Abstract
• Background and Aims The Quadrifaria group of Paspalum (Poaceae, Paniceae) comprises species native to the subtropical and temperate regions of South America. The purpose of this research was to characterize the I genomes in five species of this group and to establish phylogenetic relationships among them.
• Methods Prometaphase chromatin condensation patterns, the physical location of 5S and 45S rDNA sites by fluorescence in situ hybridization (FISH), and sequences of five chloroplast non-coding regions were analysed.
• Key Results The condensation patterns observed were highly conserved among diploid and tetraploid accessions studied and not influenced by the dyes used or by the FISH procedure, allowing the identification of almost all the chromosome pairs that carried the rDNA signals. The FISH analysis of 5S rDNA sites showed the same localization and a correspondence between the number of sites and ploidy level. In contrast, the distribution of 45S rDNA sites was variable. Two general patterns were observed with respect to the location of the 45S rDNA. The species and cytotypes Paspalum haumanii 2x, P. intermedium 2x, P. quadrifarium 4x and P. exaltatum 4x showed proximal sites on chromosome 8 and two to four distal sites in other chromosomes, while P. quarinii 4x and P. quadrifarium 2x showed only distal sites located on a variable number of small chromosomes and on the long arm of chromosome 1. The single most-parsimonious tree found from the phylogenetic analysis showed the Quadrifaria species partitioned in two clades, one of them includes P. haumanii 2x and P. intermedium 2x together with P. quadrifarium 4x and P. exaltatum 4x, while the other contains P. quadrifarium 2x and P. quarinii 4x.
• Conclusions The subdivision found with FISH is consistent with the clades recovered with cpDNA data and both analyses suggest that the Quadrifaria group, as presently defined, is not monophyletic and its species belong in at least two clades.
Keywords: Grasses, Paspalum, Quadrifaria group, cpDNA sequence analysis, molecular evolution, double-target FISH, rRNA genes, karyotype, prometaphase chromatin patterns
INTRODUCTION
The genus Paspalum (Poaceae, Paniceae) includes between 350 and 400 species most of them native to tropical and sub-tropical regions of the Americas (Chase, 1929; Clayton and Reinvoize, 1986). Several of the species are of economic importance for forage, turf and ornamental purposes (Burson and Bennett, 1971). Ploidy levels in the genus range from diploid to octoploid (De Moraes et al., 1974) and practically all Paspalum species have a basic chromosome number of x = 10 (Burton, 1940).
Based on morphological characteristics of reproductive and vegetative organs Chase (1929) subdivided the genus into informal groups. The Quadrifaria group of Paspalum was first described by Barreto (1954, 1966), and comprises at least ten species which were originally included in the Virgata group of Chase (1929). They are all native to the subtropical and temperate regions of South America (Barreto, 1966), and sexual diploids and apomictic polyploids are known for almost all of them (Norrmann et al., 1989).
Meiotic chromosome pairing in interspecific hybrids of diploid P. quadrifarium Lam., P. haumanii Parodi, P. intermedium Munro, P. quarinii Morrone & Zuloaga and P. densum Poir. showed that they all contained a particular form of the I genome (Quarín and Norrmann, 1990; Caponio and Quarín, 1993). This genome has been described in other groups such as Virgata and Dilatata in which several allotetraploid species have been assigned the genomic formula IIJJ (Burson, 1983). In contrast to the I genome, the only proposed source of the J genome is P. juergensii (Burson, 1983). Since the II genomic formula has been assigned to all the diploid Quadrifaria cytotypes analysed to date (Quarín and Norrmann, 1990; Caponio and Quarín, 1993), a species from this group may be the source of the I genome for the Dilatata and Virgata polyploids. To advance in the study of the affinities among the polyploid groups and the diploid I genome donors, the different I genomes present in the Quadrifaria species must be characterized further.
From the cytogenetic point of view, chromosome pairing is not likely to provide further insight into the phylogenetic relationships among diploid and polyploid species with I genome since some degree of multivalent formation in these polyploids is to be expected. On the other hand, the mitotic chromosomes in Paspalum are small and morphologically similar, making their karyotypic differentiation very difficult. An alternative has been the use of the prometaphase stage in which uneven condensation of the chromatin has allowed chromosome identification and karyotyping in several plant genera (Fukui and Iijima, 1991; Nakamura et al., 2001). Distinctive patterns of chromatin condensation in prometaphase were also clearly observed in Paspalum chromosome squashes allowing the construction of karyotypes in a triploid and a tetraploid cytotype of P. quadrifarium and a segmental allopolyploid origin was suggested for both (Speranza et al., 2003). Further refining of the cytogenetic approach can be achieved by analysing other chromosome markers.
Fluorescence in situ hybridization (FISH) of repetitive DNA sequences has been used as a tool for karyotype and genome analyses of a large number of plant species. The repetitive and tandemly organized ribosomal rDNA genes, 5S rDNA and 45S rDNA, are localized at one or more sites per chromosome set, and their characteristic positions provide useful markers for chromosome and genome identification. They have been used in several grasses such as Triticum and Hordeum (Jiang and Gill, 1994), Oryza (Shishido et al., 2000), Sorghum (Sang and Liang, 2000), Thinopyrum (Brasileiro-Vidal et al., 2003), Festuca (Harper et al., 2004), providing useful information about evolutionary and phylogenetic relationships between species.
A molecular phylogeny is not available for Paspalum yet, and it is likely that some of the morphological groups recognized to date, would need to be at best re-evaluated when sequence information becomes available. Because of the prevalence of polyploidy and hybridization in the genus, the use of nuclear sequences does not seem appropriate as a first approach given the technical difficulties implied in isolating them and the reticulate evolutionary pattern of the group (Sang, 2002). Several universal primers are available from the literature for highly variable, non-coding chloroplast regions (Demesure et al., 1995; Soltis and Soltis, 1998). These regions can provide a useful phylogenetic signal to analyse the relationships among the diploid species that have the different variants of the I genome.
