. In the first step, glucan chains assemble as a monomolecular glucan chain sheet using van der Waals forces and, in the next step, the glucan chain sheets assemble via hydrogen bonding to form the crystalline cellulose I microfibril. Although polymerization and crystallization are separate events, they are linked in a manner where each event influences the other. Cellulose is synthesized processively, and the growing end of the glucan chain (the non-reducing end; Koyama et al., 1997) is tightly associated with the catalytic region of cellulose synthase. Polymerization and crystallization are coupled processes in A. xylinum, and the rate of polymerization is influenced by crystallization (Benziman et al., 1980). In A. xylinum, Calcofluor disrupts the crystallization steps by binding to the glucan chain sheets, resulting in an increase in the rate of polymerization. This relationship between polymerization and crystallization may also be important for understanding increased production of non-crystalline cellulose under non-permissive conditions by the rsw1 mutant of arabidopsis (Arioli et al., 1998) and by cotton fibres in the presence of the herbicide CGA 325′615 (Peng et al., 2001). The forces generated by crystallization may be sufficient to release the glucan chain from the cellulose synthase active site during synthesis. Where crystallization of the glucan chains is affected either by a mutation in the cellulose synthase or in the presence of a herbicide such as CGA 325′615, the glucan chains remain tightly attached to the cellulose synthase. Both the mutation (rsw1) and the herbicide (CGA 325′615) result in a defect in the formation of a rosette structure, probably at different steps in the assembly of the rosette.
β-1,4-GLUCAN CHAINS ARE SYNTHESIZED BY CELLULOSE SYNTHASE
The polymerization of glucose residues into a β-1,4-linked backbone is catalysed by the enzyme cellulose synthase, which utilizes UDP-α-glucose as the substrate. In its simplest form, this is a one-step (or direct) polymerization reaction involving glycosyl transfer by inversion of configuration at the anomeric carbon. Moreover, in this type of reaction a single cellulose synthase molecule is capable of initiating, elongating and terminating a β-1,4-linked glucan chain. This mechanism implies that cellulose synthase binds directly to the substrate UDP-glucose and is capable of initiating synthesis without the requirement of a primer. Moreover, the enzyme is a processive enzyme and remains attached to the growing end without the need to attach and detach during synthesis. That cellulose synthase does indeed perform a one-step polymerization reaction is observed in vitro using cell-free extracts from A. xylinum, Agrobacterium tumefaciens, and a number of plant species. Since cellulose synthase is an integral membrane protein, in most cases these extracts are either membrane preparations, detergent-solubilized fractions, or partially purified proteins in solution or on a polyacrylamide gel. In a majority of these cases, the cellulose synthase is present with other proteins in the reaction mixture; however, relatively pure preparations of cellulose synthase from A. xylinum have also been used to demonstrate cellulose synthesis in a one-step reaction in vitro. Growth of the glucan chain in cellulose occurs at the non-reducing end by direct transfer of glucose from UDP-glucose (Koyama et al., 1997). Synthesis of β-glycan chains in a number of other polysaccharides, including chitin and hyaluronan, also occurs by direct transfer of sugar from a nucleotide sugar donor to the non-reducing end, and no requirement for a primer has been observed. More recently, it has been observed that hyaluronan synthase obtained from Xenopus laevis extends the hyaluronan chain from the non-reducing end while the enzyme obtained from Streptococcus pyogenes extends it from the reducing end (Bodevin-Authelet et al., 2005).
MULTIPLE STEPS IN POLYMERIZATION TO FORM β-1,4-GLUCAN CHAINS OF CELLULOSE?
