Abstract
• Background and Aims The long-lived and mainly outcrossing species Sarracenia purpurea has been introduced into Switzerland and become invasive. This creates the opportunity to study reactions to founder effect and how a species can circumvent deleterious effects of bottlenecks such as reduced genetic diversity, inbreeding and extinction through mutational meltdown, to emerge as a highly invasive plant.
• Methods A population genetic survey by random amplified polymorphism DNA markers (RAPD) together with historical insights and a field pollination experiment were carried out.
• Key Results At the regional scale, S. purpurea shows low structure (θst = 0·072) due to a recent founder event and important subsequent growth. Nevertheless, multivariate statistical analyses reveal that, because of a bottleneck that shifted allele frequencies, most of the variability is independent among populations. In one population (Tenasses) the species has become invasive and genetic analysis reveals restricted gene flow and family structure (θst = 0·287). Although inbreeding appears to be high (Fis > 0·410 from a Bayesian estimation), a field pollination experiment failed to detect significant inbreeding depression upon F1 seed number and seed weight fitness-traits. Furthermore, crosses between unrelated individuals produced F1 seeds with significantly reduced fitness, thus showing local outbreeding depression.
• Conclusions The results suggest that, under restricted gene flow among families, the species may not only have rapidly purged deleterious alleles, but also have undergone some form of selection for inbreeding due to co-adaptation between loci.
Keywords: Bottleneck, founder effect, introduced populations, Sarracenia purpurea, invasion, RAPD, multivariate analysis, field pollination, family structure, bi-parental purge, outbreeding depression
INTRODUCTION
Colonization often involves marked founder effects, which have an important genetic impact (Barret, 1996; Cain et al., 2000). Especially in the case of anthropogenic introduction into new areas, species populations are often founded from only a few individuals that are completely isolated from source populations and thus go through a period of considerable genetic drift. Although reduced interspecific competition and absence of co-evolved parasites are often invoked to explain the success of introduced weedy species by allowing resource re-allocation away from defence against native specialist enemies to plant size and fecundity (Kingston and Waldren, 2003), mounting evidence supports the importance of genetic attributes and rapid evolutionary events in the success of invasion (Lee, 2002; Maron et al., 2004). In that context, the reciprocal influence of genetic diversity and breeding system is of crucial importance to understanding species evolution (Holsinger, 2000). Such introduction events offer hopeful model systems for population studies on micro-evolutionary processes associated with long-distance dispersal colonization and the role of bottlenecks in evolution. They contribute to a better understanding of the invasion process as well as of the impact of natural range shifts due to climate change (e.g. Hewitt, 2000).
Genetic drift affects the allele frequency of finite populations, especially those that passed through a bottleneck or that have been founded by only a few individuals. Such populations present a relatively low genetic diversity (e.g. Leberg, 1992). Furthermore, genetic variability in such populations is a biased sample of the originating population, so that alleles, especially rare ones, are lost and heterozygosity is lowered. By increasing reproduction between relatives, bottlenecks further increase homozygosity and can thus induce inbreeding depression. This decrease of population fitness is often thought to be the consequence of the increased probability that recessive deleterious alleles are expressed in an homozygous state (Charlesworth and Charlesworth, 1999). Therefore, most species going through a bottleneck present a higher extinction risk, because of the mutational meltdown process whereby genetic diversity loss leads to inbreeding depression, which in turn leads to further genetic diversity loss and mutation fixation, and so forth (Ellestrand and Elam, 1993). An attractive hypothesis to explain how inbred populations can escape mutational meltdown is the purging of deleterious mutations (Wang et al., 1999). The probability that recessive deleterious mutations are in homozygous states is proportional to inbreeding, thereby exposing deleterious alleles to selection and leading to their progressive elimination from the population (Byers and Waller, 1999). In that context, reproduction by selfing can be viewed as adaptive.
Furthermore, bottleneck and foundation are not only able to reduce genetic variability and genetic additive variance, but can also rearrange it, possibly with adaptive changes (Carson, 1990). Since the genome is understood as a network where interaction among loci (especially epistasis) is important, a bottleneck potentially provides the opportunity for great change without great genetic variability or mutation (Carson and Templeton, 1984). In panmictic populations, loci are balanced and co-adapted in order to maximize individual fitness. If these interactions are modified by changes in the allele frequency and intense recombination, natural selection has the opportunity to reorganize genetic co-adaptation (selection realignment; Carson, 1989). Such a transformation of epistasis variance into additive variance can induce important modifications in populations by providing a new substrate for selection (e.g. Husband and Barrett, 1992).
SARRACENIA PURPUREA L.
The genus Sarracenia L. belongs to the carnivorous family Sarraceniaceae, which is thought to have had an interesting radiation and comprises nine well-recognized species originating in North America (McDaniel, 1971; Bayer et al., 1996). Sarracenia purpurea is the only species of the genus not restricted to the south of the continent, probably because of its preference for cool conditions and its outstanding tolerance to frost. This species has expanded its range, a phenomenon probably linked to post-glacial recolonization (Juniper et al., 1989) and is present naturally along the whole Atlantic seaboard of North America, stretching from Florida to the boreal zone. Sarracenia purpurea exhibits considerable morphological variability throughout its distribution and the taxon has been split into various subspecies and varieties. Godt and Hamrick (1999) have recently made an allozyme survey of the infra-specific subdivisions and concluded, as did McDaniel (1971), that only two subspecies are well supported by combined molecular and morphological data: a southern pubescent taxon, S. purpurea subsp. venosa (Raf.) Wherry and a northern glabrous taxon, S. purpurea subsp. purpurea [= S. purpurea subsp. gibbosa (Raf.) Wherry].
Sarracenia purpurea is a long-lived perennial producing a strong rhizome that allows the species to spread horizontally. Even though its life cycle has not been studied precisely, Schwaegerle and Schaal (1979) reported from eight to 15 generations in 70 years. A demographic investigation of the Swiss populations led to an approximation of a lifetime of 20 years for an individual and showed that reproduction first happens after only 3 years, while maximal flowering occurs after 10 years (P. Ecoffey, University of Lausanne, Switzerland, 1995, unpubl. res.).
The flower is protandrous and presents an unusual sophisticated morphology: the pistil has an umbrella shape with the stigma at the angles, tightly enclosed by the bracts, petaloid sepals and petals. This specialized system is thought to be an adaptation promoting allogamy by secondary pollen presentation (Schnell, 1976; Bayer et al., 1996). Although Thomas and Duncan (1986) reported from field experiments that the species seems mostly outcrossing, morphological specialization of the flower can only be viewed as a leaky mechanism to prevent autogamy; at least, selfing by geitonogamy is unavoidable. Nevertheless, self-compatibility in S. purpurea does not seem to have been investigated so far. Flowering is synchronous among individuals of the same population. Seeds measure about 2 mm and present vestigial lateral wings. Seed dispersal is thus not very efficient and long-distance dispersal could only be achieved by hydrochory (Bayer et al., 1996).