In the present study, prometaphase chromatin condensation patterns, the physical location of 5S and 45S rDNA sites by double-target FISH, and sequences of five chloroplast non-coding regions were used to characterize the I genomes of some diploid and polyploid cytotypes of five species of the Quadrifaria group of Paspalum and to establish phylogenetic relationships among them. An attempt was also made to localize the heterochromatin blocks using the base specific fluorochromes chromomycin A3 (CMA) and 4′,6-diamidino-2-phenylindole (DAPI), which bind preferentially to GC- or AT-rich chromosome regions, respectively (reviewed by Guerra, 2000).
MATERIALS AND METHODS
Plant material
Six accessions belonging to five species of the Quadrifaria group of Paspalum were used for cytogenetic analysis: P. quadrifarium Lam., P. haumanii Parodi, P. intermedium Munro, P. quarinii Morrone & Zuloaga and P. exaltatum K. Presl. Plants were obtained from collections growing at the Facultad de Agronomía, Montevideo, Uruguay, or at Embrapa-Cenargen, Brasília, Brazil (Table 1). The other known source of I genomes, Paspalum rufum Nees (Virgata group), and Axonopus rosengurttii (MCM 05, Montevideo, Uruguay) were included as outgroups for the cpDNA phylogenetic analysis.
Table 1.
Paspalum species and cytotypes analysed, with respective chromosome number, collection number, provenance, and number and position of 5S and 45S rDNA sites
| Species |
2n |
Collection no. |
Provenance |
No. and position of 5S rDNA sites* |
No. and position of 45S rDNA sites* |
|---|---|---|---|---|---|
| P. intermedium | 20 | V11802 | 18 km south from Dourados, MGS, Brazil | 2 (1p) | 4: 2 (5t) + 2 (8p) |
| P. haumanii | 20 | Q3860 | Paso Lucero, Corrientes, Argentina | 2 (1p) | 4: 2 (5t) + 2 (8p) |
| P. quadrifarium | 20 | NA2623 | R5 km 491, Rivera, Uruguay | 2 (1p) | 5: 2 (5t) + 1 (1t) + 1 (7t) + 1 (10t) |
| P. quadrifarium | 40 | NA7664 | Cañada Ibañez, Cerro Largo, Uruguay | 4 (1p) | 7: 4 (5t) + 2 (8p) + 1 (t) |
| P. quarinii** | 40 | BRA-020923 | São Miguel das Missões, RGS, Brazil | 4 (1p) | 7: 2 (both ends of one chromosome 1) + 2 (5t) + 3 (t) |
| P. exaltatum | 40 | BRA-022101 | Caseiros, RGS, Brazil | 4 (1p) | 6: 4 (8p) + 2 (t) |
The position of the site was given by the chromosome order on a monoploid complement (1–10) followed by its location as proximal (p) or terminal (t).
= P. brunneum auct. non Mez (Morrone and Zuloaga, 2000).
Pretreatment and fixation
Selected root tips were obtained from plants growing in pots and pretreated with 2 mm 8-hydroxyquinoline for 2 h at room temperature and 2 h at 4 °C. They were fixed in Carnoy 3 : 1 (ethanol : acetic acid) solution for at least 24 h at room temperature and then stored at −20 °C until used.
Chromosome preparation for karyogram analysis
The protocol used for chromosome preparations for karyotype analysis followed that of Speranza et al. (2003). Fixed root tips were rinsed in buffer (40 mm citric acid, 60 mm sodium citrate) for 15 min and digested for 4 h at 37 °C with a combination of 3 % (w/v) cellulase (Calbiochem, San Diego, CA, USA), 1 % (w/v), cellulase Onozuka RS (Yakult Pharmaceutical, Japan) and 4 % (v/v) pectinase (Sigma, St Louis, MO, USA). Root tips were squashed in 45 % acetic acid and coverslips were removed by freezing in liquid nitrogen. The slides were washed in 70 % acetic acid at 40 °C for 5–10 min to remove the remaining cytoplasm, stained with 2 % lacto-propionic (1 : 1) orcein (Sigma), and sealed. The construction of the karyograms followed Speranza et al. (2003).
Cells were photographed on an Olympus New Vanox Microscope and highly amplified conventional prints were digitized and treated with Corel Draw version 11 for processing. Idiogram construction was based on the analysis of at least five well-spread prometaphases. Measurements were made on the public domain software Image J 1.28u (http://rsb.info.nih.gov/ij/Java1.3.1_03).
Chromosome preparation for fluorochrome banding and FISH
For these procedures, chromosome preparations were performed according to Cornélio et al. (2003). Fixed root tips were washed in distilled water for 15 min and digested in 2 % cellulase Onozuka R-10 (Serva) and 20 % pectinase (Sigma) for 4 h at 37 °C. Root tips were squashed in 45 % acetic acid and coverslips were removed by freezing in liquid nitrogen and then slides were air-dried. The best slides were selected after a brief stain with DAPI/glycerol (v/v), destained in Carnoy 3 : 1 for 30 min and dehydrated in 100 % ethanol for at least 2 h. Slides were stored at −20 °C until used.
For CMA/DAPI staining the slides were aged for 3 d, stained with 0·5 mg ml−1 chromomycin A3 (CMA; Sigma) for 1 h and 2 µg ml−1 4′,6-diamidino-2-phenylindole (DAPI; Sigma) for 30 min and mounted in 1 : 1 (v/v) McIlvaine's pH 7 buffer/glycerol. Images were acquired using a Cohu-CCD video camera attached to a DMLB Leica epifluorescence microscope, and combined using the Leica Qfish software. Final processing was made using Corel Draw version 11. After image acquisition, the coverslips were removed, the slides were destained in ethanol–acetic acid (3 : 1) for 30 min, dehidratated in 70 % ethanol overnight, air dried and stored at −20 °C until used for FISH.
DNA probes and labelling
Two DNA probes, R2 and D2, were used in the FISH experiments. Probe R2 is a 6·5-kb fragment containing an 18S–5·8S–25S rDNA repeat unit (including internal transcribed spacers ITS1 and ITS2 and a short 5′ fragment of the intergenic region from Arabidopsis thaliana (Wanzenböck et al., 1997). Probe D2, is a 500-bp fragment of the 5S rDNA gene repeated unit from Lotus japonicus (Pedrosa et al., 2002). Both probes were labelled by nick translation (Gibco) with digoxigenin-11-dUTP (Roche, Molecular Biochemicals, Sussex, UK) and biotin-11-dUTP (Sigma), respectively.