That cellulose may be synthesized in multiple steps (indirect mechanism) was initially proposed by Matthysse et al. (1995a) based upon analysis of cellulose-minus mutants in A. tumefaciens. In this proposal, cellulose is synthesized through steps involving lipid intermediates and both cellulose synthase and a cellulase (an endoglucanase) is suggested to play a role in cellulose biosynthesis. Interestingly, a gene encoding the endoglucanase is present in the cellulose synthase-encoding operon in A. tumefaciens (Matthysse et al., 1995b) and a similar endoglucanase has now been observed in cellulose-synthesizing operons in a number of other bacterial species (Römling, 2002). In A. xylinum, an endoglucanase coding region is not present within the cellulose-synthesizing operon but is found adjacent to this operon, and the endoglucanase is produced as a soluble protein (Standal et al., 1994). In vitro cellulose synthesis clearly has been demonstrated using membrane proteins from A. xylinum and this rules out any role for this endoglucanase during in vitro synthesis. Whether this or any other endoglucanase may have a role during in vivo cellulose biosynthesis in A. xylinum remains to be determined. Membrane-anchored endoglucanases have also been identified in plants (Brummell et al., 1997), and mutations in some of the cellulose-deficient mutants of arabidopsis were mapped to a gene encoding a membrane-bound endoglucanase, commonly referred to as KORRIGAN (Nicol et al., 1998; Zuo et al., 2000; Lane et al., 2001; Sato et al., 2001). For some time, the endoglucanases identified in bacteria and plants were predicted to function as ‘editor/chain terminator’ during in vivo cellulose biosynthesis (Delmer, 1999) until Peng et al. (2002) proposed a model where the membrane-bound endoglucanase KORRIGAN was implicated during cellulose biosynthesis in plants. This multi-step model proposes that, in plants, sitosterol-β-glucoside (SG) serves as a primer for synthesis of sitosterol-cellodextrins (SCDs) by cellulose synthase on the cytoplasmic side of the plasma membrane. The SCDs are flipped by an unknown mechanism to the outer side of the plasma membrane where the endoglucanase KORRIGAN cleaves SCDs giving rise to SG and cellodextrins (CDs). In the next step, the CDs undergo β-1,4-glucan chain elongation catalysed by cellulose synthase proteins. This model envisages a lipid primer and a number of protein components during cellulose biosynthesis. However, evidence from in vitro cellulose synthesis using solubilized proteins from plant membranes does not support the requirement of a primer or any lipid intermediates during cellulose biosynthesis in plants (Okuda et al., 1993; Kudlicka and Brown, 1997; Lai-Kee-Him et al., 2002; Colombani et al., 2004; Somerville et al., 2004). More recently, no differences were found in the amounts of SG and SCDs in extracts of wild-type and a KORRIGAN mutant (kor1-1) of arabidopsis suggesting that the KORRIGAN endoglucanase is not involved in the recycling of the SG primer (Robert et al., 2004). Although it is difficult to provide evidence for a direct role for KORRIGAN during the polymerization step of cellulose biosynthesis, it probably affects cellulose biosynthesis indirectly during cell plate formation, cell elongation and secondary wall deposition.
BIOSYNTHESIS OF CELLULOSE IS REGULATED POST-TRANSCRIPTIONALLY
Cellulose synthase activity in A. xylinum is regulated by the allosteric activator c-di-GMP (Ross et al., 1987). This compound has also been found to be an activator of cellulose synthase activity in other bacteria, including Escherichia coli (I. M. Saxena and R. M. Brown, Jr, unpubl. res.) and A. tumefaciens (Amikam and Benziman, 1989). Genes regulating the synthesis and degradation of this novel nucleotide regulator have now been identified in a large number of bacterial species, and it appears that c-di-GMP may be involved in regulating the activity of many more pathways (Garcia et al., 2004; Paul et al., 2004; Simm et al., 2004; Tischler and Camilli, 2004). In most bacteria, genes for cellulose synthase exhibit a constitutive expression (Römling, 2002) and, although plants contain a large number of CesA genes, many of these genes are expressed throughout the plant, suggesting that regulation of cellulose synthesis in plants occurs post-transcriptionally as well (Somerville et al., 2004). As mentioned earlier, no unique regulator has been identified for modulating cellulose synthase activity in plants. On the other hand, given the large number of CesA genes in most plants, control of cellulose microfibril assembly (not necessarily polymerization) may be exercised by the interaction of different CesA sub-units in a specific orientation. In bacteria, where a single functional cellulose synthase is sufficient to form cellulose microfibrils (Saxena and Brown, 1995), an interaction between different cellulose synthase molecules makes no sense for regulating the enzyme activity.