Because of its carnivorous biology and showy morphology, S. purpurea has often been transplanted into previously unoccupied stations. Such introductions, when documented, are promising opportunities to study micro-evolutionary processes. This is precisely the reason why S. purpurea was one of the first plant species to be studied empirically in the framework of the evolutionary consequences of a bottleneck (Schwaegerle and Schaal, 1979). In that study, an American population, artificially founded by a single individual 75 years before, was shown by an allozyme survey to harbour 2·5 times less genetic diversity. Taggart et al. (1990) also studied this species in this framework after it was introduced outside of its native range into Ireland. Using the same loci as Schwaegerle and Schaal (1979), they studied a population first introduced in 1906, as well as populations founded from this stock. This study, where historical documentation is available and coupled with genetic data, offers the opportunity to estimate genetic structure in different bottleneck situations (various numbers of founders and time spans since a bottleneck, see Discussion). Furthermore, neither Schwaegerle and Schaal (1979) nor Taggart et al. (1990) found any significant heterozygote deficit, even in populations founded by one individual. Therefore, S. purpurea appears as a species able to limit the consequences of bottlenecks, at least concerning inbreeding. Nevertheless, Ellison (2001), studying seed size and germination success throughout the range of the species, has demonstrated that these parameters show high variability. As this variability was not correlated to the environmental gradients studied, it could be related to limited gene flow and inbreeding depression intensity among populations.
Every time introductions such as this have been studied, the species shows explosive demography. In Switzerland, introduced populations have shown fast population growth and aggressive behaviour enabling them to outcompete rare native peat bog species (Feldmeyer, 1985). Because of its narrow habitat requirements and since peat bogs are very fragmented in Switzerland, S. purpurea cannot properly be defined as invasive but it obviously shows weedy behaviour. This raises questions about the way an outcrossing long-lived species with limited dispersal capacities and, consequently, low gene flow survives with reduced genetic diversity and can have recurrently escaped the deleterious effect of mutational meltdown following foundation.
The present study attempts to explain (a) the impact of bottlenecks upon genetic diversity with a genome-wide perspective in a well-known spatio-temporal framework, and (b) the strategies that an expanding outcrossing species, living in isolated populations, adopts to avoid mutational meltdown and extinction. To shed light on these questions, a population genetics survey with random amplified polymorphism DNA (RAPD) markers has been carried out and a field pollination experiment performed. The results show that genetic diversity in S. purpurea is not strongly affected by bottleneck and founder effects. Although self-compatible, S. purpurea reproduces mostly by outcrossing but colonizes sites in a family structure. This probably enables this species to purge main deleterious mutations, as well as reinforcing selection to produce harmonious genomes and thus avoid extinction through mutational meltdown.
SARRACENIA PURPUREA IN SWITZERLAND: HISTORICAL INSIGHTS
Following a long botanical tradition of conducting vegetation surveys, the introduction of S. purpurea into Switzerland has been catalogued in considerable detail. Sarracenia purpurea subsp. purpurea was introduced into Switzerland by M. F. Cornu who brought seeds back from a journey in the United States at the end of the 19th century (Correvon, 1947). He first grew the species in the Jura mountains before he introduced it into peat bogs in the region of Vevey, Canton de Vaud. Jaccard (1903) reported that, ‘in the region of Vevey, the species occurs as huge clump, flowering and fructifying every year […] proving that the species is acclimating successfully’. Although the peat bogs where the species was introduced are not precisely mentioned, the species actually occurs in only two well-known peat bogs (Moret, 1992).
In the Tenasses peat bog, S. purpurea was introduced around 1900. In a careful vegetation survey, Dutoit (1924) reported the first clear mention of numerous individuals of the species at this site. Feldmeyer (1985) mentioned that S. purpurea had colonized the entire central part of the peat bog in which abiotic conditions are similar to its native habitat. At present, probably after 20 overlapping generations, the population is dense with >25 000 individuals in 2500 m2 (pers. obs.).
Around 1950, individual(s) from the Tenasses peat bog were introduced in the lowland marsh of Champ-Buet, near Lausanne. A field botanist (P. DeRham, University of Lausanne, Switzerland, 1962, unpubl. res.) reported the presence of ‘1 clump of S. purpurea growing within the acidic vegetation’ but failed to find seedlings. Around 1975, two to three individuals were present (Moret, Botanical Garden of Lausanne, Switzerland, pers. comm.) and during the present study around 120 individuals were counted. Only half of them were flowering and the density was much lower than in the Tenasse peat bog.
MATERIALS AND METHODS
Sample collection
Individuals of S. purpurea were randomly chosen to provide a representative sample of the occupied territory of the two sites. In the peat bog of Les Tenasses (T, N: 46°29′ 29″N, 6°55′16″E, 1200 m), 53 individuals (t01–t53) were collected following the bog topography, so that the population was subdivided into six groups (Ta–Tf). The last group (Tf) consisted of three isolated individuals growing in the marshes surrounding the peat bog and has not been fully analysed. In the marsh at Champ-Buet (Bu, 46°36′50″N, 6°34′50″E, 604 m), no obvious topographical subdivision was detectable and 30 individuals (b01–b30) were collected randomly. The exact location of each individual was determined using a D-GPS that provides accurate coordinates (precision approx. 1 cm). All the statistical analyses described hereafter have been performed at a regional scale (considering two populations, T and Bu) as well as at a local scale (considering the Tenasses site as topographically subdivided into six populations, giving a sample of seven groups: Ta, Tb, Tc, Td, Te, Tf and Bu). Young unopened ascidia (leaves) were collected from all 83 sampled individuals, conserved on ice and then stored at −80 °C until DNA was extracted.
DNA extraction and RAPD amplification
Following the manufacturer's instructions, DNA from 1 g of freshly iced ascidia was extracted using a Dneasy Plant Mini Kit (QIAGEN). After this procedure, DNA was resuspended in 100 µl of EB buffer (QIAGEN). To avoid the well-known reproducibility drawbacks of RAPD, sample DNA was amplified using the Ready-To-Go RAPD Analysis Kit (Amersham Pharmacia Biotech). This procedure involves lyophilized reactants and only requires the addition of 5–50 ng of DNA, 25 pmol of primer and water to a final volume of 25 µl. Thus, manipulation errors are minimized and reactant concentrations are constant, ensuring good reproductibility. Although RAPD markers have often been criticized, mostly for their lack of reproducibility (Jones et al., 1997), the procedure adopted here was checked to provide reliable genetic markers by amplifying five individuals three times independently. RAPD patterns were concordant and ambiguous fragments were discarded from analyses. PCR amplifications were all performed on the same Biometra T3 thermocycler (version 3·28T) according to the following programme: 1× (95 °C, 5 min); 45× (95 °C, 1 min; 36 °C, 1 min; 72 °C, 2 min); 1× (72 °C, 5 min). PCR products were visualized by a 5-h electrophoresis (15 V cm−1) of 8-µl aliquots on 30-cm ethidium bromide-stained 2 % agarose gels in a 1× TBE buffer system. Ten different primers furnished and named by the manufacturer were tested on a sub-sample of five individuals and three of them (P1, 5′-d[GGTGCGGGAA]-3′; P5, 5′-d[AACGCGCAAC]-3′; P6, 5′-d[CCCGTCAGCA]-3′) were chosen according to the polymorphism generated, the interpretability of the banding pattern and its reproducibility. PCR products between 400 and 2500 bp were scored for presence (1) or absence (0) from digitalized photographs and named (Px-0000) according to the primer used to generate them and the approximate band size in basepairs determined against the 100-bp ladder (Gibco-BRL 1 µg µl−1). Weak bands were often not reproducible and were all discarded from the analyses.