Double-target FISH
The in situ hybridization technique followed Heslop-Harrison et al. (1991) with the modifications introduced by Pedrosa et al. (2001). Hybridization mixture consisted of 60 % (v/v) formamide, 5 % (p/v) dextran sulfate, 2 × SSC, 0·01 % salmon sperm DNA and between 20 and 50 ng μL−1 of each labelled probe. The mixture was preheated at 75 °C for 10 min and kept on ice for 5 min. Then 30 μL of the mixture was added to each slide and chromosomes together with the probes were denatured on a hot plate at 80 °C for 10 min. Slides were then transferred to a humid chamber for overnight hybridization at 37 °C. Following hybridization, slides were given a stringent wash with 0·1× SSC at 42 °C for 5 min and washed in 2× SSC. Digoxigenin-labelled probe, R2, was detected with FITC (fluorescein isothiocyanate; Roche), and biotin-labelled probe, D2, was detected with TRITC (tetramethyl rhodamine isothiocyanate; Dako). Slides were counterstained and mounted with 2 µg mL−1 DAPI in Vectashield (Vector). The images were acquired as described before.
Sequencing
Silica-gel dried leaves of the same individuals used for cytogenetic and FISH analysis were used to extract DNA with a Sigma Genelute™ kit (Sigma-Aldrich, St Louis, MO, USA) according to the manufacturer's instructions. Universal primers (Taberlet et al., 1991) were used to amplify the trnL(UAA) intron (primers C and D of Taberlet et al., 1991), the PsbA-trnH spacer (Sang et al., 1997) and the atpB–rbcL spacer (Hodges and Arnold, 1994). Primers for the trnG(UCC) intron were obtained from Shaw et al. (2004). The trnL(UAA)–trnF(GAA) spacer could not be amplified directly from genomic DNA in any of these Paspalum species using primers E and F (Taberlet et al., 1991); therefore, E-F fragments were obtained by nested PCR with primers E and F using 1 : 1500 to 1 : 5000 dilutions of the C-F fragments as template. All PCR amplifications were carried out in 25-μL reactions containing 0·4 units of NEB Taq polymerase (New England Biolabs, Beverly, MA, USA) 1·5 mm MgCl2, 0·4 μm of each primer and 0·1 mm of each dNTP in the manufacturer's buffer. Amplification was carried out in a Biometra® T3 Thermoblock with the same programme for all fragments. The programme consisted of an initial 5 min at 95 °C, followed by one cycle of 1 min at 94 °C, 1 min at 58 °C and 2 min 30 s at 72 °C, the annealing temperature was decreased by 1 °C for six cycles and then 32 additional cycles were carried out with an annealing temperature of 52 °C followed by a final elongation step of 5 min at 72 °C. PCR products were cleaned with Wizard® SV Gel and PCR Clean-up System (Promega, Madison, WI, USA) and diluted to approx. 1 ng μL−1 before sequencing.
All regions were sequenced in both directions on a CEQ 8000 capillary sequencer (Beckman-Coulter, Fullerton, CA, USA) using one-quarter reaction volumes with the addition of 80 mm Tris and 2 mm MgCl2 (pH 9) to complete the volume of a full reaction. The same PCR primers were used for sequencing, except for primer E because a poly-A tract located 65 bp from its end prevented further sequencing in most materials. Primer E2 (5′-AAAGGAGTGCGACGAGAAC-3′) was designed starting at position 127, just internal to the poly-A tract. This primer was designed with Primer 3 (Rozen and Skaletsky, 2000) based on the sequences obtained with primer F for this region.
The sequences were edited manually using Sequencher™ (V4·1·4, Genecodes, AnnArbor, MI, USA) and all ambiguous end regions removed. The resulting partial sequences were prealigned with the Clustal-W (Thompson et al., 1994) algorithm included in BioEdit (V 5.0.6; Hall, 1997) and the resulting alignments were manually adjusted. All sequences and alignments were submitted to Genbank (Accession nos. AY941120–AY941159). Parsimony analyses for all five regions combined in a single matrix were carried out in PAUP* (V 4.0b10, Swofford) using an exhaustive search; bootstrap analysis was performed with 10 000 replicates and 100 SAR per replicate.
RESULTS
Chromatin condensation patterns and fluorochrome banding
The karyotypes of the diploid cytotypes of Paspalum intermedium, P. haumanii and P. quadrifarium are shown in Fig. 1. The three species presented symmetric karyotypes with metacentric and submetacentric chromosomes. The general features of prometaphase chromatin condensation patterns obtained after orcein staining were basically the same for the three diploid cytotypes and allowed the identification of homeologous chromosomes. CMA/DAPI double staining revealed the same condensation pattern observed after orcein staining (Fig. 2). CMA-stained chromosomes exhibited more contrasted bands than DAPI. Chromosome 1 presented an entirely homogenous chromatin condensation pattern in both arms. Chromosomes 2 and 8 showed condensed proximal and terminal regions. Condensation in the other chromosomes includes the proximal regions and a characteristic distal region of the long arm, while their short arms are entirely or almost entirely condensed. Only one submetacentric satellite–chromosome pair (chromosome 5) was observed, with the satellite located in the short arm (Fig. 1A; Fig. 2B and D). The satellites were not stained with DAPI (Fig. 2A and C). In prometaphase and metaphase, satellites are frequently found separated from the chromosomes and also became lost (Fig. 1C). In the tetraploids analysed with CMA/DAPI no secondary constriction was observed (Fig. 2H). After in situ hybridization, the DAPI banding pattern was not only carefully preserved in all species but even more contrasted than before, and it was very useful to identify the chromosomes labelled by the probes.
Fig. 1.
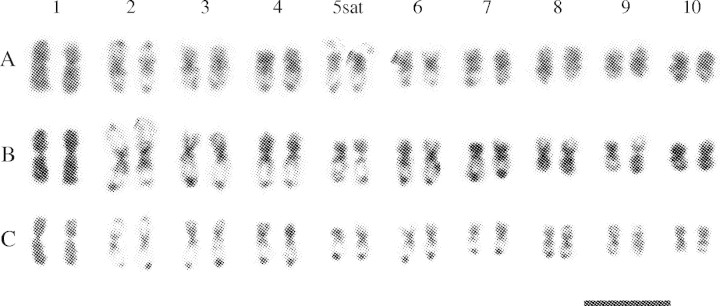
Karyograms of diploid cytotypes of (A) P. intermedium, (B) P. haumanii, and (C) P. quadrifarium based on orcein stained prometaphase chromosomes. Scale bar = 10 µm.