QUESTIONS REMAIN FOR THE STRUCTURE OF CELLULOSE SYNTHASE AS WELL AS THE MECHANISM OF POLYMERIZATION TO EXPLAIN A TWO-FOLD SYMMETRY IN THE β-1,4-GLUCAN CHAINS OF CELLULOSE
About 10 years ago, a model was proposed to explain the mechanism by which a two-fold symmetry could be obtained in the β-1,4-glucan chains of cellulose (Saxena et al., 1995). In simple terms, the model predicted two binding sites for UDP-glucose molecules in the catalytic channel of the cellulose synthase and suggested simultaneous or sequential addition of the glucose residues to the growing end of a glucan chain. In this model, two UDP-glucose molecules were positioned such that, upon addition to the growing end of the glucan chain, each glucose residue was inverted 180° with respect to its neighbouring residues. The model essentially described a mechanism to obtain a two-fold symmetry in the glucan chain using two catalytic centres within a single enzyme molecule. This model has been widely debated, and it has even been suggested that the two-fold symmetry can be obtained from a single catalytic centre as there is a fairly large degree of freedom of rotation about the β-glycosidic bond (Delmer, 1999). According to this proposal, the glucose residue added in one orientation relaxes into the native orientation after polymerization (Delmer, 1999). Other proposals have suggested that two catalytic centres may be present in two sub-units and be organized following dimerization or two different catalytic domains within the same catalytic site participate in the dual addition (Albersheim et al., 1997; Charnock et al., 2001). Cellulose synthase and other processive β-glycosyltransferases have so far resisted crystal structure determination, although the structure of a non-processive β-glycosyltransferase (SpsA from Bacillus subtilis) has been determined (Charnock and Davies, 1999). The SpsA protein lacks the conserved QXXRW motif found in the processive enzymes, and mutation analysis has indicated a role of this motif at some step in the synthesis of cellulose (Saxena et al., 2001). The structure of the globular region of the A. xylinum cellulose synthase containing all the conserved aspartic acid residues and the QXXRW motif was predicted using the genetic algorithm (Saxena et al., 2001). Based on structural and functional criteria, the location and putative functions were assigned to the conserved residues and the QXXRW motif during glucan chain polymerization. More specifically, the tryptophan residue in the QXXRW motif was suggested to be involved in the glucan chain binding. However, in the absence of a crystal structure of a protein with the conserved aspartic residues and the QXXRW motif, their exact role in the synthesis of cellulose and other β-linked polysaccharides remains to be determined.
DEPOSITION OF CELLULOSE MICROFIBRILS INFLUENCE THE DIRECTION OF PLANT CELL GROWTH: WHAT FACTORS DETERMINE DIRECTION OF MICROFIBRIL GROWTH?
Growth and development in plants follows a certain pattern dictated not only by the genes but also by a number of internal and external cues. Given the internal and external cues, how do plants determine the direction of growth? Growth is defined as an irreversible increase in volume and results from cell division and cell elongation. The direction in which growth occurs in turn is determined by the plane of cell division and the axis along which cell elongation takes place. The general role ascribed to cellulose in the cell walls of plants is to provide the necessary strength to resist the turgor pressure. However, at the cellular level, cellulose has a distinct role in maintaining the size, shape and division/differentiation potential of most plant cells. Are the signals for growth and differentiation transmitted to the cellulose-synthesizing machinery and if it is so, what are these signals and how are they sensed by the cellulose-synthesizing machinery? As these questions are considered, we have to be mindful of the role that the cellulose product may exercise in the direction and quantity in which it is incorporated in the walls of plant cells. Directional growth occurs as a result of anisotropy in the underlying cells and in plant cells it is believed to result from a directional synthesis of cellulose around the cells. Cell elongation, therefore, is presumed to occur in a direction perpendicular to the direction of synthesis of the cellulose microfibrils. A number of cellular components other than cellulose must be involved in determining the direction of cell elongation, and a common objective in a number of investigations is to identify and determine the role of these interacting components. A major component that is implicated in all this is the microtubule (Fig. 2C and D). Many explanations for the role of microtubules in determining the direction of cellulose synthesis can be found in the literature. The general view so far is that microtubules play a key role in determining the direction of microfibril growth by providing guide channels for setting up the direction of initial microfibril synthesis and also membrane flow within these channels (Mueller and Brown, 1982a, b; Giddings and Staehelin, 1991). However, recent views of microtubule/microfibril interaction suggest a reversal in the role of orientation and that cellulose microfibrils may provide cues for the orientation of the cortical microtubules (Akashi and Shibaoka, 1991; Fisher and Cyr, 1998). In an extreme case, where microtubule assembly was disrupted by a temperature-sensitive mutation (mor1-1 mutant of arabidopsis) or the drug oryzalin, it was found that cellulose microfibrils were able to self-align in the presence of adequate cellulose synthesis (Sugimoto et al., 2003). Unpublished work with freeze etch of colchicine-treated cotton fibres undergoing secondary wall formation reveals that while the microtubules are no longer present, the original bands of microfibrils formed within the channels delimited by microtubules still remain aligned (K. Okuda and R. M. Brown, Jr, unpubl. res.).