Statistical analysis of dominant markers
When analysing anonymous markers, absence of co-migration must be assumed. Each locus is supposed to be di-allelic and alleles are assumed to be induced by identical mutations. These assumptions are not testable without time-consuming segregation analyses but seem usually to provide negligible bias (Carlson et al., 1991). Statistical analysis of dominant markers is not as straightforward and powerful as the analysis of co-dominant markers. However, the lack of statistical power can be circumvented by generating numerous markers (Mariette et al., 2002). Without analysing multiple generations, the Hardy–Weinberg equilibrium must classically be postulated, which is usually unrealistic, especially in the case of small sample size and intra-population analysis. Lynch and Milligan (1994) offered statistical background and recommendations to investigate population structure with classical Fst analogues. Roughly, analysis must be limited to fragments whose frequency is less than 1 − (3/N) and Fst estimations with small sample size should be corrected following Weir and Cockerham (1984) methods of θst calculation (also see Isabel et al., 1999). These recommendations have been followed and θst have been calculated using RAPDFst software (Black, 1997). These estimates have been performed at both the regional and the local scale, and to estimate pairwise F-statistics between groups.
To avoid strict Hardy–Weinberg equilibrium postulation, it is permissible to adopt phenetic analysis, where each PCR product is assumed to represent a profile (sometimes called phenotype). The most frequently applied phenetic method to estimate population structure is certainly the use of Shannon's diversity index (Bussell, 1999, and references therein) to partition diversity among populations (Gst). Unfortunately, these estimations seem particularly biased in the case of small and unequal samples and are thus not presented here. Considering RAPD product as a phenotype, multivariate analyses based on Euclidian distances are particularly promising to reveal the structure of genetic variability. Although correspondance analysis is the most frequently used method to analyse binary data, it simultaneously maximizes inertia (multidimensional variance) among individuals (viewed as objects) and loci (viewed as descriptors). This property could potentially hide the relationships between loci (e.g. linkage). Thus, in order to provide a genome-wide variability point of view summarized in a few synthetic variables, principal component analysis based on a covariance matrix (PCAc) was preferred here. Then, between-group eigenanalysis (i.e. PCA among groups based on the PCA among individuals) can be processed to partition this variability into within- and between-group components (hereafter, called 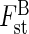 ). The statistical significance of this partition is assessed by Romesburg randomization test (9999 permutations). Multivariate analyses have been performed using ADE-4 software (Thioulouse et al., 1998).
). The statistical significance of this partition is assessed by Romesburg randomization test (9999 permutations). Multivariate analyses have been performed using ADE-4 software (Thioulouse et al., 1998).
In addition, Bayesian statistical procedures have been proposed to investigate population structure with dominant markers (Holsinger et al., 2002). This hierarchical approach aims to investigate genetic data by incorporating the effect of uncertainty about the fixation index (Fst) and the inbreeding coefficient (Fis) into the estimations of these parameters, through the use of a Markov Chain Monte Carlo (MCMC). MCMC is run 250 000 times to ensure convergence of the Markov chain to its stationary distribution and, after the initial 50 000 simulations are discarded, estimations every five steps are retained to avoid autocorrelation among samples. The deviance information criterion (DIC) is a measure that takes into account both how well the model fits the data and how many parameters are required to do it. Running different models (Fst and Fis ≠ 0; Fst = 0 and Fis ≠ 0; Fst ≠ 0 and Fis = 0) on the same dataset, DIC is a model choice criterion. Another criterion that should be taken into account is the complexity (pD; Holsinger and Wallace, 2004). Thus, because DIC and pD are lower than for other models, the full model (Fst and Fis ≠ 0) was used here (DIC = 852·551, pD = 109·882) and both most likely Fst (θB) and Fis (f) are estimated with their variance and a reliable 95 % credible interval. This procedure was achieved using Hickory v0·8 software (Holsinger et al., 2002).
Spatial analysis
For both populations separately and for topographical groups, Mantel tests have been performed to examine the relationships between geographical distance among individuals and their genetic distance from each other based on the Jaccard index:
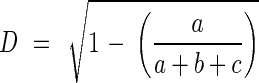 |
with a as the number of concordances 1–1, b and c as discordances 1–0 and 0–1, respectively. Statistical significance is assessed by a 9999 random permutations test. These procedures have been carried out using ADE-4 software (Thioulouse et al., 1998).
Pollination experiment and statistical measurement of inbreeding depression
Within-population estimations of inbreeding depression are often achieved by comparing fitness-traits measured on selfed (Ws) and outcrossed individuals (Wo): δ = 1 − Ws/Wo. Although inbreeding depression intensity is variable during the life cycle (with early stage fitness-traits often less strongly affected than later ones), traits such as number and weight of seeds have often been shown to provide good estimations of fitness and thus inbreeding depression (Sheridan and Karowe, 2000). Among the 53 sampled individuals of the Tenasses population, 21 individuals were manipulated to control pollinations in the field. This sub-sample was chosen to provide individuals with at least four flowers, enabling different treatments on the same individual (i.e. genet) to be applied. On each individual, three flowers were emasculated before anthesis and isolated using silk bags, while the fourth was taken as the pollen donor. A few days after emasculation, mature pollen was sampled from the fourth flower, using a cotton stick, and stored at 4 °C until the stigma was receptive for pollination. Pollinations were performed in June by applying the cotton stick loaded with pollen on the receptive stigmas. One of the three emasculated flowers was self-pollinated, while the two others were cross-pollinated by pollen from (1) an individual belonging to the same topographical group (treatment called allo1 hereafter) and (2) an individual belonging to a different topographical group (named allo2 hereafter). Then, flowers were isolated again with the silk bags until the seeds had ripened. Capsules from the 21 manipulated individuals, as well as 17 capsules from open-pollinated individuals, were sampled after reaching complete maturity in October. For each capsule, seeds were counted and individually weighed using a Mettler Toledo balance sensitive to >0·001 mg. Seed weight and seed number among the different treatments were compared (selfing vs. allo1 + allo2, selfing vs. allo1, selfing vs. allo2, allo1 vs. allo2) using appropriate statistical paired tests among individuals: paired t-tests or exact Wilcoxon signed-rank tests depending on the normality of data (all tests were performed using S-plus 2000 software; Math Soft Inc.). Furthermore, using the outcrossing pollinations (allo1 and allo2), the effects of parental geographical distance and genetic distance on seed number and seed weight were tested using standard linear regressions.