Fig. 2.
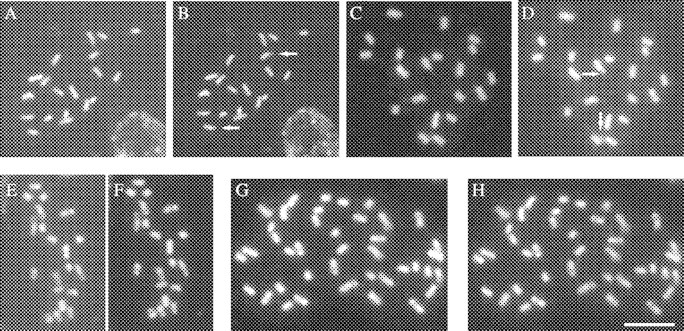
Prometaphase chromosomes of P. haumanii 2x, P. intermedium 2x, P. quadrifarium 2x and P. quadrifarium 4x after DAPI (A, C, E and G) and CMA staining (B, D, F and H). Arrows point out satellites. Scale bar = 10 µm.
Number and distribution of 5S rDNA sites
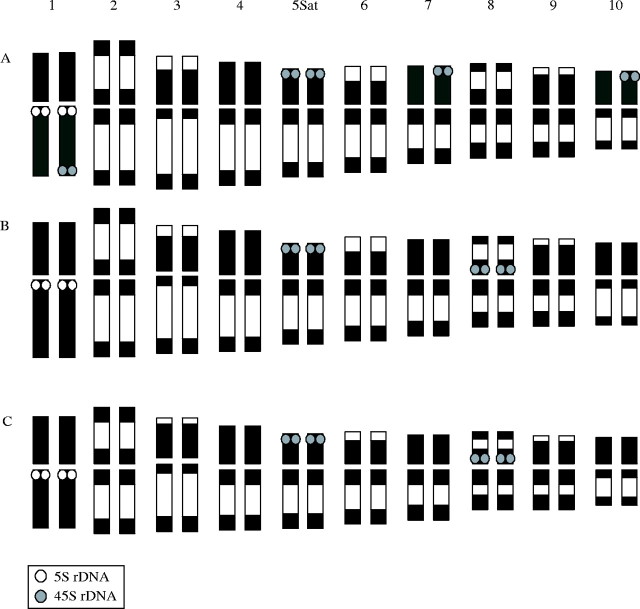
Figure 3 shows the results of the double-target in situ hybridization with the 5S rDNA and 45S rDNA probes to mitotic chromosomes of the Quadrifaria species and cytotypes. The number and localization of the rDNA sites are summarized in Table 1.
Fig. 3.
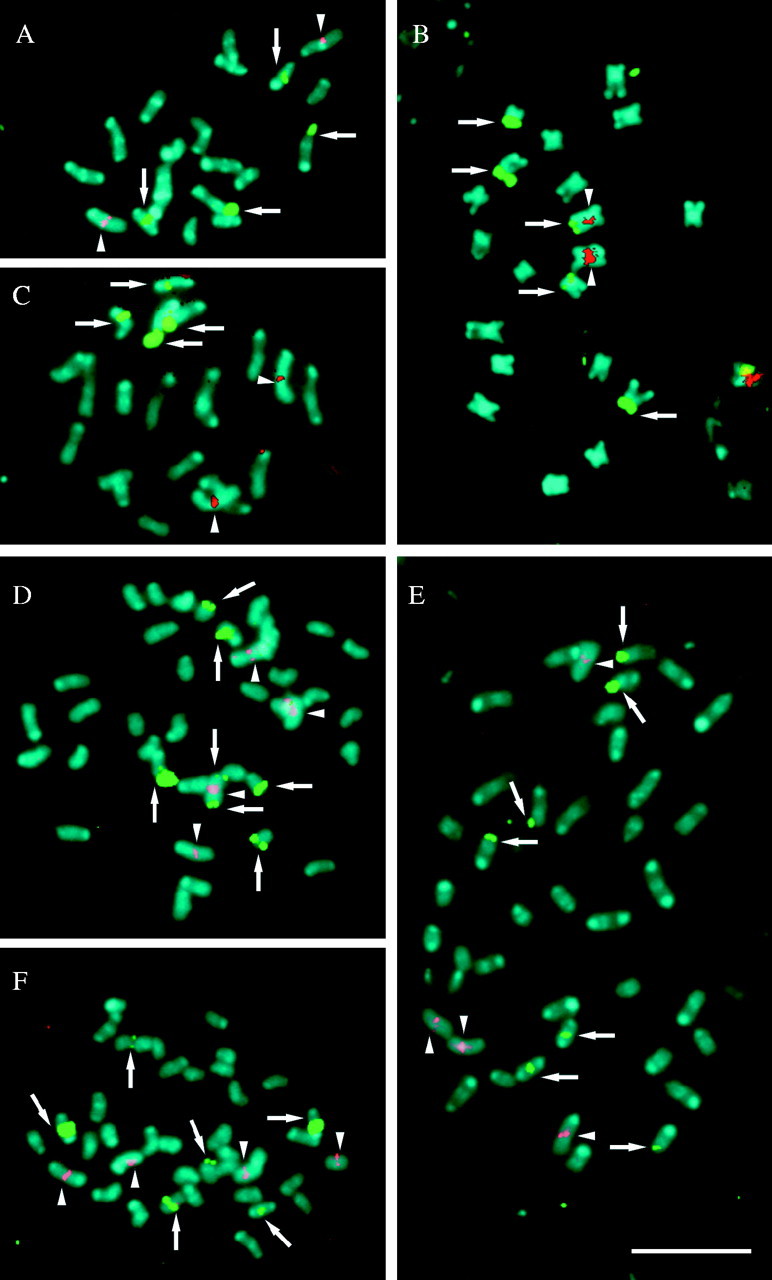
Fluorescence in situ hybridization with 45S (green) and 5S (red) rDNA probes, in prometaphase chromosomes of (A) P. haumanii 2x, (B) P. quadrifarium, (C) P. intermedium 2x, (D) P. quarinii 4x, (E) P. quadrifarium 4x, and (F) P. exaltatum 4x. Arrowheads indicate 5S rDNA sites while arrows indicate 45S rDNA sites. Note, in B and D, 45S and 5S rDNA sites located on chromosome 1. Chromosomes were counterstained with DAPI (blue). Scale bar = 10 µm.