CONCLUSIONS
A goal in many investigations is to derive or approximate general or unifying principles. The same may be true with studies of cellulose biosynthesis in various organisms. Similarities have been found in the sequence of cellulose synthases obtained from different organisms and, so far, a clear relationship is observed between these sequences (Nobles and Brown, 2004). Even though specific features are found in cellulose synthases from different organisms, it is believed that the catalytic region is conserved in all these cellulose synthases. This suggests that the different cellulose synthases catalyse synthesis of β-1,4-glucan chains in a similar manner. Furthermore, the requirement for specific cellulose microfibril dimensions in the life or growth stage of each organism has allowed selection of cellulose-synthesizing complexes and their specific arrangements. Once again the organization of these complexes follows a pattern (Roberts and Roberts, 2004). However, at this point it is clear that the paradigm of cellulose biosynthesis as exemplified in A. xylinum may not be sufficient to account for the vastly increased complexity of cellulose biosynthesis observed in vascular plants. Although a number of basic principles for cellulose synthesis are universal, requirements for cellulose synthesis are very different in A. xylinum and plants.
Acknowledgments
The authors would like to acknowledge support from the Division of Energy Biosciences, Department of Energy (Grant DE-FG03-94ER20145) and the Welch Foundation (Grant F-1217).
LITERATURE CITED
- Akashi T, Shibaoka. 1991. Involvement of transmembrane proteins in the association of cortical microtubules with the plasma membrane in tobacco BY-2 cells. Journal of Cell Science 98: 169–174. [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Staehelin LA, Varner JE. 1997. Do the structures of cell wall polysaccharides define their mode of synthesis? Plant Physiology 113: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y, Delmer DP, Benziman M. 1982. Achievement of high rates of in vitro synthesis of 1,4-β-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proceedings of the National Academy of Sciences of the USA 79: 6448–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikam D, Benziman M. 1989. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens Journal of Bacteriology 177: 6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller L, Doblin M, Barreiro R, Wang H, Niu X, Kollipara K, et al. 2004. Cellulose synthesis in maize: isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 11: 287–299. [Google Scholar]
- Arioli T, Peng L, Betzner AS, Burn J, Wittke W, Herth W, et al. 1998. Molecular analysis of cellulose biosynthesis in Arabidopsis Science 279: 717–720. [DOI] [PubMed] [Google Scholar]
- Attala RH, VanderHart DL. 1984. Native cellulose: a composite of two distinct crystalline forms. Science 223: 283–285. [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP. 1998. Polymerases and the replisome: machines within machines. Cell 92: 295–305. [DOI] [PubMed] [Google Scholar]
- Benziman M, Haigler CH, Brown Jr RM, White AR, Cooper KM. 1980. Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum Proceedings of the National Academy of Sciences of the USA 77: 6678–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodevin-Authelet S, Kusche-Gullberg M, Pummill P, DeAngelis P, Lindahl U. 2005. Biosynthesis of hyaluronan: direction of chain elongation. Journal of Biological Chemistry 280: 8813–8818. [DOI] [PubMed] [Google Scholar]
- Brown Jr RM. 1985. Cellulose microfibril assembly and orientation: recent developments. Journal of Cell Science Supplement 2: 13–32. [DOI] [PubMed] [Google Scholar]
- Brown Jr RM. 1996. The biosynthesis of cellulose. Journal of Macromolecular Science —Pure and Applied Chemistry A33: 1345–1373. [Google Scholar]
- Brown Jr RM. 2004. Cellulose structure and biosynthesis: what is in store for the 21st century? Journal of Polymer Science. Part A. Polymer Chemistry 42: 487–495. [Google Scholar]