RESULTS
Population genetics
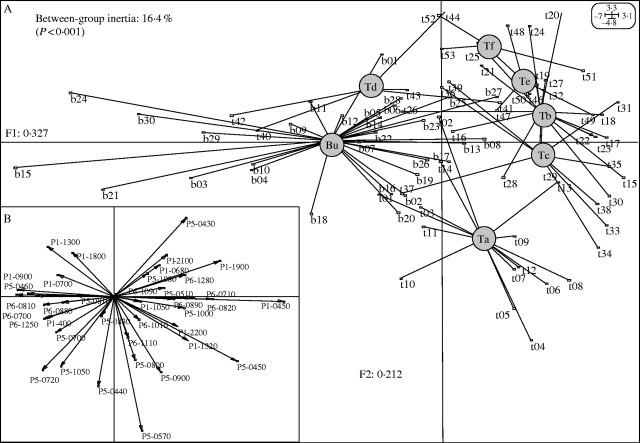
Phenotypes among the 83 sampled individuals were distinguished. At the regional scale, comparing the two populations taken as a whole (Bu and T), between-group eigenanalysis (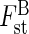 ) shows that 7·2 % of the variance was partitioned between the populations (P < 0·001). θst gave a very similar result (0·076). When the same analyses were undertaken considering the Tenasses population as a topographically subdivided sample (Ta–Tf), genetic structure was higher: θst (0·287) and
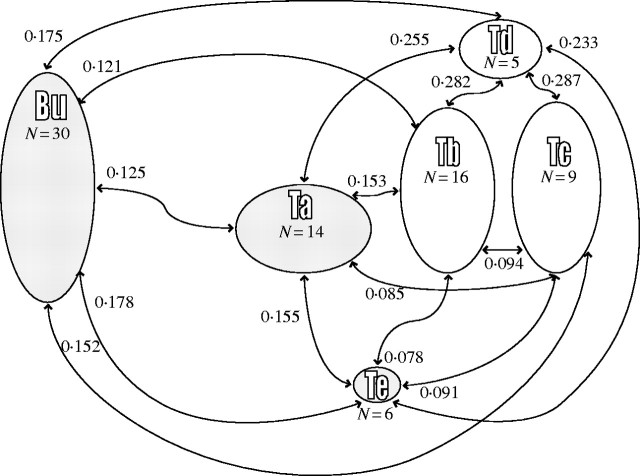
) shows that 7·2 % of the variance was partitioned between the populations (P < 0·001). θst gave a very similar result (0·076). When the same analyses were undertaken considering the Tenasses population as a topographically subdivided sample (Ta–Tf), genetic structure was higher: θst (0·287) and 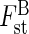 (0·164, P < 0·001). A graphical representation of the between analysis is presented in Fig. 1A. The genetic variability is summarized on the first two principal components, representing 54 % of the total inertia. The loci contribution to the F1 and F2 axes are presented in Fig. 1B and show typical star-like patterns, suggesting that the loci do not systematically covariate and thus mainly segregate randomly. While the groups of the Tenasses population differentiated along the F2 axis, the individuals of Champ-Buet principally did so along the F1 axis. Individuals belonging to the Td group had intermediate positions. Pairwise-θst values between topographical groups are also presented in Fig. 2. Once again, the Td group was clearly differentiated from the others. The Bayesian statistics gave a very similar partition with a mean θB = 0·267 (variance of 0·031 and 95 % credible interval from 0·217 to 0·319), but also allowed a rough estimate of the mean Fis = 0·802 (variance of 0·165 and 95 % credible interval from 0·410 to 0·992).
(0·164, P < 0·001). A graphical representation of the between analysis is presented in Fig. 1A. The genetic variability is summarized on the first two principal components, representing 54 % of the total inertia. The loci contribution to the F1 and F2 axes are presented in Fig. 1B and show typical star-like patterns, suggesting that the loci do not systematically covariate and thus mainly segregate randomly. While the groups of the Tenasses population differentiated along the F2 axis, the individuals of Champ-Buet principally did so along the F1 axis. Individuals belonging to the Td group had intermediate positions. Pairwise-θst values between topographical groups are also presented in Fig. 2. Once again, the Td group was clearly differentiated from the others. The Bayesian statistics gave a very similar partition with a mean θB = 0·267 (variance of 0·031 and 95 % credible interval from 0·217 to 0·319), but also allowed a rough estimate of the mean Fis = 0·802 (variance of 0·165 and 95 % credible interval from 0·410 to 0·992).
Fig. 1.
(A) Representation of the between-group eigenanalysis among the seven topological groups of individuals (tii and bii) of Sarracenia purpurea delimited in the Tenasses population (Ta to Tf) and Champ-Buet population (Bu). Between-group genetic inertia (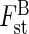 ) represents 16·4 % of the total inertia (P < 0·001). F1 and F2 represent the first two principal components that maximize inertia in the ordination-reduced space and summarize 54 % of the total genetic inertia. (B) Contribution of the 40 RAPD loci to the first two principal components (F1 and F2). The star-like pattern indicates that most loci randomly segregate.
) represents 16·4 % of the total inertia (P < 0·001). F1 and F2 represent the first two principal components that maximize inertia in the ordination-reduced space and summarize 54 % of the total genetic inertia. (B) Contribution of the 40 RAPD loci to the first two principal components (F1 and F2). The star-like pattern indicates that most loci randomly segregate.
Fig. 2.
Pairwise-θst between topographical groups of Sarracenia purpurea delimited in the Tenasses populations (Ta to Te) and Champ-Buet population (Bu). Size of the schematic group is proportional to the patch size and N is the number of individuals sampled. Groups shaded in grey present significant Mantel test results between geographical and genetic distances. The Tenasses population taken as a whole also shows a significant Mantel test result.
The Mantel test shows significant correlation between individual geographical distance and genetic distance as calculated by the Jaccard index in the case of the Tenasses population taken as a whole (RM = 0·128, P = 0·04) and the Champ-Buet population (RM = 0·286, P = 0·005). When the Tenasses population was topographically subdivided, only the Ta and the Te groups showed significant association between geographical and genetic distances [RM = 0·487 (P < 0·001) and RM = 0·836 (P = 0·012), respectively].