The three diploid species, P. quadrifarium, P. haumanii and P. intermedium, have one pair of 5S rDNA sites (Fig. 3A–C) and polyploid plants were found to have the expected number of these loci (Table 1). Thus tetraploid cytotypes of P. quarinii, P. quadrifarium and P. exaltatum had four hybridization sites (Fig. 3D–F). The signals in all species and cytotypes were detected in a proximal location on the short arm of chromosome 1, which is characterized by its homogeneous chromatin condensation pattern in prometaphase.
Number and distribution of 45S rDNA sites
The in situ hybridization analysis revealed four sites for the 45S rDNA in the species P. haumanii and P. intermedium: two strong large-sized signals were localized at the end of the short arms of SAT chromosome 5 and a minor pair of sites was visible at the condensed proximal region of chromosome 8 (Figs 3A and C and 4B and C). For the diploid cytotype of P. quadrifarium, a different organization was observed. Five 45S rDNA sites were detected and all of them were located on the terminal end of the short arms of the chromosomes. Strong signals were localized at the end of the short arm of both homologous SAT chromosomes 5, and three minor sites on the short arms of only one member of chromosome pairs 1, 7 and 10. The signals in the homologous chromosome of these pairs were not detected (Figs 3B and 4A).
Fig. 4.
Idiograms of (A) Paspalum quadrifarium 2x, (B) P. haumanii 2x, and (C) P. intermedium 2x. Distribution of 5S rDNA sites (white dots) and 45S rDNA sites (grey dots) is indicated.
In the tetraploid cytotype of P. quarinii, a total of seven 45S rDNA sites were observed (Fig. 3D). Two of these were located at both terminal regions of only one member of chromosome pair 1. It is interesting to note that these two sites were unequal in size; the site at the end of the long arm was larger than the one found at the shorter one. Two other sites were observed at the terminal regions of two chromosomes that resemble chromosome 5 in diploid species. The other three sites were distributed at terminal regions of three small chromosomes.
The tetraploid individual of P. exaltatum had six 45S rDNA sites (Fig. 3F): four major sites were located in the condensed proximal regions of chromosomes that present a chromatin pattern similar to chromosome pair 8 of the diploid species, while the other two minor signals were located at the terminal region of two small chromosomes. No signal was detected at the chromosome corresponding to the number 5 of diploid species.
For the tetraploid cytotype of P. quadrifarium seven signals were observed and its hybridization pattern did not represent the exact doubling of the diploid cytotype of P. quadrifarium (Fig. 3E). Four 45S rDNA sites were located on the distal region of the short arms of two chromosome pairs corresponding to chromosome 5 of diploids, a minor site was observed on the terminal region of a small chromosome, and two signals were detected in the proximal region of chromosome 8.
cpDNA sequences
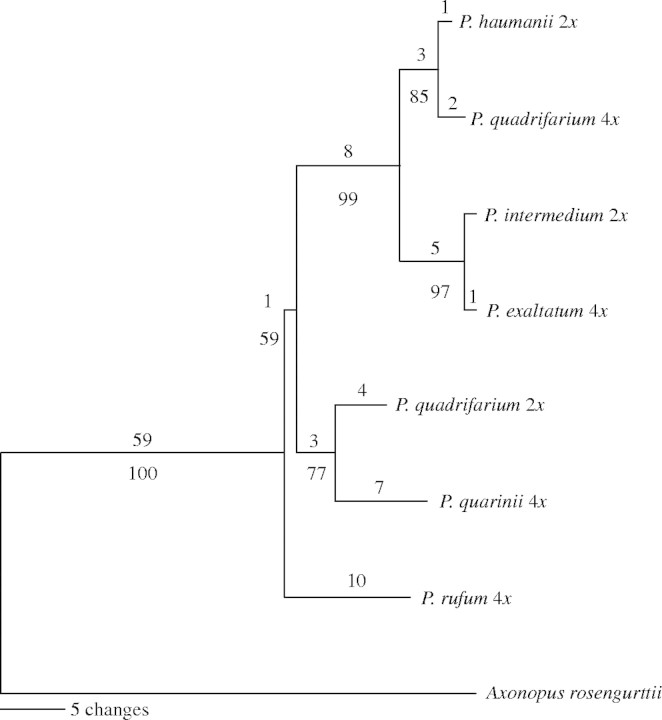
Separate phylogenetic analyses were conducted for each sequenced region and no conflicts were found for supported branches. Each gene supported a subset of the nodes; however, the trnL–trnF spacer sequence matrix alone produced almost all the relationships found for the combined data set, except with bootstrap supports ranging from 60 to 94 %. All the data were combined in a single matrix for further analysis. The resulting combined matrix consisted of a total length of 2367 bp positions, 74 of which were variable and 23 parsimony informative. A single most-parsimonious tree of length 104 (CI = 0·9327, RC = 0·7772) was found for this set of sequences (Fig. 5). Bootstrap values for all nodes ranged between 77 and 100 % with one short branch showing 59 %. The tree shows the Quadrifaria species analysed here partitioned in two clades, one of them includes P. haumanii and P. intermedium together with the tetraploid cytotypes of P. quadrifarium and P. exaltatum, while the other contains the diploid P. quadrifarium and the tetraploid P. quarinii.
Fig. 5.
The single most-parsimonious tree found using an exhaustive search from the phylogenetic analysis of five chloroplast non-coding regions (see text). Numbers above the branches indicate the number of base substitutions and numbers below the branches are bootstrap support percentages out of 10000 replicates.