- Brown Jr RM, Montezinos D. 1976. Cellulose microfibrils: visualization of biosynthetic and orienting complexes in association with the plasma membrane. Proceedings of the National Academy of Sciences of the USA 73: 143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Catala C, Lashbrook CC, Bennett AB. 1997. A membrane-anchored E-type endo-1,4-beta-glucanase is localized on Golgi and plasma membranes of higher plants. Proceedings of the National Academy of Sciences of the USA 94: 4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. 2002. Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiology 129: 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Davies GJ. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry 38: 6380–6385. [DOI] [PubMed] [Google Scholar]
- Charnock SJ, Henrissat B, Davies GJ. 2001. Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiology 125: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani A, Djerbi S, Bessueille L, Blomqvist K, Ohlsson A, Berglund T, Teeri TT, Bulone V. 2004.In vitro synthesis of (1→3)-β-D-glucan (callose) and cellulose by detergent extracts of membranes from cell suspension cultures of hybrid aspen. Cellulose 11: 313–327. [Google Scholar]
- Cousins SK, Brown Jr RM. 1995. Cellulose I microfibril assembly: computational molecular mechanics energy analysis favours bonding by van der Waals forces as the initial step in crystallization. Polymer 36: 3885–3888. [Google Scholar]
- Cousins SK, Brown Jr RM. 1997. X-ray diffraction and ultrastructural analyses of dye-altered celluloses support van der Waals forces as the initial step in cellulose crystallization. Polymer 38: 897–902. [Google Scholar]
- Cousins SK, Brown Jr RM. 1997. Photoisomerization of a dye-altered β-1, 4 glucan sheet induces the crystallization of a cellulose-composite. Polymer 38: 903–912. [Google Scholar]
- Delmer DP. 1999. Cellulose biosynthesis: exciting times for a difficult field of study. Annual Reviews of Plant Physiology and Plant Molecular Biology 50: 245–276. [DOI] [PubMed] [Google Scholar]
- Djerbi S, Aspeborg H, Nilsson P, Sundberg B, Mellerowicz E, Blomqvist K, et al. 2004. Identification and expression analysis of genes encoding putative cellulose synthases (CesA) in the hybrid aspen, Populus tremula (L.)×P. tremuloides (Michx.). Cellulose 11: 301–312. [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. 2002. Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiology 43: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, et al. 2000. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12: 2409–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DD, Cyr RJ. 1998. Extending the microtubule/microfibril paradigm. Plant Physiology 116: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B, Latasa C, Solano C, Portillo FG, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Molecular Microbiology 54: 264–277. [DOI] [PubMed] [Google Scholar]
- Giddings Jr TH, Staehelin LA. 1991. Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In: Lloyd CW, ed. The cytoskeletal basis of plant growth and form. London: Academic Press, 85–99. [Google Scholar]
- Giddings Jr TH, Brower DL, Staehelin LA. 1980. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. Journal of Cell Biology 84: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L. 1958. The synthesis of cellulose in cell-free extracts of Acetobacter xylinum Journal of Biological Chemistry 232: 627–636. [PubMed] [Google Scholar]
- Grimson, MJ, Haigler CH, Blanton, RL. 1996. Cellulose microfibrils, cell motility, and plasma membrane protein organization change in parallel during culmination in Dictyostelium discoideum Journal of Cell Science 109: 3079–3087. [DOI] [PubMed] [Google Scholar]
- Ha M-A, Apperley DC, Evans BW, Huxham IM, Jardine WG, Viëtor RJ, et al. 1998. Fine structure in cellulose microfibrils: NMR evidence from onion and quince. The Plant Journal 16: 183–190. [DOI] [PubMed] [Google Scholar]
- Haigler CH, Brown Jr RM. 1986. Transport of rosettes from the Golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma 134: 111–120. [Google Scholar]
- Haigler CH, Brown Jr RM, Benziman M. 1980. Calcofluor White ST alters the in vivo assembly of cellulose microfibrils. Science 210: 903–906. [DOI] [PubMed] [Google Scholar]
- Hamann T, Osborne E, Youngs HL, Misson J, Nussaume L, Somerville C. 2004. Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11: 279–286. [Google Scholar]