Pollination experiment
Careful pollinations have been achieved by saturating stigmas with a large amount of pollen. Nevertheless, artificial pollinations produced significantly less seeds than natural ones (t-test, P < 0·001). Then, in order to compare the effect of the types of pollination (i.e. selfing vs. allo1 vs. allo2) on seed fitness-traits, seed weight and seed numbers were shown to be unaffected by different confounding factors (data not shown). Thus, seed traits were shown to be independent of the individual measured (i.e. maternal investment measured by the number of flowers produced), as well as the group origin. The weight of seeds was also shown to be unaffected by seed number. In this case, seed traits measured are thus independent and reliable markers of early inbreeding depression. Seed numbers ranged from 0 to 649 for selfed flowers, from 0 to 852 for the allo1 treatment and from 0 to 366 for the allo2 treatment. Selfed and outcrossed flowers did not produce significantly different amounts of seeds (P = 0·506, d.f. = 61). Neither allo1 nor allo2 produced significantly different numbers of seeds than selfed flowers [P = 0·294 (d.f. = 20) and P = 0·893 (d.f. = 20), respectively]. There were no differences either among the outcrossing treatments (allo1 vs. allo2; P = 0·375, d.f. = 20). Finally, no effect of geographical or genetic distances among parents was detected (rho = 0·19, P = 0·219 and rho = 0·15, P = 0·346, respectively).
Seed weight ranged from 0·023 to 0·841 mg. Given the sample size of 21 treated individuals, a seed weight difference of 0·067 mg (>15 %) among treatments should be detected without type-I error. No significant differences have been detected among treatments on seed weight (d.f. = 10; selfing vs. allo1: P = 0·896; selfing vs. allo2: P = 0·206; allo1 vs. allo2: P = 0·279). Although not in a significant proportion, seeds produced by selfing often show a bimodal weight distribution, with about one-quarter of seeds lighter than others (data not shown).
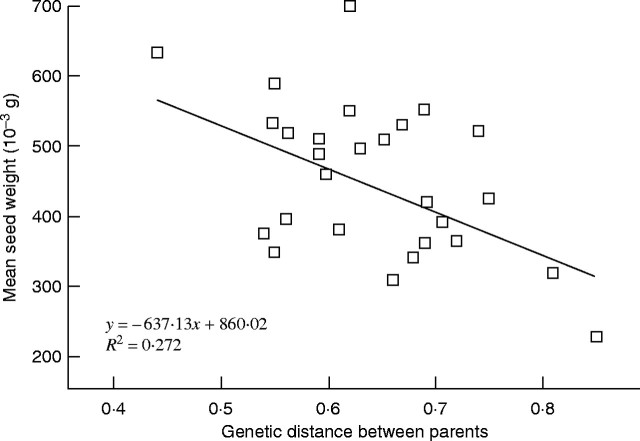
Linear regression failed to explain an effect of the geographical distance between parents on the seed weight they produce (P = 0·054). More interestingly, the effect of the genetic distance among parents using the Jaccard index was shown by linear regression to have a significant negative effect on the weight of produced seeds (Fig. 3; R2 = 0·272, slope: −637·14, P = 0·004 and intercept: 860·02, P < 0·001).
Fig. 3.
Linear regression of the mean F1 seed weight on the genetic distance among parents used to achieve the outcross pollinations. Genetic distances have been estimated using the Jaccard index on the RAPD phenotype of parents. Genetic distance explains 27·24 % of the seed weight variance (R2) and both slope (−637·13) and intercept (860·02) of the linear model are significant (P = 0·004 and P < 0·001, respectively).
DISCUSSION
The three RAPD primers used in this study gave similar fragment numbers and together provided 40 clear, reproducible and polymorph fragments. RAPD thus provided reliable markers to assess genetic variability in this study (also see Etisham-Ul-Haq et al., 2001; Rossello et al., 2002).
Founder effect and genetic variability
At the regional scale, although the Tenasses and Champ-Buet populations are >30 km apart and thus cannot be connected by extensive gene flow, genetic differentiation was weak (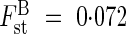 ). Considering that inferences from RAPD and allozymes are comparable (Isabel et al., 1995), this differentiation is characteristic of long-lived perennials presenting outbreeding mating systems whose mean Gst is 0·094 (Hamrick and Godt, 1996). The inferred dispersal rate between these populations nevertheless seems irrelevant in this situation, because recurrent gene flow between the Tenasses peat bog and the Champ-Buet marsh was certainly very low and close to zero as no seed is able to travel the distance by natural means. Furthermore, gene flow by pollen does not seem to provide a better explanation because the usual pollinators of S. purpurea (Bombus spp., Syrphidae and Halictidae; Thomas and Duncan, 1986; pers. obs.) do not usually travel such distances (Richards, 1997). Thus, the weak differentiation between the Tenasses and Champ-Buet populations must be viewed as a consequence of the perennial habit of the species, together with common ancestry and recent isolation (<50 years).
). Considering that inferences from RAPD and allozymes are comparable (Isabel et al., 1995), this differentiation is characteristic of long-lived perennials presenting outbreeding mating systems whose mean Gst is 0·094 (Hamrick and Godt, 1996). The inferred dispersal rate between these populations nevertheless seems irrelevant in this situation, because recurrent gene flow between the Tenasses peat bog and the Champ-Buet marsh was certainly very low and close to zero as no seed is able to travel the distance by natural means. Furthermore, gene flow by pollen does not seem to provide a better explanation because the usual pollinators of S. purpurea (Bombus spp., Syrphidae and Halictidae; Thomas and Duncan, 1986; pers. obs.) do not usually travel such distances (Richards, 1997). Thus, the weak differentiation between the Tenasses and Champ-Buet populations must be viewed as a consequence of the perennial habit of the species, together with common ancestry and recent isolation (<50 years).
Such a low genetic structure is surprising for populations that have been affected by a bottleneck. Although Schwaegerle and Schaal (1979) sampled their reference populations amongst what is now recognized by Godt and Hamrick (1999) as two subspecies, they reported a much higher Gst (0·528) than the present estimation for a population founded by only one individual. When turning to the estimations of Taggart et al. (1990) for populations introduced into Ireland, only populations founded by three individuals or more showed similar genetic differentiation. More precisely, Taggart et al. (1990) have studied a similar situation to that found in Switzerland, with populations founded by individuals taken from the population initially introduced into Ireland in 1906. Two populations known to have been founded by three and four individuals 30 years before their study showed Gst values of 0·106 and 0, respectively. From these estimations, the Champ-Buet population would have been founded by at least three individuals. This estimation derived from genetic data is in contradiction with the historical one that describes the foundation of the Champ-Buet population by only one individual.