DISCUSSION
Karyograms based on the differential prometaphase chromatin condensation pattern
The prometaphase chromosome condensation patterns observed here allowed the identification of most homologous chromosomes and the karyotype construction of the diploid cytotypes of Paspalum haumanii, P. intermedium and P. quadrifarium. This banding pattern was also highly conserved in the tetraploid accessions investigated here and in the polyploid cytotypes of P. quadrifarium reported by Speranza et al. (2003). These patterns were not influenced by the dyes used (CMA, DAPI or orcein) or by the FISH procedure, therefore the bands are not necessarily related to heterochromatin or to GC- and AT-rich regions. They most probably represent the longitudinal variation in the chromosome condensation at the prophase and prometaphase (for a discussion of heterochromatin and condensation patterns, see Guerra, 1988). Eventually, this uneven chromatin condensation allowed the identification of almost all the chromosome pairs that carried the rDNA signals in the species analysed here.
FISH pattern of the 5S rDNA sites
The number and location of the 5S rDNA sequence appeared to be conserved among the Quadrifaria species and cytotypes. As seen for other plant genera (D'Hont et al., 1998; Adams et al., 2000; de Melo and Guerra, 2003), a correspondence between the number of 5S rDNA loci and ploidy level was found for the tetraploid specimens of P. quadrifarium, P. quarinii and P. exaltatum, which all exhibit the four expected signals. Such a conservation of the 5S rDNA loci is not a common feature in grasses, where variability in number, location or both has been reported (Thomas et al., 1996; Shishido et al., 2000; Taketa et al., 2001; Harper et al., 2004).
Variability in the number and location of 45S rDNA sites among species
In contrast to the 5S rDNA, several differences in the location of 45S rDNA loci were detected. The large number of heterozygous pairs found in the diploid individual of P. quadrifarium is particularly remarkable. This unexpected variation may be attributed to the fact that diploid cytotypes of P. quadrifarium are sexual and self-incompatible (Norrmann et al., 1989). The population from which this individual was collected most probably is polymorphic for the presence of 45S rDNA sites on chromosomes 1, 7 and 10, and provides evidence of the high mobility of the 45S rDNA within a population. Polymorphisms for 45S rDNA sites have been also reported in other grasses such as Lolium (Thomas et al., 1996) and Hordeum (Taketa et al., 1999, 2001). In the Quadrifaria case, the general structural similarity among the karyotypes of all the species studied suggested that major chromosome rearrangements are not frequent in this group. Therefore, mechanisms such as transposon mobility and amplification of cryptic minor rDNA sites by unequal crossing-over rather than chromosomal rearrangements may be involved in the origin of the variation of the 45S rDNA. The possibility that nucleolar organizer regions (NORs) in Paspalum may have moved via magnification of minor loci consisting of few repeats and deletion of major inactive sites, as proposed by Dvořák (1989) and Dubcovsky and Dvořák (1995) for the Triticeae, may be a serious concern regarding the use of ITS sequences for phylogeny reconstruction (Dubcovsky and Dvořák, 1995; Adams et al., 2000).
Two general patterns were observed in the Quadrifaria accessions with respect to the location of the 45S rDNA. The species and cytotypes P. haumanii 2x, P. intermedium 2x, P. quadrifarium 4x and P. exaltatum 4x showed proximal sites on chromosome 8 and a variable number of distal sites, while P. quarinii 4x and P. quadrifarium 2x showed only distal sites located on a variable number of small chromosomes and on the long arm of chromosome 1.
cpDNA sequence analysis
The subdivision between the Quadrifaria species found with the FISH analysis is consistent with the two clades recovered with cpDNA data. These clades, in turn, are congruent with those found in a preliminary cpDNA sequence analysis of about 70 Paspalum species which includes the ones studied in this paper (P. Speranza and G. Rua, unpubl. res.). This preliminary analysis also shows that diploid P. quadrifarium is sister to a clade containing P. quarinii and a wide array of species including the Paniculata group, a proposed source of the J genomes to the Dilatata group (Burson, 1978). From the morphological point of view, the Quadrifaria group includes species with very similar characteristics. Differences among P. quadrifarium, P. exaltatum and P. haumanii for example, are mostly quantitative (Barreto, 1966); however, P. haumanii and P. intermedium are extremely large bunchgrasses often reaching heights of 2–3 m, while the species included in the other clade never reach such size (Barreto, 1966). The information presented here suggests that the Quadrifaria group, as presently defined, is not monophyletic and its species belong in at least two clades which may in turn be paraphyletic to other morphological groups. More species need to be analysed in order to identify stable synapomorphies for the clades that make up the Quadrifaria group.
Origin of the polyploid cytotypes
The tetraploid plant of P. quadrifarium used here is likely to be a hybrid between species belonging to the two main clades into which the Quadrifaria group appears to be divided. The presence of two distinct genomes reported by Speranza et al. (2003) in the same individual, suggested a segmental allopolyploid origin. That conclusion, based on the lack of multivalent formation during meiosis and prometaphase chromosome analysis, is further supported here by the 45S rDNA hybridization pattern of this plant – only two signals in the proximal region of chromosome 8. Moreover, this plant morphologically resembles diploid P. quadrifarium but its chloroplast genome is more similar to that of P. haumanii. This provides additional support for an allopolyploid origin for this cytotype.
The chloroplast haplotype of the tetraploid individual of P. exaltatum used here, was closely related to that of P. intermedium; however, P. exaltatum is morphologically very similar to P. haumanii, which also suggests a hybrid origin for this plant. The presence of four 45S rDNA proximal signals on chromosome 8 in this tetraploid P. exaltatum indicates that it most probably originated by hybridization between two species with a karyotype similar to those of P. intermedium and P. haumanii.
Differences between the expected and observed number of 45S rDNA loci was also detected in the polyploid cytotypes of P. quadrifarium and P. quarinii. There is significant evidence for genetic alteration of the genome following polyploidization (Wendel, 2000; Levy and Feldman, 2004). The loss of ribosomal genes after allopolyploidization has been seen in many plant genera (Vaughan et al., 1993; Thomas et al., 1997; Mishima et al., 2002). Thus, unlike the 5S rDNA sites, some 45S rDNA loci of the Paspalum accessions investigated here appear to have been eliminated after polyploidization.