- Herth W. 1983. Arrays of plasma-membrane ‘rosettes’ involved in cellulose microfibril formation of Spirogyra Planta 159: 347–356. [DOI] [PubMed] [Google Scholar]
- Holland N, Holland D, Helentjaris T, Dhugga KS, Xoconostle-Cazares B, Delmer DP. 2000. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiology 123: 1313–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Brown Jr RM. 1984. The assembly of cellulose microfibrils in Valonia macrophysa Kutz. Planta 160: 372–381. [DOI] [PubMed] [Google Scholar]
- Itoh T, Brown Jr RM. 1988. Development of cellulose synthesizing complexes in Boergesenia and Valonia Protoplasma 144: 160–169. [Google Scholar]
- Jarvis M. 2003. Cellulose stacks up. Nature 426: 611–612. [DOI] [PubMed] [Google Scholar]
- Kiermayer O, Sleytr UB. 1979. Hexagonally ordered ‘rosettes’ of particles in the plasma membrane of Micrasterias denticulata Bréb. and their significance for microfibril formation and orientation. Protoplasma 101: 133–138. [Google Scholar]
- Kimura S, Itoh T. 1996. New cellulose synthesizing complexes (terminal complexes) involved in animal cellulose biosynthesis in the tunicate Metandrocarpa uedai Protoplasma 194: 151–163. [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui X, Linder CR, Brown Jr RM. 1999. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis Plant Cell 11: 2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Shoda S. 1995. Chemical synthesis of cellulose and cello-oligomers using a hydrolysis enzyme as a catalyst. International Journal of Biological Macromolecules 17: 373–379. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Kashiwa K, Kawasaki T, Shoda S. 1991. Novel method for polysaccharide synthesis using an enzyme: the first in vitro synthesis of cellulose via a nonbiosynthetic path utilizing cellulase as catalyst. Journal of the American Chemical Society 113: 3079–3084. [Google Scholar]
- Kondo T, Togawa E, Brown Jr RM. 2001. Nematic ordered cellulose: a concept of glucan chain association. Biomacromolecules 2: 1324–1330. [DOI] [PubMed] [Google Scholar]
- Koyama M, Helbert W, Imai R, Sugiyama J, Henrissat B. 1997. Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proceedings of the National Academy of Sciences of the USA 94: 9091–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Brown Jr RM. 1997. Cellulose and callose biosynthesis in higher plants. I. Solubilization and separation of (1→3)- and (1→4)-β-glucan synthase activities from mung bean. Plant Physiology 115: 643–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Brown Jr RM, Li L, Lee JH, Shin H, Kuga S. 1995. β-glucan synthesis in the cotton fiber. IV. In vitro assembly of the cellulose I allomorph. Plant Physiology 107: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicka K, Wardrop A, Itoh T, Brown Jr RM. 1987. Further evidence from sectioned material in support of the existence of a linear terminal complex in cellulose synthesis. Protoplasma 136: 96–103. [Google Scholar]
- Kuga S, Takagi S, Brown Jr RM. 1993. Native folded-chain cellulose II. Polymer 34: 3293–3297. [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. 2002. Dimerization of cotton fiber cellulose synthase catalytic sub-units occurs via oxidation of the zinc-binding domains. Proceedings of the National Academy of Sciences of the USA 99: 11109–11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Kee-Him J, Chanzy H, Müller M, Putaux J-L, Imai T, Bulone V. 2002.In vitro versus in vivo cellulose microfibrils from plant primary wall synthases: structural differences. Journal of Biological Chemistry 277: 36931–36939. [DOI] [PubMed] [Google Scholar]
- Lane DR, Wiedemeier A, Peng L, Höfte H, Vernhettes S, Desprez T, et al. 2001. Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiology 126: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Brown Jr RM, Kuga S, Shoda S, Kobayashi S. 1994. Assembly of synthetic cellulose I. Proceedings of the National Academy of Sciences of the USA 91: 7425–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Brown Jr RM. 1993. β-Glucan synthesis in the cotton fiber. II. Regulation and kinetic properties of β-glucan synthases. Plant Physiology 101: 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FC, Brown Jr RM. 1989. Purification of cellulose synthase from Acetobacter xylinum In: Schuerch C, ed. Cellulose and wood—chemistry and technology. New York: John Wiley and Sons, 473–492. [Google Scholar]