Because 92·8 % of the genetic variability is partitioned within populations at the regional scale, the analysis of genetic structure at a local scale (i.e. using population's topographic subdivision) is of great interest to gain insights about bottlenecks and the processes shaping the observed genetic variability. Therefore, it appears that a great part of the genetic variability is partitioned between groups (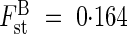 and θst = 0·287), reinforcing the idea of limited gene flow in S. purpurea and explaining the isolation by distance detected by the Mantel test (Tenasses, RM = 0·128; Champ-Buet, RM = 0·286). Furthermore, when looking at how genetic variability is partitioned among groups (Fig. 1), the differences between the Tenasses and Champ-Buet groups become obvious. It appears that most of the genetic variability within the Champ-Buet population is independent of that found within the Tenasses population (i.e. between the Tenasses groups). The pattern of PCAc on individual RAPD phenotypes is reflecting the common ancestry as well as the isolation of the two populations; some individuals of Champ-Buet show similar RAPD patterns to those found in the Tenasses population, but the whole-population genetic variability is clearly differentiated. This pattern can be viewed as the consequence of a bottleneck that shifted allele frequencies. Even more so, when multivariate analyses on RAPD phenotypes are viewed as a way of revealing genomic tendencies, the independence of genomic patterns showed by Fig. 1 indicates that the founder effect has greatly affected the genomic composition. The Champ-Buet population occurs in a lowland acidic marsh with dense vegetation, while the Tenasses population occurs in an acidic peat bog at high altitude. This sharp genetic pattern (Fig. 1) may thus also be partly related to the contrasting ecological conditions at work in these two sites and the strong effect of selection upon bottlenecked populations (Carson and Templeton, 1984). Pairwise-θst values between the Champ-Buet group and the Tenasses subpopulations are almost equivalent (Fig. 2). It is thus impossible to define precisely the origin of the founder individual transplanted to Champ-Buet and a foundation by several individuals with different genetic composition could be an explanation of this pattern. Nevertheless, historical insights, as well as the differentiation of genomic composition previously discussed, seem to be in accordance with a severe bottleneck and an important founder effect. Therefore, it is more plausible that the Champ-Buet population has been founded by a single (or very few) individual(s), but had a subsequent important growth rate (see the historical insights section) that preserved most of the genetic variability present in the Tenasses population (Grant et al., 2001). This variability was nevertheless rearranged.
and θst = 0·287), reinforcing the idea of limited gene flow in S. purpurea and explaining the isolation by distance detected by the Mantel test (Tenasses, RM = 0·128; Champ-Buet, RM = 0·286). Furthermore, when looking at how genetic variability is partitioned among groups (Fig. 1), the differences between the Tenasses and Champ-Buet groups become obvious. It appears that most of the genetic variability within the Champ-Buet population is independent of that found within the Tenasses population (i.e. between the Tenasses groups). The pattern of PCAc on individual RAPD phenotypes is reflecting the common ancestry as well as the isolation of the two populations; some individuals of Champ-Buet show similar RAPD patterns to those found in the Tenasses population, but the whole-population genetic variability is clearly differentiated. This pattern can be viewed as the consequence of a bottleneck that shifted allele frequencies. Even more so, when multivariate analyses on RAPD phenotypes are viewed as a way of revealing genomic tendencies, the independence of genomic patterns showed by Fig. 1 indicates that the founder effect has greatly affected the genomic composition. The Champ-Buet population occurs in a lowland acidic marsh with dense vegetation, while the Tenasses population occurs in an acidic peat bog at high altitude. This sharp genetic pattern (Fig. 1) may thus also be partly related to the contrasting ecological conditions at work in these two sites and the strong effect of selection upon bottlenecked populations (Carson and Templeton, 1984). Pairwise-θst values between the Champ-Buet group and the Tenasses subpopulations are almost equivalent (Fig. 2). It is thus impossible to define precisely the origin of the founder individual transplanted to Champ-Buet and a foundation by several individuals with different genetic composition could be an explanation of this pattern. Nevertheless, historical insights, as well as the differentiation of genomic composition previously discussed, seem to be in accordance with a severe bottleneck and an important founder effect. Therefore, it is more plausible that the Champ-Buet population has been founded by a single (or very few) individual(s), but had a subsequent important growth rate (see the historical insights section) that preserved most of the genetic variability present in the Tenasses population (Grant et al., 2001). This variability was nevertheless rearranged.
Multivariate analysis of genetic phenotype, especially PCA which takes into account the interaction among loci, thus appears as a powerful inference method for dominant markers. Obviously, it provides insights about processes that would not be detected by classical F-statistics. In this case, discordance between historical and genetic insights could not have been analysed without multivariate analysis (PCA).
Family structure and mutational meltdown avoidance
As a whole, the important local genetic structure indicates great isolation between groups, reinforcing the view of a very restricted gene flow in S. purpurea, even on a local scale (mean seed dispersal, 12·8 cm; Ellison and Parker, 2002). It thus appears that even geographically small populations such as the Tenasses one (i.e. <60 m in diameter) have not reached equilibrium. Although the Tenasses population as a whole shows isolation by distance, no group but the marginal ones (Ta and Te) show such a pattern. This is a strong indication of an expanding population colonizing the space into a family, which is easily explained by restricted seed dispersal (Ellison and Parker, 2002). Each mound can thus be viewed as an island where the founder individual shares the space and reproduces with its own offspring, unable to disperse further recurrently. Such a family structure can explain the apparent paradox of a perennial species that principally outcrosses, easily avoids extinction through mutational meltdown and presents an invasive behaviour after a bottleneck. When colonizing a site in such a family structure, most mating occurs between close relatives and bi-parental inbreeding is increased. Individuals would thus first suffer from bi-parental inbreeding depression but the genetic load would decrease with time by favouring bi-parental purge (Byers and Waller, 1999).
Bayesian estimation of the population structure indicates a strong inbreeding in the introduced populations of S. purpurea (mean Fis = 0·802). Although the Bayesian inferences about inbreeding coefficient with dominant markers are only indicative and should be taken with great caution (Holsinger et al., 2002), such high inbreeding is not uncommon for populations affected by founder effect. Furthermore, the 95 % credible interval indicates that the inbreeding coefficient is clearly above 0·410. Such an inbred population is expected to show inbreeding depression, with a fitness decrease reaching at least 20–40 % (Keller and Waller, 2002). Sheridan and Karowe (2000) have shown in a sister species (Sarracenia flava) that the F1 seed weight is related to inbreeding and affects the subsequent germination rate (also see Ellison, 2001). Thus, even though the F1 seed weight measured here can be used as a reliable marker (Simons and Johnston, 2000; Tremayne and Richards, 2000), S. purpurea in the Tenasses population does not show early inbreeding depression. Although sample size is powerful enough to allow the detection of <15 % seed weight decrease, the comparison among treatments used to detect inbreeding depression failed to be significant. The only indication of decreased fitness for inbred seeds is a trend to bimodal distribution for seed weight produced by selfing (data not shown). In some cases, about one-quarter of the selfed seeds were lighter than others and were also lighter than seeds produced by outcrossing, which are certainly individuals expressing a deleterious allele in the homozygous state. This pattern is in accordance with an efficient purge that has already eliminated most of the deleterious alleles from the population, at least at early stages of development. Furthermore, it reinforces the demographic observations of P. Ecoffey (unpubl. res.), who was unable to explain current density and demography in the Tenasses peat bog by a constant growth rate.