Cytogenetic differentiation
All the diploid species analysed here have been assigned the II genome formula. Quarín and Norrmann (1990) found that for crosses between pairs of diploid cytotypes belonging to the same cpDNA clade (P. intermedium × P. haumanii and P. quadrifarium × P. quarinii), the mean number of bivalents of each hybrid was nearly 10. In the same paper it is reported that for inter-clade crosses that number was close to 8·5 and for hybrids of all four species to P. rufum (Virgata group), the only diploid source of I genome outside the Quadrifaria group, it was approx. 7. The mean number of bivalents in the hybrids reported by Quarín and Norrmann (1990) is then roughly proportional to the phylogenetic distances estimated in this paper for their parents based on cpDNA. However, for Seberg and Petersen (1998) chromosome pairing between two species may be a poor indicator of phylogenetic relatedness, since it has been shown to be under genetic control, and even influenced by environmental conditions (Jahuar and Joppa, 1996; Seberg and Petersen, 1998). Thus, more phylogenetic and cytogenetic data are required to assess whether chromosome pairing reflects phylogenetic distances in the genus Paspalum both among and within its major clades.
The evidence presented here sharply contrasts with the implicit interpretation of meiotic studies that autopolyploidy had played a major role in the evolution of the polyploid cytotypes of the Quadrifaria group (Quarín and Lombardo, 1986; Quarín and Norrmann, 1987). Instead, it appears that at least some of the polyploid apomictic cytotypes of the Quadrifaria group are part of a vast hybrid complex.
Acknowledgments
The authors are thankful to Dr G. Rua (Argentina) for providing the Paspalum haumanii and P. rufum specimens. This research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico and Fundacão de Amparo a Ciencia e Tecnología do Estado de Pernambuco (Brazil), and Comisión Sectorial de Investigación Científica (UDELAR, Uruguay). M.V. was supported by a grant from the Red Latinoamericana de Botánica (RLB-03-P5).
LITERATURE CITED
- Adams SP, Leitch IJ, Bennett MD, Chase MW, Leitch AR. 2000. Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). American Journal of Botany 87: 1578–1583. [PubMed] [Google Scholar]
- Barreto IL. 1954. Las especies afines a Paspalum virgatum en la América del Sur. Revista Argentina de Agronomía 23: 53–70. [Google Scholar]
- Barreto IL. 1966. Las especies afines a Paspalum quadrifarium (Gramineae) en la América del Sur de clima sub-tropical y templado. Darwiniana 14: 130–155. [Google Scholar]
- Brasileiro-Vidal AC, Cuadrado A, Brammer SP, Zanatta AC, Prestes AM, Moraes-Fernandes M, Guerra M. 2003. Chromosome characterization in Thinopyrum ponticum (Triticeae, Poaceae) using in situ hybridization with different DNA sequences. Genetics and Molecular Biology 26: 505–510. [Google Scholar]
- Burson BL. 1978. Genome relations between Paspalum conspersum and two diploid Paspalum species. Canadian Journal of Genetics and Cytology 20: 365–372. [Google Scholar]
- Burson BL. 1983. Phylogenetic investigations of Paspalum dilatatum and related species. In: Norman MJT, ed. Proceedings of the XIV International Grassland Congress. Lexington, KY, June 1983. Boulder, CO: Westview Press, 170–173. [Google Scholar]
- Burson BL, Bennett HW. 1971. Chromosome numbers, microsporogenesis, and mode of reproduction of seven Paspalum species. Crop Science 11: 292–294. [Google Scholar]
- Burton GW. 1940. A cytological study of some species in the genus Paspalum Journal of Agronomical Research 60: 193–197. [Google Scholar]
- Caponio I, Quarín CL. 1993. Cytology and reproduction of Paspalum densum and its genomic relationship with P. intermedium and P. urvillei Journal of Heredity 84: 220–222. [Google Scholar]
- Chase A. 1929. North American species of Paspalum Contributions from the US National Herbarium, Vol 28, Part 1. Washington: US Government Printing Office. [Google Scholar]
- Clayton WD, Renvoize SA. 1986. Genera Graminum, grasses of the world. Kew Bulletin, Additional Series 13. [Google Scholar]
- Cornélio MTMN, Figueirôa ARS, Santos KGB, Carvalho R, Soares Filho WS, Guerra M. 2003. Chromosomal relationships among cultivars of Citrus reticulate Blanco, its hybrids and related species. Plant Systematics and Evolution 240: 149–161. [Google Scholar]
- de Melo NF, Guerra M. 2003. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Annals of Botany 92: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moraes MI, Barreto IL, Salzano FM, Sacchet OF. 1974. Cytological and evolutionary relationships in Brazilian forms of Paspalum Caryologia 27: 455–465. [Google Scholar]
- Demesure B, Sodzi N, Petit RJ. 1995. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Molecular Ecology 4: 129–131. [DOI] [PubMed] [Google Scholar]
- D'Hont A, Paget-Goy A, Escoute J, Carrel F. 1998. The interspecific genome structure of cultivated banana, Musa spp. revealed by genomic DNA in situ hybridization. Theoretical Applied Genetics 100: 177–183. [Google Scholar]
- Dubcovsky J, Dvořák J. 1995. Ribosomal RNA multigene loci: nomads of the Triticeae genomes. Genetics 140: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák J. 1989. Organization and evolution of multigene families: inferences from ribosomal RNA gene loci of wheat and related species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS, eds. Plant population genetics, breeding, and genetic resources. Sunderland, MA: Sinauer Associates, 83–97. [Google Scholar]
- Fukui K, Iijima K. 1991. Somatic chromosome map of rice by imaging methods. Theoretical Applied Genetics 81: 589–596. [DOI] [PubMed] [Google Scholar]
- Guerra M. 1988. Characterization of different types of condensed chromatin in Costus (Zingiberaceae). Plant Systematics and Evolution 158: 107–115. [Google Scholar]
- Guerra M. 2000. Patterns of heterochromatin distribution in plant chromosomes. Genetics and Molecular Biology 23: 1029–1041. [Google Scholar]