- Matthysse AG, Thomas DL, White AR. 1995. Mechanism of cellulose synthesis in Agrobacterium tumefaciens Journal of Bacteriology 177: 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse AG, White S, Lightfoot R. 1995. Genes required for cellulose synthesis in Agrobacterium tumefaciens Journal of Bacteriology 177: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta S, Brown Jr RM. 1992. High resolution analysis of the formation of cellulose synthesizing complexes in Vaucheria hamata Protoplasma 166: 187–199. [Google Scholar]
- Mueller SC, Brown Jr RM. 1980. Evidence for an intramembrane component associated with a cellulose microfibril synthesizing complex in higher plants. Journal of Cell Biology 84: 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Brown Jr RM. 1982. The control of cellulose microfibril deposition in the cell wall of higher plants. I. Planta 154: 489–500. [DOI] [PubMed] [Google Scholar]
- Mueller SC, Brown Jr RM. 1982. The control of cellulose microfibril deposition in the cell wall of higher plants. II. Planta 154: 501–515. [DOI] [PubMed] [Google Scholar]
- Nakashima J, Laosinchai W, Cui X, Brown Jr RM. 2003. New insight into the mechanism of cellulose and callose biosynthesis: proteases may regulate callose biosynthesis upon wounding. Cellulose 10: 369–389. [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. 1998. A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO Journal 17: 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Sugiyama J, Chanzy H, Langan P. 2003. Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. Journal of the American Chemical Society 125: 14300–14306. [DOI] [PubMed] [Google Scholar]
- Nobles Jr DR, Brown Jr RM. 2004. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose 11: 437–448. [Google Scholar]
- Okuda K. 2002. Structure and phylogeny of cell coverings. Journal of Plant Research 115: 283–288. [DOI] [PubMed] [Google Scholar]
- Okuda K, Li L, Kudlicka K, Kuga S, Brown Jr RM. 1993. β-Glucan synthesis in the cotton fiber. I. Identitification of β-1,4- and β-1,3-glucans synthesized in vitro Plant Physiology 101: 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Sekida S, Yoshinaga S, Suetomo Y. 2004. Cellulose-synthesizing complexes in some chromophyte algae. Cellulose 11: 365–376. [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, et al. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes and Development 18: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. 1996. Higher plants contain homologs of the bacterial celA genes encoding the catalytic sub-unit of cellulose synthase. Proceedings of the National Academy of Sciences of the USA 93: 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D. 2002. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295: 147–150. [DOI] [PubMed] [Google Scholar]
- Peng L, Xiang F, Roberts E, Kawagoe Y, Greve LC, Kreuz K, et al. 2001. The experimental herbicide CGA 325′615 inhibits synthesis of crystalline cellulose and causes accumulation of non-crystalline β-1,4-glucan associated with CesA protein. Plant Physiology 126: 981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM. 2001. Cellulose: how many cellulose synthases to make a plant? Current Biology 11: R213–R216. [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. 2000. The cellulose synthase superfamily. Plant Physiology 124: 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Mouille G, Höfte H. 2004. The mechanism and regulation of cellulose synthesis in primary walls: lessons from cellulose-deficient Arabidopsis mutants. Cellulose 11: 351–364. [Google Scholar]
- Roberts AW, Roberts E. 2004. Cellulose synthase (CesA) genes in algae and seedless plants. Cellulose 11: 419–435. [Google Scholar]
- Römling U. 2002. Molecular biology of cellulose production in bacteria. Research in Microbiology 153: 205–212. [DOI] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281. [DOI] [PubMed] [Google Scholar]