In addition, the genetic distances between parents used for controlled pollination have a significant negative effect on the F1 seed weight and, in turn, on offspring fitness (Fig. 3). This is an indication of outbreeding depression rather than inbreeding depression. Because geographical distance failed to explain seed weight differences and because homogenous ecological conditions can be assumed within the species range in the Tenasses peat bog, the outbreeding depression pattern observed in the F1 is likely to be caused by a genetic mechanism (Lynch, 1991). Epistasic and dominant interactions at different loci within the genome cause co-adaptation of genes, so that unrelated individuals do not have similar gene combinations. These intrinsic incompatibilities, reinforced by family structure and drift, lead to a decrease of F1 fitness independent of the environment when genetic co-adaptations (heterozygote × heterozygote interactions) are disrupted by gene flow between distantly related individuals (Edmands, 2002). In other words, in the absence of inbreeding depression in the F1, the bottleneck could have generated genetic additive variance mainly from the epistasic variance component (Naciri-Graven and Goudet, 2003) and to a lesser extent from dominance (Willis and Orr, 1993). The lack of inbreeding depression in the F1 generation and the outbreeding depression expressed in the F1 suggest that the species has not only purged the main deleterious alleles, but may also have undergone some forms of selection for inbreeding due to co-adaptation between loci. Family structure and bi-parental inbreeding is a convenient alternative to selfing to purge the genetic load. What is more, as S. purpurea shows self-compatibility but is morphologically unexpected to self, individuals that produce more than one flower would be able to produce selfed offspring by geitonogamy and would thus avoid mutational meltdown more easily through an efficient purge. Although not studied precisely (M. J. Godt, University of Georgia, USA, pers. comm.), individuals of S. purpurea inside its native range are often described with one or two scapes and thus one or two flowers (Britton and Brown, 1913; Schwaegerle and Schaal, 1979), but in the populations studied here up to 15 scapes and flowers are produced (pers. obs.). Due to the stenoecious ecological requirement of S. purpurea and the lack of obvious habitat differences between native and introduced habitat, such an increase could easily be achieved by resource reallocation from defence to fecundity and the transformation of epistasic variance into additive variance related to foundation.
A likely scenario is that the founding event and subsequent genetic drift would have driven allele frequencies toward extreme values, exposing previously rare homozygotes to selection and would have altered the selective values of alleles, thereby producing selection for a new system of co-adapted genes (Cheverud et al., 1999). Such a scenario should explain how S. purpurea could avoid extinction through mutational meltdown even after a severe decrease of genetic diversity by bottlenecks. This is because under restricted gene flow and limited seed dispersal, populations rapidly structure into family breeding units that allow an efficient bi-parental purge of deleterious alleles and selection for genomic systems of co-adapted genes.
Acknowledgments
We thank Lise Bavaud, Guillaume Besnard, John Gaskin, Mélanie Glaettli, Jérôme Goudet, Jean-Louis Moret and Christophe Randin for their help during this work.
LITERATURE CITED
- Barrett SCH. 1996. The reproductive biology and genetics of island plants. Philosophical Transactions of the Royal Society of London B 351: 725–733. [Google Scholar]
- Bayer RJ, Hufford L, Soltis DE. 1996. Phylogenetic relationship in Sarraceniaceae based on rbcL and ITS sequences. Systematic Botany 21: 121–134. [Google Scholar]
- Black B. 1997. RAPDFST – a FORTRAN program to estimate F(ST) and effective migration rates among subpopulations using RAPD-PCR files. Fort Collins, CO: Colorado State University (wcb4@lamar.colostate.edu). [Google Scholar]
- Britton NL, Brown A. 1913.An illustrated flora of the northern United States, Canada and British Possessions, Vol. II. New York: Charles Scribner & Sons. [Google Scholar]
- Bussell JD. 1999. The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petrea (Lobeliaceae). Molecular Ecology 8: 775–789. [Google Scholar]
- Byers DL, Waller DM. 1999. Do plant populations purge their genetic load? Effects of populations size and mating history on inbreeding depression. Annual Review in Ecology and Systematics 30: 479–513. [Google Scholar]
- Cain ML, Milligan BG, Strand AE. 2000. Long-distance seed dispersal in plant population. American Journal of Botany 87: 1217–1227. [PubMed] [Google Scholar]
- Carlson JE, Tulsieram LK, Glaubitz JC, Luk VWK, Kauffeldt C, Rutledge R. 1991. Segregation of random amplified DNA markers in F1 progeny of conifers. Theoretical and Applied Genetics 83: 194–200. [DOI] [PubMed] [Google Scholar]
- Carson HL. 1989. Genetic imbalance, realigned selection and the origin of species. In: Giddings LV, Kaneshiro KY, Anderson WW, eds. Genetics, speciation and the founder principle. Oxford: Oxford University Press, 345–362. [Google Scholar]
- Carson HL. 1990. Increased genetic variance after a population bottleneck. Trends in Ecology and Evolution 5: 228–230. [DOI] [PubMed] [Google Scholar]
- Carson HL, Templeton AR. 1984. Genetic revolutions in relation to speciation phenomena: the founding of new populations. Annual Review in Ecology and Systematics 15: 97–131. [Google Scholar]
- Charlesworth D, Charlesworth B. 1999. The genetic basis of inbreeding depression. Genetical Research 74: 329–340. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Vaughin TT, Pletscher LS, King-Ellison K, Bailiff J, Adams E, Erickson C, Bonislawski A. 1999. Epistasis and the evolution of additive genetic variance in populations that pass through a bottleneck. Evolution 53: 1009–1018. [DOI] [PubMed] [Google Scholar]
- Correvon H. 1947.Fleurs des eaux et des marais. Neuchâtel: Delachaux et Niestlé. [Google Scholar]
- Dutoit D. 1924.Les associations végétales des Sous-Alpes de Vevey (Suisse). PhD Thesis, University of Lausanne, Switzerland. [Google Scholar]
- Edmands S. 2002. Does parental divergence predict reproductive compatibility? Trends in Ecology and Evolution 17: 520–527. [Google Scholar]
- Ellestrand NC, Elam DR. 1993. Population genetic consequences of small population size: implcation for plant conservation. Annual Review in Ecology and Systematics 24: 217–242. [Google Scholar]
- Ellison AM. 2001. Interspecific and intraspecific variation in seed size and germination requirements of Sarracenia (Sarraceniaceae). American Journal of Botany 88: 429–437. [PubMed] [Google Scholar]