- Hall T. 1997. BioEdit: Biological Sequence Alignment Editor for Windows 95/98/NT, 4.8.6 edn. Raleigh: North Carolina State University. [Google Scholar]
- Harper JA, Thomas ID, Lovatt JA, Thomas HM. 2004. Physical mapping of rDNA sites in possible diploid progenitors of polyploid Festuca species. Plant Systematics and Evolution 245: 163–168. [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T, Anamthawat-Jónsson K, Leitch AR, Shi M, Leitch IJ. 1991.In situ hybridization with automated chromosome denaturation. Technique 3: 109–115. [Google Scholar]
- Hodges SA, Arnold ML. 1994. Columbines, a geographically spread species flock. Proceedings of the National Academy of Sciences of the USA 91: 5129–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahuar PP, Joppa LR. 1996. Chromosome pairing as a tool in genome analysis: merits and limitations. In: Jahuar PP, ed. Methods of genome analysis in plants. Boca Raton, FL: CRS Press, 9–37. [Google Scholar]
- Jiang J, Gill BS. 1994. Nonisotopic in situ hybridization and plant genome mapping: the first 10 years. Genome 37: 717–725. [DOI] [PubMed] [Google Scholar]
- Levy AA, Feldman M. 2004. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society 82: 607–613. [Google Scholar]
- Mishima M, Ohmido N, Fukui K, Yahara T. 2002. Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110: 550–558. [DOI] [PubMed] [Google Scholar]
- Morrone O, Zuloaga FO. 2000.Paspalum quarinii Morrone & Zuloaga (Gramineae-Paniceae), una nueva especie de Paraguay, sur de Brasil y Argentina. Candollea 55: 311–314. [Google Scholar]
- Nakamura R, Kitamura S, Inoue M, Ohmido N, Fukui K. 2001. Karyotype analysis of Nicotiana kawakamii Y. Ohashi using DAPI banding and rDNA FISH. Theoretical and Applied Genetics 102: 810–814. [Google Scholar]
- Norrmann GA, Quarín CL, Burson BL. 1989. Cytogenetics and reproductive behaviour of different chromosome races in six Paspalum species. Journal of Heredity 80: 24–28. [Google Scholar]
- Pedrosa A, Jantsch MF, Moscone E, Ambros PF, Schweizer D. 2001. Characterisation of pericentromeric and sticky intercalary heterochromatin in Ornithogalum longibracteatum (Hyacinthaceae). Chromosoma 110: 203–213. [DOI] [PubMed] [Google Scholar]
- Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A. 2002. Chromosomal map of the model Lotus japonicus. Genetics 161: 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarín CL, Lombardo IP. 1986. Niveles de ploidía y distribución geográfica de Paspalum quadrifarium (Gramineae). Mendeliana 7: 101–107. [Google Scholar]
- Quarín CL, Norrmann GA. 1987. Relaciones entre el número de cromosomas, su comportamiento en la meiosis y el sistema reproductivo del género Paspalum. Anales del IV Congreso Latinoamericano de Botánica 3: 25–34. [Google Scholar]
- Quarín CL, Norrmann GA. 1990. Interspecific hybrids between five Paspalum species. Botanical Gazette 151: 366–369. [Google Scholar]
- Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, eds. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press, 365–386. [DOI] [PubMed] [Google Scholar]
- Sang T. 2002. Utility of low-copy nuclear gene sequences in plant phylogenies. Critical Review in Biochemistry and Molecular Biology 37: 121–147. [DOI] [PubMed] [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120–1136. [PubMed] [Google Scholar]
- Sang Y, Liang GH. 2000. Comparative physical mapping of the 18S-5·8S-26S rDNA in three sorghum species. Genome 43: 22–28. [PubMed] [Google Scholar]
- Seberg O, Petersen G. 1998. A critical review of concepts and methods used in classical genome analysis. Botanical Review 64: 372–417. [Google Scholar]
- Shaw J, Lickey EB, Beck JT, Farmer SS, Liu W, Miller J, et al. 2004. The tortoise and the hare. II. Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany 92: 142–196. [DOI] [PubMed] [Google Scholar]
- Shishido R, Sano Y, Fukui K. 2000. Ribosomal DNAs: an exception to the conservation of gene order in rice genomes. Molecular and General Genetics 263: 586–591. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. 1998. Choosing an approach and an appropriate gene for phylogenetic analysis. In: Soltis DE, Soltis PS, Doyle JJ, eds. Molecular systematics of plants. II. Boston, MA: Kluwer Academic Publishers, 1–42. [Google Scholar]
- Speranza P, Vaio M, Mazzella C. 2003. Karyotypes of two cytotypes of Paspalum quadrifarium Lam. (Poaceae). An alternative technique for small chromosomes in plants. Genetics and Molecular Biology 26: 499–503. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Taketa S, Harrison GE, Heslop-Harrison JS. 1999. Comparative physical mapping of the 5S and 18S-25S rDNA in nine wild Hordeum species and cytotypes. Theoretical Applied Genetics 98: 1–9. [Google Scholar]
- Taketa S, Ando H, Takeda K, Von Bothmer R. 2001. Physical locations of 5S and 18S-25S rDNA in Asian and American diploid Hordeum species with the I genome. Heredity 86: 522–530. [DOI] [PubMed] [Google Scholar]
- Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E, King IP. 1996. Comparison of ribosomal DNA sites in Lolium species by fluorescence in situ hybridization. Chromosome Research 4: 486–490. [DOI] [PubMed] [Google Scholar]
- Thomas HM, Harper JA, Meredith MR, Morgan WG, Thomas ID, Timms E, King IP. 1997. Physical mapping of ribosomal DNA sites in Festuca arundinaceae and related species by in situ hybridization. Genome 40: 406–410. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan HE, Jamilena M, Ruiz Rejón C, Parker JS, Garrido-Ramos MA. 1993. Loss of nucleolar-organizer regions during polyploid evolution in Scilla autumnalis Heredity 71: 574–580. [Google Scholar]
- Wanzenböck EM, Schöfer D, Schweizer D, Bachmair A. 1997. Ribosomal transcription units integrated via T-DNA transformation associate with the nucleolus and do not require upstream repeat sequences for activity in Arabidopsis thaliana The Plant Journal 11: 1007–1016. [DOI] [PubMed] [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249. [PubMed] [Google Scholar]