- Sato S, Kato T, Kakegawa K, Ishii T, Liu YG, Awano T, et al. 2001. Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana Plant Cell Physiology 42: 251–263. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown Jr RM. 1995. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum Journal of Bacteriology 177: 5276–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Brown Jr RM, Dandekar T. 2001. Structure–function characterization of cellulose synthase: relationship to other glycosyltransferases. Phytochemistry 57: 1135–1148. [DOI] [PubMed] [Google Scholar]
- Saxena IM, Brown Jr RM, Fevre M, Geremia RA, Henrissat B. 1995. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. Journal of Bacteriology 177: 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Kudlicka K, Okuda K, Brown Jr RM. 1994. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. Journal of Bacteriology 176: 5735–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Lin FC, Brown Jr RM. 1990. Cloning and sequencing of the cellulose synthase catalytic sub-unit gene of Acetobacter xylinum Plant Molecular Biology 15: 673–683. [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Eshed R, Richmond T, Delmer D, Somerville C. 2001. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proceedings of the National Academy of Sciences of the USA 98: 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Molecular Microbiology 53: 1123–1134. [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, et al. 2004. Towards a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Standal R, Iversen TG, Coucheron DH, Fjaervik E, Blatny JM, Valla S. 1994. A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. Journal of Bacteriology 176: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Himmelspach R, Williamson RE, Wasteneys GO. 2003. Mutation or drug-dependent microtubule disruption causes radial swelling without altering parallel cellulose microfibril deposition in Arabidopsis root cells. Plant Cell 15: 1414–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama J, Harada H, Fujiyoshi Y, Uyeda N. 1985. Lattice images from ultrathin sections of cellulose microfibrils in the cell wall of Valonia macrophysa Kutz. Planta 166: 161–168. [DOI] [PubMed] [Google Scholar]
- Sugiyama J, Persson J, Chanzy H. 1991. Combined infrared and electron diffraction study of the polymorphism of native celluloses. Macromolecules 24: 2461–2466. [Google Scholar]
- Tanaka K, Murata K, Yamazaki M, Onosato K, Miyao A, Hirochika H. 2003. Three distinct rice cellulose synthase catalytic sub-unit genes required for cellulose synthesis in the secondary wall. Plant Physiology 133: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. 2003. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proceedings of the National Academy of Sciences of the USA 100: 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Molecular Microbiology 53: 857–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsekos I. 1999. The sites of cellulose synthesis in algae: diversity and evolution of cellulose-synthesizing enzyme complexes. Journal of Phycology 35: 635–655. [Google Scholar]
- Tsekos I, Reiss HD. 1992. Occurrence of the putative microfibril-synthesizing complexes (linear terminal complexes) in the plasma membrane of the epiphytic marine red alga Erythrocladia subintegra Rosenv. Protoplasma 169: 57–67. [Google Scholar]
- Tsekos I, Okuda K, Brown Jr RM. 1996. The formation and development of cellulose-synthesizing linear terminal complexes (TCs) in the plasma membrane of the marine red alga Erythrocladia subintegra Rosenv. Protoplasma 193: 33–45. [Google Scholar]
- Tsekos I, Orologas N, Herth W. 1999. Cellulose microfibril assembly and orientation in some bangiophyte red algae: relationship between synthesizing terminal complexes and microfibril structure, shape, and dimensions. Phycologia 38: 217–224. [Google Scholar]
- Willison JHM, Brown Jr RM. 1978. Cell wall structure and deposition in Glaucocystis Journal of Cell Biology 77: 103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HC, Fear AL, Calhoon RD, Eichinger GH, Mayer R, Amikam D, et al. 1990. Genetic organization of the cellulose synthase operon in Acetobacter xylinum Proceedings of the National Academy of Sciences of the USA 87: 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH. 2000. KORRIGAN, an Arabidopsis endo-1,4-beta-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]