- Ellison AM, Parker JN. 2002. Seed dispersal and seedling establishment of Sarracenia purpurea (Sarraceniaceae). American Journal of Botany 89: 1024–1026. [DOI] [PubMed] [Google Scholar]
- Etisham-Ul-Haq M, Allnut TR, Smith-Ramirez C, Gardner MF, Armesto JJ, Newton AC. 2001. Patterns of genetic variation in in and ex situ populations of the threatened Chilean vine Berberidopsis corallina, detected using RAPD markers. Annals of Botany 87: 813–821. [Google Scholar]
- Feldmeyer E. 1985. Étude phyto-écologique de la tourbière des Tenasses. Botanica Helvetica 95: 99–115. [Google Scholar]
- Godt MJW, Hamrick JL. 1999. Genetic divergence among infraspecific taxa of Sarracenia purpurea Systematic Botany 23: 427–438. [Google Scholar]
- Grant PR, Grant BR, Petren K. 2001. A population founded by a single pair of individuals: establishment, expansion, and evolution. Genetica 112–113: 359–382. [PubMed] [Google Scholar]
- Hamrick JL, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London B 351: 1291–1298. [Google Scholar]
- Hewitt GM. 2000. The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. [DOI] [PubMed] [Google Scholar]
- Holsinger KE. 2000. Reproductive systems and evolution in vascular plants. Proceedings of the National Academy of Sciences of the USA 97: 7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Wallace LE. 2004. Bayesian approaches for the analysis of population genetic structure: an example from Platanthera leucophea (Orchidaceae). Molecular Ecology 13: 887–894. [DOI] [PubMed] [Google Scholar]
- Holsinger KE, Lewis PO, Dey DK. 2002. A Bayesian approach to inferring population structure from dominant markers. Molecular Ecology 11: 1157–1164. [DOI] [PubMed] [Google Scholar]
- Husband BC, Barrett SCH. 1992. Effective population size and genetic drift in tristylous Eichornia paniculata (Pontederiaceae). Evolution 46: 1875–1890. [DOI] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Bousquet J. 1995. Complete congruence between gene diversity estimates derived from genotypic data at enzyme and Random Amplified Polymorphic DNA loci in black spruce. Proceedings of the National Academy of Sciences of the USA 92: 6369–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel N, Beaulieu J, Thériault P, Bousquet J. 1999. Direct evidence for biased gene diversity estimates from dominant random amplified polymorphic DNA (RAPD) fingerprints. Molecular Ecology 8: 477–483. [Google Scholar]
- Jaccard P. 1903. Procès-verbal de la séance du 5 novembre 1902 de la SVSN. Bulletin de la Societé Vaudoise des Sciences Naturelles 39: 105. [Google Scholar]
- Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Van der wiel C, Bredemeijer G, Vosman B, Matthes D, Daly A, Brettschneider R, et al. 1997. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Molecular Breeding 3: 381–390. [Google Scholar]
- Juniper BE, Robins RJ, Joel DM. 1989.The carnivorous plants. London: Academic Press. [Google Scholar]
- Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends in Ecology and Evolution 17: 230–241. [Google Scholar]
- Kingston N, Waldren S. 2003. The plant communities and environmental gradients of Pitcairn Island: The significance of invasive species and the need for conservation management. Annals of Botany 92: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberg PL. 1992. Effects of population bottlenecks on genetic diversity as measured by allozyme electrophoresis. Evolution 46: 477–494. [DOI] [PubMed] [Google Scholar]
- Lee CE. 2002. Evolutionary genetics of invasive species. Trends in Ecology and Evolution 17: 386–391. [Google Scholar]
- Lynch M. 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45: 622–629. [DOI] [PubMed] [Google Scholar]
- Lynch M, Milligan BG. 1994. Analysis of population genetic structure with RAPD markers. Molecular Ecology 3: 91–99. [DOI] [PubMed] [Google Scholar]
- McDaniel S. 1971. The genus Sarracenia (Sarraceniaceae). Bulletin of the Tall Timber Research Station 9: 1–36. [Google Scholar]
- Mariette S, Le Corre V, Austerlitz F, Kremer A. 2002. Sampling within the genome for measuring within-population diversity: trade-offs between markers. Molecular Ecology 11: 1145–1156. [DOI] [PubMed] [Google Scholar]
- Maron JL, Vila M, Bommarco R, Elmendorf S, Beardsley P. 2004. Rapid evolution of an invasive plant. Ecological Monographs 74: 261–280. [Google Scholar]
- Moret JL. 1992. La Sarracenie dans le canton de Vaud. Bulletin du Cercle Vaudois de Botanique 21: 55–57. [Google Scholar]
- Naciri-Graven Y, Goudet J. 2003. The additive genetic variance after bottlenecks is affected by the number of loci involved in epistatic interactions. Evolution 57: 706–716. [DOI] [PubMed] [Google Scholar]
- Richards AJ. 1997.Plant breeding systems, 2nd edn. London: Chapman & Hall. [Google Scholar]
- Rossello JA, Cebrian MC, Mayol M. 2002. Testing taxonomic and biogeographical relationships in a narrow Mediterranean endemic complex (Hippocrepis balearica) using RAPD markers. Annals of Botany 89: 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell DE. 1976.Carnivorous plants of the United States and Canada. North Carolina: Winston-Salem. [Google Scholar]
- Schwaegerle KE, Schaal BA. 1979. Genetic variability and founder effect in the pitcher plant Sarracenia purpurea Evolution 33: 1210–1218. [DOI] [PubMed] [Google Scholar]
- Sheridan PM, Karowe DN. 2000. Inbreeding, outbreeding and heterosis in the yellow pitcher plant, Sarracenia flava (Sarraceniaceae), in Virginia. American Journal of Botany 87: 1628–1633. [PubMed] [Google Scholar]
- Simons AM, Johnston MO. 2000. Variation in seed traits of Lobelia inflata (Campanulaceae): sources and fitness consequences. American Journal of Botany 87: 124–132. [PubMed] [Google Scholar]
- Taggart JB, McNally SF, Sharp PM. 1990. Genetic variability and differentiation among founder population of the pitcher plant (Sarracenia purpurea L.) in Ireland. Heredity 64: 177–183. [Google Scholar]
- Thioulouse J, Chessel D, Doledec S, Olivier JM. 1998. ADE-4. University of Lyon 1. Villeurbanne, France (http://pbil.univ-lyon1.fr/ADE-4/ADE-4.html). [Google Scholar]
- Thomas K, Duncan MC. 1986. Pollination and fertilization in the pitcher plant (Sarracenia purpurea L). American Journal of Botany 73: 678. [Google Scholar]
- Tremayne MA, Richards AJ. 2000. Seed weight and seed number affect subsequent fitness in outcrossing and selfing Primula species. New Phytologist 148: 127–142. [DOI] [PubMed] [Google Scholar]
- Wang J, Hill WG, Charlesworth D, Charlesworth B. 1999. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genetical Research 74: 165–178. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population-structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Willis JH, Orr AH. 1993. Increased heritable variation following population bottlenecks: the role of dominance. Evolution 47: 949–957. [DOI] [PubMed] [Google Scholar]